ABSTRACT
Russula is the largest genus in the Russulales and is widespread throughout the world. Almost all Russula species are known to be ectomycorrhizal with high ecological and edible values, and some are lethal poisonous. In this study, four new species belonging to the subgenus Russula crown clade are identified based on morphological and phylogenetic evidence from the Xizang Autonomous Region and other provinces of China. Morphologically, Russula paragraveolens (sect. Polychromae, subsect. Xerampelinae) is mainly characterised by a cherry red to blood red pileus centre, a reddish orange pileus margin; R. pseudograveolens (sect. Polychromae, subsect. Xerampelinae) is characterised by a violet brown to brownish red pileus centre, a pale red to pastel red pileus margin and short basidia; R. shigatseensis (sect. Flavisiccantes, subsect. Lepidinae) is characterised by a brownish orange to madder red pileus centre, pinkish red pileus margin, and having lateral branches or branches of hyphal terminations in pileipellis; R. yadongensis (sect. Tenellae, subsect. Laricinae) is characterised by a dark purplish red pileus centre with brownish purple tints and having isolated to clustered spines of spore ornamentations. Their distinct taxonomic status is confirmed by the positions of the four new species in both the ITS and 4-locus (nucLSU, mtSSU, rpb2, tef1) phylogenetic trees.
KEYWORDS: Agaricomycetes, morphology, phylogeny, Russulales, taxonomy
1. Introduction
Russula Pers. belongs to the Basidiomycota, Agaricomycetes, Russulales, Russulaceae, and is the type genus of Russulaceae (Kirk et al. 2008; He et al. 2019). Russula species are widespread throughout the world, from the tundra of Greenland in the north to the broad-leaved forest of New Zealand in the south (Looney et al. 2015). They form ectomycorrhizae with gymnosperms, such as Abies, Larix, Picea, Pinus, and Pseudotsuga, and angiosperms, such as Fabaceae, Fagales, Malpighiales, and Myrtaceae. They increase plant resistance, promote root growth and nutrient uptake, and closely correspond to the plant communities (Looney et al. 2018; Sarwar et al. 2020). Many species of Russula are important wild edible mushrooms, rich in nutritional elements, with anti-tumour, antioxidant, cholesterol-lowering, blood lipid-lowering, and blood sugar-lowering effects, such as Russula cyanoxantha (Schaeff.) Fr. and R. griseocarnosa X.H. Wang, Zhu L. Yang & Knudsen (Chen and Zhang 2010; Kaewnarin et al. 2016; Khatua et al. 2021; Liu et al. 2023). However, there are also toxic species such as R. emetica (Schaeff.) Pers. and R. senecis S. Imai, which often cause gastroenteritis-type poisoning when accidentally consumed. Russula subnigricans Hongo is a highly toxic species that has caused many deaths in food poisoning incidents (Chen et al. 2014; Cho and Han 2016; Matsuura et al. 2016).
In recent years, due to the rapid development of molecular biology techniques, the taxonomic research of Russula has been promoted. Phylogenetic relationships of infrageneric taxa have been more objectively elucidated. The latest classification system shows that the genus comprises eight subgenera, Archaeae Buyck & V. Hofst., Brevipedum Buyck & V. Hofst., Compactae (Fr.) Bon, Crassotunicatae Buyck & V. Hofst., Glutinosae Buyck & X.H. Wang, Heterophyllidiae Romagn., Malodorae Buyck & V. Hofst., Russula Buyck & V. Hofst (Looney et al. 2015; Buyck et al. 2018, 2020; Adamčík et al. 2019). Among them, subg. Russula consists of two major clades: The Russula core clade and the Russula crown clade. Since 2006, at least 68 new species have been published for the crown clade of subg. Russula, 48 of which are from Asia, and 23 new species have been described for this clade from China, demonstrating the extraordinary species richness of this clade of the Russula crown clade (Li et al. 2012, 2013a, 2013b, 2015, 2016; Jiang et al. 2018; Li et al. 2018a, 2018b, 2021; Caboň et al. 2019; Song et al. 2021; Li 2022; Zhou et al. 2022). Members of Russula crown clade mostly have unchanging, yellowing, browning, reddening, greying, or blackening of context when bruised, sometimes with distinct disagreeable to agreeable smell, mild to strongly acrid tasted context, white to yellow spore print (Buyck et al. 2018). Four new species of this clade were also identified in our macrofungal diversity survey, and these four new species were elucidated by morphological and phylogenetic analyses in this study.
2. Materials and methods
2.1. Morphological study
During collection, the macro-morphological characteristics of the specimens and the habitat were photographed with a camera (Canon EOS 80D), and fresh specimens were wrapped in tin foil in order to avoid mixing or crushing. After recording the macro-morphological characteristics in the field, the fresh specimens were dehydrated in a drying oven at 55 °C for 12 h in the laboratory. The dried specimens were deposited in the Mycological Herbarium, Institute of Microbiology, Chinese Academy of Sciences (HMAS). The description followed the criteria for Russula morphology observation (Adamčík et al. 2019). The colour designation refers to HTML Color Codes (https://htmlcolorcodes.com).
Specimens were rehydrated with 5% KOH solution. Observation of spores was made in Melzer’s reagent. The morphology of hymenial cystidia and pileocystidia were stained with 1% aqueous Congo red solution (Heilmann-Clausen et al. 2000). Tissues of the pileus were also examined in Cresyl Blue solution to verify the presence of a metachromatic reaction (Buyck 1989). Carbolfuchsin was used to observe incrustations on primordial hyphae (Singer 1968). Sulfovanillin was used to observe the colouring of cystidia contents (Caboň et al. 2017).
2.2. DNA extraction, PCR and sequencing
DNA was extracted from 5 to 20 mg tissue of dried specimen with the Broad-spectrum Plant Rapid Genomic DNA Kit (Biomed, Beijing) following the manufacturer’s instructions.
In this study, five loci were amplified and sequenced: ITS was amplified using primer pair ITS1-F/ITS4 (White et al. 1990); ribosomal nuclear large subunit (nucLSU) was amplified by LROR/LR5 (Moncalvo et al. 2000); ribosomal mitochondrial small subunit (mtSSU) was amplified by MS1/MS2 (White et al. 1990); second largest subunit of the RNA polymerase II (rpb2) was amplified by RPB2-6F/RPB2–7.1R (Matheny 2005); translation elongation factor 1-alpha (tef1) was amplified by EF1-983F/EF1-1567R (Rehner and Buckley 2005). The amplified PCR products were detected by electrophoresis and sent to BGI Genomics Co., Ltd. (Beijing) for purification and sequencing.
2.3. Molecular phylogenetic study
Newly generated and reference sequences obtained from GenBank are listed in Tables 1 and 2. The sequences were aligned in AliView v1.19 and manually adjusted to eliminate poorly aligned or ambiguous regions. The six partitions were assembled in PhyloSuite v1.2.2 (Zhang et al. 2020), in the order of six loci (nucLSU, mtSSU, rpb2 exons, rpb2 introns, tef1 exons, tef1 introns). The final alignments have been deposited in TreeBASE (study no. 30337, 30338).
Table 1.
Sequences used in phylogenetic analysis based on ITS.
| Taxon | Voucher specimen | Location | ITS accession number |
|---|---|---|---|
| Gymnomyces abietis | - | USA | AY239347 |
| Gymnomyces abietis | - | USA | AY239348 |
| Gymnomyces californicus | - | USA | AY239312 |
| Gymnomyces californicus | - | USA | AY239308 |
| Gymnomyces foetens | - | USA | AY239316 |
| Gymnomyces monticola | - | USA | AY239313 |
| Gymnomyces monticola | - | USA | AY239314 |
| Gymnomyces setigerus | - | USA | AY239317 |
| Gymnomyces subalpinus | - | USA | AY239309 |
| Gymnomyces subalpinus | - | USA | AY239311 |
| Macowanites vinaceodorus | - | Spain | AJ438034 |
| Macowanites vinaceodorus | 46374 (AH) | Spain | MK105695 |
| Russula abbottabadensis | FH00304589 (holotype) | Pakistan | MG386704 |
| Russula abietiphila | HCCN14799 (type) | South Korea | MN130060 |
| Russula abietiphila | HCCN18498 | South Korea | MN130061 |
| Russula adwanitekae | AG 16-1430 (type) | India | MN263242 |
| Russula adwanitekae | AG 16-1435 | India | MN263243 |
| Russula amethystina | LAH35058 (holotype) | Pakistan | KT953613 |
| Russula amoenipes | 309IS77 | USA | AY061656 |
| Russula aurantioflammans | r3245 | Slovakia | KU928167 |
| Russula brunneovinacea | RITF 2242 (holotype) | China | KY114148 |
| Russula brunneoviolacea | MC01-507 | Denmark | AM113956 |
| Russula brunneoviolacea | PRM 922,557 | Canada | MG687327 |
| Russula buyckii | CUHAM277 (holotype) | India | KT962833 |
| Russula cessans | CR19 | Canada | KP406550 |
| Russula changbaiensis | HMAS262369 (holotype) | China | KC412162 |
| Russula claroflava | 224IS76 | USA | AY061665 |
| Russula claroflava | FH12212 | USA | KT933997 |
| Russula clavatohyphata | CAL1756 (holotype) | India | MG934209 |
| Russula clavipes | SAV:F-1327 | Slovakia | KU205292 |
| Russula coronaspora | GDGM79711 (holotype) | China | MN275689 |
| Russula coronaspora | GDGM79712 | China | MN275690 |
| Russula cremeirosea | BPL289 | USA | KT933983 |
| Russula cremeoavellanea | SAV F-2125 | Slovakia | KY582695 |
| Russula cuprea | FH12250 | USA | KT934010 |
| Russula curtipes | 1123IS77 | USA | AY061668 |
| Russula curtipes | FH12206 | USA | KT933995 |
| Russula decolorans | - | Germany | AF418637 |
| Russula decolorans | FH12196 | USA | KT933992 |
| Russula dhakuriana | CUHAM343 | India | MK414576 |
| Russula emeticicolor | FH12253 | USA | KT934011 |
| Russula faginea | SAV F-1337 | France | KU205289 |
| Russula faginea | SAV F-997 | Slovakia | KU205286 |
| Russula favrei | SAV F-1333 | Finland | KU205311 |
| Russula favrei | UPS UE06.09.2003-9 | Sweden | KU205272 |
| Russula firmula | AT2004142 | Sweden | DQ422017 |
| Russula flavobrunnescens | TLXM (AK5024) (type) | Mexico | MN130082 |
| Russula fontqueri | FH12223 | USA | KT934003 |
| Russula gnathangensis | CAL1733 (holotype) | India | MK253441 |
| Russula graveolens | SAV F-1342 | Slovakia | KU205302 |
| Russula graveolens | SAV:F-1338 | Belgium | KU205306 |
| Russula griseocarnosa | KUN F51839 (holotype) | China | EF627042 |
| Russula griseocarnosa | - | China | EF627043 |
| Russula guangxiensis | HMAS267867 (holotype) | China | KT286852 |
| Russula hakkae | HMAS267765 (holotype) | China | KT286848 |
| Russula heilongjiangensis | HMAS255142 (holotype) | China | MG719932 |
| Russula hookeri | CUHAM275 (holotype) | India | KP713777 |
| Russula integra | FH12172 | USA | KT933984 |
| Russula jilinensis | HMAS194253 (holotype) | China | GU966632 |
| Russula katarinae | BB03.159 (PC) (holotype) | USA | KP966377 |
| Russula katarinae | BB03.159 (holotype) | USA | NR153255 |
| Russula kewzingensis | CAL1636 (holotype) | India | MG674302 |
| Russula khinganensis | HMAS278895 (holotype) | China | MG719928 |
| Russula laeta | SAV F-3949 | Slovakia | KY582708 |
| Russula laricina | BB 08.681 | Italy | JN944008 |
| Russula laricina | E Watling 25556 | Europe | AY061685 |
| Russula lepida | - | Germany | AF418641 |
| Russula madrensis | TLXM (AK3422) (type) | Mexico | MN130093 |
| Russula magica | GENT (FH12-061) (type) | Thailand | MN130096 |
| Russula messapica var. messapica | ALV1991 | Spain | MK105669 |
| Russula messapica var. messapicoides | JL1493 | Spain | MK105674 |
| Russula minor | GDGM79686 (holotype) | China | MN275666 |
| Russula minor | GDGM79687 | China | MN275665 |
| Russula nauseosa | F30315 | Canada | KJ748441 |
| Russula nauseosa | F30317 | Canada | KJ748443 |
| Russula nuoljae | HMJAU37320 | China | KY357333 |
| Russula nuoljae | SAV:F-3092 | Norway | KU205350 |
| Russula nympharum | FH11121505 (holotype) | Spain | KU928157 |
| Russula odorata | BB 07.186 | Slovakia | JN944010 |
| Russula olivaceohimalayensis | CAL 1659 (AG 17-1447) (type) | India | MN130097 |
| Russula olivaceohimalayensis | CAL 1664 (AG 15-910) | India | MN130098 |
| Russula paragraveolens | HMAS281158 (holotype) | China | OQ871504 |
| Russula paragraveolens | HMAS279574 | China | OQ871506 |
| Russula paragraveolens | HMAS279575 | China | OQ871505 |
| Russula pascua | IB:1998/0124 | Germany | KU205314 |
| Russula peckii | BPL270 | USA | KT933970 |
| Russula pseudograveolens | HMAS279577 | China | OQ871507 |
| Russula pseudograveolens | HMAS279579 | China | OQ871508 |
| Russula pseudograveolens | HMAS287385 | China | OQ871497 |
| Russula pseudograveolens | HMAS287384 (holotype) | China | OQ871496 |
| Russula pseudotsugarum | UBC:F33077 | Canada | MF908478 |
| Russula pseudotsugarum | UWBM:WTU-F-038562 (type) | USA | KX813578 |
| Russula puellaris | nl1372 (TUB) | Germany | AF418628 |
| Russula puellula | SAVF 3107 | Slovakia | KY582704 |
| Russula purpureoverrucosa | GDGM32902 (holotype) | China | MG214692 |
| Russula purpureozonate | KD 18-003 (type) | India | MN267570 |
| Russula purpureozonate | KD 18-15 | India | MN269951 |
| Russula pusilla | BPL267 | USA | KT933968 |
| Russula rosea | BB 07.780 | France | JN944003 |
| Russula rubra | SAV F-914 | Slovakia | KY582723 |
| Russula rugulosa | BPL654 | USA | KY848516 |
| Russula rutila | SAV F-1504 | Slovakia | KY582687 |
| Russula sancti-pauli | PC (BB 06.494) (type) | Mexico | MN130101 |
| Russula sancti-pauli | PC (BB 06.499) | Mexico | MN130102 |
| Russula sapinea | PA38 | Latvia | KR019818 |
| Russula seperina | GENT (Verbeken 2000-135) | Italy | MN130109 |
| Russula seperina | SAV F-3156 (type) | Slovakia | MN130108 |
| Russula shigatseensis | HMAS287390 | China | OQ871502 |
| Russula shigatseensis | HMAS287389 (holotype) | China | OQ871501 |
| Russula shigatseensis | HMAS287391 | China | OQ871503 |
| Russula sichuanensis | HKAS 53885 | China | JX391968 |
| Russula sichuanensis | HKAS53792 (holotype) | China | JX391969 |
| Russula sichuanensis | HMAS 255316 | China | MG786566 |
| Russula solaris | BB 07.282 | Slovakia | JN944007 |
| Russula sp. | FLAS-F-61146 | USA | MH211767 |
| Russula sp. | FLAS-F-61609 | USA | MH211995 |
| Russula sp. | HMAS:260700 | China | KX441055 |
| Russula sp. | JLF6993 | USA | MN263039 |
| Russula sp. | LM1553 | UK | KM576511 |
| Russula sp. | S.D. Russell 439 | USA | MK397035 |
| Russula sp. | SR48-10MX | Mexico | KT697966 |
| Russula sp. | F30324 | Canada | KJ748450 |
| Russula sp. | S.D. Russell 7799 | USA | MK532803 |
| Russula subrutilans | RITF1874 (holotype) | China | KJ868237 |
| Russula subsulphurea | F18743 | Europe | KF810135 |
| Russula subsulphurea | TENN:F18743 | USA | NR153231 |
| Russula subtilis | SAV F-3805 (type) | USA | KY509504 |
| Russula subtilis | TENN-F-067624 (BPL666) (type) | USA | KY509511 |
| Russula tengii | HMAS262728 (holotype) | China | MG386708 |
| Russula tinctipes | SAVF-2494 | Slovakia | KY582698 |
| Russula uttarakhandia | CAL 1537 (holotype) | India | KY873997 |
| Russula velenovskyi | 526IS77 | USA | AY061721 |
| Russula versatilis | PRM 922558 | Czech Republic | MG687329 |
| Russula versicolor | BB 07.288 | Slovakia | JN944009 |
| Russula veternosa | SAV F-2588 | Slovakia | KY582699 |
| Russula vidalii | JC100508BT01, JMV800688 (BCN) | Spain | MK105693 |
| Russula vidalii | JMV20160517-1 (BCN) | Spain | MK105694 |
| Russula vinosa | nl1386 | Germany | AF418638 |
| Russula vinosa | SAV F-20024 | Slovakia | KY582692 |
| Russula vinosobrunneola | HMAS 278885 | China | MG719925 |
| Russula vinosobrunneola | HMAS281138 (holotype) | China | MG719927 |
| Russula violaceoincarnata | O73136 | Netherland | GU234047 |
| Russula xerampelina | DG05-28 | UK | JQ888204 |
| Russula xerampelina | UPS:UE14.09.2004-3 | Sweden | KU205279 |
| Russula yadongensis | HMAS287387 | China | OQ871499 |
| Russula yadongensis | HMAS287386 (holotype) | China | OQ871498 |
| Russula yadongensis | HMAS287388 | China | OQ871500 |
| Russula zelleri | NYGB-761009 (type) | USA | KX812833 |
| Russula zelleri | OSC 5610 | USA | MK169364 |
| Russula zvarae | BB 08.639 | Italy | JN944004 |
| Outgroup | |||
| Russula emetica | lw81 (TUB) | Germany | AF418619 |
| Russula emetica | UE05.10.2003-11 | Sweden | DQ421997 |
Newly generated sequences are shown in bold.
Table 2.
Sequences used in phylogenetic analysis based on 4-locus data.
| Taxon | Voucher specimen | Location | nucLSU | mtSSU | rpb2 | tef1 |
|---|---|---|---|---|---|---|
| R. aff. fucosa | 27/BB 06.596 | Canada | KU237457 | KU237301 | KU237743 | KU237892 |
| R. aff. subdensifolia | 552/BB 07.158 | USA | KU237544 | KU237390 | KU237830 | KU237974 |
| R. aff. turci | 433/BB 07.325 | Slovakia | KU237497 | KU237342 | KU237783 | KU237927 |
| R. aff. viscidula | 73/BB 06.049 | Madagascar | KU237469 | KU237313 | KU237755 | – |
| R. aff. xerampelina | 592/DM fl07-14 | USA | KU237576 | KU237424 | KU237862 | KU238004 |
| R. amara | 532/BB 07.782 | France | KU237524 | KU237370 | KU237810 | KU237954 |
| R. amethystina | 529/BB 07.314 | Slovakia | KU237521 | KU237367 | KU237807 | KU237951 |
| R. aurata | 547/BB 07.211 | Slovakia | KU237539 | KU237385 | KU237825 | KU237969 |
| R. azurea | 537/BB 08.668 | Italy | KU237529 | KU237375 | KU237815 | KU237959 |
| R. badia | 587/BB 07.324 | Slovakia | KU237571 | KU237419 | KU237857 | KU237999 |
| R. burlinghamiae | 548/BB 05.108 | USA | KU237540 | KU237386 | KU237826 | KU237970 |
| R. carminipes | 531/BB 07.192 | Slovakia | KU237523 | KU237369 | KU237809 | KU237953 |
| R. carpini | 551/BB 07.262 | Slovakia | KU237543 | KU237389 | KU237829 | KU237973 |
| R. cf. aurantioflammans | 241/BB 06.603 | Canada | KU237488 | KU237332 | KU237774 | KU237917 |
| R. cf. brunneoviolacea | 524/BB 06.606 | Canada | KU237516 | KU237362 | KU237802 | KU237946 |
| R. cf. decipiens | 231/BB 06.521 | Mexico | KU237482 | KU237326 | KU237768 | KU237911 |
| R. cf. katarinae | 30/BB 06.617 | Canada | KU237460 | KU237304 | KU237746 | KU237895 |
| R. cf. odorata | 525/BB 07.219 | Slovakia | KU237517 | KU237363 | KU237803 | KU237947 |
| R. cf. olivobrunnea | 240/BB 06.505 | Mexico | KU237487 | KU237331 | KU237773 | KU237916 |
| R. cf. sejuncta | 557/BB 08.143 | Madagascar | KU237547 | KU237393 | KU237833 | – |
| R. cf. sesenagula | 84/BB 06.129 | Madagascar | KU237473 | KU237317 | KU237759 | KU237904 |
| R. cf. vinosobrunea | 533/BB 07.231 | Slovakia | KU237525 | KU237371 | KU237811 | KU237955 |
| R. citrinolutea sp. ined. | 29/BB 06.611 | Canada | KU237459 | KU237303 | KU237745 | KU237894 |
| R. corallina | 229/BB 06.324 | USA | KU237481 | KU237325 | KU237767 | KU237910 |
| R. cuprea | 565/BB 07.233 | Slovakia | KU237555 | KU237401 | KU237841 | KU237984 |
| R. decipiens | 585/BB 07.178 | Slovakia | KU237569 | KU237417 | KU237855 | KU237997 |
| R. decolorans | 549/BB 07.322 | Slovakia | KU237541 | KU237387 | KU237827 | KU237971 |
| R. discopus | 48/BB 06.027 | Madagascar | KU237467 | KU237311 | KU237753 | KU237900 |
| R. echinospermatinae sp. ined. | 736/BB 09.173 | New Caledonia | KU237589 | KU237437 | KU237874 | KU238016 |
| R. flavisiccans | 236/BB 06.336 | Mexico | KU237485 | KU237329 | KU237771 | KU237914 |
| R. gigasperma | 438/BB 07.280 | Slovakia | KU237501 | KU237346 | KU237787 | KU237931 |
| R. globispora | 436/BB 07.243 | Slovakia | KU237499 | KU237344 | KU237785 | KU237929 |
| R. heinemannianus | 594/CS s.n. | Zimbabwe | KU237577 | KU237425 | KU237863 | KU238005 |
| R. integra | 518/BB 07.198 | Slovakia | KU237513 | KU237359 | KU237799 | KU237943 |
| R. laeta | 519/BB 07.267 | Slovakia | KU237514 | KU237360 | KU237800 | KU237944 |
| R. laricina | 575/BB 08.681 | Italy | KU237560 | KU237408 | KU237846 | KU237991 |
| R. lepida | 437/BB 07.189 | Slovakia | KU237500 | KU237345 | KU237786 | KU237930 |
| R. lilacea | 435/BB 07.213 | Slovakia | KU237498 | KU237343 | KU237784 | KU237928 |
| R. melliolens | 554/BB 07.194 | Slovakia | KU237545 | KU237391 | KU237831 | KU237975 |
| R. musaecolor sp. ined. | 558/BB 08.063 | Madagascar | KU237548 | KU237394 | KU237834 | KU237977 |
| R. nauseosa | 588/BB 07.285 | Slovakia | KU237572 | KU237420 | KU237858 | KU238000 |
| R. nothofagineae sp. ined. | 723/BB 09.044 | New Caledonia | KU237583 | KU237431 | – | KU238010 |
| R. nothofagineae sp. ined. | 725/BB 09.068 | New Caledonia | KU237584 | KU237432 | KU237869 | KU238011 |
| R. nothofagineae sp. ined. | 726/BB 09.069 | New Caledonia | KU237585 | KU237433 | KU237870 | KU238012 |
| R. nothofagineae sp. ined. | 732/BB 09.124 | New Caledonia | KU237586 | KU237434 | KU237871 | KU238013 |
| R. nothofagineae sp. ined. | 733/BB 09.125 | New Caledonia | KU237587 | KU237435 | KU237872 | KU238014 |
| R. obscurosordida sp. ined. | 591/BB 06.564 | Canada | KU237575 | KU237423 | KU237861 | KU238003 |
| R. odorata | 526/BB 07.186 | Slovakia | KU237518 | KU237364 | KU237804 | KU237948 |
| R. olivacea | 426/BB 07.223 | Slovakia | KU237492 | KU237336 | KU237778 | KU237921 |
| R. olivascens | 530/BB 08.663 | Italia | KU237522 | KU237368 | KU237808 | KU237952 |
| R. paludosa | 442/BB 07.330 | Slovakia | KU237505 | KU237350 | KU237791 | KU237935 |
| R. paragraveolens | HMAS281158 | China | OQ875223 | OQ878262 | OQ933792 | OQ948122 |
| R. paragraveolens | HMAS279574 | China | OQ875224 | OQ878263 | OQ933793 | OQ948123 |
| R. pelargonia | 586/BB 07.169 | Slovakia | KU237570 | KU237418 | KU237856 | KU237998 |
| R. pseudograveolens | HMAS279577 | China | OQ875227 | OQ878266 | OQ933794 | OQ948121 |
| R. pseudograveolens | HMAS287385 | China | OQ875226 | OQ878265 | OQ933796 | OQ948120 |
| R. pseudograveolens | HMAS287384 | China | OQ875225 | OQ878264 | OQ933795 | OQ948119 |
| R. puellaris | 523/BB 07.311 | Slovakia | KU237515 | KU237361 | KU237801 | KU237945 |
| R. romellii | 427/BB 07.202 | Slovakia | KU237493 | KU237337 | KU237779 | KU237922 |
| R. rosea | 430/BB 07.780 | France | KU237496 | KU237340 | KU237782 | KU237925 |
| R. roseinae sp. ined. | 735/BB 09.172 | New Caledonia | KU237588 | KU237436 | KU237873 | KU238015 |
| R. shigatseensis | HMAS287390 | China | OQ875231 | OQ878270 | OQ933788 | OQ948115 |
| R. shigatseensis | HMAS287389 | China | OQ875230 | OQ878269 | OQ933787 | OQ948114 |
| R. shigatseensis | HMAS387391 | China | OQ875232 | OQ878271 | OQ933789 | OQ948116 |
| R. sichuanensis | HMAS255316 | China | MG786572 | MG792323 | – | MG812160 |
| R. sichuanensis | HMAS268888 | China | KX441372 | KX441619 | – | MF893457 |
| R. solaris | 559/BB 07.282 | Slovakia | KU237549 | KU237395 | KU237835 | KU237978 |
| R. subtilis | 536/BB 05.107 | USA | KU237528 | KU237374 | KU237814 | KU237958 |
| R. tlaxcalae | 33/BB 06.542 | Mexico | KU237463 | KU237307 | KU237749 | KU237897 |
| R. turci | 528/BB 07.328 | Slovakia | KU237520 | KU237366 | KU237806 | KU237950 |
| R. versicolor | 589/BB 07.288 | Slovakia | KU237573 | KU237421 | KU237859 | KU238001 |
| R. vinosobrunneola | HMAS278885 | China | MG786570 | MG792321 | – | MG812158 |
| R. vinosobrunneola | HMAS281138 | China | MG786569 | MG792320 | – | MG812157 |
| R. vinosobrunneola | HMAS278896 | China | MG786567 | MG792318 | – | MG812155 |
| R. vinosobrunneola | HMAS278960 | China | MG786568 | MG792319 | – | MG812156 |
| R. yadongensis | HMAS287386 | China | OQ875228 | OQ878267 | OQ933790 | OQ948117 |
| R. yadongensis | HMAS287388 | China | OQ875229 | OQ878268 | OQ933791 | OQ948118 |
| R. zvarae | 538/BB 08.639 | Italy | KU237530 | KU237376 | KU237816 | KU237960 |
| Outgroup | ||||||
| R. raoultii | 561/BB 08.674 | Italy | KU237551 | KU237397 | KU237837 | KU237980 |
| R. viscida | 425/BB 07.298 | Slovakia | KU237491 | KU237335 | KU237777 | KU237920 |
Newly generated sequences are shown in bold.
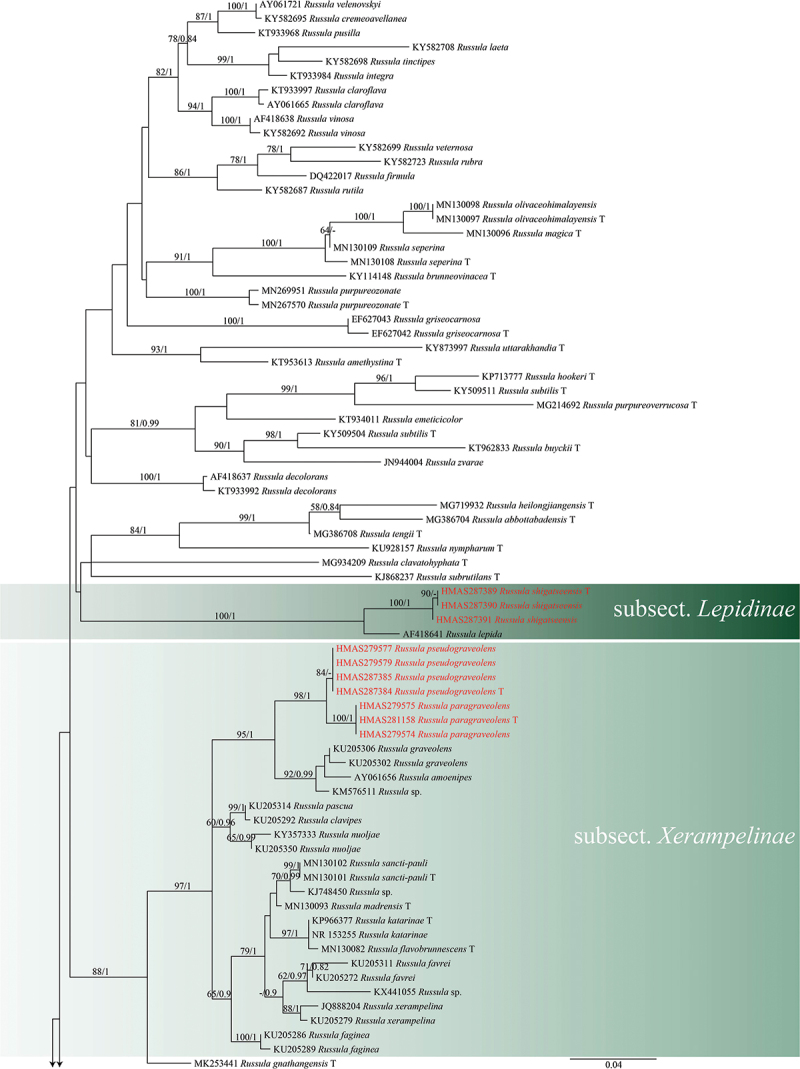
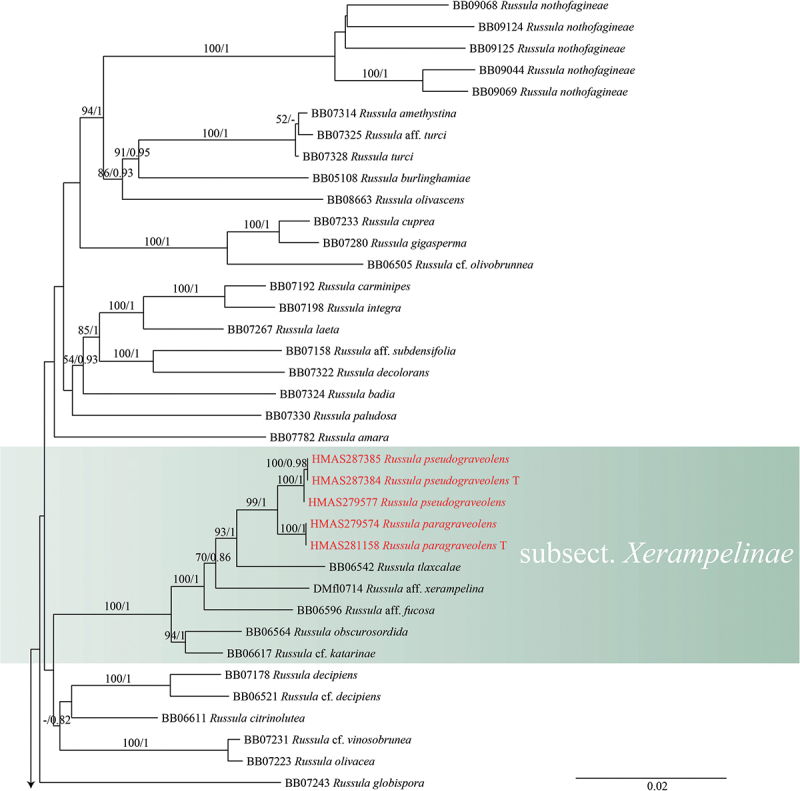
The ITS-based phylogenetic analysis consisted of 152 sequences representing 109 species, with R. emetica as the outgroup. A total of 80 concatenated sequences representing 66 species were used for multigene phylogenetic analyses, with R. raoultii Quél. and R. viscida Kudřna as the outgroups. The ITS and concatenated sequences were analysed using RAXMLGUI 1.3.1 (Silvestro and Michalak 2012) with the GTRGAMMAI model and 1,000 rapid bootstrap (BS) replicates. The best-fit model for ITS was selected using ModelFinder in PhyloSuite v1.2.2 (Kalyaanamoorthy et al. 2017). The best partitioning scheme and evolutionary models for six predefined partitions were selected using PartitionFinder2 in PhyloSuite v1.2.2 (Lanfear et al. 2017). Bayesian inference (BI) analyses was performed using MrBayes 3.2.6 (Ronquist et al. 2012) under the best model. Four Markov chains were run for 2 million generations, stopping when the average standard deviation of split frequencies fell below 0.01. Trees were sampled every 100th generation. The initial 25% of sampled data were discarded as burn-in.
The resulting file after tree construction was used to view the phylogenetic tree using FigTree 1.4.3 (Andrew 2016). Bootstrap Support (BS) ≥70% considered significantly supported. Bayesian Posterior Probability (PP) ≥95% was regarded as significant.
3. Results
3.1. Phylogeny
The best evolutionary model was selected using ModelFinder, with the BIC criterion: HKY+F+I+G4 for ITS. The best partitioning scheme and evolutionary models for six predefined partitions were selected using PartitionFinder2 (Lanfear et al. 2017), with greedy algorithm and AICc criterion: GTR+I+G for nucLSU, GTR+I+G for mtSSU, SYM+I+G for rpb2 exons, HKY+G for rpb2 introns, SYM+I+G for tef1 exons, GTR+I+G for tef1 introns.
Maximum likelihood and Bayesian analyses were performed on the ITS and 4-locus data sets, and both maximum likelihood and Bayesian analyses yielded the same topology. The four proposed new species, R. paragraveolens, R. pseudograveolens, R. shigatseensis, and R. yadongensis, are all nested in the Russula crown clade in both ITS and 4-locus (nucLSU-mtSSU-rpb2-tef1) trees, and are clearly separated from known species (Figures 1 and 2).
Figure 1.
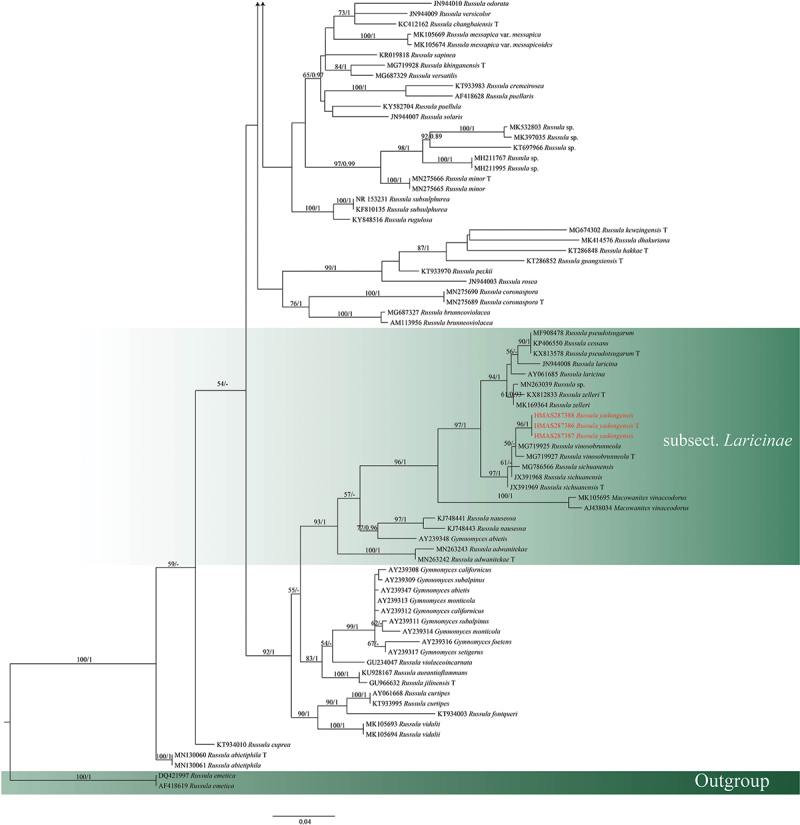
(Continued).
Figure 2.
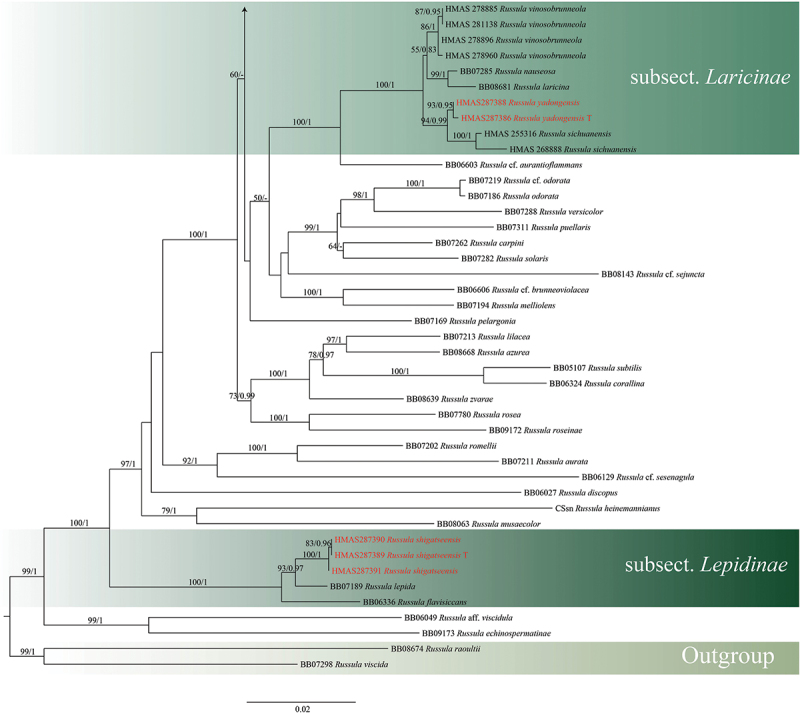
(Continued).
Figure 1.
Maximum likelihood tree of subgen. Russula crown clade based on ITS sequences, bootstrap values higher than 70% were displayed around nodes. Accession numbers of the four species are shown in red. “T” refers to a type specimen.
Figure 2.
Maximum likelihood tree of subgen. Russula crown clade based on 4-locus (nucLSU-mtSSU-rpb2-tef1) combined sequences, bootstrap values higher than 70% were displayed around nodes. Collections of the two novel species are shown in red. “T” refers to a type specimen.
3.2. Taxonomy
Russula paragraveolens S.H. Wang, G.J. Li, R.L. Zhao & B. Cao, sp. nov., Figures 3a, 4a, 4b, 5, 6, 13a
Figure 3.

Basidiomata in the field. (a) Russula paragraveolens (HMAS281158, holotype). (b) R. pseudograveolens (HMAS287384, holotype). (c–d) R. pseudograveolens (HMAS279577). (e–f) R. pseudograveolens (HMAS279579). (g–h) R. yadongensis (HMAS287387). (i) R. yadongensis (HMAS287386, holotype). (j) R. yadongensis (HMAS287388). (k–l) R. shigatseensis (HMAS287390). (m) R. shigatseensis (HMAS287389, holotype). (n–o) R. shigatseensis (HMAS287391).
Figure 4.

Basidiospores under scanning electron microscope. (a–b) Russula paragraveolens (HMAS281158, holotype). (c–d) R. pseudograveolens (HMAS287384, holotype). (e–f) R. yadongensis (HMAS287386, holotype). (g–h) R. shigatseensis (HMAS287389, holotype).
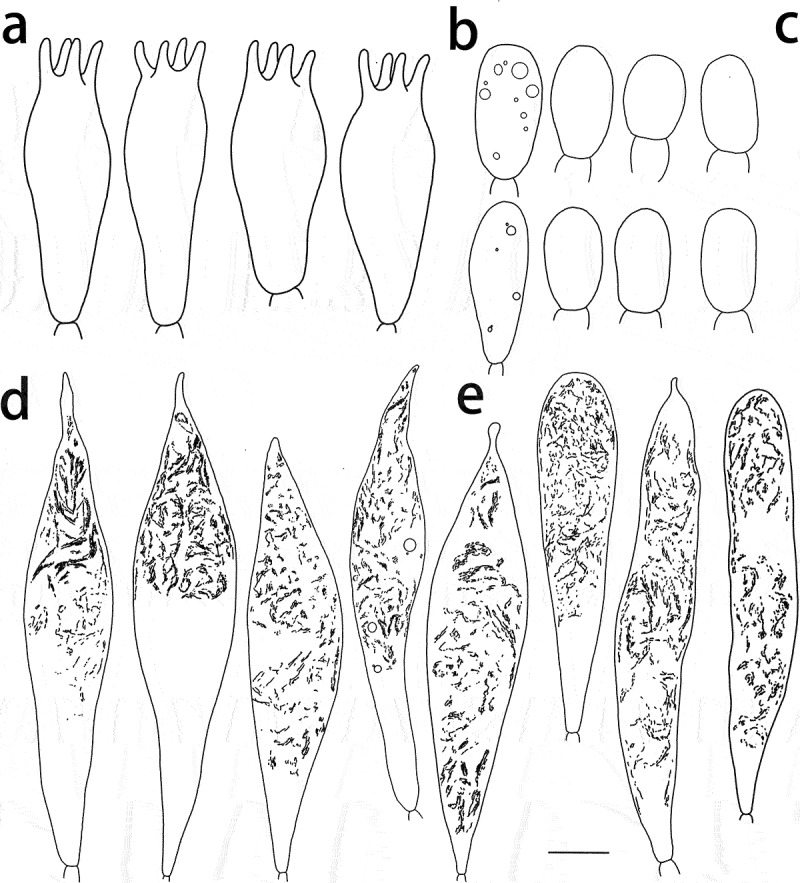
Figure 5.
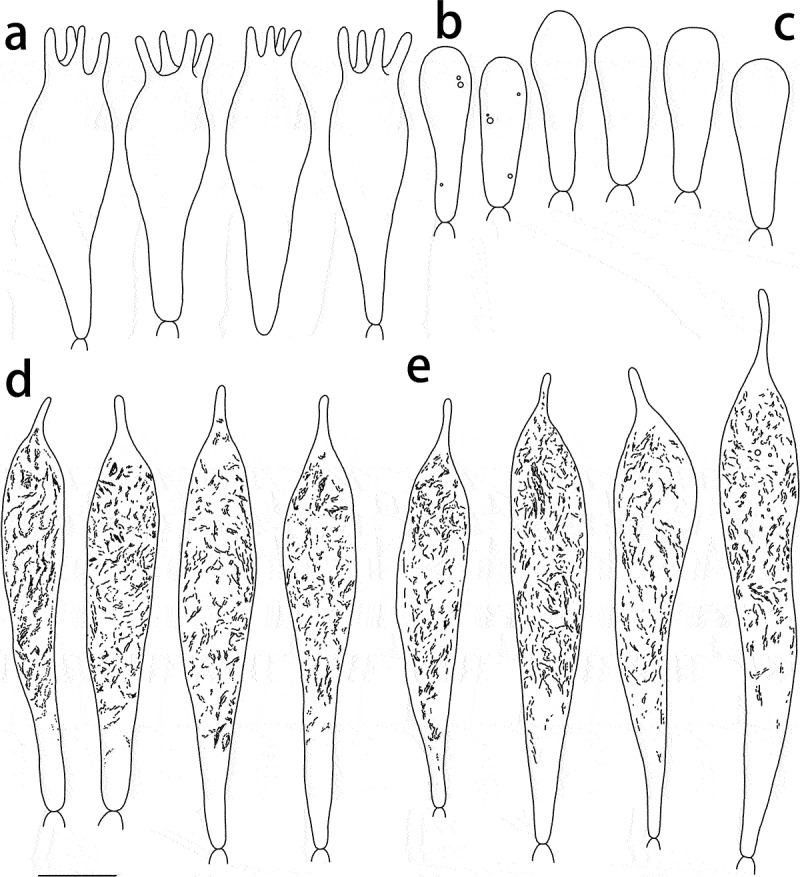
Russula paragraveolens (HMAS281158, holotype), hymenium. (a) Basidia. (b) Basidiola. (c) Marginal cells on the lamella edges. (d) Hymenial cystidia near the lamella sides. (e) Hymenial cystidia on the lamella edges. Cystidia with contents as observed in Congo red. Scale bar = 10 μm.
Figure 6.
Russula paragraveolens (HMAS281158, holotype), pileipellis. (a) Pileocystidia near the pileus centre. (b) Hyphal terminations near the pileus centre. (c) Pileocystidia near the pileus margin. (d) Hyphal terminations near the pileus margin. Cystidial contents as observed in Congo red. Scale bar = 10 μm.
Figure 13.

Spore drawings showing ornamentation in Melzer reagent. (a) Russula paragraveolens (HMAS281158, holotype). (b) R. pseudograveolens (HMAS287384, holotype). (c) R. shigatseensis (HMAS287389, holotype). (d) R. yadongensis (HMAS287386, holotype). Scale bar = 2 μm.
Fungal Names: FN571273.
Typification CHINA. Jilin Province, Yanbian Korean Autonomous Prefecture, Wangqing County, Mantianxing National Forest Park, N 43°18′ E 129°45′, 381 m asl, 22 July 2016, Ming-Zhe Zhang, Xu-Ming Bai, Rong-Chun Dai, Guo-Jie Li, ZRL20160546 (holotype HMAS281158). GenBank: OQ871504 (ITS), OQ875223 (nucLSU), OQ878262 (mtSSU), OQ933792 (rpb2), OQ948122 (tef1).
Etymology Named after its similarity to the species R. graveolens Romell.
Diagnosis Pileus medium-sized, with bright red tinge; lamellae dense, with yellowish white to pale yellow colour; stipe 25–40 × 10–17 mm, cylindrical and slightly thick near the base; spores (5.0–)5.5–5.9–6.3(−6.6) × (4.0–)4.6–5.0–5.4(−5.6) μm, broadly ellipsoid, large, with isolated or occasionally fused, prominent spines; basidia (31–)31–36–41(−50) × (10–)10–11–12(−14) μm, clavate; hymenial cystidia (42–)53–59–64(−71) × (8–)9–10(−11) μm, mainly clavate or fusiform, apically acute; hyphal terminations near the pileus margin occasionally narrow, thin-walled, terminal cells mainly cylindrical, apically obtuse or slightly constricted.
Pileus medium-sized, 40–70 mm diam., applanate with depressed centre; margin crenulate; cuticle smooth and shiny, peeling to 3/4 of the radius, bright red tinge (#AE1B0C), near the margin reddish orange (#CE4E24), towards the centre cherry red (#FE7C4D) to blood red (#6E1705), pale red (#FEB190) and pastel red (#F46733) when young, becoming cherry red (#FE7C4D) when mature. Lamellae 2–3 mm deep, adnate, dense, white (#F9F6E7) when young, becoming yellowish white (#F3F0CF) to pale yellow (#E4D39C) when mature; furcations absent, unequal with a few lamellulae of different lengths; edges concolorous and even. Stipe 25–40 × 10–17 mm, cylindrical or slightly thicker near the base, white (#F9F6E7), staining yellowish brown (#F5DD93) when bruised; medulla stuffed and becoming hollow when mature. Context 2–3 mm thick in a half of the pileus radius, pale yellow (#E4D39C) to yellow (#F3E392), yellow (#F3E392) to yellowish brown (#F5DD93) when bruised; odour fishy; taste mild. Spore print ochre.
Spores (5.0–)5.5–5.9–6.3(−6.6) × (4.0–)4.6–5.0–5.4(−5.6) μm, broadly ellipsoid, Q = (1.1–)1.13–1.18–1.23(−1.3); ornamentation of large, moderately distant [5–6(−6) in a 3 μm diam. circle] amyloid spines or warts, which are (0.7–)0.9–1.3(−1.4) μm high, isolated or fused in pairs or short chains [0–1(−2) fusions in the circle]; line connections absent or dispersed; suprahilar spot large, amyloid. Basidia (31–)31–36–41(−50) × (10–)10–11–12(−14) μm, clavate, 4-spored; basidiola first cylindrical, then clavate, 8.5–10.5 μm wide. Hymenial cystidia dispersed to moderately numerous, 300–1,100/mm2, (42–)53–59–64(−71) × (8–)9–10(−11) μm, mainly clavate or fusiform, apically acute, mucronate with a 1–9 μm long appendage, originating in subhymenium, thin-walled; contents completely heteromorphous crystalline, turning pale yellow-brown in sulfovanillin; abundant near the lamellae edges, (55–)58–62–65(−66) × (8–)9–10–11(−11) μm, similar to those on the sides. marginal cells (17–)20–23–26(−33) × (7–)8–9–10(−11) μm, undifferentiated. Pileipellis orthochromatic in Cresyl blue, sharply delimited from the underlying context, 220–450 μm deep, with a well-defined, strongly gelatinised, 70–120 μm deep suprapellis composed of ascending to erect hyphal terminations; subpellis 130–360 μm deep, composed of horizontally oriented, dense, intricate and narrow hyphae. Acid-resistant incrustations absent. Hyphal terminations near the pileus margin occasionally narrow, thin-walled; terminal cells (25–)33–47–60(−67) × (3–)3–4–5(−5) μm, cylindrical, apically obtuse or slightly constricted; subterminal cells usually equally long and wide, but often also shorter and wider, 1–2 μm wide. Hyphal terminations near the pileus centre similar, terminal cells even narrower, (38–)40–46–51(−58) × (3–)3–4(−4) μm; subterminal cells unbranched and embedded in intricate hyphae of the subpellis. Pileocystidia near the pileus margin very abundant, typically 1-celled, sometimes 2-celled, usually clavate, occasionally slightly flexuous, thin-walled, terminal cells variable in length, (59–)61–64–68(−72) × (5–)6–7–9(−10) μm, mostly subcylindrical or narrowly clavate, apically mainly obtuse, occasionally subacute, contents heteromorphous, usually dense and crystalline-granulose, turning grey-brown to black in sulfovanillin. Pileocystidia near the pileus centre slightly smaller; terminal cells (46–)50–60–70(−77) × (5–)5–6–8(−8) μm, mostly subclavate, cylindrical or fusiform, apically obtuse but occasionally also subacute to constricted. Cystidioid hyphae in subpellis and context dispersed, with heteromorphous granulose contents, oleiferous hyphae frequent in the lower part of subpellis and context.
Habit and habitat Solitary on soil in secondary broad-leaved forest (dominated by Quercus mongolica) at 300–500 m.
Other specimens examined CHINA. Jilin Province, Yanbian Korean Autonomous Prefecture, Wangqing County, Mantianxing National Forest Park, N 43°18′ E 129°45′, 381 m asl, 22 July 2016, Ming-Zhe Zhang, Xu-Ming Bai, Rong-Chun Dai, Guo-Jie Li, ZRL20162647 (HMAS279575); Jilin Province, Yanbian Korean Autonomous Prefecture, Wangqing County, Mantianxing National Forest Park, N 43°18′ E 129°45′, 381 m asl, 21 July 2016, Ming-Zhe Zhang, Xu-Ming Bai, Rong-Chun Dai, Guo-Jie Li, ZRL20162503 (HMAS279574).
Notes: According to the ITS phylogenetic tree (Figure 1), R. paragraveolens is phylogenetically related to R. amoenipes and R. graveolens. However, there are clear morphological differences, with R. paragraveolens having smaller spores and a brighter red pileus colour (Romagnesi 1967, 1985; Sarnari 1998; Sarnari and Redeuilh 2005). The new species R. paragraveolens is sister to R. pseudograveolens, both species belong to subsect. Xerampelinae of sect. Polychromae, but is morphologically distinct. Russula pseudograveolens is clearly distinguished from R. paragraveolens by having shorter and slender basidia [basidia of R. pseudograveolens (27.0–)28.5–30.4–32.3(−35.1) × (8.5–)9.1–9.7–10.2(−10.7) μm], longer projections of hymenial cystidia on lamellae sides [hymenial cystidia of R. pseudograveolens (41.3–)43.4–49.6–55.8(−64.1) × (5.4–)7.8–9.2–10.6(−10.3) μm], more lateral branches of hyphal terminations in pileipellis.
Russula pseudograveolens S.H. Wang, G.J. Li, R.L. Zhao & B. Cao, sp. nov., Figures 3b–f, 4 c–d, 7, 8, 13b
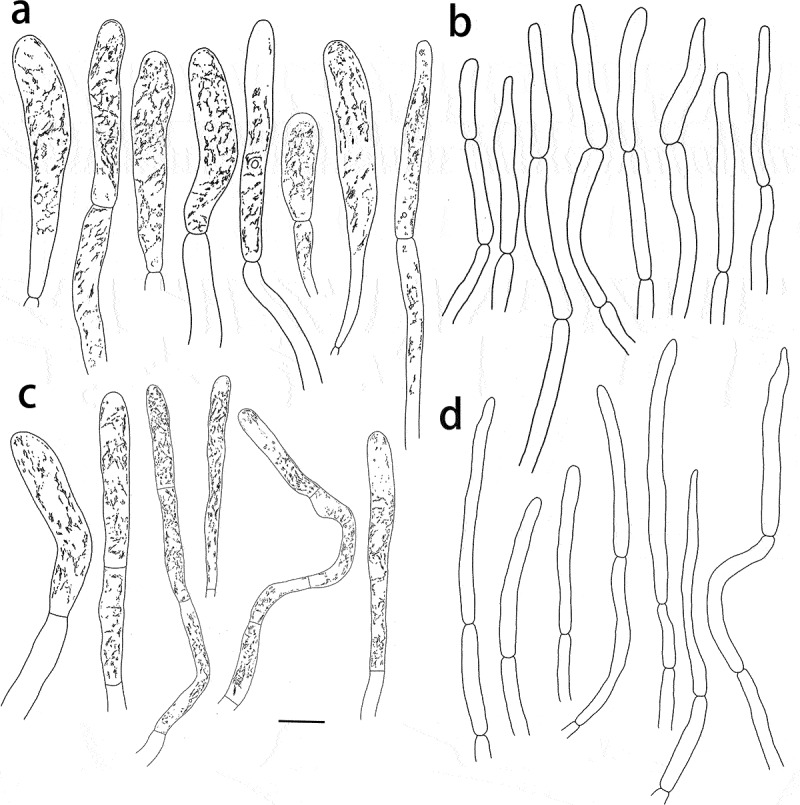
Figure 7.
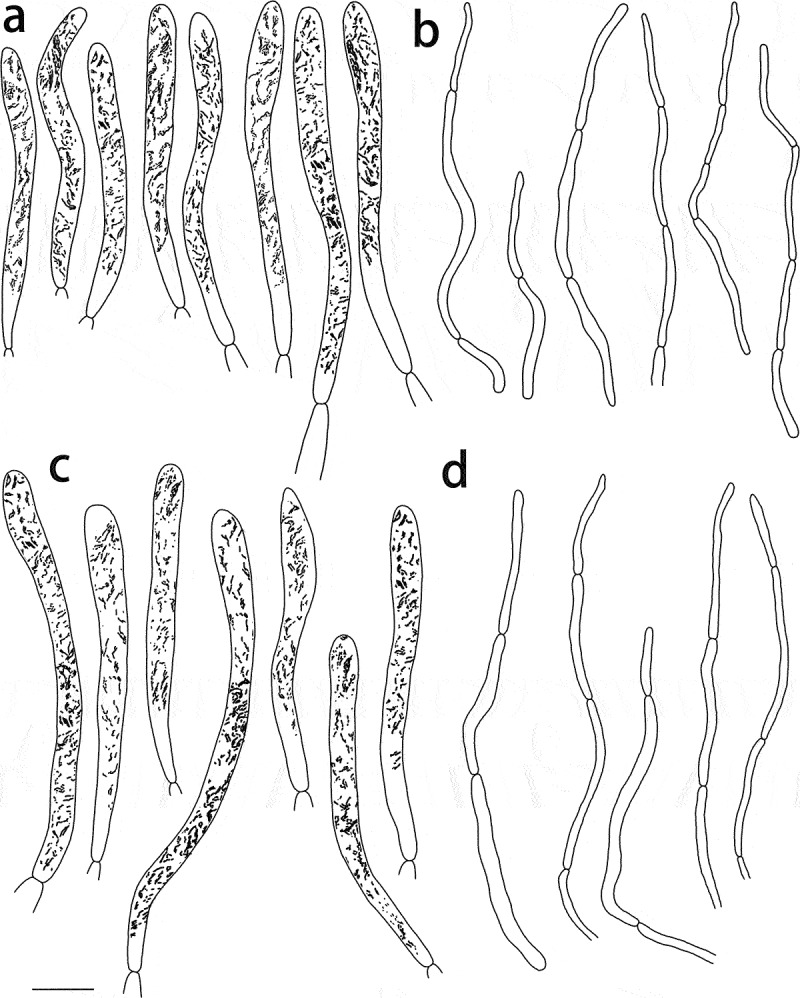
Russula pseudograveolens (HMAS287384, holotype), hymenium. (a) Basidia. (b) Basidiola. (c) Marginal cells on the lamella edges. (d) Hymenial cystidia near the lamella sides. (e) Hymenial cystidia on the lamella edges. Cystidia with contents as observed in Congo Red. Scale bar = 10 μm.
Figure 8.
Russula pseudograveolens (HMAS287384, holotype), pileipellis. (a) Pileocystidia near the pileus centre. (b) Hyphal terminations near the pileus centre. (c) Pileocystidia near the pileus margin. (d) Hyphal terminations near the pileus margin. Cystidial contents as observed in Congo Red. Scale bar = 10 μm.
Fungal Names: FN571275.
Typification CHINA. Chongqing City, Chengkou County, Daba Mountain, Beiping Mountain, N 32°0′ E 108°44′, 1,570 m asl, 14 September 2021, Xin-Yu Zhu, Ming-Zhe Zhang, Yang Liu, Chen-Hao Li, ZRL20211703 (holotype HMAS287384). GenBank: OQ871496 (ITS), OQ875225 (nucLSU), OQ878264 (mtSSU), OQ933795 (rpb2), OQ948119 (tef1).
Etymology Named after its similarity to the species R. graveolens.
Diagnosis Pileus medium-sized, with violet brown to brownish red colour; lamellae with yellowish white to pale cream colour; stipe 22–24 × 8–10 mm, cylindrical, mainly pale brown; spores (5.5–)6.0–6.3–6.6(−6.9) × (4.7–)5.0–5.2–5.5(−5.7) μm, broadly ellipsoid, large; basidia (27–)28–30–32(−35) × (8.5–)9–10(−11) μm, clavate; hymenial cystidia (41–)43–50–56(−64) × (5–)8–9–11(−11) μm, mainly clavate or fusiform, apically acute; hyphal terminations near the pileus margin occasionally narrow, thin-walled, terminal cells mainly cylindrical, apically obtuse or slightly constricted. subterminal cells branched and embedded in intricate hyphae of the subpellis.
Pileus medium-sized, 40–47 mm diam., hemispherical when young, becoming applanate when mature, margin undulate, typically cracking when mature; near the centre darker and more brown (#56464F), violet brown (#6B5D6E) to brownish red (#623C47), near the margin pale red (#BD8791) to pastel red (#905361), cuticle dry, matt, minutely pruinose or encrusted with granulose tufts over entire surface which are darker than the background. Lamellae 2 mm deep, pale yellow (#C9CEC8), yellowish white (#F6FCFF) when young, yellowish white (#F6FCFF) to pale cream (#D0D3C8), adnate, dense, lamellulae and furcations absent; edges concolorous and even. Stipe 22–24 × 8–10 mm, cylindrical, mainly pale brown (#777671), near the lamellae yellowish white (#F6FCFF) to white (#C1CEED), medulla stuffed and becoming hollow when mature. Context 1 mm thick in a half of the pileus radius, yellowish white (#F6FCFF) to cream (#CDCDC3), yellow (#D2D5CA) to yellowish brown (#A29D83) when bruised; no distinct odour first, somewhat fishy when dry; taste mild. Spore print not observed.
Spores (5.5–)6.0–6.3–6.6(−6.9) × (4.7–)5.0–5.2–5.5(−5.7) μm, broadly ellipsoid, Q = (1.1–)1.18–1.20–1.23(−1.3); ornamentation of large, dense [7–8(−8) in a 3 μm diam. circle] amyloid spines or warts, which are (0.6–)0.7–0.9(−1.0) μm high, isolated or fused in pairs or short chains [0–1(−2) fusions in the circle]; line connections absent or dispersed; suprahilar spot large, amyloid. Basidia (27–)28–30–32(−35) × (8.5–)9–10(−11) μm, clavate, 4-spored; basidiola first cylindrical, then clavate, 6.8–8.6 μm wide. Hymenial cystidia dispersed to moderately numerous, 300–1,100/mm2, (41–)43–50–56(−64) × (5–)8–9–11(−11) μm, mainly clavate or fusiform, apically acute, mucronate with a 6–10 μm long appendage, originating in subhymenium, thin-walled; contents completely heteromorphous crystalline, turning pale yellow-brown or pale greyish brown in sulfovanillin; abundant near the lamellae edges, (42–)42–48–54(−63) × (6–)6–7(−8) μm, similar to those on the sides but usually smaller. Marginal cells (13–)13–15–18(−19) × (5–)5–6–7(−8) μm, undifferentiated. Pileipellis orthochromatic in Cresyl blue, sharply delimited from the underlying context, 200–320 μm deep, with a well-defined, strongly gelatinised, 80–120 μm deep suprapellis composed of ascending to erect hyphal terminations; subpellis 110–200 μm deep, composed of horizontally oriented, dense, intricate and narrow hyphae. Acid-resistant incrustations absent. Hyphal terminations near the pileus margin occasionally narrow, thin-walled; terminal cells (36–)37–53–69(−83) × (1.6–)2.0–2.4–2.9(−3.0) μm, cylindrical, apically obtuse or slightly constricted; subterminal cells usually equally long and wide, but often also shorter and wider, 1–2 μm wide, branched or not. Hyphal terminations near the pileus centre similar, terminal cells even narrower, (28–)32–37–42(−43) × (1.5–)1.9–2.7–3.4(−3.7) μm; subterminal cells branched and embedded in intricate hyphae of the subpellis. Pileocystidia near the pileus margin very abundant, typically 1-celled, sometimes 2-celled, usually clavate, occasionally slightly flexuous, thin-walled, terminal cells variable in length, (48–)49–54–59(−60) × (4.2–)4.7–5.3–5.9(−5.9) μm, mostly subcylindrical or narrowly clavate, apically mainly obtuse, occasionally subacute, contents heteromorphous, usually dense and crystalline-granulose, turning grey-brown to black in sulfovanillin. Pileocystidia near the pileus centre slightly smaller; terminal cells (43–)47–51–55(−58) × (3.4–)4.0–4.8–5.6(−5.7) μm, mostly subclavate, cylindrical or fusiform, apically obtuse but occasionally also subacute to constricted. Cystidioid hyphae in subpellis and context dispersed, with heteromorphous granulose contents, oleiferous hyphae frequent in the lower part of subpellis and context.
Habit and habitat Solitary on soil in mixed coniferous and broad-leaved forest (dominated by Fagaceae spp. of Castanopsis, Lithocarpus, and Quercus, intermixed with Pinus yunnanensis var. tenuifolia) at 500–2,200 m.
Other specimens examined CHINA. Chongqing City, Chengkou County, Daba Mountain, Beiping Mountain, N 32°0′ E 108°44′, 1,570 m asl, 14 September 2021, Xin-Yu Zhu, Ming-Zhe Zhang, Yang Liu, Chen-Hao Li, ZRL20211685 (HMAS287385); Guangxi Zhuang Autonomous Region, Baise City, Leye County, Yachang Orchid National Nature Reserve, N 24°44′ E 106°19′, 1,211 m asl, 6 August 2017, Rui-Lin Zhao, GX20170438 (HMAS279577); Guangxi Zhuang Autonomous Region, Baise City, Leye County, Yachang Orchid National Nature Reserve, N 24°44′ E 106°19′, 1,211 m asl, 6 August 2017, Hui-Jun Wang, GX20170498 (HMAS279579).
Notes: According to the ITS phylogenetic tree (Figure 1), R. pseudograveolens is phylogenetically related to R. graveolens. However, there are clear morphological differences, with R. pseudograveolens having smaller spores, shorter basidia, and a brighter red pileus colour (Romagnesi 1967, 1985; Sarnari 1998; Sarnari and Redeuilh 2005). The new species R. pseudograveolens is sister to R. paragraveolens. However, differences not only in morphology, but also in host plants and geographical distribution. Russula paragraveolens grows solitarily on the ground in secondary broadleaved forest (dominated by Quercus mongolica) at 300–500 m, while R. pseudograveolens grows at higher altitudes and has more complex host plants at 500–2,200 m. Furthermore, R. paragraveolens and R. pseudograveolens belong to different temperature zones, the former being a temperate species and the latter a subtropical species.
Russula shigatseensis S.H. Wang, R.L. Zhao & B. Cao, sp. nov., Figures 3k–o, 4g–h, 9, 10, 13c
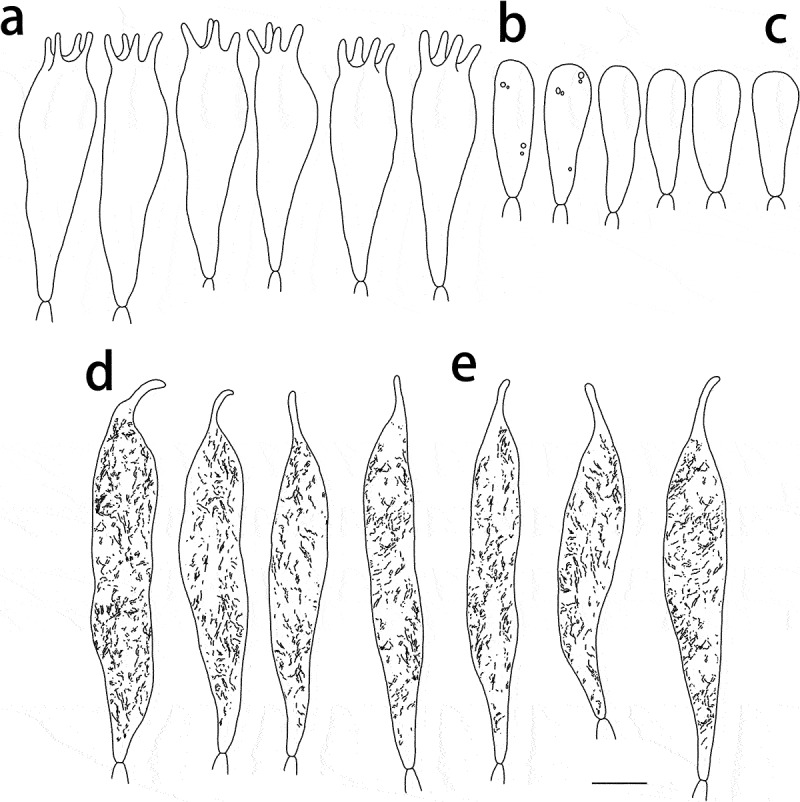
Figure 9.
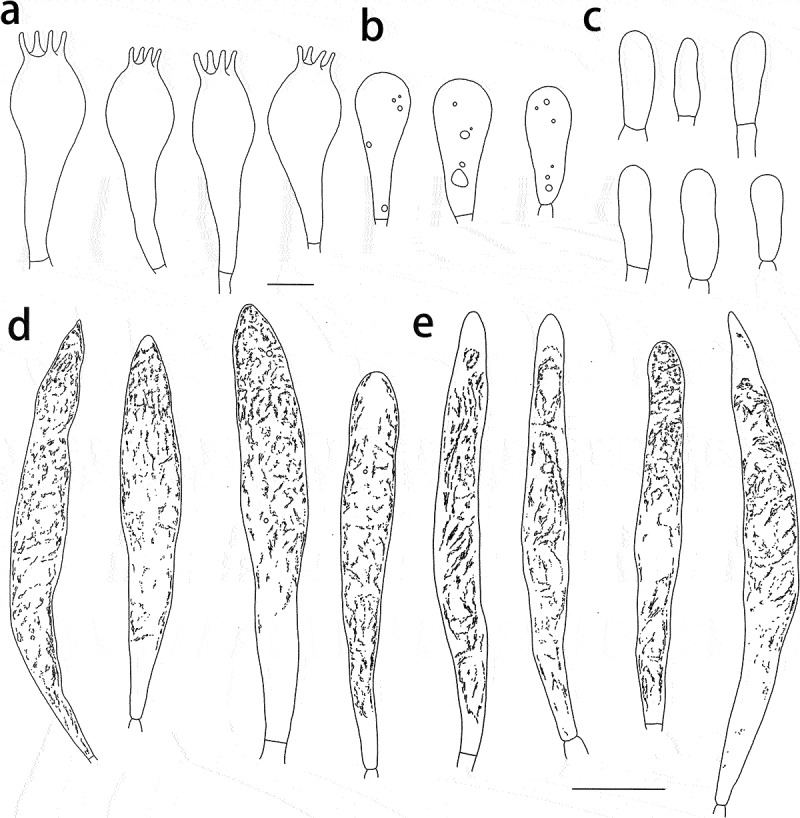
Russula shigatseensis (HMAS287389, holotype), hymenium. (a) Basidia. (b) Basidiola. (c) Marginal cells on the lamella edges. (d) Hymenial cystidia near the lamella sides. (e) Hymenial cystidia on the lamella edges. Cystidia with contents as observed in Congo Red. Scale bar = 10 μm.
Figure 10.
Russula shigatseensis (HMAS287389, holotype), pileipellis. (a) Pileocystidia near the pileus centre. (b) Hyphal terminations near the pileus centre. (c) Pileocystidia near the pileus margin. (d) Hyphal terminations near the pileus margin. Cystidial contents as observed in Congo Red. Scale bar = 10 μm.
Fungal Names: FN571577.
Typification CHINA. Xizang Autonomous Region, Shigatse Municipality, Yadong County, Lower Yadong Township, N 27°25′ E 88°56′, 3,024 m asl, 26 July 2022, Rui-Lin Zhao, Xin-Yu Zhu, Jia-Xin Li, ZRL20220194 (holotype HMAS287389). GenBank: OQ871501 (ITS), OQ875230 (nucLSU), OQ878269 (mtSSU), OQ933787 (rpb2), OQ948114 (tef1).
Etymology Refers to Shigatse Municipality, the locality of the type specimen.
Diagnosis Pileus medium-sized, with brownish orange to madder red colour; lamellae with yellowish white to pale cream colour; stipe 45–104 × 12–18 mm, cylindrical and slightly thick near the base; spores (5.6–)6.0–6.4–6.9(−7.5) × (4.8–)5.2–5.7–6.2(−6.7) μm, subglobose, medium-sized; basidia (24–)31–35–39(−39) × (8–)10–12–13(−13) μm, clavate; hymenial cystidia (64–)69–78–87(−97) × (5–)6–7–8(−8) μm, mainly clavate or fusiform, apically acute; hyphal terminations near the pileus margin occasionally narrow, thin-walled, terminal cells mainly cylindrical, apically obtuse or slightly constricted. subterminal cells branched and embedded in intricate hyphae of the subpellis.
Pileus medium-sized, 59–80 mm diam., hemispherical when young, becoming applanate with depressed centre when mature, applanate with depressed centre; margin smooth; cuticle dry to viscid, smooth, peeling to 1/2 of the radius, brownish orange (#B66747) to madder red (#C43925) in the centre, pinkish red (#E68A99) towards the margin. Lamellae 1–7 mm deep, yellowish white (#A19D87) to white (#CED0CD) when young, yellowish white (#A19D87) to pale cream (#CAD0C9), adnate, dense, lamellulae and furcations absent; edges concolorous and even. Stipe 45–104 × 12–18 mm, cylindrical and slightly thick near the base, white (#CED0CD), often with pinkish flush or pink areas, medulla stuffed and becoming hollow when mature. Context 1–2 mm thick in a half of the pileus radius, yellowish white (#A19D87) to cream (#C1BDBA). Spore print not observed
Spores (5.6–)6.0–6.4–6.9(−7.5) × (4.8–)5.2–5.7–6.2(−6.7) μm, subglobose, Q = (1.0–)1.0–1.13–1.18(−1.3); ornamentation of medium-sized, moderately distant to dense [6–8 (−12) in a 3 μm diam. circle] amyloid spines or warts, which are (0.5–)0.5–0.7(−0.8) μm high, isolated or fused in pairs or short chains [0–1(−2) fusions in the circle]; line connections absent or dispersed; suprahilar spot large, amyloid. Basidia (24–)31–35–39(−39) × (8–)10–12–13(−13) μm, clavate, 4-spored; basidiola first cylindrical, then clavate, 7.0–10.7 μm wide. Hymenial cystidia dispersed to moderately numerous, 300–1,100/mm2, (64–)69–78–87(−97) × (5–)6–7–8(−8) μm, mainly clavate or fusiform, apically acute, mucronate with a 0–5 μm long appendage, originating in subhymenium, thin-walled; contents completely heteromorphous crystalline, turning grey-brown to black in sulfovanillin; abundant near the lamellae edges, (49–)50–58–65(−77) × (3–)5–6–7(−8) μm, similar to those on the sides but usually smaller. Marginal cells (9–)8–12–16(−21) × (4–)3–5–6(−9) μm, undifferentiated. Pileipellis orthochromatic in Cresyl blue, sharply delimited from the underlying context, 140–330 μm deep, with a well-defined, strongly gelatinised, 50–150 μm deep suprapellis composed of ascending to erect hyphal terminations; subpellis 90–180 μm deep, composed of horizontally oriented, dense, intricate and narrow hyphae. Acid-resistant incrustations absent. Hyphal terminations near the pileus margin occasionally narrow, thin-walled; terminal cells (44–)57–86–116(−142) × (2.5–)3–4–5(−5) μm, cylindrical, apically obtuse or slightly constricted; subterminal cells usually equally long and wide, but often also shorter and wider, 1–2 μm wide, branched or not. Hyphal terminations near the pileus centre similar, terminal cells even narrower, (26–)29–38–46(−56) × (1.0–)1.3–1.9–2.5(−3.2) μm; subterminal cells branched and embedded in intricate hyphae of the subpellis. Pileocystidia near the pileus margin very abundant, typically 1–2-celled, seldom 3–4-celled, usually clavate, frequently slightly flexuous, thin-walled, terminal cells variable in length, (36–)40–46–52(−55) × (2.5–)2.9–3.4–4.0(−4.4) μm, mostly subcylindrical or narrowly clavate, apically mainly obtuse, occasionally subacute, contents heteromorphous, usually dense and crystalline-granulose, turning pale yellow-brown or pale greyish brown in sulfovanillin. Pileocystidia near the pileus centre slightly smaller; terminal cells (25–)28–35–41(−46) × (2.5–)2.7–3.2–3.7(−3.8) μm, mostly subclavate, cylindrical or fusiform, apically obtuse but occasionally also subacute to constricted. Cystidioid hyphae in subpellis and context dispersed, with heteromorphous granulose contents, oleiferous hyphae frequent in the lower part of subpellis and context.
Habit and habitat: Scattered in coniferous forests.
Other specimens examined CHINA. Xizang Autonomous Region, Shigatse Municipality, Yadong County, Lower Yadong Township, N 27°25′ E 88°56′, 3,024 m asl, 26 July 2022, Mao-Qiang He, Bin Cao, ZRL20220072 (HMAS287390); Xizang Autonomous Region, Shigatse Municipality, Yadong County, Lower Yadong Township, N 27°25′ E 88°56′, 3,024 m asl, 26 July 2022, Mao-Qiang He, Bin Cao, ZRL20220207 (HMAS287391).
Notes: According to the ITS and multigene phylogeny, R. shigatseensis belonging to sect. Flavisiccantes, subsect. Lepidinae, is represented in a well-supported clade (BS = 100%) by three specimens from the Shigatse region of Xizang, China. The species is closely related to R. lepida Fr., but R. shigatseensis has smaller spores [(5.6–)6.0–6.4–6.9(−7.5) × (4.8–)5.2–5.7–6.2(−6.7) μm]. Compared to R. lepida [2–9 × 4–7 μm], the spores of R. shigatseensis have spore ornamentations of more or less isolated spines (Sarnari and Redeuilh 2005). R. flavisiccans Bills also appeared close to the present species in multigene phylogeny but the former differs from newly proposed species in having adnate to notched, forked, or anastomosing lamellae with scattered lamellulae, larger spores [7–8.5(−9) × 6.5–8(−8.5) μm] (Bills 1989).
Russula yadongensis S.H. Wang, R.L. Zhao & B. Cao, sp. nov., Figures 3g–j, 4e–f, 11, 12, 13d
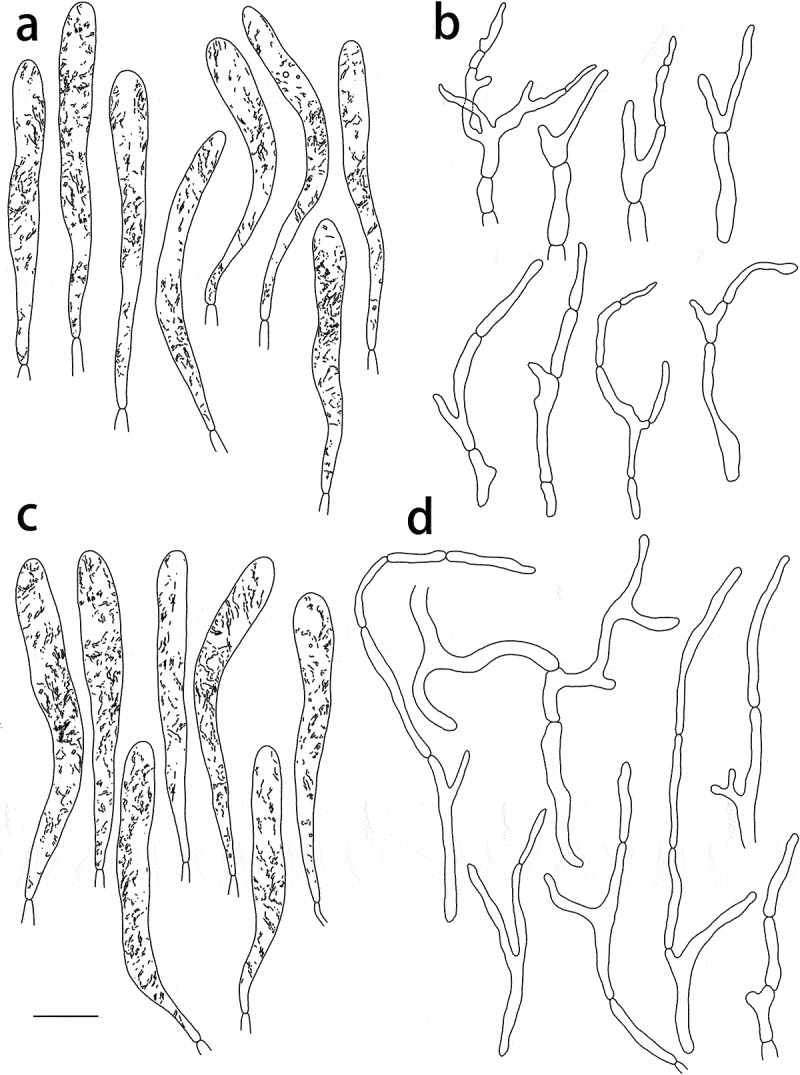
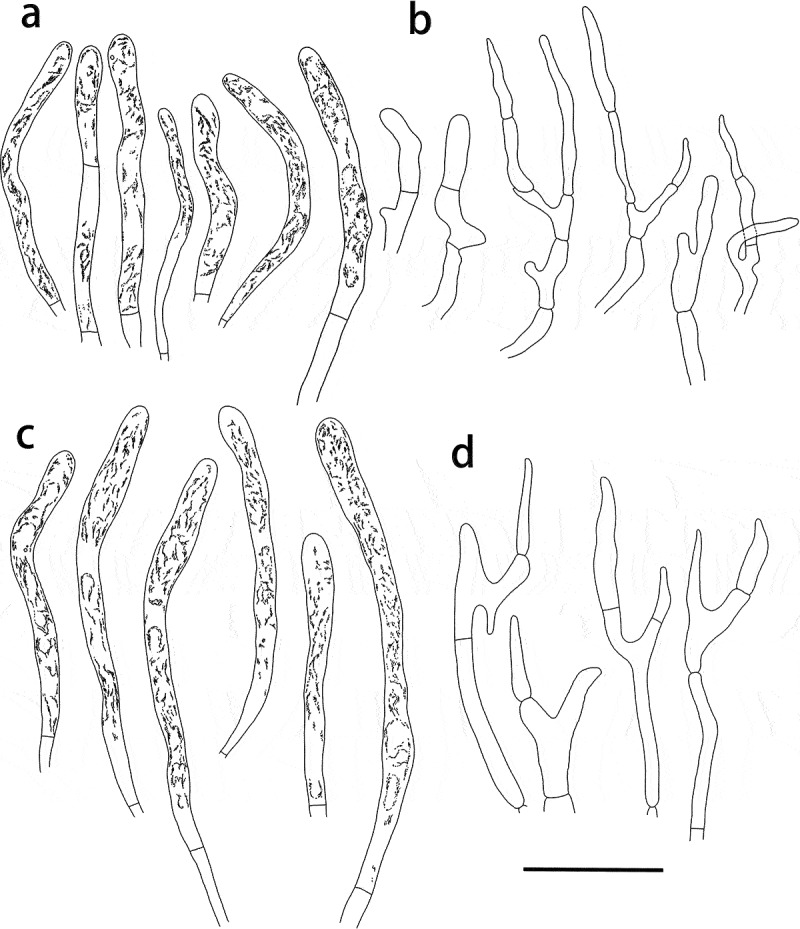
Figure 11.
Russula yadongensis (HMAS287386, holotype), hymenium. (a) Basidia. (b) Basidiola. (c) Marginal cells on the lamella edges. (d) Hymenial cystidia near the lamella sides. (e) Hymenial cystidia on the lamella edges. Cystidia with contents as observed in Congo Red. Scale bar = 10 μm.
Figure 12.
Russula yadongensis (HMAS287386, holotype), pileipellis. (a) Pileocystidia near the pileus centre. (b) Hyphal terminations near the pileus centre. (c) Pileocystidia near the pileus margin. (d) Hyphal terminations near the pileus margin. Cystidial contents as observed in Congo Red. Scale bar = 10 μm.
Fungal Names: FN571578.
Typification CHINA. Xizang Autonomous Region, Shigatse Municipality, Yadong County, Lower Yadong Township, N 27°25′ E 88°56′, 26 July 2022, 3,024 m asl, Mao-Qiang He, Bin Cao, ZRL20220204 (holotype HMAS287386). GenBank: OQ871498 (ITS), OQ875228 (nucLSU), OQ878267 (mtSSU), OQ933790 (rpb2), OQ948117 (tef1).
Etymology Refers to Yadong County, the locality of the type specimen.
Diagnosis Pileus medium-sized, light purple to purple, dark purplish red with brownish purple tints in the centre; lamellae with yellowish white to pale yellow colour; stipe 37–66 × 5–15 mm, cylindrical and slightly thick near the base; spores (6.0–)6.3–6.6–7.0(−7.2) × (4.5–)5.0–5.4–5.8(−6.2) μm, broadly ellipsoid, large; basidia (21–)22–24–26(−28) × (10–)10–11(−11) μm, clavate; hymenial cystidia (40–)35–41–46(−58) × (7–)7–8–9(−11) μm, mainly clavate or fusiform, apically acute; Hyphal terminations near the pileus margin occasionally narrow, thin-walled; terminal cells cylindrical, apically obtuse or slightly constricted; subterminal cells unbranched.
Pileus medium-sized, 32–46 mm diam., applanate with depressed centre; margin smooth or slightly striate; cuticle smooth and shiny, peeling to 1/3 of the radius, light purple (#CAB5B1) when young, purple (#996F7F) when mature, dark purplish red (#7B5E6A) with brownish purple tints in the centre. Lamellae 2–4 mm deep, adnate to free, dense, white (#D0D6E7) when young, becoming yellowish white (#D1DCD8) to pale yellow (#EBF6FA) when mature; lamellulae and furcations absent; edges concolorous and even. Stipe 37–66 × 5–15 mm, cylindrical and slightly thick near the base, white (#D0D6E7); medulla stuffed and becoming hollow when mature. Context 1–2 mm thick in a half of the pileus radius, white (#D0D6E7), yellowish white (#D1DCD8) to cream (#D7E0DF) when mature. Spore print not observed.Spores (6.0–)6.3–6.6–7.0(−7.2) × (4.5–)5.0–5.4–5.8(−6.2) μm, broadly ellipsoid, Q = (1.1–)1.15–1.23–1.3(−1.4); ornamentation of large, moderately distant [5–6(−7) in a 3 μm diam. circle] amyloid spines or warts, which are (0.7–)0.9–1.2(−1.3) μm high, isolated or fused in pairs or short chains [0–1(−2) fusions in the circle]; line connections absent or dispersed; suprahilar spot large, amyloid. Basidia (21–)22–24–26(−28) × (10–)10–11(−11) μm, clavate, 4-spored; basidiola first cylindrical, then clavate, 7.2–9.1 μm wide. Hymenial cystidia dispersed to moderately numerous, 300–1,100/mm2, (40–)35–41–46(−58) × (7–)7–8–9(−11) μm, mainly clavate or fusiform, apically acute, mucronate with a 0–6 μm long appendage, originating in subhymenium, thin-walled; contents completely heteromorphous crystalline, turning pale yellow-brown or pale greyish brown in sulfovanillin; abundant near the lamellae edges, (30–)44–53–61(−58) × (5–)7–9–10(−11) μm, similar to those on the sides. Marginal cells (9–)12–15–17(−18) × (6–)6–8–9(−10) μm, undifferentiated. Pileipellis orthochromatic in Cresyl blue, sharply delimited from the underlying context, 160–360 μm deep, with a well-defined, strongly gelatinised, 60–140 μm deep suprapellis composed of ascending to erect hyphal terminations; subpellis 100–220 μm deep, composed of horizontally oriented, dense, intricate and narrow hyphae. Acid-resistant incrustations absent. Hyphal terminations near the pileus margin occasionally narrow, thin-walled; terminal cells (26–)25–41–56(−81) × (2–)2–3(−3) μm, cylindrical, apically obtuse or slightly constricted; subterminal cells usually equally long and wide, but often also shorter and wider, 1–2 μm wide. Hyphal terminations near the pileus centre similar, terminal cells even narrower, (25–)27–34–40(−45) × (1.2–)1.5–2.0–2.4(−2.6) μm; subterminal cells unbranched and embedded in intricate hyphae of the subpellis. Pileocystidia near the pileus margin very abundant, typically 1–2-celled, sometimes 2–3-celled, usually clavate, occasionally slightly flexuous, thin-walled, terminal cells variable in length, (31–)33–56–78(−103) × (3.5–)3.7–4.9–6.2(−7.8) μm, mostly subcylindrical or narrowly clavate, apically mainly obtuse, occasionally subacute, contents heteromorphous, usually dense and crystalline-granulose, turning grey-brown to black in sulfovanillin. Pileocystidia near the pileus centre slightly smaller; terminal cells (30–)33–44–55(−63) × (2.6–)3.7–4.6–5.6(−6.5) μm, mostly subclavate, cylindrical or fusiform, apically obtuse but occasionally also subacute to constricted. Cystidioid hyphae in subpellis and context dispersed, with heteromorphous granulose contents, oleiferous hyphae frequent in the lower part of subpellis and context.
Habit and habitat: Scattered in coniferous forests.
Other specimens examined CHINA. Xizang Autonomous Region, Shigatse Municipality, Yadong County, Lower Yadong Township, N 27°22′ E 88°58′, 2,872 m asl, 26 July 2022, Rui-Lin Zhao, Xin-Yu Zhu, Jia-Xin Li, ZRL20220152 (HMAS287387); Xizang Autonomous Region, Shigatse Municipality, Yadong County, Lower Yadong Township, Yanqinggang Village, N 27°25′ E 88°55′, 3,254 m asl, 27 July 2022, Mao-Qiang He, Bin Cao, ZRL20220377 (HMAS287388).
Notes: Russula yadongensis belonging to subsect. Laricinae of sect. Tenellae, is represented by three specimens from the Shigatse region of Xizang, China in a well-supported clade (BS = 97%), and is phylogenetically related to R. sichuanensis G.J. Li & H.A. Wen and R. vinosobrunneola G.J. Li & R.L. Zhao (Figure 1). However, there are clear morphological differences, with R. yadongensis having smaller spores [(6.0–)6.3–6.6–7.0(−7.2) × (4.5–)5.0–5.4–5.8(−6.2) μm], a deeper purple pileus colour. Compared with R. vinosobrunneola and R. sichuanensis, the spores of R. yadongensis have spore ornamentations of more or less isolated spines (Li et al. 2018a). Russula yadongensis is also closely related to R. nauseosa (Pers.) Fr. and R. laricina Fr. in the multigene phylogeny (Figure 2). Compared to the closely related species R. nauseosa, the spores of R. yadongensis are smaller [spores of R. nauseosa 7,8–10 × 6,6–7,8 μm] and the hymenial cystidia are slightly smaller [hymenial cystidia of R. nauseosa 45–80 × 9–13 μm] (Sarnari and Redeuilh 2005). Compared to the closely related species R. laricina, the spores of R. yadongensis are smaller [spores of R. laricina 6,5–9,5 × 6–8 μm] and have a deeper purple pileus colour (Sarnari and Redeuilh 2005).
4. Discussion
The ITS phylogenetic analyses are most commonly used for the practical identification of Russula species (Li et al. 2019). However, it is often difficult to distinguish between closely related species based on ITS phylogenetic analysis alone. Multi-locus phylogenetic analyses have become the preferred technique for revealing relationships within the Russula genus in recent years (Li et al. 2019; Buyck et al. 2020). In this study, the topological structure of the ITS and multi-locus phylogenetic analyses are basically similar, but the Bayesian posterior probability values and maximum likelihood bootstrap were higher in the multi-locus phylogenetic analyses.
In this study, four new species belong to three different subsections within the crown clade, namely subsect. Laricinae (R. yadongensis), subsect. Lepidinae (R. shigatseensis), subsect. Xerampelinae (R. paragraveolens and R. pseudograveolens). In China, some new species of these three subsections have previously been reported, such as R. cessans, R. faginea, R. laricina, R. lepida, R. nauseosa, R. nuoljae, R. pascua, R. sichuanensis, R. vinosobrunneola, R. xerampelina (Li 2014; Li et al. 2018b; Cao et al. 2019).
Species of subsect. Laricinae generally have a purple pileus, white stipe, and a yellow spore print, and mainly grow in coniferous forests (Romagnesi 1967). Many species of this subsection have previously been reported, namely R. adwanitekae A. Ghosh, K. Das & Buyck, R. cessans A. Pearson, R. curtipes F.H. Møller & Jul. Schäff., R. laricina, R. nauseosa, R. obscurozelleri Bazzic., D. Mill. & Buyck, R. pseudotsugarum Bazzic., D. Mill. & Buyck, R. sichuanensis, R. vidalii Trappe & T.F. Elliott, R. vinaceodora (Calonge & J.M. Vidal) Trappe & T.F. Elliott, R. vinosobrunneola, R. zelleri Burl (Ghosh et al. 2021). In this study, ITS phylogenetic analyses showed significant support (BS = 93%) for R. yadongensis with other species in this subsection, while morphological and phylogenetic results could distinguish this species well from other known species within this subsection.
Subsection Lepidinae is characterised by a velvety pileus surface, hard context, the cystidia of the pileus, and hymenium not reacting to sulfovanillin, mild taste (Sarnari and Redeuilh 2005). Many species of subsect. Lepidinae have previously been reported, namely R. amarissima Romagn. & E.-J. Gilbert, R. Baniyakundensis A. Ghosh, K. Das & D. Chakr., R. flavisiccans, R. indoarmeniaca A. Ghosh, K. Das & R.P. Bhatt, R. lepida, R. ochroleucoides Kauffman (Kauffman 1917; Bills 1989; Sarnari and Redeuilh 2005; Ghosh et al. 2016, 2021). In this study, ITS phylogenetic analyses showed significant support (BS = 100%) for R. shigatseensis with other species in this subsection, while morphological and phylogenetic results could distinguish this species well from other known species within this subsection.
Species of the subsect. Xerampelinae can be easily recognised in field by following characters: tardily fishy context smell, mild taste, context slowly turning brownish when bruised or old, lamellae turning red in aniline, absence of acid-resistant incrustation on pileocystidia which turn grey in sulphovanillin (Adamczík and Marhold 2000; Adamcik 2002, 2003; Adamčík 2004; Adamčik and Knudsen 2004; Sarnari and Redeuilh 2005; Buyck and Adamcík 2013; Adamčík et al. 2016). Many species in this subsection have previously been reported, such as R. faginea Romagn., R. favrei M.M. Moser, R. clavipes Velen., R. graveolens, R. nuoljae Kühner, R. pascua (F.H. Møller & Jul. Schäff.) Kühner, R. subrubens (J.E. Lange) Bon, and R. xerampelina (Schaeff.) Fr (Adamčík et al. 2016). In this study, ITS phylogenetic analyses showed significant support (BS = 97%) for R. paragraveolens and R. pseudograveolens with other species in this subsection, while morphological and phylogenetic results could distinguish this species well from other known species within this subsection.
Acknowledgments
We thank Chun-Rong Dai, Hui-Jun Wang, Xu-Ming Bai, Yan-Lei Ding, Zhi-Lin Ling, Hu-Sheng Ma, Guang-Fu Mou, Chen-Hao Li, and Yang Liu for assistance in specimen collecting.
Funding Statement
This project was supported by the Survey of Wildlife Resources in Key Areas of Tibet (ZL202203601), the National Natural Science Foundation of China (31961143010, 31500013, 30770013), the Beijing Innovation Consortium of Agriculture Research System (BAIC03-01), the Talent Introduction Scientific Research Special Project of Hebei Agricultural University (YJ201849), and the Projects of Science and Technology Programs of Tibet (XZ202202YD0031C).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Adamcik S. 2002. Taxonomy of the Russula xerampelina group. Part 2. Taxonomic and nomenclatural study of Russula xerampelina and R. erythropoda. Mycotaxon. 82:241–267. [Google Scholar]
- Adamcik S. 2003. Russula faginea and similar taxa. Czech Mycol. 55:177–192. doi: 10.33585/cmy.54306. [DOI] [Google Scholar]
- Adamčík S. 2004. Studies on Russula lavipes and related taxa of Russula section Xerampelinae with a predominantly olivaceous pileus. Persoonia. 18:393–409. [Google Scholar]
- Adamčik S, Knudsen H. 2004. Red-capped species of Russula sect. Xerampelinae associated with dwarf scrub. Xerampelinae Assoc Dwarf Scrub Mycol Res. 108(12):1463–1475. doi: 10.1017/S0953756204000875. [DOI] [PubMed] [Google Scholar]
- Adamčík S, Looney B, Caboň M, Jančovičová S, Adamčíková K, Avis PG, Barajas M, Bhatt RP, Corrales A, Das K. 2019. The quest for a globally comprehensible Russula language. Fungal Divers. 99:369–449. doi: 10.1007/s13225-019-00437-2. [DOI] [Google Scholar]
- Adamčík S, Slovák M, Eberhardt U, Ronikier A, Jairus T, Hampe F, Verbeken A. 2016. Molecular inference, multivariate morphometrics and ecological assessment are applied in concert to delimit species in the Russula clavipes complex. Mycologia. 108:716–730. doi: 10.3852/15-194. [DOI] [PubMed] [Google Scholar]
- Adamczík S, Marhold K. 2000. Taxonomy of the Russula xerampelina group I. Morphometric study of the Russula xerampelina group in Slovakia. Mycotaxon. 76:463–479. [Google Scholar]
- Andrew R 2016. Tree drawing tool version 1.4.3. Edinburgh, UK: Institute of Evolutionary Biology, University of Edinburgh. http://tree.bio.ed.ac.uk/software/figtree/. [Google Scholar]
- Bills GF. 1989. Southern Appalachian Russulas. IV. Mycologia. 81:57–65. doi: 10.1080/00275514.1989.12025626. [DOI] [Google Scholar]
- Buyck B. 1989. Valeur taxonomique du bleu de crésyl pour le genre Russula [Taxonomic value of cresyl blue for the genus Russula]. Bull Soc Mycol Fr. 105:1–6. [Google Scholar]
- Buyck B, Adamcík S. 2013. The Russula xerompelina complex Russulales, Agaricomycotina in North America. Scr Bot Belg. 51:117–131. [Google Scholar]
- Buyck B, Wang X, Adamcíková K, Cabon M, Jancovicova S, Hofstetter V, Adamcik S. 2020. One step closer to unravelling the origin of Russula: subgenus glutinosae subg. nov. Mycosphere. 11(1):285–304. doi: 10.5943/mycosphere/11/1/6. [DOI] [Google Scholar]
- Buyck B, Zoller S, Hofstetter V. 2018. Walking the thin line… ten years later: the dilemma of above- versus below-ground features to support phylogenies in the Russulaceae (Basidiomycota). Fungal Divers. 89(1):267–292. doi: 10.1007/s13225-018-0397-5. [DOI] [Google Scholar]
- Caboň M, Eberhardt U, Looney B, Hampe F, Kolařík M, Jančovičová S, Verbeken A, Adamčík S. 2017. New insights in Russula subsect. Rubrinae: phylogeny and the quest for synapomorphic characters. Mycol Prog. 16(9):877–892. doi: 10.1007/s11557-017-1322-0. [DOI] [Google Scholar]
- Caboň M, Li GJ, Saba M, Kolařík M, Jančovičová S, Khalid AN, Moreau PA, Wen HA, Pfister DH, Adamčík S. 2019. Phylogenetic study documents different speciation mechanisms within the Russula globispora lineage in boreal and arctic environments of the Northern Hemisphere. IMA Fungus. 10:1–16. doi: 10.1186/s43008-019-0003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B, Li GJ, Zhao RL. 2019. Species diversity and geographic components of Russula from the greater and Lesser Khinggan Mountains. Biodiversity Sci. 27:854. doi: 10.17520/biods.2019040. [DOI] [Google Scholar]
- Chen XJ, Zhang YJ. 2010. Polysaccharide extract from Russula and its role of lowering blood glucose and lipid. Food Sci. 31:255–258. [Google Scholar]
- Chen Z, Zhang P, Zhang Z. 2014. Investigation and analysis of 102 mushroom poisoning cases in Southern China from 1994 to 2012. Fungal Divers. 64:123–131. doi: 10.1007/s13225-013-0260-7. [DOI] [Google Scholar]
- Cho JT, Han JH. 2016. A case of mushroom poisoning with Russula subnigricans: development of rhabdomyolysis, acute kidney injury, cardiogenic shock, and death. J Korean Med Sci. 31:1164–1167. doi: 10.3346/jkms.2016.31.7.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Das K, Adhikari S, Bhatt R. 2016. A novel species of Russula (Russulaceae) from Indian Himalaya. Mycosphere. 7(6):778–785. doi: 10.5943/mycosphere/7/6/8. [DOI] [Google Scholar]
- Ghosh A, Das K, Buyck B. 2021. Two new species in the Russula (Russulaceae, Basidiomycota) crown clade from Indian Himalaya. Eur J Taxon. 782:157–172. doi: 10.5852/ejt.2021.782.1595. [DOI] [Google Scholar]
- Ghosh A, Das K, Chakraborty D. 2021. Morphology and molecular approach reveal a new species of the genus Russula subsect. Lepidinae (Russulaceae) from India. Lepidiane Russ India Phytox. 483(3):244–254. doi: 10.11646/phytotaxa.483.3.4. [DOI] [Google Scholar]
- Heilmann-Clausen J, Verbeken A, Vesterholt J, Leonard P. 2000. The genus Lactarius: book review. Field Mycol. 1(1):6–6. doi: 10.1016/S1468-1641(10)60005-9. [DOI] [Google Scholar]
- He MQ, Zhao RL, Hyde KD, Begerow D, Kemler M, Yurkov A, Mckenzie E, Raspe O, Kakishima M, Sanchez-Ramrez S. 2019. Notes, outline and divergence times of Basidiomycota. Fungal Divers. 99(1):105–367. doi: 10.1007/s13225-019-00435-4. [DOI] [Google Scholar]
- Jiang XM, Li YK, Liang JF, Wu JR. 2018. Russula brunneovinacea sp nov., from northeastern China. Mycotaxon. 132(4):789–797. doi: 10.5248/132.789. [DOI] [Google Scholar]
- Kaewnarin K, Suwannarach N, Kumla J, Lumyong S. 2016. Phenolic profile of various wild edible mushroom extracts from Thailand and their antioxidant properties, anti-tyrosinase and hyperglycaemic inhibitory activities. J Funct Foods. 27:352–364. doi: 10.1016/j.jff.2016.09.008. [DOI] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, WongThomas KF, Haeseler V, Jermiin A, Lars S. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman CH. 1917. Tennessee and Kentucky fungi. Mycologia. 9:159–166. doi: 10.1080/00275514.1917.12018913. [DOI] [Google Scholar]
- Khatua S, Gupta SS, Ghosh M, Tripathi S, Acharya K. 2021. Exploration of nutritional, antioxidative, antibacterial and anticancer status of Russula alatoreticula: towards valorization of a traditionally preferred unique myco-food. J Food Sci Technol. 58:2133–2147. doi: 10.1007/s13197-020-04723-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk PM, Cannon PF, Minter DW, Stalpers JA. 2008. Ainsworth & Bisby's Dictionary of the Fungi. 10th ed. Wallingford: CABI. [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34:772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- Li GJ. 2014. The Taxonomy of Russula in China. Beijing: University of Chinese Academy of Sciences. [Google Scholar]
- Li GJ. 2022. Russula rubiginosus sp. nov. In Russula subsect. maculatinae from Heilongjiang Province, Northeast China. Phytotaxa. 575(2):140–148. doi: 10.11646/phytotaxa.575.2.3. [DOI] [Google Scholar]
- Li GJ, Hyde KD, Zhao RL, Hongsanan S, Abdel-Aziz FA, Abdel-Wahab MA, Alvarado P, Alves-Silva G, Ammirati JF, Ariyawansa HA, et al. 2016. Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 78:1–237. doi: 10.1007/s13225-016-0366-9. [DOI] [Google Scholar]
- Li GJ, Li SF, Liu XZ, Wen HA. 2012. Russula jilinensis sp. nov. (Russulaceae) from Northeast China. Mycotaxon. 120:49–58. doi: 10.5248/120.49. [DOI] [Google Scholar]
- Li F, Li GJ, Zhang J, Gao H, Shi RS, Deng CY. 2021. Russula fanjing, a new species of Russula subsect. Russula (Russulaceae, Russulales) from Guizhou province, China. Phytotaxa. 480:139–151. doi: 10.11646/phytotaxa.480.2.3. [DOI] [Google Scholar]
- Li YK, Xin Z, Yuan Y, Cao Z, Liang JF. 2015. Morphological and molecular evidence for a new species of Russula (Russulaceae) from southern China. Phytotaxa. 202(2):94–102. doi: 10.11646/8466. [DOI] [Google Scholar]
- Li GJ, Zhang CL, Zhao RL, Lin FC. 2018a. Two new species of Russula from Northeast China. Mycosphere. 9(3):431–443. doi: 10.5943/mycosphere/9/3/1. [DOI] [Google Scholar]
- Li GJ, Zhang C, Zhao RL, Lin FC. 2018b. Hypogeous gasteroid lactarius sulphosmus sp. nov. and agaricoid Russula vinosobrunneola sp. nov. (Russulaceae) from China. Mycosphere. 9(4):838–858. doi: 10.5943/mycosphere/9/4/9. [DOI] [Google Scholar]
- Li GJ, Zhao D, Li SF, Yang HJ, Liu XZ. 2013a. Russula changbaiensis sp. nov. From northeast China. Mycotaxon. 124:269–278. doi: 10.5248/124.269. [DOI] [Google Scholar]
- Li GJ, Zhao RL, Zhang CL, Lin FC. 2019. A preliminary DNA barcode selection for the genus Russula (Russulales, Basidiomycota). Mycology. 10:61–74. doi: 10.1080/21501203.2018.1500400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GJ, Zhao Q, Zhao D, Yue SF, Li SF, Wen HA, Liu XZ. 2013b. Russula atroaeruginea and R. sichuanensis spp. nov. From southwest China. Mycotaxon. 124(1):173–188. doi: 10.5248/124.173. [DOI] [Google Scholar]
- Liu L, Liu Z, Y Q, Sui Z, Peng N, Y Y, Zang Y, Wang X, Zhou L, Y Q, et al. 2023. Research progress on nutritional function and bioactivities of Russula. Sci Technol Food Ind. 44:447–453. doi: 10.13386/j.issn1002-0306.2022030358. [DOI] [Google Scholar]
- Looney BP, Meidl P, Piatek MJ, Miettinen O, Martin FM, Matheny PB, Labbé JL. 2018. Russulaceae: a new genomic dataset to study ecosystem function and evolutionary diversification of ectomycorrhizal fungi with their tree associates. New Phytol. 218:54–65. doi: 10.1111/nph.15001. [DOI] [PubMed] [Google Scholar]
- Looney BP, Ryberg M, Hampe F, Sánchez‐García M, Matheny PB. 2015. Into and out of the tropics: global diversification patterns in a hyperdiverse clade of ectomycorrhizal fungi. Mol Ecol. 25(2):630–647. doi: 10.1111/mec.13506. [DOI] [PubMed] [Google Scholar]
- Matheny PB. 2005. Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Mol Phylogenet Evol. 35:1–20. doi: 10.1016/j.ympev.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Matsuura M, Kato S, Saikawa Y, Nakata M, Hashimoto K. 2016. Identification of cyclopropylacetyl-(R)-carnitine, a unique chemical Marker of the fatally toxic mushroom Russula subnigricans. Chem Pharm Bull. 64(6):602–608. doi: 10.1248/cpb.c15-01033. [DOI] [PubMed] [Google Scholar]
- Moncalvo JM, Lutzoni FM, Rehner SA, Johnson J, Vilgalys R. 2000. Phylogenetic relationships of agaric fungi based on nuclear large subunit ribosomal DNA sequences. Syst Biol. 49:278–305. doi: 10.1093/sysbio/49.2.278. [DOI] [PubMed] [Google Scholar]
- Rehner SA, Buckley E. 2005. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 97(1):84–98. doi: 10.1080/15572536.2006.11832842. [DOI] [PubMed] [Google Scholar]
- Romagnesi H. 1967. Les russules d’Europe et d’Afrique du Nord [The Russula of Europe and North Africa]. Paris: Bordas. [Google Scholar]
- Romagnesi H. 1985. Les russules d’Europe et d’Afrique du Nord [The Russula of Europe and North Africa]. Lehre: Vaduz. [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP.. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic. 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnari M. 1998. Monografia illustrata del genere Russula in Europa [Illustrated monograph of the genus Russula in Europe]. Trento: Fondazione centro studi micologici. [Google Scholar]
- Sarnari M, Redeuilh G. 2005. Monografia illustrata del genere Russula in europa [illustrated monograph of the genus Russula in Europe]. Trento: Associazione Micologica Bresadola. [Google Scholar]
- Sarwar S, Aziz T, Hanif M, Ilyas S, Saba M, Khalid S, Fiaz M. 2020. Plectological and molecular identification of economically important wild Russulales mushrooms from Pakistan and their antifungal potential against food pathogenic fungus Aspergillus niger. Bangl J Plant Taxon. 27(1):67–77. doi: 10.3329/bjpt.v27i1.47568. [DOI] [Google Scholar]
- Silvestro D, Michalak I.. 2012. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12:335–337. doi: 10.1007/s13127-011-0056-0. [DOI] [Google Scholar]
- Singer R. 1968. Les Russules d’Europe et d’Afrique du Nord [Russula of Europe and North Africa]. Paris: Jstor. [Google Scholar]
- Song Y, Xie XC, Buyck B. 2021. Two novel species of subgenus Russula crown clade (Russulales, Basidiomycota) from China. Mus Natl Hist Nat. 775:15–33. doi: 10.5852/ejt.2021.775.1543. [DOI] [Google Scholar]
- White T, Bruns T, Lee S, Taylor F, White T, Lee SH, Taylor L, Shawetaylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications. Mycologia. 18(1):315–322. [Google Scholar]
- Zhang D, Gao F, Jakovli I, Zou H, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- Zhou H, Cheng GQ, Wang QT, Guo MJ, Zhuo L, Yan HF, Li GJ, Hou CL. 2022. Morphological characteristics and phylogeny reveal six new species in Russula Subgenus Russula (Russulaceae, Russulales) from Yanshan Mountains, North China. J Fungi. 8(12):1283. doi: 10.3390/jof8121283. [DOI] [PMC free article] [PubMed] [Google Scholar]


