ABSTRACT
Vibrio alginolyticus, an emergent species of Vibrio genus, exists in aquatic and marine environments. It has undergone genetic diversification, but its detailed genomic diversity is still unclear. Here, we performed a multi-dimensional comparative genomic analysis to explore the population phylogeny, virulence-related genes and potential drug resistance genes of 184 V. alginolyticus isolates. Although genetic diversity is complex, we analysed the population structure using three sub-datasets, including the subdivision for three lineages into sublineages and the distribution of strains in the marine ecological niche. Accessory genes, most of which reclassified V. alginolyticus genomes as different but with relatively close affinities, were nonuniformly distributed among these isolates. We demonstrated that the spread of some post-evolutionary isolates (mainly L3 strains isolated from Chinese territorial seas) was likely to be closely related to human activities, whereas other more ancestral strains (strains in the L1 and L2) tended to be locally endemic and formed clonal complex groups. In terms of pathogenicity, the potential virulence factors were mainly associated with toxin, adherence, motility, chemotaxis, and the type III secretion system (T3SS). We also found five types of antibacterial drug resistance genes. The prevalence of β-lactam resistance genes was 100%, which indicated that there may be a potential risk of natural resistance to β-lactam drugs. Our study reveals insights into genomic characteristics, evolution and potential virulence-associated gene profiles of V. alginolyticus.
KEYWORDS: Vibrio alginolyticus, whole genome sequencing, population structure, virulence-related factors, antibiotic resistance genes
Introduction
Vibrio alginolyticus, a halophilic bacterium in the genus of Vibrio, is widely distributed in marine and estuarine habitats such as seawaters, estuaries, ocean sediment, and aquaculture settings [1]. V. alginolyticus was previously listed as a biotype of V. parahaemolyticus in the 8th edition of Bergey's Manual of Systematic Bacteriology, and became a separate species in the 9th edition of the manual [2]. V. alginolyticus is an emerging opportunistic pathogen to humans that causes foodborne diarrhoea in coastal areas [3]. Additionally, it can cause otitis media [4], soft tissue infections [5], conjunctivitis, and sepsis [3,6,7]. This bacterium also has pathogenic effects on marine organisms, including farm fish, molluscs, shellfish, and shrimp [8], and represents a sustained threat to aquaculture industries and ecosystems around the world [9].
Climate change and marine traffic lead to changing Vibrio species communities in the oceans [10]. As a mesophilic bacterium, pathogenic V. alginolyticus could become significant even in temperate zones such as the coastal areas of northern China and Europe. In our recent investigation, the increasing incidence of V. alginolyticus infection was found to threaten human health in coastal areas in China [11,12]. The major virulence factors of V. alginolyticus are motility, biofilm, extracellular proteases, type III secretion systems (T3SS), and type VI secretion systems (T6SS) [13–15]. V. alginolyticus shares relatively high similarities with closely related species in the Parahaemolyticus clade of the genus Vibrio [16]. V. alginolyticus and V. parahaemolyticus have similar genotypes [17], niche adaptations [18], and biomedical applications [19]. Flagellar motility plays an important role in the lifestyle of V. alginolyticus, both in the aquatic environment and in host colonization [20], and is associated with movement, colonization, adhesion, biofilm formation, and virulence. The regulation of adhesion in V. alginolyticus can be attributed in several chemotactic factors (such as the genes mcp, cheB, and cheV) [21,22].
To gain a comprehensive understanding of the genetic mechanisms shaping the epidemic dynamics of V. alginolyticus in the marine environment, we performed multi-dimensional comparative genomics research in this study. The rapid development of high-throughput sequencing technology in recent years has allowed the accumulation of genome data, which has provided opportunities for the genomics research on these pathogenic bacteria. Genome sequences of some V. alginolyticus strains have been reported in recent years [23,24], but the genomic characters were only presented for a single strain. To date, genomic variation characteristics related to horizontal gene transfer (HGT) through mobile genetic elements (MGEs) of V. alginolyticus has been described [25]. However, there is still a lack of genome studies related to the evolution and clonal structure of the V. alginolyticus population. Additionally, the phylogenetic relationships among V. alginolyticus strains isolated from marine environment and patients remain unclear. Little is known about the abundance, pathogenicity, and ecology of V. alginolyticus.
In this study, 184 strains from different regions and different host origins were genetically investigated to obtain the core- and pan-genome features, virulence-related genes, drug resistance genes, and transmission characteristics of V. alginolyticus. Here, we not only revealed unprecedented details of the population structure in V. alginolyticus, but also provided an overall analysis of virulence-related factors.
Materials and methods
Sampling, isolation and DNA extraction
Free-living, plankton-attached and shellfish-associated V. alginolyticus were collected between May 2008 and January 2020 in China. A total of 133 V. alginolyticus strains were isolated from seawater, seafood, freshwater, and other samples. Strains were cultured on 1% NaCl lysogeny broth (LB, w/v) agar at 35°C for 18 h. All strains collected in this study are listed in Supplementary Table S1.
Genomic DNA with no contamination or DNA degradation was prepared by using the QIAamp DNA Mini Kit (Qiagen, Dusseldorf, Germany). Then, a NanoDrop spectrophotometer was used to measure the concentration and purity of DNA genomes. Subsequently, the DNA integrity was checked by agarose gel electrophoresis. The quality requirements were concentration ≥ 100 ng/μL and total amount > 20 μg.
Whole genome sequencing
The genomes of 133 V. alginolyticus strains were sequenced using the Illumina HiSeq 4000 platform (Illumina Inc., San Diego, CA, USA) at the Beijing Genomics Institute (BGI, Shenzhen, China) with a depth of 200× coverage. Library construction (paired-end read sizes of 150 bp), genome sequencing, and data pipelining were performed in accordance with the manufacturer’s protocols.
Genome assembly and annotation
FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was applied to evaluate the raw sequence data, and the low-quality reads were filtered. The data filtering criteria were removal of contaminated adapters, removal of repeated reads, and removal of low-quality reads (Phred score ≤ 20 bases exceeding 50% of the read length). Then, the clean data were assembled into contigs using SPAdes v3.8.2 with the default parameters [26]. After removing contigs with lengths < 500 bp, QUAST v5.0.1 was applied to evaluate the genome assembly [27]. Open reading frame prediction and annotation were performed by Prodigal v2.6.3 [28] and Prokka v1.13.3 [29].
V. alginolyticus genomes from NCBI
In addition to 133 laboratory isolates, there were 61 whole genomic sequences available at the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/) as of March 2021. Because of the existence of mislabelled species, species-level identification was clarified by average nucleotide identity (ANI) analysis. High-quality sequenced genomes were screened by using the criteria of 4.5 Mb ≤ genome size ≤ 6.5 Mb and contigs < 200. A total of 51 strains from online databases were screened out and listed in Supplementary Table S2.
Average nucleotide identity (ANI) analysis
A Perl script based on the previous algorithm [30] was used to compute the ANIs for the draft sequences from the V. alginolyticus strains. The ANI was evaluated between the query genome and the reference genome, and the mean identity of all BLASTn matches was computed as the ANI value. The computation required that the BLASTn matches should have ≥ 30% sequence identity and ≥ 70% length coverage.
Pan-genome analysis
After obtaining the annotated assemblies in GFF3 format (produced by Prokka software), the pan genome matrix of V. alginolyticus was calculated using Roary v1.12 [31] (https://sanger-pathogens.github.io/Roary), a pan genome pipeline, with the identity parameter set to 90%. The heatmap of the presence/absence of pan genes was drawn in R with the gplots 3.1.1 package.
Phylogenetic tree construction
Coding sequences (CDSs) were predicted using Prodigal v2.6.3 in the V. alginolyticus genome sequences. A non-redundant homologous gene set was done by CD-hit v4.6.6 [32]. Then BLAST+ was applied to search for the homologous genes of the CDS of each strain in the non-redundant homologous gene set. Each homologous gene contained in all strains with a single copy was considered a core gene. Next, core SNPs were called using snippy v4.3.6 (https://github.com/tseemann/snippy) from all the predicted core genes against the reference genome sequence of strain ATCC 17749T (BioSample accession: SAMN02603463). Gubbins v2.4.1 was used as a recombination-removal tool to analyse the clonal recombination relationships [33]. The fastBAPS algorithm (https://github.com/gtonkinhill/fastbaps/) was used to rapidly identify genomic lineages or genetic clusters [34]. The maximum-likelihood tree was reconstructed using PhyML v3.1 based on core SNPs with 1000 bootstraps.
Virulence-related gene searching
The Virulence Factor Database (VFDB, http://www.mgc.ac.cn/VFs/) was used to predict and annotate the virulence genes. All the protein sequences from the V. alginolyticus strains were searched against the VFDB using BLASTp with an E-value of 1e-5. The virulence genes were determined by using a relatively relaxed cutoff (identity ≥ 80%, length coverage ≥ 50%) and a stricter cutoff (identity ≥ 90%, length coverage ≥ 80%). Sequence alignment analysis and phylogenetic construction of a neighbor-joining (NJ) tree were performed with 1000 bootstrap replicates.
Antibiotic resistance gene searching
The antibiotic resistance genes were detected using the ResFinder tool (https://cge.cbs.dtu.dk/services/ResFinder/) and were cross-compared with the Comprehensive Antibiotic Resistance Database (CARD, http://arpcard.mcmaster.ca). The BLASTn cutoff values were chosen as follows: E-value of 1e-5, identity ≥ 90%, and length coverage ≥ 60%.
Type III secretion system (T3SS) identification
The CDSs of T3SS from V. alginolyticus strain E06333 (Accession No.: CP071058.1) were used as references for comparison with genomes using BLAST+ . For a single CDS, the alignments of more than half the length and identity > 50% were considered positive matches, and the e-value cutoff was set to a lower cutoff, 1e-2. A concatenated segment of more than three regions positively mapped to the reference CDS was considered a potential T3SS segment. This screening process was conducted by an automatic pipeline.
In vitro cytotoxicity assays
Bacterial infection was performed using the human epidermoid carcinoma cell line Hep-2 (CCC0068; Peking Union Medical College Cell Resource Center). Cytotoxicity assays were performed in 96-well plates at a density of 1 × 105 cells/well. The lactate dehydrogenase (LDH) released by cells was detected using a Cytotox 96 kit (Promega, Madison, USA). All experiments were performed three times in duplicate.
Motility and biofilm formation assays
Strains were diluted to OD600 1.0, and then spotted on LBS medium containing 0.35% agar. The swimming ability was recorded by measuring the colony diameter after 12 h at 30°C. For biofilm formation assay, strains were cultured overnight at 30°C in 1% NaCl-LB broth. The cultures (50 µL) were diluted in 5 mL LBS in glass tubes and incubated at 30°C without shaking for 72 h. The bacterial incubation was transferred to a clean cuvette and the optical density of was measured at 600 nm. Total biofilm formation was measured by 1% (w/v) crystal violet staining at room temperature for 30 min. Crystal violet that stained biofilm was solubilized with dimethylsulphoxide (DMSO) and then measured at an optical density of 570 nm using Multiskan Spectrum (Thermo Scientific, USA). Finally, we calculated the relative biofilm formation as 1000 × OD570 / (OD600 × V1 × V2), where V1 was the volume of bacterial incubation and V2 was the volume of DMSO.
Data availability
The genome sequences of the V. alginolyticus strains sequenced in this study have been deposited at GenBank/DDBJ/ENA under the BioProject ID no. PRJNA836558.
Results
Phylogeny and population structure of V. alginolyticus
The genome sizes of the 184 V. alginolyticus strains ranged from 5.00 (strains Va62, Va116, and Va119) to 5.86 Mb (strain UCD-9C), with an average of 5.20 Mb. The GC contents ranged from 44.2 mol% (strain UCD-9C) to 44.8 mol% (strain 2014V-1011), with an average of 44.56 mol%. Approximately 4344–5303 CDSs per genome were predicted (Supplementary Table S2 and Table S3). ANI calculation showed the vast majority of strains had ANI values between 98.13% and 100% compared with the V. alginolyticus type strain ATCC 17749T (Supplementary Figure S1), which was greater than the species boundaries of 95%−96% [35].
Among the 184 V. alginolyticus genomes, the core genome size was estimated to be 3.1 Mb, with an average GC content of 46.02 mol%, which was slightly higher than the average of the 184 genomes (44.56 mol%). The core genome was searched against the COG database to infer gene functions. Aside from the core genes mainly related to the fundamental physiological activities and biological processes, eight mobile genetic elements (MGEs) related genes were also identified using RAST Annotation Server against the core genome sequences, including one integrase and seven phage/ prophage-related factors, which indicates that they were acquired early in the emergence of V. alginolyticus and that they are stable in the genome.
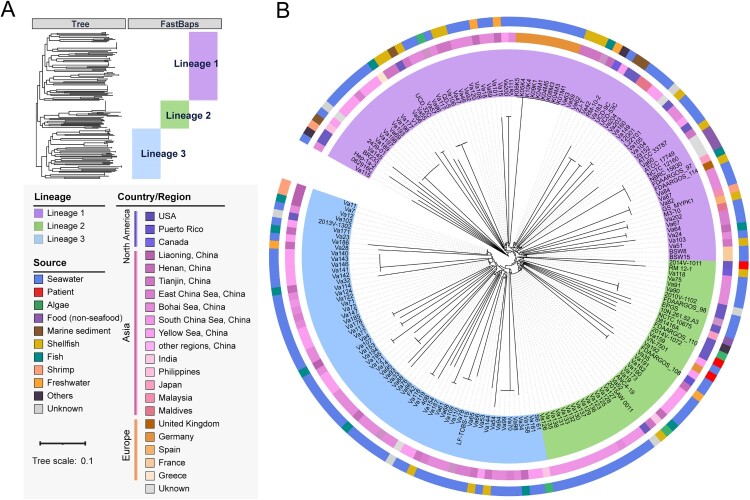
A total of 230,751 SNPs were identified in the V. alginolyticus core genome. Compared with the complete genome ATCC 17749T as the reference sequence, the number of core genome SNPs (cgSNPs) ranged from 29,652 (strain Va46) to 34,424 (both strains Va109 and Va110). A phylogenetic tree was constructed based on the cgSNPs, and three major lineages could be inferred, named Lineage 1 (L1) to Lineage 3 (L3) (Figure 1(A)). L1 showed deeper evolutionary roots and contained more isolates (43.5%, 80/184), which indicated they were introduced into the local region earlier than the isolates in L2 and L3. The phylogenetic analysis showed that some of the strains from China seas (like the Yellow Sea and South China Sea) were clustered together. However, regardless of isolation region or isolation source, there was no clear correlation between single isolation source and evolutionary cluster found in the phylogenetic tree (Figure 1(B)). Therefore, the various V. alginolyticus strains showed substantial evolutionary differences in geographic distribution or host source, indicating a high genetic diversity even in a local region.
Figure 1.
Population structure and phylogenetic analyses of V. alginolyticus. (A) Phylogenetic tree of V. alginolyticus constructed based on de-recombinational cgSNPs. Colours indicate the isolation region and host source of V. alginolyticus strains. Bootstrap values were calculated by 1000 replications. The horizontal bar indicates 0.1 estimated nucleotide substitutions per site. (B) Evolutionary tree divided into 3 main lineages using FastBaps R package with an optimized BAPS prior. Lineages 1–3 are indicated in purple, green and blue, respectively.
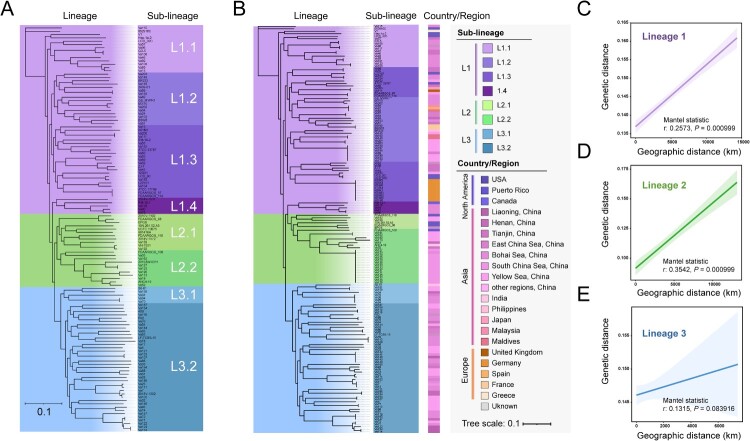
We further divided the total genomic dataset into several hierarchical sub-datasets containing different numbers of genomes. In the first sub-dataset, we removed the same clone obtained from a single sampling to form a sub-dataset of 123 isolates for analysis of more refined population structure of V. alginolyticus. Hierarchical Bayesian clustering based the phylogenetic tree was reconstructed for these V. alginolyticus isolates, and SNP-based genotyping showed that, among the L1 strains, four sub-lineages (named L1.1, L1.2, L1.3, and L1.4, respectively) could be further differentiated. Both L2 and L3 were further divided into two sub-lineages each (Figure 2(A)).
Figure 2.
Phylogenetic analysis of three hierarchical sub-datasets containing different number of V. alginolyticus. (A) Phylogenetic tree of V. alginolyticus eliminated from a single sampling belonging to the same clonal group. (B) Phylogenetic tree constructed based on marine habitat-related strains. The colours indicate sublineages and isolation regions of V. alginolyticus strains. (C) Correlation analysis between genetic distance and geographical distances within L1 strains (Mantel statistic r: 0.2573, P value = 0.000999). (D) Correlation analysis between genetic distance and geographical distances within L2 strains (Mantel statistic r: 0.3542, P value = 0.000999). (E) Correlation analysis between genetic distance and geographical distances within L3 strains (Mantel statistic r: 0.1315, P value = 0.083916).
Sub-dataset-2 contained 164 strains associated with marine habitats, with isolated sources including seawater, marine sediment, fishes, and seagrass. This sub-dataset, from which clinical and food-derived strains were removed, was ideal for characterizing the distribution of V. alginolyticus populations in marine ecological niches. Strains from Europe and North America showed a scattered distribution; for example, strains isolated from the USA were nested within the L1 and L2 population but clustered in different sub-lineages with restricted diversification (Figure 2(B)). The evolutionary tree indicated that, even for the same marine niche, V. alginolyticus populations did not show clear geographical aggregation. However, strains from Asia (mainly Chinese isolates) were found throughout in the L1, L2, and L3 lineages (Figure 2(B)). We speculated that the genetic diversity of V. alginolyticus may be related to some non-geographic separation factors, such as human activities.
To test this hypothesis, we then singled out 163 strains with well-defined isolation sites as a third sub-dataset. By calculating the genetic distance and the actual geographical distance among V. alginolyticus strains, we found that the L3 strains were more affected by non-geographic separation factors (Mantel statistic r: 0.1315, P = 0.083916, Figure 2(C)), whereas L1 and L2 were less affected (both had a Mantel statistic P = 0.000999, Figures 2(D) and (E)). This evidence suggested that the spread of L3 (mainly including strains isolated from Chinese territorial seas) was likely to be closely related to human activities, whereas L1 and L2 tended to be locally endemic and formed clonal complex groups.
Pan-genome diversification
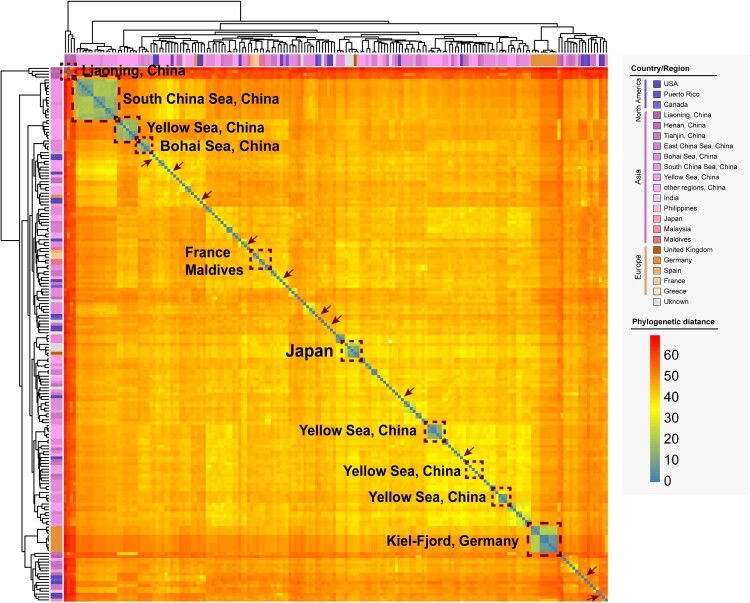
All genome sequences were analysed by two-dimensional hierarchical clustering analysis based on the presence or absence of CDSs to preliminarily assess the consistency of these V. alginolyticus genomes. From a global perspective, there were certain differences in the genome composition of V. alginolyticus, and the geographical distribution was complex and diverse. Some strains from China territorial seas formed a separate cluster that distinguished strains from other regions, such as strains from the South China Sea and the Bohai Sea, whereas the strains from the Yellow Sea showed a multiple-clustering trend (Figure 3). In the Chinese Mainland, only the strains from Liaoning Province formed a single cluster, whereas the strains from Henan Province and Tianjin Municipality were scattered in various clusters. The same was true for the clustering of global strains. For example, several strains from Japan, Germany, France, and Maldives formed separate clusters despite a very limited number of strains and isolation sources; however, strains from other geographical locations, represented by the United States strains, were widely distributed, as highlighted in the arrows in Figure 3.
Figure 3.
Two-dimensional hierarchical clustering based on the whole genome sequences of V. alginolyticus. Isolation regions or countries are presented in the corresponding dashed boxes, whereas strains from the USA are marked with brown arrows. Colours from blue to red represent increasing genomic distances.
Next, to calculate the genomic differences of strains from different geographical origins, the core genome and accessory genome of these sequenced V. alginolyticus strains were analysed in depth. Approximately 24,725 genes were predicted and annotated that were considered to be the pan-genome of the V. alginolyticus strains in this study (Figure S2A). The core-genome curve (Figure S2B) showed a flat slope, which indicated that it had achieved a full representation of this species core diversity. As the number of strains increased, the number of pan genes was growing rapidly while the number of the core genes gradually stabilized. Altogether there were 3323 (13.4%) genes identified as core genes (99% ≤ strains ≤ 100%), and the number of pan genes was equal to 7.4 times that of core genes. And there were also 422 soft core genes (95% ≤ strains < 99%) in the pan-genome.
In addition, the analysis with protein identity set to 90% resulted in partial strains with similar accessory genome content gathering to form several groups, such as the three typical groups in the red box (Figure S2A), named Groups 1–3. Exploring the gene set that was exclusive for these three groups, 147 genes were found to be unique for and present in all nine isolates from the Kiel-Fjord, Germany in Group 1 (Supplementary Table S4). In the same way, 71 and 80 unique gene clusters were identified in Group 2 and Group 3, comprising 14 South China Sea-derived strains and 7 partial Yellow Sea-derived strains, respectively (Supplementary Tables S5 and S6). The annotation of these three groups remains incomplete because the majority of genes (more than 2/3) were only annotated as hypothetical proteins despite being mapped against the Gene Ontology (GO) database, Cluster of Orthologous Groups of proteins (COG), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Swiss-Prot protein database. The annotation information for the remaining genes of Group 1 mainly included aspects of molecular function and biological process, and included the specific functions such as lipid transport and metabolism, transcription, energy production and conversion, and multi-drug resistance (Supplementary Table S4). The unique accessory genes of the strains from the South China Sea (Group 2) were mainly responsible for carbohydrate transport and metabolism, and cellular component organization or biogenesis (Supplementary Table S5). For partial Yellow Sea-derived strains (Group 3), the genes with more detailed functional information were involved in signal transduction and transport and metabolism (Supplementary Table S6). These accessory genome-related evidence reclassified V. alginolyticus as a relatively closer affinity, thereby indicating substantial genomic differences with respect to other groups.
Distribution characteristics of virulence-related genes
Based on the annotation of the VFDB database via the relatively relax cutoff (identity ≥ 80%, length coverage ≥ 50%), the virulence components harboured by 184 V. alginolyticus strains were identified and categorized as 8 virulence-related classes involved in 14 virulence-related factors and 85 potential virulence genes (Table 1). The number of virulence genes carried by each strain ranged from 50 (strain Va52) to 88 (strain 062916C), with an average of 74 ± 4 (Supplementary Table S7). The functions of these virulence genes were identified as cell chemotaxis, motility, T3SS, haemolysin, lipopolysaccharide synthesis, and immune evasion. The positivity rate of virulence factors related to T3SS reached 99.5% (183/184) in these strains. It is worth mentioning that all V. alginolyticus strains carried the thermolabile haemolysin gene (tlh). Only one strain isolated from Canada, RM-12-1, carried the haemolysin hlyD gene (Supplementary Table S7).
Table 1.
Function and pathogenic role of the virulence factors of Vibrio alginolyticus.
| VFclass | Virulence factors | Function or Pathogenic Role |
|---|---|---|
| Adherence | Multivalent adhesion molecule | Cell–cell adhesion |
| Chemotaxis and motility | Flagella | Flagellum biogenesis, motor activity, pathogenesis |
| Quorum sensing | Autoinducer-2 | Autoinducer synthesis, quorum sensing |
| Secretion system | T3SS1 | Translocated effector, protein secretion |
| T6SS | T6SS structual component | |
| T4SS | T4SS effector | |
| Toxin | Haemolysin/cytolysin | Protein transport, pathogenesis |
| Thermolabile haemolysin | Lipid metabolic process, pathogenesis | |
| Endotoxin | LOS | Multifunctional enzyme |
| LPS | Lipopolysaccharide biosynthesis | |
| Immune evasion | Capsule | Polysaccharide biosynthesis, chemotaxis |
| immunogenic lipoprotein | Membrane component | |
| Others | O-antigen | Lipopolysaccharide biosynthesis |
| elongation factor | Protein biosynthesis |
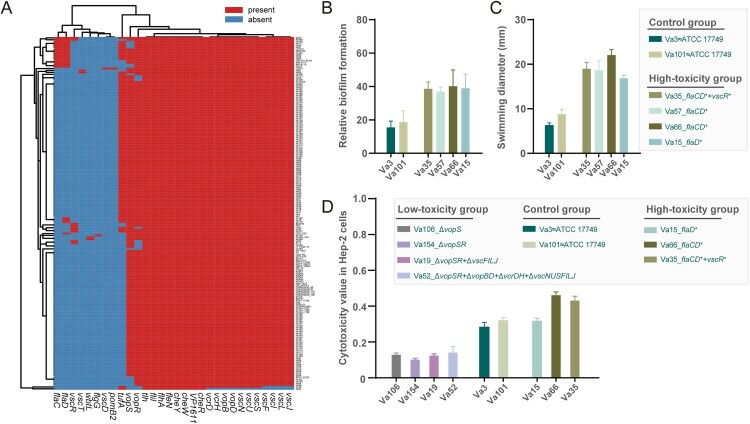
Then we used a more conservative and stricter cutoff for protein sequence comparison. Consequently, we identified 30 virulence genes with known functions (Figure 4(A)). All these V. alginolyticus strains had 8 highly conserved virulence genes, with the biological functions including thermolabile haemolysin (tlh), adherence factor (VP1611), and flagella synthesis and cell chemotaxis (fliI, flhA, fleN, cheR, cheW, cheY). The heatmap of virulence-related factors showed that most of the V. alginolyticus harboured almost an equal number of virulence genes, which indicated that these strains shared a similar virulence spectrum (Figure 4(A)). An important finding was that there did not appear to be any clear distinctions in (potential) virulence determinants found in China vs. global isolates. By linking virulence-related genes with isolation regions or isolation hosts, no relevant distribution characteristics were observed between the V. alginolyticus strains.
Figure 4.
Virulence analysis of V. alginolyticus. (A) Heatmap for virulence-related factors of V. alginolyticus. Red represents the presence of genes, while blue represents the absence of genes. (B) Relative biofilm formation of six representative strains. The vertical coordinate represents the relative amount of biofilm formation measured by 1% (w/v) crystal violet (w/v) staining. (C) Swimming ability of six representative strains. The vertical coordinate represents the colony diameter after 12 h of incubation. Strains Va3 and Va101 were selected as the control group based on the same virulence spectrum to ATCC 17749T, the type strain of V. alginolyticus. In the high-toxicity group, four strains (Va15, Va35, Va57, and Va66) with 1–3 additional flaC (flagella), flaD (flagella), and vscR (T3SS) genes were compared with ATCC 17749T. (D) Cytotoxicity values of nine representative strains. Higher values indicate higher cytotoxicity. The high-toxicity group and the control group were as described above. For the low-toxicity group, compared with ATCC 17749T, another four strains (Va19, Va52, Va106, and Va154) were chosen with natural deletion of one or more virulence genes (the gene family including vopSR, vopBD, vcrDH, and vscNUSFILJ).
Additionally, the genes of cholera toxin (ctxA/ctxB) of V. cholerae O1/O139, thermostable direct haemolysin (tdh) and TDH related haemolysin (trh) of V. parahaemolyticus were not detected. The thermolabile haemolysin gene (tlh) was subsequently compared with the reference sequence V. parahaemolyticus ATCC 33846 (GenBank accession No.: GU971655.1). Sequence comparison showed that the DNA sequence similarity was 84.6% – 85.7%, but the amino acid sequence similarity reached 92.8% – 93.7%. The tlh gene sequences were used to construct the split network tree (Supplementary Figure S3). Consequently, the tlh gene carried by V. alginolyticus is likely to have contributed to pathogenicity because it encoded a phospholipase A2 protein in V. parahaemolyticus when its expression was upregulated under conditions mimicking the human intestine [36,37]. Moreover, it was found that the cheR gene sequence NJ tree roughly classified these V. alginolyticus isolates into four groups (Supplementary Figure S4).
Subsequently, the strains in the high-toxicity group had flagella-related genes (flaCD) exhibiting superior biofilm formation ability and swimming motility compared with the control group (Figures 4(B) and (C)). In addition, we also selected four representative strains missing the T3SS-related vopBDSR and vscNUSFILJ genes as the low-toxicity group. Strains lacking vopS showed a significant reduction in cytotoxic infectivity (Figure 4(D)). Genes such as vopBDR and vscNUSFILJ had minimal impact on cytotoxicity, whereas flaCD genes enhanced cytotoxic effects (Figure 4(D)).
Type III secretion system
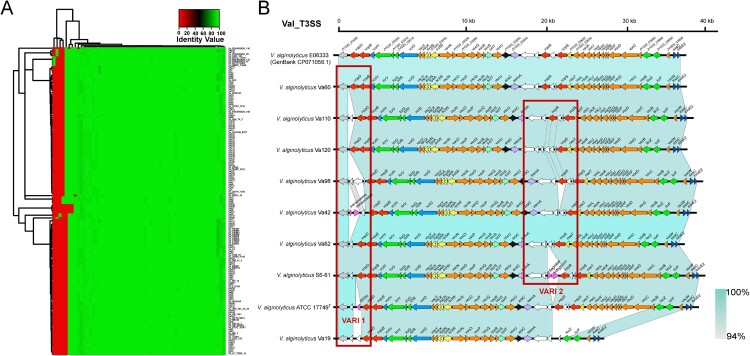
Numerous T3SS-associated genes were identified in this study. T3SS was of particular interest, and there were indeed some doubts about the distribution and composition of T3SS in V. alginolyticus. First, T3SS was initially identified by the potential T3SS segment composed of CDSs. As a result, we obtained a total of 165 positive T3SS sequences from each genome (the remaining 19 incomplete T3SS sequences were discarded). Then, all the potential T3SS sequences were manually checked and determined based on the locus of these segments and the existence of known conserve genes of T3SS.
After an extraction of each CDS in the T3SS sequences, a clustering heatmap based on comparison of the similarity of each CDS was constructed (Figure 5(A)). According to the identified CDS arrangement, these T3SS sequences could be classified into five subtypes. The subtype represented by strain Va110 accounted for the highest proportion (76/165, 46.1%), followed by the subtype represented by Va60 (75/165, 45.5%). The remaining three subtypes only appeared in a few strains. Among them, the acquired sequence length ranged from 36,924 to 38,742 bp with the variant number of CDSs ranging from 46 to 50. The annotation information of the five types of T3SS gene clusters is presented in Supplementary Table S8. The CDS components of these five T3SS types were presented in Figure 5(B). Their backbone had a similar structure, but there were two variable regions in the red box representing some small differences involving one to four CDSs, due to the inserted sequence (i.e. IS200 in the T3SS of V. alginolyticus ATCC 17749T) or other genetic events (i.e. the deletion or insertion of several hypothetical proteins).
Figure 5.
Type III secretion system of V. alginolyticus. (A) Heatmap for 165 available T3SS sequences. Red indicates low identity values, whereas green indicates high identity values. (B) Gene cluster distribution of five different T3SS genotypes in V. alginolyticus. The two identified variable regions (VARI1 and VARI2) are marked with red boxes. The colours represent different gene families, and the length of the arrow indicates the size of each CDS.
Antibiotic resistance genes
We found that five classes of antibiotic resistance-related genes were present in the V. alginolyticus genomes (Table 2), named β-lactams (blaCARB-42), quinolones (qnrVC5), aminoglycosides [aph(3’)-Ia, aph(3'’)-Ib, aph(6)-Id], sulphonamides (dfrA15, dfrA31, sul1, sul2), and amide alcohols (floR). Among them, the prevalence of β-lactam resistance genes was 100% and included a single genotype (blaCARB-42). This finding indicated that V. alginolyticus may be naturally resistant to β-lactam drugs. The sulphonamide resistance genes ranked secondly and were distributed in 11 of 184 strains (6.0%). When considering multi-drug resistance, only a handful of strains (n = 11, 6.0%) carried multiple drug resistance genes. For example, the strain ZJ-T from China carried four different kinds of resistance genes, and it had carried the largest number of resistance genes. Further investigation revealed that these drug-resistance genes of strain ZJ-T were located in a large gene cluster on the same scaffold. After comparison with the BLASTn tool, this fragment was very similar to the integrated conjugative elements (ICEs, ICEVal056-1, accession No.: KR231688.1) of V. alginolyticus, with coverage and identity of 70% and 99.98%, respectively. This indicated that the strain ZJ-T may have obtained a large fragment carrying multiple drug-resistance genes through HGT.
Table 2.
Distribution and positive rate of antibiotic resistance genes.
| Class of antibiotics | Genotype | Number of strains | Positive rate (%) |
|---|---|---|---|
| β-lactams | blaCARB-42 | 184 | 100 |
| Quinolones | qnrVC5 | 1 | 0.5 |
| Aminoglycosides | aph(3’)-Ia,aph(3'’)-Ib,aph(6)-Id | 9 | 4.9 |
| Sulphonamides | dfrA15,dfrA31,sul1,sul2 | 11 | 6.0 |
| Amido alcohols | floR | 7 | 3.8 |
Discussion
A variety of marine microorganisms can spread across oceans, such as in the case of the global spread of pathogenic clones of V. parahaemolyticus and V. cholerae. However, human activity is an important factor in the spread of these pathogenic clones. For example, human activity is associated with a cholera outbreak in Haiti [38]. We still do not have a clear understanding of the divergence, prevalence, and spread of V. alginolyticus. In this study, comparative genomic analysis has revealed several different dimensions of evolutionary characteristics in V. alginolyticus genomes on the basis of a systematic description of the population structure features for 184 isolates from different regions and sources and investigation of the distribution of virulence genes and drug resistance in the V. alginolyticus strains.
Pan-genome analysis helps us understand the core-/pan-genes and unique gene clusters of this species, whereas the core genome is usually composed of essential genes, such as those that encode for the major redox reactions [39]. The accessory genome was found to be related to V. alginolyticus niche adaptation and biological characteristics, including genes related to virulence and drug resistance. The number of pan-genes was equal to 7.4 times that of the core genes from the pan-genome analysis (Figure 4(A,B)); this indicates that V. alginolyticus had a strong adaptability to the external environment and the variable host habitats, with a rich and diverse pan gene pool. Based on these results, it is speculated that human activities, such as shipping, tourism, and global-seafood trade, may promote to transoceanic transmission of V. alginolyticus in seawater. This allows strains in different sea areas to frequently exchange genes (e.g. HGT mediated by MGEs [40]), which ultimately affects the V. alginolyticus population structure and evolution. For example, the V. alginolyticus strains from the Kiel-Fjord (Germany) have evolved from clonal expansion following HGT-driven niche-adaptation [25].
The phylogenetic tree based on the cgSNPs provided a clear interpretation of the evolutionary relationship between 184 V. alginolyticus strains. It is generally accepted that strains in the same clade are more closely related to each other and may have a common evolutionary ancestor. However, the phylogenetic tree (Figure 1) showed that V. alginolyticus were classified into the same clade and were isolated from different times and different geographic regions. The strains Va115, 062916C, and Hep-1a-2 formed a single evolutionary branch respectively, which was distinct from other strains, and these strains were ancient from an evolutionary perspective. This finding indicates that the V. alginolyticus isolated from China and the USA between 2014 and 2016 had evolved and spread. We used a hierarchical Bayesian clustering algorithm implemented in fastBAPS. It helped to split phylogenetic trees into three lineages and eight sublineages (Figures 1 and 2) using a population genomic model. We further divided the total genomic dataset of V. alginolyticus into three sub-datasets. However, because of the uneven number of strains in the isolation area, the isolation region of V. alginolyticus did not cover the global geographic location well, which posed a challenge for analysing the global population structure.
The core genome evolutionary relationships were not completely consistent with those of the pan-genome. The accessory genes in bacteria may have been derived in different stages of evolution in order to adapt to various hard-condition living situations, the accessory genome plays an inescapable role in evolution of V. alginolyticus for its environmental adaptability. The phylogenetic tree not only appeared little interspecies relation between V. alginolyticus strains isolated from different host sources and geographical regions but indicated that there was a highly genetic diversity among the strains.
The creatures that inhabit the Earth can fully utilize resources if there is not any competition or other predators. However, because of the existence of interspecies competition, what a creature can occupy is nothing but the ecological niche within the scope of viability [41,42]. Over a long period of time, bacteria have evolved a competition mechanism to compete for limited resources, which included the evolution of virulence genes and antibiotic resistance genes. This mechanism allows Vibrio species to infect humans and aquatic animals, and to expand their ecological niches. This study found that there was a rich diversity of virulence factors in V. alginolyticus populations, which are mainly associated with chemotaxis, motility, and secretion system. Consistent with a previous study [43], more than 100 acetylation-modified proteins were predicted as virulence factors in V. alginolyticus. Another study in Mexico found that several genes encoding virulence factors were present in environmental strains of V. alginolyticus, including tdh, proA, wza, vopD, hcp, vasH and vgrG genes [44].
Additionally, flagella-related genes (like fleN, flhA, fliL) have roles in flagellum biogenesis and motor activity, which have been proven to be closely related to biological and cellular processes such as chemotaxis, biofilm formation, and colonization for bacteria of the genus Vibrio [21,45,46]. As for cheW gene, an important downstream gene in the bacterial chemotaxis pathway, was composed of V. cholerae chemotaxis cluster III system which was regulated by RpoS and quorum sensing in response to polar localization [47]. V. parahaemolyticus RIMI 2210633 possesses two sets of T3SSs on chromosomes 1 and 2, designated T3SS1 and T3SS2, respectively [48]. Likewise, V. alginolyticus required its T3SS to cause rapid death of infected host cells such as fish and shrimp, and the T3SS is a highly conserved apparatus among these V. alginolyticus strains (Figure 5).
Many other studies have explored the infection mechanism of T3SS in V. alginolyticus [49,50]; it can deliver the effectors directly into cells of hosts across bacterial and host membranes. Vibrio T3SS initiates a multifaceted process leading to the death of mammalian cell lines, starting with autophagy, followed by cell rounding, and culminating in caspase-independent cell death [51,52]. The effector protein VopS plays a crucial role in inducing cell rounding and disrupting the actin cytoskeleton by inhibiting Rho GTPases [53]. Zhao et al. demonstrated that VopS from V. alginolyticus not only contributes to fish cell rounding through Rho GTPase inhibition but also plays an essential role in apoptosis induction. A deletion mutant lacking VopS exhibited severe impairment in its ability to induce these two cellular responses, whereas transfection of VopS into the fish cell line FHM alone is sufficient to trigger both cell rounding and apoptosis [50]. Those findings are consistent with the results of this study.
There is currently a lack of worldwide surveillance information regarding V. alginolyticus infections, as this pathogen is not taken seriously in many countries, which probably underestimate the clinical infectivity of this pathogen. Unfortunately, our efforts to link the virulence factors to the isolation sources under a variety of conditions have been unsuccessful. The divergence of virulence genes among environmental strains is so subtle that the minimal variation in a certain virulence gene sequence or SNP-based locus is not powerful enough to establish the relationship between strains and pathogenicity. Therefore, the pathogenicity of V. alginolyticus may be affected by multiple factors. The pathogenicity may depend on the complex interaction among the multiplexing systems, such as the host species, the host’s niche, and the pathogenic bacteria itself.
Nowadays, exposure to seawater environments through recreational (e.g. surfing, snorkelling, speed boating, jet skiing) or occupational activities (e.g. sailing, crabbing, fishing) increases the risk of disease caused by V. alginolyticus, especially when an individual’s immunity is weak. This suggests that V. alginolyticus is a foundational condition rather than a decisive factor for human diseases, and the pathogenic progress may be accelerated and deteriorated by the synergistic effect of multiple factors. The functions of these potential virulence genes deserve more attention.
The formation of flagella confers motility to bacteria, enabling them to colonize both abiotic and biotic surfaces, optimize growth and survival, and evade desiccation in hostile host environments. Vibrio genomes harbour five or six polar flagellin genes located at two distinct genomic locations [54]. The expression of these genes are tightly linked to Vibrio virulence and host colonization [55]. Mutation in flaC within V. vulnificus leads to a significant reduction in motility, adhesion, and cytotoxicity [56]. Mobley et al. introduced a nonpolar mutation into the flaD gene and evaluated its impact on virulence in vivo. The hpmA flaD mutant exhibited approximately 100-fold lower recovery compared with the hmpA mutant or wild-type parent strain, which indicates clear attenuation of virulence potential [57].
More importantly, several studies have demonstrated that the antibiotic resistance genes are physically linked to MGEs and could potentially propagate to other bacterial species through HGT [58]. We found that 184 strains all carried the β-lactam resistance gene blaCARB-42; therefore, V. alginolyticus may be naturally resistant to β-lactam drugs. Similarly, another study also found that more than 90% of the V. alginolyticus strains were resistant to β-lactams antibiotics, and other antibiotics including cephalotin, amikacin, cephotaxime and pefloxacin [44]. Another survey in Korea showed that V. alginolyticus isolates were mainly resistant to ampicillin, vancomycin, and cephalothin [59]. In our study, the only strain ZJ-T (one out of 184 strains), isolated from diseased Epinephelus coioides in Guangdong Province, China [60], could serve as a representative with carrying several potential drug resistance genes. However, another study found that the prevalence of multi-resistance to four or more antimicrobials in V. alginolyticus (11.1%) was higher than that in V. parahaemolyticus (5%) [61]. Overall, these investigations of V. alginolyticus antimicrobial resistance should play an early-warning role in V. alginolyticus monitoring and treatment of clinical infection.
Our comprehensive analyses of V. alginolyticus genomes have further enhanced the understanding of their virulence factors, drug resistance genes, population structure and evolutionary characters. We demonstrated the detailed genetic diversity of the V. alginolyticus genomes. This study provided a framework for systematic characterization of the V. alginolyticus genomes, which may be broadly applicable to diverse Vibrionaceae microorganisms and amenable to comparative analysis.
Supplementary Material
Acknowledgements
We thank Mallory Eckstut, PhD, from Liwen Bianji (Edanz) for editing the English text of a draft of this manuscript.
Funding Statement
This work was supported by the National Science and Technology Major Project (2018ZX10714002, 2017ZX10303405-002).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Grimes DJ. The vibrios: scavengers, symbionts, and pathogens from the Sea. Microb Ecol. 2020;80(3):501–506. doi: 10.1007/s00248-020-01524-7 [DOI] [PubMed] [Google Scholar]
- 2.Rockstroh T. [Changes in the nomenclature of bacteria after the 8th edition of Bergey's Manual of the Determinative Bacteriology]. Z Arztl Fortbild (Jena). 1977;71(11):545–550. [PubMed] [Google Scholar]
- 3.Baker-Austin C, Oliver JD, Alam M, et al. Vibrio spp. infections. Nat Rev Dis Primers. 2018;4(1):8. doi: 10.1038/s41572-018-0005-8 [DOI] [PubMed] [Google Scholar]
- 4.Feingold MH, Kumar ML.. Otitis media associated with Vibrio alginolyticus in a child with pressure-equalizing tubes. Pediatr Infect Dis J. 2004;23(5):475–476. doi: 10.1097/01.inf.0000126592.19378.30 [DOI] [PubMed] [Google Scholar]
- 5.Howard RJ, Pessa ME, Brennaman BH, Ramphal R (1985) Necrotizing soft-tissue infections caused by marine vibrios. Surgery 98(1):126-130. [PubMed] [Google Scholar]
- 6.Jacobs Slifka KM, Newton AE, Mahon BE.. Vibrio alginolyticus infections in the USA, 1988-2012. Epidemiol Infect. 2017;145(7):1491–1499. doi: 10.1017/S0950268817000140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janda JM, Powers C, Bryant RG, et al. Current perspectives on the epidemiology and pathogenesis of clinically significant Vibrio spp. Clin Microbiol Rev. 1988;1(3):245–267. doi: 10.1128/CMR.1.3.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Q, Ji W, Xu Z.. Current use and development of fish vaccines in China. Fish Shellfish Immunol. 2020;96:223–234. doi: 10.1016/j.fsi.2019.12.010 [DOI] [PubMed] [Google Scholar]
- 9.Ruwandeepika HA, Defoirdt T, Bhowmick PP, et al. In vitro and in vivo expression of virulence genes in Vibrio isolates belonging to the Harveyi clade in relation to their virulence towards gnotobiotic brine shrimp (Artemia franciscana). Environ Microbiol. 2011;13(2):506–517. doi: 10.1111/j.1462-2920.2010.02354.x [DOI] [PubMed] [Google Scholar]
- 10.Oberbeckmann S, Wichels A, Maier T, et al. A polyphasic approach for the differentiation of environmental vibrio isolates from temperate waters. FEMS Microbiol Ecol. 2011;75(1):145–162. doi: 10.1111/j.1574-6941.2010.00998.x [DOI] [PubMed] [Google Scholar]
- 11.Kuang SF, Xiang J, Chen YT, et al. Exogenous pyruvate promotes gentamicin uptake to kill antibiotic-resistant Vibrio alginolyticus. Int J Antimicrob Agents. 2024;63(1):107036. doi: 10.1016/j.ijantimicag.2023.107036 [DOI] [PubMed] [Google Scholar]
- 12.Zhou K, Tian KY, Liu XQ, et al. Characteristic and otopathogenic analysis of a Vibrio alginolyticus strain responsible for chronic otitis externa in China. Front Microbiol. 2021;12:750642. doi: 10.3389/fmicb.2021.750642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu D, Liu H, Yang Z, et al. Chromatin immunoprecipitation sequencing technology reveals global regulatory roles of low-cell-density quorum-sensing regulator AphA in the pathogen vibrio alginolyticus. J Bacteriol. 2016;198(21):2985–2999. doi: 10.1128/JB.00520-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu C, Zhao Z, Liu Y, et al. Type III secretion 1 effector gene diversity among vibrio isolates from coastal areas in China. Front Cell Infect Microbiol. 2020;10:301. doi: 10.3389/fcimb.2020.00301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheng L, Lv Y, Liu Q, et al. Connecting type VI secretion, quorum sensing, and c-di-GMP production in fish pathogen Vibrio alginolyticus through phosphatase PppA. Vet Microbiol. 2013;162(2-4):652–662. doi: 10.1016/j.vetmic.2012.09.009 [DOI] [PubMed] [Google Scholar]
- 16.Huang Z, Yu K, Fang Y, et al. Comparative genomics and transcriptomics analyses reveal a unique environmental adaptability of vibrio fujianensis. Microorganisms. 2020;8(4):555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montieri S, Suffredini E, Ciccozzi M, et al. Phylogenetic and evolutionary analysis of Vibrio parahaemolyticus and Vibrio alginolyticus isolates based on toxR gene sequence. New Microbiol. 2010;33(4):359–372. [PubMed] [Google Scholar]
- 18.Yingkajorn M, Sermwitayawong N, Palittapongarnpimp P, et al. Vibrio parahaemolyticus and its specific bacteriophages as an indicator in cockles (Anadara granosa) for the risk of V. parahaemolyticus infection in Southern Thailand. Microb Ecol. 2014;67(4):849–856. doi: 10.1007/s00248-014-0382-9 [DOI] [PubMed] [Google Scholar]
- 19.Salamone M, Nicosia A, Ghersi G, et al. Vibrio proteases for biomedical applications: modulating the proteolytic secretome of V. alginolyticus and V. parahaemolyticus for improved enzymes production. Microorganisms. 2019;7(10):387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu S, Nishikino T, Hu B, et al. Molecular architecture of the sheathed polar flagellum in Vibrio alginolyticus. Proc Natl Acad Sci U S A. 2017;114(41):10966–10971. doi: 10.1073/pnas.1712489114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan F, Tabassum N, Anand R, et al. Motility of Vibrio spp.: regulation and controlling strategies. Appl Microbiol Biotechnol. 2020;104(19):8187–8208. doi: 10.1007/s00253-020-10794-7 [DOI] [PubMed] [Google Scholar]
- 22.Echazarreta MA, Klose KE.. Vibrio flagellar synthesis. Front Cell Infect Microbiol. 2019;9:131. doi: 10.3389/fcimb.2019.00131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu XF, Cao Y, Zhang HL, et al. Complete genome sequence of Vibrio alginolyticus ATCC 17749 T. Genome Announc. 2015;3(1):45–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang P, Wen Z, Li B, et al. Complete genome sequence of Vibrio alginolyticus ATCC 33787(T) isolated from seawater with three native megaplasmids. Mar Genomics. 2016;28:45–47. doi: 10.1016/j.margen.2016.05.003 [DOI] [PubMed] [Google Scholar]
- 25.Chibani CM, Roth O, Liesegang H, et al. Genomic variation among closely related Vibrio alginolyticus strains is located on mobile genetic elements. BMC Genomics. 2020;21(1):354. doi: 10.1186/s12864-020-6735-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gurevich A, Saveliev V, Vyahhi N, et al. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. doi: 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyatt D, Chen GL, Locascio PF, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 30.Richter M, Rosselló-Móra R.. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106(45):19126–19131. doi: 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Page AJ, Cummins CA, Hunt M, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3693. doi: 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu L, Niu B, Zhu Z, et al. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28(23):3150–3152. doi: 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Croucher NJ, Page AJ, Connor TR, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43(3):e15. doi: 10.1093/nar/gku1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tonkin-Hill G, Lees JA, Bentley SD, et al. Fast hierarchical Bayesian analysis of population structure. Nucleic Acids Res. 2019;47(11):5539–5549. doi: 10.1093/nar/gkz361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jain C, Rodriguez RL, Phillippy AM, et al. High throughput ANI analysis of 90 K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9(1):5114. doi: 10.1038/s41467-018-07641-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein SL, Gutierrez West CK, Mejia DM, et al. Genes similar to the Vibrio parahaemolyticus virulence-related genes tdh, tlh, and vscC2 occur in other vibrionaceae species isolated from a pristine estuary. Appl Environ Microbiol. 2014;80(2):595–602. doi: 10.1128/AEM.02895-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Broberg CA, Calder TJ, Orth K.. Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 2011;13(12-13):992–1001. doi: 10.1016/j.micinf.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lantagne D, Balakrish Nair G, Lanata CF, et al. The cholera outbreak in Haiti: where and how did it begin? Curr Top Microbiol Immunol. 2014;379:145–164. [DOI] [PubMed] [Google Scholar]
- 39.Falkowski PG, Fenchel T, Delong EF.. The microbial engines that drive Earth's biogeochemical cycles. Science. 2008;320(5879):1034–1039. doi: 10.1126/science.1153213 [DOI] [PubMed] [Google Scholar]
- 40.Aminov RI. Horizontal gene exchange in environmental microbiota. Front Microbiol. 2011;2:158. doi: 10.3389/fmicb.2011.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicholson E, Keith DA, Wilcove DS.. Assessing the threat status of ecological communities. Conserv Biol. 2009;23(2):259–274. doi: 10.1111/j.1523-1739.2008.01158.x [DOI] [PubMed] [Google Scholar]
- 42.Harrisson KA, Pavlova A, Gonçalves da Silva A, et al. Scope for genetic rescue of an endangered subspecies though re-establishing natural gene flow with another subspecies. Mol Ecol. 2016;25(6):1242–1258. doi: 10.1111/mec.13547 [DOI] [PubMed] [Google Scholar]
- 43.Pang H, Li W, Zhang W, et al. Acetylome profiling of Vibrio alginolyticus reveals its role in bacterial virulence. J Proteomics. 2020;211:103543. doi: 10.1016/j.jprot.2019.103543 [DOI] [PubMed] [Google Scholar]
- 44.Hernández-Robles MF, Álvarez-Contreras AK, Juárez-García P, et al. Virulence factors and antimicrobial resistance in environmental strains of Vibrio alginolyticus. Int Microbiol. 2016;19(4):191–198. [DOI] [PubMed] [Google Scholar]
- 45.Syed KA, Beyhan S, Correa N, et al. The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J Bacteriol. 2009;191(21):6555–6570. doi: 10.1128/JB.00949-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takekawa N, Isumi M, Terashima H, et al. Structure of vibrio FliL, a new stomatin-like protein that assists the bacterial flagellar motor function. mBio. 2019;10(2):e00292–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ringgaard S, Hubbard T, Mandlik A, et al. RpoS and quorum sensing control expression and polar localization of Vibrio cholerae chemotaxis cluster III proteins in vitro and in vivo. Mol Microbiol. 2015;97(4):660–675. doi: 10.1111/mmi.13053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Makino K, Oshima K, Kurokawa K, et al. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae . Lancet. 2003;361(9359):743–749. doi: 10.1016/S0140-6736(03)12659-1 [DOI] [PubMed] [Google Scholar]
- 49.Zhao Z, Chen C, Hu CQ, et al. The type III secretion system of Vibrio alginolyticus induces rapid apoptosis, cell rounding and osmotic lysis of fish cells. Microbiology (Reading). 2010;156(Pt 9):2864–2872. doi: 10.1099/mic.0.040626-0 [DOI] [PubMed] [Google Scholar]
- 50.Zhao Z, Liu J, Deng Y, et al. The Vibrio alginolyticus T3SS effectors, Val1686 and Val1680, induce cell rounding, apoptosis and lysis of fish epithelial cells. Virulence. 2018;9(1):318–330. doi: 10.1080/21505594.2017.1414134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burdette DL, Yarbrough ML, Orvedahl A, et al. Vibrio parahaemolyticus orchestrates a multifaceted host cell infection by induction of autophagy, cell rounding, and then cell lysis. Proc Natl Acad Sci U S A. 2008;105(34):12497–12502. doi: 10.1073/pnas.0802773105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burdette DL, Yarbrough ML, Orth K.. Not without cause: Vibrio parahaemolyticus induces acute autophagy and cell death. Autophagy. 2009;5(1):100–102. doi: 10.4161/auto.5.1.7264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yarbrough ML, Li Y, Kinch LN, et al. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science. 2009;323(5911):269–272. doi: 10.1126/science.1166382 [DOI] [PubMed] [Google Scholar]
- 54.McCarter LL. Polar flagellar motility of the Vibrionaceae. Microbiol Mol Biol Rev. 2001;65(3):445–462. table of contents. doi: 10.1128/MMBR.65.3.445-462.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jung YC, Lee MA, Kim HS, et al. Role of DegQ in differential stability of flagellin subunits in Vibrio vulnificus . NPJ Biofilms Microbiomes. 2021;7(1):32. doi: 10.1038/s41522-021-00206-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liang H, Xia L, Wu Z, et al. Expression, characterization and immunogenicity of flagellin FlaC from Vibrio alginolyticus strain HY9901. Fish Shellfish Immunol. 2010;29(2):343–348. doi: 10.1016/j.fsi.2010.04.003 [DOI] [PubMed] [Google Scholar]
- 57.Mobley HL, Belas R, Lockatell V, et al. Construction of a flagellum-negative mutant of Proteus mirabilis: effect on internalization by human renal epithelial cells and virulence in a mouse model of ascending urinary tract infection. Infect Immun. 1996;64(12):5332–5340. doi: 10.1128/iai.64.12.5332-5340.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verma J, Bag S, Saha B, et al. Genomic plasticity associated with antimicrobial resistance in Vibrio cholerae. Proc Natl Acad Sci U S A. 2019;116(13):6226–6231. doi: 10.1073/pnas.1900141116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kang CH, Shin Y, Jang S, et al. Antimicrobial susceptibility of Vibrio alginolyticus isolated from oyster in Korea. Environ Sci Pollut Res Int. 2016;23(20):21106–21112. doi: 10.1007/s11356-016-7426-2 [DOI] [PubMed] [Google Scholar]
- 60.Chang C, Jin X, Chaoqun H.. Phenotypic and genetic differences between opaque and translucent colonies of Vibrio alginolyticus. Biofouling. 2009;25(6):525–531. doi: 10.1080/08927010902964578 [DOI] [PubMed] [Google Scholar]
- 61.Oh EG, Son KT, Yu H, et al. Antimicrobial resistance of Vibrio parahaemolyticus and Vibrio alginolyticus strains isolated from farmed fish in Korea from 2005 through 2007. J Food Prot. 2011;74(3):380–386. doi: 10.4315/0362-028X.JFP-10-307 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.