Abstract
Azotobacter vinelandii produces two polymers: the extracellular polysaccharide alginate and the intracellular polyester poly-β-hydroxybutyrate (PHB). A cosmid clone (pSMU588) from an A. vinelandii gene library diminished alginate production by A. vinelandii mucoid strain ATCC 9046. The nucleotide sequence and predicted amino acid sequence of the locus responsible for the mucoidy suppression revealed 65% identity to Pseudomonas GacS, a transmembrane sensor kinase of the two-component regulators, whose cognate response regulator, GacA, is a global activator regulating several products and virulence factors. Plasmid pMC15, harboring gacS, and a strain carrying a gacS nonpolar mutation were constructed. Either pMC15 or the gacS mutation significantly reduced alginate production and transcription of algD, the gene coding for the key enzyme GDP-mannose dehydrogenase of the alginate biosynthetic pathway. We found that the gacS mutation also reduced PHB accumulation and impaired encystment. Taken together, these data indicate that in A. vinelandii the gacSA global system regulates polymer synthesis.
Azotobacter vinelandii is a nitrogen-fixing soil bacterium that undergoes differentiation to form desiccation-resistant cysts and produces two polymers of industrial importance: alginate and poly-β-hydroxybutyrate (PHB). Both polymers are involved in the encystment process; alginate is a component of the cyst capsule (28), and PHB accumulation correlates with the frequency of cyst formation (30).
A. vinelandii has been shown to possess an alginate biosynthetic gene cluster (2, 16, 20, 25, 32), organized in three operons, one of which transcribes algD encoding a GDP-mannose dehydrogenase, which converts GDP-mannose to GDP-mannuronic acid, the substrate for alginate polymerization. The algUmucABCD cluster controls alginate production. AlgU is a ςE homolog (19). The mucA and mucB genes code for negative regulators of AlgU activity. In strain ATCC 9046, transcription of the algD gene is initiated at three sites, one of which is AlgU dependent (2). AlgU activity was shown to be involved in the encystment process independent of its role in alginate synthesis (23).
Three enzymes are involved in PHB biosynthesis in A. vinelandii: a β-ketothiolase, an acetoacetyl-coenzyme A reductase and a PHB synthetase (18). PHB synthesis in A. vinelandii was shown to be regulated at the level of the β-ketothiolase activity (18). The genes encoding the enzymes participating in PHB synthesis in A. vinelandii have not been reported.
Members of our group have previously reported the identification of cosmid pSMU588 from a gene bank of nonmucoid strain UW136, which reduced alginate production in A. vinelandii ATCC 9046 (19). The characterization of pSMU588 reported here allowed us to identify a regulatory gene homologous to Pseudomonas gacS, coding for a sensor kinase of the two-component regulatory systems. This study also provides evidence for GacS playing a role as a regulator of alginate and PHB synthesis in A. vinelandii.
Identification of the locus responsible for the suppression of mucoidy.
Mini-Tn5-lacZ1 (6) mutagenesis of plasmid pSMU588 was carried out to identify the locus responsible for the reduction of alginate. A pSMU588::mini-Tn5-lacZ1 derivative that no longer suppressed mucoidy was isolated and named pSMU588-21. Alginate production of ATCC 9046 carrying this plasmid was determined and the results are shown in Table 1.
TABLE 1.
Polymer production and encystment in A. vinelandii strainsa
| Strain | Alginate production (μg/mg of protein)
|
PHB production (μg/mg of protein) | Encystment (%) | ||
|---|---|---|---|---|---|
| BSb
|
BBc | ||||
| Liquid | Solid | ||||
| ATCC 9046 | 1,667 ± 11 | 1,246 ± 15 | 1,717 ± 11 | 413.1 ± 17.8 | 7.2 ± 1.5 |
| ATCC 9046/pSMU588 | 666 ± 39 | NDd | ND | ND | ND |
| ATCC 9046/pSMU588-21 | 1,320 ± 68 | ND | ND | ND | ND |
| ATCC 9046/pMC15 | 16 ± 1.8 | ND | ND | 48.5 ± 0.5 | ND |
| JM1 | 19 ± 1.5 | 411 ± 25 | 38 ± 2.8 | ND | <0.0001 |
| JM2 | 36 ± 2.3 | 465 ± 53 | 29 ± 3.5 | 49.6 ± 0.9 | ND |
Alginate production was determined as previously described (21); all determinations were done in triplicate. PHB production was determined as previously described (29), in cells grown for 48 h in Burk's liquid medium supplemented with 2% sucrose. Values are means ± standard errors of the means.
BS, Burk's medium supplemented with 2% sucrose as the carbon source.
BB, Burk's medium supplemented with 0.2% n-butanol as the carbon source.
ND, not determined.
We determined 3 kb of nucleotide sequence around the mini-Tn5-lacZ1 insertion. DNA sequence analysis revealed one open reading frame encoding a polypeptide of 905 amino acid residues, sharing similarity with transmembrane sensor kinases belonging to signal transduction proteins of the family of two-component regulators (12). Among those with the highest identity (65%) was GacS (previously called LemA), which is present in the following Pseudomonas species: P. syringae (13), P. viridiflava (15), P. fluorescens (5), P. aureofaciens (3), and P. tolaasii (11). In P. syringae, GacS and its cognate response regulator, GacA, are required for the production of the toxin syringomycin and for extracellular proteases (27); in P. viridiflava, GacS controls protease, pectate lyase and alginate production (15); and in P. fluorecens, GacS controls antibiotic production (5). A gacS homolog, rpfA, which regulates the production of virulence factors, has been reported in the phytopathogenic bacterium Erwinia carotovora (8). In P. aeruginosa and P. aureofaciens, GacA, the GacS cognate response regulator, has been shown to be a global activator regulating several products and virulence factors via acyl-l-homoserine lactones (3, 26).
The A. vinelandii GacS amino acid sequence contains specific motifs typical of the ArcB subfamily, histidine protein kinases that are characterized by containing, in addition to the two transmembrane domains and the orthodox transmitter H1 domain, a response regulator D1 domain and a second phosphorylatable histidine, H2 (9, 14). The exact location of the mini-Tn5-lacZ1 mutation within gacS was mapped between codons encoding amino acids 735 and 736.
Plasmid pMC15.
To rule out the possibility that plasmid pSMU588-21 no longer suppressed alginate production due to a polar effect of the gacS::mini-Tn5-lacZ1 insertion rather than to the lack of the gacS gene product, oligonucleotides gacS1 (5′-AAGCGGAGCTCGAGCCGTCAGG-3′) and gacS2 (5′-ACGGTGCCGTCTCGAGTTTCCGCTC-3′) were used to isolate, by PCR, a fragment containing the gacS gene flanked by the ATG codon (200 bp upstream) and the stop codon (20 bp downstream); this fragment was cloned into plasmid pKT230 (1). The resultant plasmid, pMC15, was found to have a much stronger effect on alginate production (Table 1), confirming the negative effect of gacS on alginate production and ruling out the possibility of a polar effect.
Construction and characterization of gacS mutants.
Sensor kinases of the two-component regulatory systems usually act as positive regulators by phosphorylating the cognate response regulator. To investigate a positive role of gacS on alginate biosynthesis, ATCC 9046 derivatives carrying gacS mutations were constructed: in A. vinelandii the insertion of Ω cassettes into genes with the same orientation as the direction of transcription produces nonpolar mutations which allow transcription of the downstream genes in the same operon (21, 24). Plasmid pMC5, a pBluescript KS(+) plasmid which carries a 2.5-kb SmaI DNA fragment including gacS, was used to construct gacS::Ω-Sp mutations. A 2-kb fragment containing a spectinomycin resistance gene (Ω-Sp) from plasmid pHP45Ω-Sp (7) was inserted into the unique EcoRI site present within gacS. Plasmids pMC7 and pMC8 with the cassette inserted in both orientations were selected and introduced into strain ATCC 9046. Spr transformants were selected. Strains JM1 and JM2 were isolated and were shown, by Southern blot analysis, to carry the gacS::Ω-Sp nonpolar and polar mutations, respectively (data not shown). RNA isolated from strains JM1, JM2, and ATCC 9046 were hybridized with a 700-bp fragment corresponding to the 3′ end of gacS. Hybridization of RNA from the wild type with that of the gacS nonpolar JM1 mutant but not with that of the JM2 polar mutant was observed (data not shown). Both gacS mutant strains produced threefold less alginate than the parental strain, ATCC 9046, when grown on solid medium (Table 1). When cultures of these mutants were grown on liquid medium, a 30- to 60-fold reduction in alginate production was detected. These data indicate that the negative effect on alginate production observed is due to the lack of the gacS gene product and not to a polar effect.
GacS also controls PHB production.
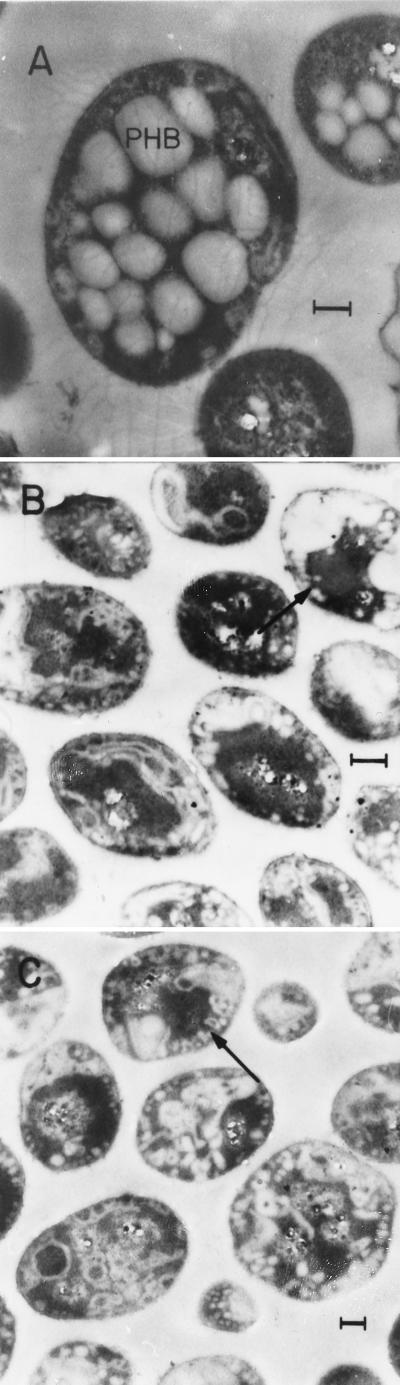
PHB granules were visible in cells of wild-type ATCC 9046 under a light microscope; however, no PHB granules were observed in cells of strains JM1 or JM2. PHB accumulation in these strains was determined. As shown in Table 1, either a gacS mutation or plasmid pMC15 caused a significant reduction in PHB accumulation. Electron microscopic examination of cultures of ATCC 9046, JM2, and ATCC 9046/pMC15 was carried out as described previously (21), and the results are shown in Fig. 1. In contrast to the wild-type ATCC 9046, where big PHB granules are observed, strains JM2 and ATCC 9046/pMC15 appear to contain numerous very small PHB granules and disorganized or amorphic white structures that appear to contain PHB. These observations indicate that GacS also regulates PHB accumulation.
FIG. 1.
Electron micrographs of A. vinelandii cells grown for 48 h in Burk's medium supplemented with 2.0% sucrose. (A) Strain ATCC 9046; (B) strain JM2; (C) strain ATCC 9046/pMC15. Arrows indicate small PHB granules. Bars, 0.4 μm.
The effect of the gacS mutation on alginate and PHB production indicates that GacS plays a positive role in the regulation of polymer synthesis in A. vinelandii. This role is likely due to the kinase activity of GacS that results in phosphorylation of GacA, leading to activation of alginate and PHB genes. On the other hand, in the presence of the gacS gene cloned into the pKT230 vector, with a copy number of 15 to 20 (4), the synthesis of these polymers is significantly reduced. A similar observation was reported for the Escherichia coli histidine protein kinase RcsC, which, together with RcsA, positively regulates synthesis of colanic acid, a component of the capsule conferring resistance to desiccation in E. coli (31). Cells carrying a multicopy rcsC plasmid were shown to reduce the level of colanic acid (10). However, the negative effects observed when two-component regulatory systems are present on multicopy plasmids do not necessarily reflect a natural situation; thus, the reduced alginate level may represent an artifact.
The algD gene is a target of signal transduction by GacS.
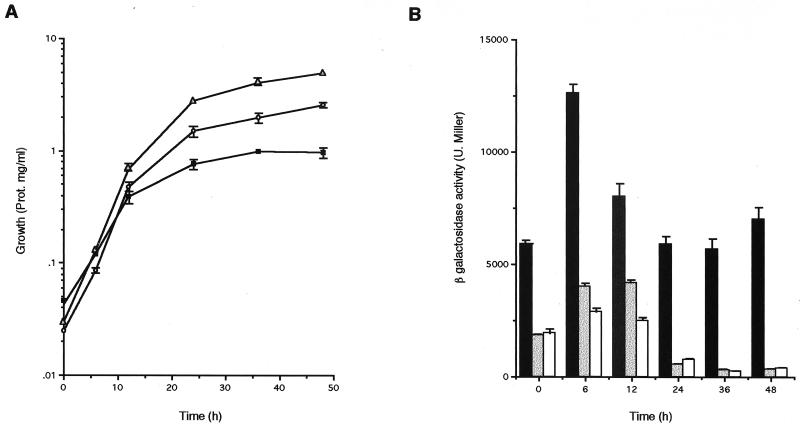
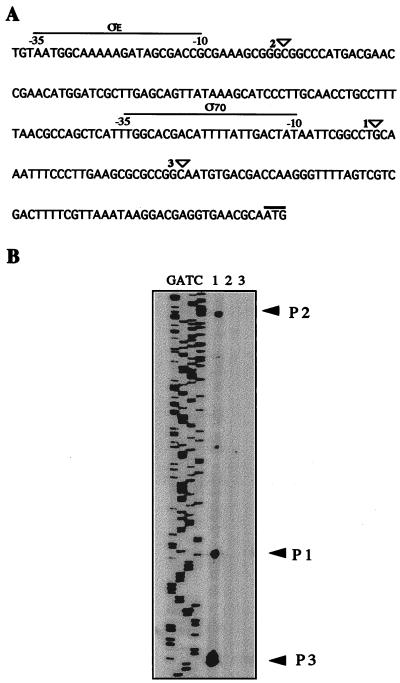
algD, the gene encoding GDP-mannose dehydrogenase, the key enzyme in the alginate biosynthetic pathway, seems to be highly regulated, since its transcription can initiate from three different sites: p1, a ς70 type of promoter (2); p2, controlled by ςE (23); and p3. We determined whether the effect of GacS on alginate biosynthesis was exerted on the transcription of the algD gene by measuring the β-galactosidase activity of strain WI12, an ATCC 9046 derivative carrying an algD::lacZ gene fusion (in the presence and absence of plasmid pMC15), as well as of strain WI12-1, a WI12 derivative carrying the gacS::Ω-Sp nonpolar mutation. The β-galactosidase activity was measured as reported by Miller (22); 1 U corresonds to 1 nmol of O-nitrophenyl-β-d-galactoside hydrolyzed per min per μg of protein. Protein was determined by the Lowry method (17). All measurements were done in triplicate. The β-galactosidase activity was measured during growth on Burk's medium supplemented with 2% sucrose (Fig. 2A). In both the WI12-1 and WI12/pMC15 strains the β-galactosidase activity was reduced (Fig. 2B). We determined which of the algD promoters was regulated by GacS. Primer extension of algD on RNA isolated from strains JM1 and ATCC 9046/pMC15 was performed as previously described (2). No primer extension products were detected with RNA from these strains grown for 48 h in liquid Burk's medium supplemented with 2% sucrose (Fig. 3). This result is in agreement with the low β-galactosidase activity level detected under the same conditions. These data show that, in regulating alginate synthesis GacS exerts influence on transcription of algD from its three promoters. Similarly, in Pseudomonas fluorecens GacS and GacA regulate gene expression by influencing the ςS levels in addition to being required for expression of genes not regulated by ςS (33). Thus, the GacSA system controls expression from promoters recognized by different sigma factors. In this regard, it is possible that the putative ς70 algD promoter, P1, may instead be a ςS-dependent promoter.
FIG. 2.
Growth (A) and β-galactosidase activity (B) on Burk's medium supplemented with 2% sucrose are shown for the strains WI12 (squares and black bars), WI12-1 (triangles and grey bars), and WI12/pMC15 (circles and white bars). Prot., protein.
FIG. 3.
Primer extension analysis of algD transcription. (A) DNA sequence of the 5′ end of algD. Triangles indicate the start sites of algD transcription. The ATG initiation codon is overlined. (B) Primer extension of the algD gene in strains ATCC 9046 (lane 1), JM2 (lane 2), and ATCC 9046/pMC15 (lane 3).
GacA must mediate signal transduction between GacS and algD. Members of our group have identified in the A. vinelandii chromosome a gene homologous to gacA (M. Castañeda, J. Sánchez, and G. Espín, unpublished results). Whether GacA directly interacts with the algD promoter region remains to be determined. In P. aureofaciens, GacA functions upstream of the PhzR regulator, in the signal transduction pathway that regulates antibiotic synthesis via acyl-homoserine lactones (3); thus, it is possible that in A. vinelandii other regulatory proteins mediate signal transduction between GacS, GacA, and algD.
It is difficult to speculate on the target of signal transduction by GacS in PHB synthesis, since regulation of the A. vinelandii phb biosynthetic genes is presently unknown. The A. vinelandii gacS mutation caused a PHB leaky phenotype and affected the size of the PHB granules. Similarly, in Ralstonia eutropha, phaP, a locus causing a PHB leaky phenotype, encodes a PHB-binding protein that determines the size of polyhydroxyalkanoic acid granules (34). Thus, gacS could be involved in the control of a gene homologous to phaP.
Effect of the gacS mutation on encystment.
Alginate has been shown to be essential for the formation of mature cysts (2). As shown above, strain JM1 produces some alginate on plates of Burk's medium supplemented with 2% sucrose, and strains producing similar or lower levels of alginate are still able to form cysts (23); we analyzed the encysting capacity of strain JM1, measuring desiccation resistance of cultures induced for encystment as previously described (2). A reduction of more than 1,000-fold in encystment frequency was observed in the gacS mutant JM1 (Table 1). However under encysting conditions, i.e., in n-butanol plates, alginate production by strain JM1 was very low. Thus, as is the case with other mutants impaired in alginate production, strain JM1 is unable to form desiccation-resistant cysts. GacS may affect this differentiation process exclusively via its effect on alginate biosynthesis; however, whether this global regulator is required for expression of other genes involved in encystment remains to be investigated.
Nucleotide sequence accession number.
The nucleotide sequence determined in this study has the GenBank accession no. AF197912.
Acknowledgments
This work was supported by grant 27767 from CONACyT. M. Castañeda thanks CONACyT and PADEP-UNAM for financial support during his Ph.D. studies.
We thank G. Soberón, D. Segura, and C. Núñez for helpful discussions. We acknowledge R. Nájera for technical support.
REFERENCES
- 1.Bagdasarian M, Lurz R, Ruckert B, Franklin F C H, Bagdasarian M M, Frey J, Timmis K N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 2.Campos M E, Martínez-Salazar J M, Lloret L, Moreno S, Núñez C, Espín G, Soberón-Chávez G. Characterization of the gene coding for GDP-mannose dehydrogenase (algD) from Azotobacter vinelandii. J Bacteriol. 1996;178:1793–1799. doi: 10.1128/jb.178.7.1793-1799.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chancey S T, Wood D W, Pierson L S., III Two-component transcriptional regulation of N-acyl-homoserine lactone production in Pseudomonas aureofaciens. Appl Environ Microbiol. 1999;65:2294–2299. doi: 10.1128/aem.65.6.2294-2299.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christopher F, Franklin K. Broad host range cloning vectors for gram negative bacteria. In: Glover D M, editor. DNA cloning. Vol. 1. Washington, D.C.: IRL Press; 1985. pp. 165–184. [Google Scholar]
- 5.Corbell N, Loper J E. A global regulator of secondary metabolite production in Pseudomonas fluorescens Pf5. J Bacteriol. 1995;177:6230–6236. doi: 10.1128/jb.177.21.6230-6236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Lorenzo V, Herrero M, Jakubzik V, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 8.Frederick R D, Chiu J, Bennetzen J L, Handa A K. Identification of a pathogenicity locus, rpfA, in Erwinia carotovora that encodes a two component sensor-regulator protein. Mol Plant-Microbe Interact. 1997;7:455–463. doi: 10.1094/MPMI.1997.10.3.407. [DOI] [PubMed] [Google Scholar]
- 9.Georgellis D, Linch A S, Lin E C C. In vitro phosphorylation study of the Arc two component system of Escherichia coli. J Bacteriol. 1997;179:5429–5435. doi: 10.1128/jb.179.17.5429-5435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gervais F G, Phoenix P, Drapeau G R. The rcsB gene, a positive regulator of colanic acid biosynthesis in Escherichia coli, is also an activator of ftsZ expression. J Bacteriol. 1992;174:3964–3971. doi: 10.1128/jb.174.12.3964-3971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han B, Arnab P, Johnstone K. Spontaneous duplication of 661 bp element within a two component sensor regulator gene causes phenotypic switching in colonies of Pseudomonas tolaasii, cause of brown blotch disease of mushrooms. Mol Microbiol. 1997;25:211–218. doi: 10.1046/j.1365-2958.1997.4411811.x. [DOI] [PubMed] [Google Scholar]
- 12.Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. [Google Scholar]
- 13.Hrabak E M, Willis D K. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J Bacteriol. 1992;174:3011–3020. doi: 10.1128/jb.174.9.3011-3020.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishige K, Nagasawa S, Tokishita S, Mizuno T. A novel device of bacterial signal transducers. EMBO J. 1994;13:5195–5202. doi: 10.1002/j.1460-2075.1994.tb06850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao C H, MacCullus D E, Fett W F. Molecular characterization of two gene loci required for production of the key pathogenicity factor pectate lyase in Pseudomonas viridiflava. Mol Plant-Microbe Interact. 1994;7:391–400. doi: 10.1094/mpmi-7-0391. [DOI] [PubMed] [Google Scholar]
- 16.LLoret L, Barreto R, León R, Moreno S, Martínez-Salazar J, Espín G, Soberón-Chávez G. Genetic analysis of the transcriptional arrangement of Azotobacter vinelandii alginate biosynthetic genes: identification of two independent promoters. Mol Microbiol. 1996;21:449–457. doi: 10.1111/j.1365-2958.1996.tb02554.x. [DOI] [PubMed] [Google Scholar]
- 17.Lowry O H, Rosebrough N J, Farr A L, Randall J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 18.Manchak J, Page W J. Control of polyhydroxyalkanoate synthesis in Azotobacter vinelandii strain UWD. Microbiology. 1994;140:953–963. [Google Scholar]
- 19.Martínez-Salazar J M, Moreno S, Nájera R, Boucher J C, Espín G, Soberón-Chávez G, Deretic V. Characterization of the genes coding for the putative sigma factor AlgU and its regulators MucA, MucB, MucC, and MucD in Azotobacter vinelandii and evaluation of their roles in alginate biosynthesis. J Bacteriol. 1996;178:1800–1808. doi: 10.1128/jb.178.7.1800-1808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mejía-Ruíz H, Guzmán J, Moreno S, Soberón-Chávez G, Espín G. The Azotobacter vinelandii alg8 and alg44 genes are essential for alginate synthesis and can be transcribed from an algD-independent promoter. Gene. 1997;199:271–277. doi: 10.1016/s0378-1119(97)00380-6. [DOI] [PubMed] [Google Scholar]
- 21.Mejía-Ruíz H, Moreno S, Guzmán J, Nájera R, León R, Soberón-Chávez G, Espín G. Isolation and characterization of an Azotobacter vinelandii algK mutant. FEMS Microbiol Lett. 1997;156:101–106. doi: 10.1111/j.1574-6968.1997.tb12712.x. [DOI] [PubMed] [Google Scholar]
- 22.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. pp. 431–435. [Google Scholar]
- 23.Moreno S, Guzmán J, Nájera R, Soberón-Chávez G, Espín G. Role of the alternative ς factor AlgU in encystment of Azotobacter vinelandii. J Bacteriol. 1998;180:2766–2769. doi: 10.1128/jb.180.10.2766-2769.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mouncey N J, Mitchell L A, Pau R. Mutational analysis of genes of the mod locus involved in molybdenum transport, homeostasis, and processing in Azotobacter vinelandii. J Bacteriol. 1995;177:5294–5302. doi: 10.1128/jb.177.18.5294-5302.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rehm H A, Ertesvag H, Valla S. A new A. vinelandii mannuronan C-5 epimerase gene (algG) is part of an alg gene cluster physically organized in a manner similar to that in P. aeruginosa. J Bacteriol. 1996;178:5884–5889. doi: 10.1128/jb.178.20.5884-5889.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A, Haas D. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol Microbiol. 1997;24:309–319. doi: 10.1046/j.1365-2958.1997.3291701.x. [DOI] [PubMed] [Google Scholar]
- 27.Rich J J, Kinscherf T G, Kitten T, Willis D K. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J Bacteriol. 1994;176:7468–7475. doi: 10.1128/jb.176.24.7468-7475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sadoff H L. Encystment and germination in Azotobacter vinelandii. Bacteriol Rev. 1975;39:516–539. doi: 10.1128/br.39.4.516-539.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segura D, Espín G. Mutational inactivation of a gene homologous to Escherichia coli ptsP affects poly-β-hydroxybutyrate accumulation and nitrogen fixation in Azotobacter vinelandii. J Bacteriol. 1998;180:4790–4798. doi: 10.1128/jb.180.18.4790-4798.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevenson L H, Socolofsky M D. Cyst formation and poly-hydroxybutyric acid accumulation in Azotobacter. J Bacteriol. 1966;91:304–310. doi: 10.1128/jb.91.1.304-310.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stout V, Gottesman S. RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J Bacteriol. 1990;172:659–669. doi: 10.1128/jb.172.2.659-669.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vázquez A, Moreno S, Guzmán J, Alvarado A, Espín G. Transcriptional organization of the Azotobacter vinelandii algGXLIVFA genes: characterization of algF mutants. Gene. 1999;232:217–222. doi: 10.1016/s0378-1119(99)00119-5. [DOI] [PubMed] [Google Scholar]
- 33.Whistler C A, Corbell N A, Sarniguet A, Ream W, Loper J E. The two component regulators GacS and GacA influence accumulation of the stationary-phase sigma factor sigmaS and the stress response in Pseudomonas fluorescens Pf-5. J Bacteriol. 1998;180:6635–6641. doi: 10.1128/jb.180.24.6635-6641.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wieczorek R, Pries A, Steinbüchel A, Mayer F. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J Bacteriol. 1995;177:2425–2435. doi: 10.1128/jb.177.9.2425-2435.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]