Abstract
Coenzyme M (CoM) (2-mercaptoethanesulfonic acid) biosynthesis is shown to be coordinately regulated with the expression of the enzymes of alkene and epoxide metabolism in the propylene-oxidizing bacteria Xanthobacter strain Py2 and Rhodococcus rhodochrous strain B276. These results provide the first evidence for the involvement of CoM in propylene metabolism by R. rhodochrous and demonstrate for the first time the inducible nature of eubacterial CoM biosynthesis.
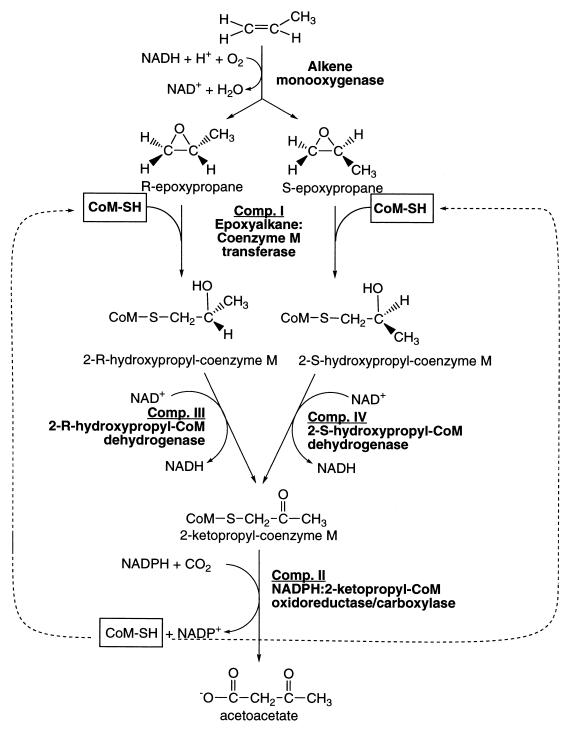
Recent studies of propylene metabolism in Xanthobacter strain Py2, a gram-negative proteobacterium, and Rhodococcus rhodochrous strain B276, a gram-positive actinomycete, have demonstrated the presence of a conserved bacterial pathway that involves oxidation to epoxypropane followed by carboxylation to acetoacetate (2, 4, 19). The latter transformation occurs in a sequence of reactions that use coenzyme M (CoM) (2-mercaptoethanesulfonic acid) as a nucleophile to open epoxide rings and as the carrier of intermediates ultimately cleaved and carboxylated to form acetoacetate (see Fig. 1) (1). The role of CoM as a C3 carrier in the epoxide carboxylation pathway is only the second defined function for this cofactor and its first observed usage in a eubacterial system. The other defined function of CoM is as an ubiquitous methyl group carrier in the pathway of methanogenesis in methanogenic archaea, where in the reductive cleavage of methyl-CoM results in the production of methane (25, 28). The newly discovered role for CoM in bacterial olefin metabolism raises interesting questions regarding the evolutionary relationships of bacterial and archaeal metabolism, especially in light of recent studies demonstrating the presence and usage of other methanogenic cofactors, specifically tetrahydromethanopterin and coenzyme F420, in eubacteria (9, 12, 15, 27).
FIG. 1.
Pathway of propylene metabolism in Xanthobacter strain Py2 and R. rhodochrous B276. Comp. I, epoxide carboxylase component I.
To date, CoM has been positively identified as an essential cofactor and as the C3 carrier in epoxide metabolism only in Xanthobacter strain Py2, which is a facultative methylotrophic bacterium (1). This observation may be significant, since the bacteria in which tetrahydromethanopterin has been identified are either obligate or facultative methylotrophs (27). Given the high degree of biochemical similarity between the epoxide carboxylation systems of Xanthobacter strain Py2 and R. rhodochrous (4, 6), it seems logical to predict that CoM is used as the C3 carrier in R. rhodochrous as well. These considerations also raise the questions of how widely CoM might be distributed within the bacterial domain, what additional functions it might play, and under what growth conditions it is produced.
In order to address the questions and considerations raised above, we have expressed the epoxyalkane:CoM transferase from Xanthobacter strain Py2 in Escherichia coli, an organism not thought to produce CoM (7). As purified from Xanthobacter strain Py2, the transferase contains tightly bound CoM (1). The addition of epoxypropane results in the stoichiometric reaction of the bound CoM and epoxypropane, which then dissociate from the transferase as the 2-hydroxypropyl-CoM adduct (see Fig. 1) (1). In this article we show that the transferase can be expressed in E. coli as a fully active enzyme that completely lacks CoM. We have exploited this property to develop a convenient bioassay for CoM that relies on recycling of CoM from various sources in epoxide carboxylation assays using the recombinant transferase with the additional complementing native components. These studies reveal that CoM biosynthesis is coordinately regulated with expression of the alkene- and epoxide-utilizing enzymes in both Xanthobacter strain Py2 and R. rhodochrous B276.
Growth of bacteria.
Xanthobacter strain Py2 and R. rhodochrous B276 (ATCC 31338) were grown as described previously (4). The carbon source for cell growth was one of the following: propylene (10% [vol/vol] gas phase), propane (30% [vol/vol] gas phase), acetate (20 mM), glucose (10 g/liter), acetone (40 mM), or isopropanol (40 mM). E. coli JM109 and E. coli BL21(DE3)/pLysS were grown in Luria-Bertani (LB) broth aerobically at 30°C and on LB agar aerobically at 37°C. Antibiotic concentrations used for selection and maintenance of plasmids were 100 μg of ampicillin per ml and 50 μg of chloramphenicol per ml.
Plasmid construction and purification of recombinant epoxyalkane:CoM transferase.
Frozen Xanthobacter Py2 cell paste (propylene grown) was thawed in 10 volumes of mineral salts medium containing 1% glycine. After the cell paste was thawed, the cell suspension was incubated at 30°C for 3 h in a sealed shaking flask containing 10% propylene. The cells were pelleted and resuspended in 1 volume of TEG (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 50 mM glucose) containing 1.5% sodium dodecyl sulfate (SDS) and incubated at 37°C for 20 min. High-molecular-weight DNA was then isolated by standard protocols (17). The gene encoding epoxyalkane:CoM transferase (herein after referred to xecA) was amplified by PCR using the primers 5′-CATATGCTGATCCGAGGGGAAGACG-3′ and 5′-GGATCCTCAGGCCGCCTGCTTGGCCT-3′. DNA amplification was performed in a Perkin-Elmer GeneAmp PCR Model 2400 with 30 cycles, with 1 cycle consisting of 30 s at 95°C, 30 s at 60°C, and 30 s at 72°C. The PCR product was digested with NdeI and BamHI and inserted into the expression vector T7-7 (21, 23). Ligated product was used to transform E. coli JM109, and plasmids were isolated using plasmid minipreps (Promega) and screened by single and double digestion with restriction endonucleases BamHI and NdeI. The properly ligated expression vector was labeled pJK1. E. coli BL21(DE3)/pLysS was transformed with pJK1 and grown in 2.5 liters of LB broth containing ampicillin and chloramphenicol at 30°C. Recombinant protein expression was induced with the addition of α-lactose to a final concentration of 1%. After a 5-h induction period, cells were harvested by centrifugation. Recombinant epoxyalkane:CoM transferase was subsequently purified using the protocol for the wild-type enzyme (5).
Sources of CoM for epoxide carboxylation assays.
Epoxide carboxylation assays (see below) were performed either with or without a supplemental source of CoM added. The supplemental source of CoM was either the commercially obtained compound (Sigma Chemicals), a preparation prepared from bacterial cell extract, or a preparation prepared from bacterial spent media. Cell extracts were prepared by thawing cells in 2 volumes of 100 mM Tris-HCl (pH 8.2), followed by lysis in a French pressure cell as described previously (4). The cell extract was clarified by ultracentrifugation at 137,000 × g for 45 min at 4°C. Clarified cell extract was boiled for 30 min and centrifuged at 14,000 × g for 20 min. A portion (1 ml) of the supernatant was lyophilized and resuspended in 300 μl of 100 mM Tris-HCl (pH 8.2) and added to the epoxide carboxylation assay mixture. Samples of authentic CoM at concentrations believed to be similar to that of cell extracts were treated identically in order to determine whether this treatment would damage the CoM (e.g., due to irreversible disulfide formation or other oxidative processes). It was found that the boiling-lyophilization treatment had no effect on subsequent usage of CoM in the assays. Spent media were prepared by retaining the supernatant obtained after centrifugation (14,000 × g for 30 min) of cultures of actively growing bacteria (A600 between 1 and 2). A portion (1 ml) of the supernatant was then lyophilized, resuspended in 300 μl of 100 mM Tris-HCl (pH 8.2), and added to the epoxide carboxylation assay mixture.
Assay of epoxide carboxylation reactions.
Epoxide carboxylase components I to IV (i.e., the four enzymes of Fig. 1) were purified from Xanthobacter strain Py2 as described previously (3, 5). The individual epoxide carboxylase protein components were assayed according to their respective reactions shown in Fig. 1 using the assays and conditions described previously (5). The complete epoxide carboxylation reaction, using racemic epoxypropane as the substrate, requires all four enzyme components shown in Fig. 1 in addition to CO2, NADPH, NAD+, and CoM according to equation 1 as follows:
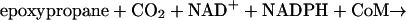 |
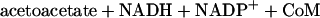 |
The complete assays (1-ml total volume) were performed as described previously (5), with the following modifications. Unless indicated otherwise, the amounts of the four proteins used in the assay were as follows: recombinant or native epoxyalkane:CoM transferase, 400 μg; NADPH:2-ketopropyl-CoM oxidoreductase/carboxylase, 5 mg; 2-R-hydroxypropyl-CoM dehydrogenase, 100 μg; and 2-S-hydroxypropyl-CoM dehydrogenase, 100 μg. These conditions were designed such that CoM, when added at low concentrations (i.e., low micromolar range), was the limiting component of the coupled assay. Supplemental CoM or a potential source of CoM (from cell extract or spent medium; see above) were either excluded from or included in individual assay mixtures as elaborated in the figure legends. Epoxypropane degradation and acetoacetate formation were monitored over time as described previously (1, 5).
Estimation of CoM concentrations from biological sources.
The concentrations of commercial CoM were 0 to 200 μM in epoxide carboxylation assay mixtures prepared as described above and using recombinant transferase as the source of component I. The rates of epoxypropane degradation were plotted against CoM concentration and fit to the equation for a rectangular hyperbola (i.e., the Michaelis-Menten equation) using the program SigmaPlot. The standard curve and equation were used to calculate CoM concentrations from initial rate data for assays where the CoM was from spent media.
Induction of CoM biosynthesis.
Shaking flask cultures of Xanthobacter strain Py2 and R. rhodochrous that had been grown with propylene were subcultured into shaking flasks containing medium in which acetate replaced propylene as the carbon source. After reaching stationary phase, the cells were subcultured again into acetate-containing medium, and this process was repeated two additional times. When the third subcultures reached an A600 of 0.9, propylene was added as overpressure to a final concentration of 10% (vol/vol) gas phase. The air and propylene in the flasks were subsequently replenished at 8-h intervals. At various times, cells were removed from cultures and assayed for epoxypropane degradation activity as described previously (3). Spent media were also prepared from the cultures at various times by the procedure described above. For controls, cultures were treated identically to those induced by addition of propylene but lacked propylene during the course of the experiment.
Heterologous expression of CoM-free epoxypropane:CoM transferase.
The high specificity of CoM as the nucleophilic thiolate for epoxide carboxylation (1) and its ability to be indefinitely recycled for additional epoxide to β-ketoacid conversions provide a potentially sensitive assay for assessing the presence or absence of this cofactor in different organisms. In order for this assay to be effective, CoM must be completely absent from the source of transferase used in the assay. One means to accomplish this would be to express the transferase in and purify it from an organism that does not contain CoM. Accordingly, the gene encoding the transferase was overexpressed in E. coli, which is believed not to contain CoM, based on the extensive studies performed by Balch and Wolfe (7).
Epoxalkane:CoM transferase was expressed at high levels (∼5% of cellular protein) from a T7-7 expression vector in lactose-induced E. coli cultures. The purified recombinant CoM (rCoM) transferase appeared identical to the native enzyme on SDS-polyacrylamide gels, migrated identically to the native enzyme on nondenaturing polyacrylamide gels, and eluted identically when chromatographed on a Superose 12 size exclusion column. Metal analysis of rCoM transferase showed the presence of 1.2 zinc molecules per monomeric unit, a value similar to that reported previously for the native enzyme (0.85 Zn/monomer). Together, these results suggest that the quaternary structure (i.e., α6) and cofactor complement are identical for the native and rCoM transferase.
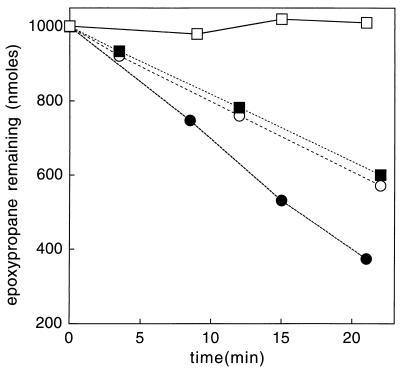
Figure 2 shows the time courses for epoxypropane degradation in complete epoxide carboxylation assay mixtures (equation 1) where the source of transferase was either the native or recombinant protein. In the absence of exogenously added CoM, no detectable activity was observed in the assay mixture containing rCoM transferase, while a low and sustained rate of epoxide degradation was observed in the assay mixture containing the same amount of the native transferase. As shown in Fig. 2, the addition of 5 μM CoM to the assay mixture containing rCoM transferase resulted in a linear rate of epoxide degradation that was nearly identical to the rate observed for the native transferase to which no CoM was added. Thus, it would appear that approximately 5 μM CoM was associated with the sample of native transferase used in this experiment. For this particular experiment, 400 μg of transferase was included in the assay mixtures, an amount that corresponds to a concentration of 9.6 μM transferase monomers. This suggests a complement of 0.5 CoM bound/native transferase active site for this enzyme preparation. The addition of 5 μM exogenous CoM to the native transferase stimulated the rate of epoxypropane degradation by 1.7-fold over the nonsupplemented rate. This result demonstrates that, under these assay conditions, CoM is sufficiently limiting that the rate of epoxide carboxylation can be nearly doubled by doubling the amount of available CoM.
FIG. 2.
Requirement of exogenous CoM for epoxide carboxylation in assays using recombinant epoxyalkane:CoM transferase. Assays were performed in duplicate, and the measurements were averaged. Experiments were done with recombinant (open symbols) or native (closed symbols) epoxyalkane:CoM transferase and with no exogenous CoM (squares) or 5 μM exogenous CoM added (circles).
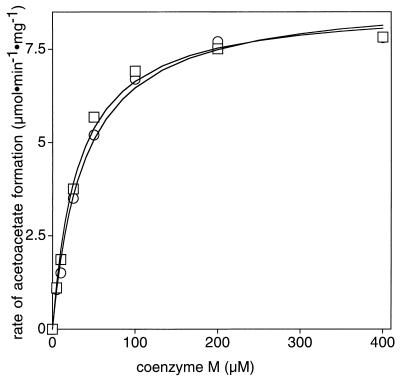
Figure 3 shows the effect of CoM concentration on the rate of epoxypropane carboxylation using native or recombinant transferase under conditions where the transferase is the rate-limiting protein component of the assay. The saturation curves for the two transferases are indistinguishable. Together, the results presented in Fig. 2 and 3 demonstrate that CoM is not essential for the synthesis of, stability of, and incorporation of zinc into the epoxyalkane:CoM transferase of Xanthobacter strain Py2.
FIG. 3.
Saturation of epoxide carboxylase activity by CoM. Assays were performed in duplicate as described in the text but with the following amounts of proteins: native or recombinant epoxyalkane:CoM transferase, 5 μg; NADPH-2-ketopropyl-CoM oxidoreductase/carboxylase, 250 μg; 2-R-hydroxypropyl-CoM dehydrogenase, 40 μg; 2-S-hydroxypropyl-CoM dehydrogenase, 25 μg. Native (squares) or recombinant (circles) epoxyalkane:CoM transferase was used.
The sensitivity of the CoM recycling assay used here is sufficient for quantifying CoM at concentrations in the low-concentration range (0.5 μM and higher). While this sensitivity is suitable for the present purposes, a bioassay described by Balch and Wolfe, in which potential sources of CoM are used to stimulate growth of the CoM auxotroph Methanobacterium ruminantium, is apparently capable of detecting CoM concentrations as low as 6 nM (7). This growth-based bioassay provided estimations of CoM concentrations in methanogenic biomass that have recently been corroborated by high pressure liquid chromatography-based determinations (13).
Identification of CoM in cell extracts and culture media.
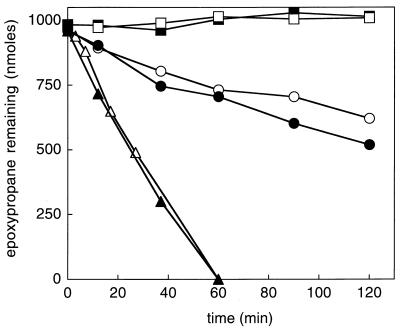
The CoM recycling assay described above was used to determine whether CoM could be detected in cell extracts and spent culture media from Xanthobacter strain Py2 and R. rhodochrous B276. The ability of the cell extract or spent medium preparation to substitute for commercial CoM in epoxide carboxylation assay mixtures containing rCoM transferase was used as the diagnostic for the presence or absence of CoM. As shown in Fig. 4, CoM was detected in extracts and spent media prepared from propylene-grown cultures of both bacteria. Importantly, no CoM was detected in cell extracts or culture media prepared from Xanthobacter strain Py2 or R. rhodochrous grown with glucose as the carbon source (Fig. 4). These results suggest that the expression of the CoM biosynthetic genes is not constitutive but induced in response to a specific need.
FIG. 4.
CoM is present in cells and culture media of propylene-grown Xanthobacter strain Py2 and R. rhodochrous. Extract or media from Xanthobacter strain Py2 (closed symbols) or R. rhodochrous (open symbols) were used. Symbols: squares, cell extract prepared from glucose-grown cells; circles, cell extract prepared from propylene-grown cells; triangles, spent media prepared from propylene-grown cells.
CoM synthesis is coordinately regulated with synthesis of alkene monooxygenase and epoxide carboxylation enzymes.
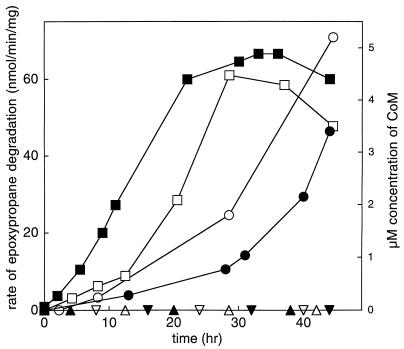
The alkene monooxygenase and epoxide carboxylation proteins (Fig. 1) of Xanthobacter strain Py2 and R. rhodochrous are inducible enzymes that are not expressed when cells are cultured in the absence of alkenes or epoxides (4, 14). The lack of detectable CoM in glucose-grown cultures of Xanthobacter strain Py2 and R. rhodochrous suggests that CoM biosynthesis is coordinately regulated with the expression of the alkene monooxygenase and epoxide carboxylation enzymes. This possibility was further investigated by growing the bacteria under conditions where the alkene and epoxide genes are repressed and then inducing their expression by addition of propylene. As shown in Fig. 5, cultures of Xanthobacter strain Py2 and R. rhodochrous grown with acetate as the source of carbon did not contain detectable levels of epoxypropane degradation activity or CoM prior to addition of propylene. The addition of propylene resulted in simultaneous increases in epoxide carboxylation activity and CoM accumulation in the culture media for both bacteria. Thus, CoM biosynthesis is induced by propylene in both bacteria.
FIG. 5.
Simultaneous induction of CoM biosynthesis and epoxide carboxylase activity in Xanthobacter strain Py2 and R. rhodochrous. Symbols: closed symbols, Xanthobacter strain Py2; open symbols, R. rhodochrous; squares, whole cell rates of epoxypropane degradation in cells exposed to propylene at t = 0 h; circles, estimated concentrations of CoM accumulating in the spent media of cells exposed to propylene at t = 0 h; triangles, whole cell rates of epoxypropane degradation in cells not exposed to propylene; inverted triangle, estimated concentrations of CoM accumulating in the spent media of cells not exposed to propylene.
Xanthobacter strain Py2 and R. rhodochrous cell extracts were prepared from cells grown with propylene, glucose, acetone, isopropanol, and in the case of R. rhodochrous, propane as the carbon source and then screened for the presence of CoM. Only those cultures grown with propylene as the carbon source contained detectable levels of CoM. Cell extract from propylene-grown Xanthobacter provided CoM in an amount that stimulated activity to a rate of 0.23 ± 0.02 nmol of epoxypropane degraded min−1 mg of protein−1 in the clarified extract (prior to boiling) that was used as the source of CoM. The specific activity of the R. rhodochrous cell extract was nearly identical (0.18 ± 0.01 nmol of epoxypropane degraded min−1 mg−1) suggesting that similar amounts of CoM accumulate in the two different bacteria. With regard to this screen of growth substrates, the absence of CoM in cell extracts prepared from cultures of R. rhodochrous grown with propane, a saturated hydrocarbon, is particularly noteworthy. The details of the pathway of bacterial propane metabolism have not been fully elucidated but may involve sequential oxidation to isopropanol and acetone as the first intermediates (26). The pathway of acetone metabolism has been elucidated for both Xanthobacter strain Py2 and R. rhodochrous and shown to involve carboxylation of acetone to acetoacetate (10, 18). Although acetone and epoxypropane are isomeric, acetone carboxylation differs significantly from epoxypropane carboxylation in that a single enzyme (acetone carboxylase) catalyzes the carboxylation in a reaction coupled to nucleoside triphosphate hydrolysis (10, 18). The lack of CoM in propane-, isopropanol-, and acetone-grown cells confirms the highly specialized role for CoM in unsaturated hydrocarbon catabolism.
Implications of these studies.
Prior to the identification of CoM as the C3 carrier in the reactions of aliphatic epoxide carboxylation, the only known function for CoM was as the methyl group carrier in archaeal methanogenesis (25, 28). Since methane formation is essential to the metabolism of all methanogens under all growth conditions, CoM must be available at all times and, accordingly, the CoM biosynthetic genes are probably constitutively expressed. Taylor et al. have isolated a methanogen, M. ruminantium, that is a CoM auxotroph, suggesting that it has a mutation in one or more of the genes involved in CoM biosynthesis (24). To date, the pathway of CoM biosynthesis has not been elucidated, although a plausible pathway beginning from phosphoenolpyruvate and bisulfite has been proposed (11).
The present work showing inducible CoM biosynthesis in nutritionally versatile heterotrophic bacteria warrants examining CoM biosynthesis in these bacteria as a model for methanogenic CoM synthesis. Both Xanthobacter and Rhodococcus are genetically tractable (16, 22, 29), suggesting that a combined genetic-biochemical approach could be used to identify the genes and enzymes involved in CoM biosynthesis. The availability of complete genome sequences of two methanogens, Methanobacterium thermoautotrophicum and Methanococcus jannaschii (8, 20), might allow the methanogenic genes to be deduced from sequence comparisons.
The above discussion raises the questions of how Xanthobacter strain Py2 and R. rhodochrous, bacteria that are phylogenetically quite distinct, acquired the genes that synthesize CoM and why CoM was chosen as the C3 carrier for epoxide carboxylation. With regard to how the genes were acquired, it is probable that they are of methanogenic origin, given the highly specialized usage of CoM in methanogens. One consideration that may be significant to this discussion is the observation that cultures of Xanthobacter strain Py2 repeatedly subcultured (i.e., for a period of several weeks) with glucose or acetone as the carbon source lose the ability to grow with propylene as the source of carbon (S. Ensign, unpublished results). In contrast, the ability of Xanthobacter strain Py2 to grow with a variety of other carbon sources (i.e., acetate, isopropanol, acetone, propylene glycol, CO2 and H2, n-propanol, and glucose) is not affected by repeated subculturing on another carbon source (S. Ensign, unpublished results). These results suggest the possibility that the genes encoding alkene monooxygenase, the epoxide carboxylation enzymes, and/or accessory proteins (e.g., CoM biosynthetic enzymes) may reside on an extrachromosomal element. Of possible relevance to this idea, Saeki and colleagues recently showed that the genes encoding the alkene monooxygenase of R. rhodochrous reside on a 185-kb linear megaplasmid, one of four such plasmids in the bacterium (16). It will be interesting to determine whether a similar situation exists in Xanthobacter strain Py2, in particular with respect to the epoxide carboxylation and CoM biosynthetic genes. It will also be interesting to determine whether a similar situation exists in Xanthobacter strain Py2, in particular with respect to the epoxide carboxylation and CoM biosynthetic genes. It will also be interesting to determine the molecular mechanisms involved in propylene sensing and the associated coordinated regulation of alkene and epoxide metabolism and CoM biosynthesis.
Acknowledgments
This work was supported by National Institutes of Health grant GM51805.
REFERENCES
- 1.Allen J R, Clark D D, Krum J G, Ensign S A. A role for coenzyme M (2-mercaptoethansulfonic acid) in a bacterial pathway of aliphatic epoxide carboxylation. Proc Natl Acad Sci USA. 1999;96:8432–8437. doi: 10.1073/pnas.96.15.8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen J R, Ensign S A. Carboxylation of epoxides to β-keto acids in cell extracts of Xanthobacter strain Py2. J Bacteriol. 1996;178:1469–1472. doi: 10.1128/jb.178.5.1469-1472.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen J R, Ensign S A. Characterization of three protein components required for functional reconstitution of the epoxide carboxylase multienzyme complex from Xanthobacter strain Py2. J Bacteriol. 1997;179:3110–3115. doi: 10.1128/jb.179.10.3110-3115.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen J R, Ensign S A. Identification and characterization of epoxide carboxylase activity in cell extracts of Nocardia corallina B276. J Bacteriol. 1998;180:2072–2078. doi: 10.1128/jb.180.8.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen J R, Ensign S A. Purification to homogeneity and reconstitution of the individual components of the epoxide carboxylase multiprotein enzyme complex from Xanthobacter strain Py2. J Biol Chem. 1997;272:32121–32128. doi: 10.1074/jbc.272.51.32121. [DOI] [PubMed] [Google Scholar]
- 6.Allen J R, Ensign S A. Two short-chain dehydrogenases confer stereoselectivity for enantiomers of epoxypropane in the multiprotein epoxide carboxylating systems of Xanthobacter strain Py2 and Nocardia Corallina B276. Biochemistry. 1999;38:247–256. doi: 10.1021/bi982114h. [DOI] [PubMed] [Google Scholar]
- 7.Balch W E, Wolfe R S. Specificity and biological distribution of coenzyme M (2-mercaptoethansulfonic acid) J Bacteriol. 1978;137:256–263. doi: 10.1128/jb.137.1.256-263.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J D, Geoghagen N S, Weidman J F, Fuhrmann J L, Nguyen D T, Utterback T, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Hurst M A, Roberts K M, Kaine B B, Borodovsky M, Klenk H P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 9.Chistoserdova L, Vorholt J A, Thauer R K, Lidstrom M E. C-1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic Archaea. Science. 1998;281:99–102. doi: 10.1126/science.281.5373.99. [DOI] [PubMed] [Google Scholar]
- 10.Clark D D, Ensign S A. Evidence for an inducible nucleotide-dependent acetone carboxylase in Rhodococcus rhodochrous B276. J Bacteriol. 1999;181:2752–2758. doi: 10.1128/jb.181.9.2752-2758.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiMarco A A, Bobik T A, Wolfe R S. Unusual coenzymes of methanogenesis. Annu Rev Biochem. 1990;59:355–394. doi: 10.1146/annurev.bi.59.070190.002035. [DOI] [PubMed] [Google Scholar]
- 12.Ebert S, Rieger P G, Knackmuss H J. Function of coenzyme F420 in aerobic catabolism of 2,4,6-trinitrophenol and 2,4-dinitrophenol by Nocardioides simplex FJ2-1A. J Bacteriol. 1999;181:2669–2674. doi: 10.1128/jb.181.9.2669-2674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elias D A, Krumholz L R, Tanner R S, Suflita J M. Estimation of methanogen biomass by quantitation of coenzyme M. Appl Environ Microbiol. 1999;65:5541–5545. doi: 10.1128/aem.65.12.5541-5545.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ensign S A. Aliphatic and chlorinated alkenes and epoxides as inducers of alkene monooxygenase and epoxidase activities in Xanthobacter strain Py2. Appl Environ Microbiol. 1996;62:61–66. doi: 10.1128/aem.62.1.61-66.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purwantini E, Daniels L. Molecular analysis of the gene encoding F420-dependent glucose-6-phosphate dehydrogenase from Mycobacterium smegmatis. J Bacteriol. 1998;180:2212–2219. doi: 10.1128/jb.180.8.2212-2219.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saeki H, Akira M, Keizo F, Averhoff B, Gottschalk G. Degradation of trichloroethene by a linear-plasmid-encoded alkene monooxygenase in Rhodococcus corallinus (Nocardia corallina) B-276. Microbiology. 1999;145:1721–1730. doi: 10.1099/13500872-145-7-1721. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 18.Sluis M K, Ensign S A. Purification and characterization of acetone carboxylase from Xanthobacter strain Py2. Proc Natl Acad Sci USA. 1997;94:8456–8461. doi: 10.1073/pnas.94.16.8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Small F J, Ensign S A. Carbon dioxide fixation in the metabolism of propylene and propylene oxide by Xanthobacter strain Py2. J Bacteriol. 1995;177:6170–6175. doi: 10.1128/jb.177.21.6170-6175.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H-M, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicare R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nolling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 22.Swaving J, Weijers C A, van Ooyen A J, deBont J A M. Complementation of Xanthobacter Py2 mutants defective in epoxyalkane degradation, and expression and nucleotide sequence of the complementing DNA fragment. Microbiology. 1995;141:477–484. doi: 10.1099/13500872-141-2-477. [DOI] [PubMed] [Google Scholar]
- 23.Tabor S. Expression using the T7 RNA polymerase/promoter system. New York, N.Y: Greene Publishing and Wiley Interscience; 1990. [Google Scholar]
- 24.Taylor C D, McBride B C, Wolfe R S, Bryant M P. Coenzyme M, essential for growth of a rumen strain of Methanobacterium ruminatium. J Bacteriol. 1974;120:974–975. doi: 10.1128/jb.120.2.974-975.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thauer R K. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology. 1998;144:2377–2406. doi: 10.1099/00221287-144-9-2377. [DOI] [PubMed] [Google Scholar]
- 26.Vestal J R, Perry J J. Divergent metabolic pathways for propane and propionate utilization by a soil isolate. J Bacteriol. 1969;99:216–221. doi: 10.1128/jb.99.1.216-221.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vorholt J A, Chistoserdova L, Stolyar S M, Thauer R K, Lidstrom M E. Distribution of tetrahydromethanopterin-dependent enzymes in methylotrophic bacteria and phylogeny of methenyl tetrahydromethanopterin cyclohydrolases. J Bacteriol. 1999;181:5750–5757. doi: 10.1128/jb.181.18.5750-5757.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolfe R S. My kind of biology. Annu Rev Microbiol. 1991;45:1–35. doi: 10.1146/annurev.mi.45.100191.000245. [DOI] [PubMed] [Google Scholar]
- 29.Zhou N Y, Chion C K, Leak D J. Cloning and expression of the genes encoding the propene monooxygenase from Xanthobacter Py2. Appl Microbiol Biotechnol. 1996;44:582–588. [Google Scholar]