Abstract
Studies of Bacillus subtilis RNases that are involved in mRNA degradation reveal a different pattern from that of Escherichia coli. A strain lacking polynucleotide phosphorylase, the major 3′-to-5′ exoribonuclease activity in cell extracts, is viable. Here, we show that the B. subtilis yvaJ gene encodes a second 3′-to-5′ exoribonuclease. A strain lacking both of these RNases grows slowly but is viable. The existence of another, as yet unknown, 3′-to-5′ exoribonuclease in B. subtilis is suggested.
The short half-life of bacterial mRNAs is thought to be required for rapid changes in gene expression programs. In Escherichia coli, mRNA exonucleolytic decay is accomplished by two 3′-to-5′ exoribonucleases, RNase II and polynucleotide phosphorylase (PNPase). The essential nature of mRNA turnover is inferred from the inviability of an RNase IIts PNPase− double mutant at the nonpermissive temperature (5).
While hydrolytic decay by RNase II constitutes the major 3′-to-5′ exonuclease activity in E. coli, Deutscher and Reuven (4) reported that there was no RNase II-like activity in Bacillus subtilis extracts. Instead, phosphorolytic degradation catalyzed by a putative PNPase of B. subtilis was primarily responsible for poly(A) decay. A minor, Mn2+-stimulated hydrolytic activity was also detectable in the B. subtilis extract.
We cloned the B. subtilis PNPase gene (pnpA) and constructed a pnpA deletion strain (10). This strain displayed several phenotypes: slightly slower growth at 37°C, cold sensitivity, competence deficiency, tetracycline sensitivity, and altered mRNA processing (1, 8, 10). If 3′-to-5′ exonucleolytic decay of mRNA is essential for B. subtilis, the isolation of a PNPase− B. subtilis strain suggested that another 3′-to-5′ exonuclease must fulfill this function. Analysis of decay of labeled substrates in the pnpA deletion strain extract indicated an Mn2+-stimulated hydrolytic activity (10).
We initiated a biochemical study to purify the “other” 3′-to-5′ exoribonuclease from B. subtilis, using decay of 3H-labeled total RNA as the assay (10). The pnpA deletion strain, BG119, was used to prepare extracts for purification of the Mn2+-stimulated RNase activity. After growth in YT medium (0.8% yeast extract, 0.8% tryptone, 0.25% NaCl) to 250 to 300 Klett units, cells were collected, washed with buffer A (20 mM Tris-HCl [pH 7.75], 100 mM KCl, 10% glycerol, 0.2 mM EDTA), and frozen at −80°C. Cell pellets were washed in 200 ml of buffer A (without glycerol) containing 1 mM phenylmethylsulfonyl fluoride and 1 mM dithiothreitol. Lysozyme was added to 0.2 mg/ml. After incubation at 37°C for 20 min, cells were passed through a French press and cell debris was pelleted at 12,000 × g. The supernatant from this low-speed spin was used as the “extract.”
For further purification, the low-speed-spin supernatant was pelleted at 200,000 × g, and nucleic acid-binding proteins were precipitated by addition of streptomycin sulfate (130 mg/ml), followed by centrifugation at 27,000 × g. More than 90% of the Mn2+-stimulated activity coprecipitated with the bulk nucleic acid. The pellet was dissolved in buffer A, dialyzed overnight against the same buffer, and diluted to a concentration of 0.2 mg of nucleic acid/ml. Nucleic acids were digested by addition of micrococcal nuclease (Worthington Biochemical) to 1.5 U per μg of nucleic acid and incubation for 20 min at 30°C in the presence of 1 mM CaCl2. After dialysis in a 20-fold volume of buffer A containing 1 mM CaCl2, protein was loaded onto a DEAE-Sepharose CL-6B (Sigma) column, which was equilibrated with buffer A. The column was washed with 10 bed volumes of buffer A, and protein was eluted with a linear gradient of KCl (100 to 500 mM). This protocol yielded a >50-fold purification of the activity, assayed using 3H-labeled total RNA.
In the course of these studies, Cheng et al. reported that the vacB gene of E. coli encodes the 3′-to-5′ exonuclease RNase R (2) and proposed that the vacB gene be named rnr. Since the yvaJ gene of B. subtilis showed significant homology (37% identity) with the E. coli rnr gene, we investigated possible RNase activity encoded by yvaJ. A strain was constructed (BG295) in which about half of the yvaJ coding sequence was replaced with a spectinomycin resistance gene. In addition, a pnpA yvaJ double mutant strain (BG296) was constructed. The growth rates of these strains in Luria-Bertani medium are shown in Table 1. Disruption of the yvaJ gene had no effect on the growth rate. However, the pnpA yvaJ strain grew even slower than the pnpA strain. Thus, the effect on growth rate conferred by the absence of PNPase was exacerbated by the absence of the yvaJ gene product. Northern blot analysis of mRNA encoded by the erythromycin resistance gene ermC showed no difference in the decay pattern between the pnpA and pnpA yvaJ strains (data not shown). The rate of bulk mRNA decay may be affected in the pnpA yvaJ strain, but this has not been measured.
TABLE 1.
Growth rate of RNase mutant strains
| Strain | Genotype | Doubling time (min)a |
|---|---|---|
| BG1 | Wild type | 19.5 ± 1.0 |
| BG119 | pnpA | 27.0 ± 0.0 |
| BG295 | yvaJ | 20.0 ± 0.7 |
| BG296 | pnpA yvaJ | 34.0 ± 1.7 |
Data are averages of measurements from at least four independent growth experiments.
Extracts of BG1 (wild type), BG119, and BG295 strains were prepared to assess RNase activity. For the RNA decay assay, 100-μl reaction mixtures contained 0.2 to 12 μg of protein, 10 to 40 nmol of RNA (1,000 to 4,000 μCi/mmol), 50 mM Na-Tricine (pH 8.0), and 100 mM KCl, with or without addition of 1 mM MgCl2 or MnCl2. After 20 min at 37°C, 300 μl of 0.5-mg/ml E. coli tRNA and 400 μl of 20% trichloroacetic acid were added. Samples were incubated on ice for 10 min and then centrifuged in the cold for 10 min. Four hundred microliters of the supernatant was removed and counted in 5 ml of Ecoscint A (National Diagnostics).
Addition of Mn2+ to BG1 or BG119 extracts resulted in a twofold increase in activity, relative to the activity in the presence of Mg2+ (Table 2). Activity in the BG295 extract in the presence of Mg2+ was only 11% of the wild type (Table 2). Thus, disruption of the yvaJ gene caused a substantial loss of RNase activity. Interestingly, activity in the BG295 extract in the presence of Mn2+ was 60% that of the wild type. This suggested that an Mn2+-stimulated RNase activity was still present in the yvaJ mutant strain.
TABLE 2.
RNA degradative activity in protein extractsa
| Divalent cation | Activity for strain:
|
||
|---|---|---|---|
| BG1 (wild type) | BG119 (pnpA) | BG295 (yvaJ) | |
| None | 0.4 | 0.6 | 0.9 |
| Mg2+ | 14.8 | 18.0 | 1.7 |
| Mn2+ | 25.6 | 35.5 | 15.7 |
Decay of 3H-labeled total RNA was measured in the presence of 1 mM MgCl2 or 1 mM MnCl2. Values are nt released in nanomoles per minute per milligram of protein. Values are from a single experiment (average of duplicate samples), using 22 nmol of labeled RNA per assay, but similar results were obtained from numerous experiments using different concentrations of labeled RNA.
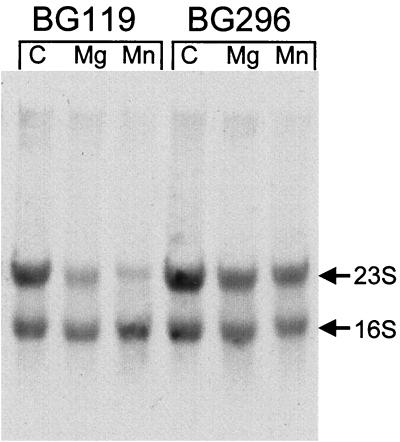
Earlier (10), we had assumed that the bulk of the 3H-labeled RNA used to assay RNase activity was mRNA, since the RNA was isolated from a late-logarithmic-phase culture following a 10-min pulse of [3H]uridine. However, when the labeled RNA was run on a morpholinopropanesulfonic acid-formaldehyde gel to directly observe RNase activity, fluorography (Autoflour; National Diagnostics) revealed that the bulk of this labeled RNA was rRNA (Fig. 1). 23S rRNA was degraded upon incubation with the most active DEAE fraction from BG119, but to a much lesser degree with the corresponding fraction extract from BG296 (pnpA yvaJ) (Fig. 1). This suggested that the major activity detected in our assays was an RNase that could degrade rRNA. Such an activity was reminiscent of E. coli RNase R, so named due to its nonspecific degradation of rRNA (3, 7).
FIG. 1.
Decay of 3H-labeled total RNA. Migration of 23S and 16S rRNAs is indicated. Substrate RNA was incubated in the presence of the most active BG119 fraction from the DEAE-Sepharose column and the corresponding BG296 fraction. Incubation at 37°C for 80 min was in the absence of divalent cation (C) and in the presence of 1 mM Mg2+ or 1 mM Mn2+.
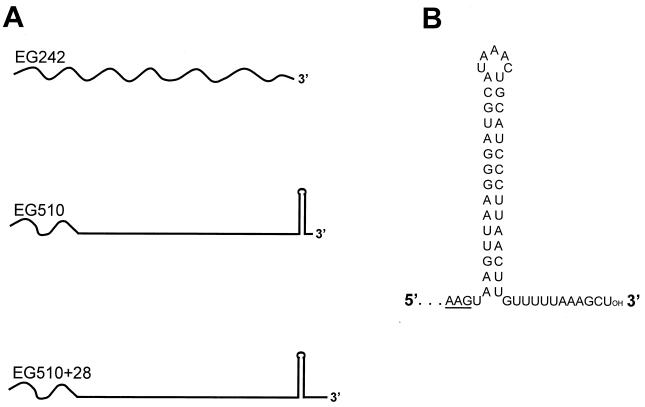
Three 32P-labeled RNA substrates (shown schematically in Fig. 2A) were prepared, as described previously (6), to test for 3′-to-5′ exoribonuclease activity. The 187-nucleotide (nt) EG242 RNA contained sequences of SP82 bacteriophage, as described previously (9). In EG510 RNA (222 nt), the 5′ terminal 42 nt were the same as in EG242 RNA, followed by the sequence of the 3′ end of ermC mRNA, which contains a strong terminator structure (predicted free energy of −20.9 kcal/mol) located 12 nt from the 3′ end (Fig. 2B). EG510+28 RNA was prepared from the same template as EG510 RNA, but such that the transcribed product contained an additional 28 nt at the 3′ end.
FIG. 2.
(A) Schematic diagram of 32P-labeled RNAs used to test for 3′-sensitive exoribonucleolytic decay. Wavy lines are SP82 sequences; straight lines are ermC sequences. (B) Predicted secondary structure of the ermC terminator sequence, which is present at the end of EG510 RNA. The ermC coding sequence ends with the AAG codon (underlined), followed by a UAA stop codon.
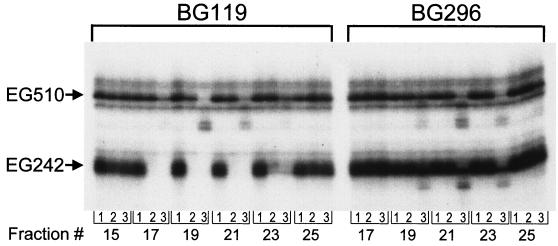
Peak fractions from the DEAE-Sepharose column were assayed for degradation of 32P-labeled EG242 and EG510 RNAs in the presence of 1 mM Mg2+ or Mn2+ (Fig. 3). For the BG119 fractions in the presence of Mg2+ or Mn2+, complete degradation of EG242 RNA was observed in fractions 17, 19, and 21, with partial degradation in fraction 23. EG510 RNA was not degraded in the presence of Mg2+ but was cleaved in the presence of Mn2+. The latter activity is likely due to nonspecific B. subtilis RNase III endonuclease activity, which copurifies in these fractions (D. H. Bechhofer, unpublished results). The differential activity on substrates with (EG510) and without (EG242) a 3′-proximal stem-loop structure suggested that the RNase activity was degrading from the 3′ end.
FIG. 3.
Decay of 32P-labeled RNAs. EG242 RNA and EG510 RNA were incubated with DEAE-Sepharose fractions from BG119 (pnpA) and BG296 (pnpA yvaJ). The samples contained either no divalent cation (lane 1), 1 mM Mg2+ (lane 2), or 1 mM Mn2+ (lane 3). The products were electrophoresed on a high-resolution 6% denaturing polyacrylamide gel. Bands other than the major ones indicated by “EG242” and “EG510” are minor transcription products.
Corresponding fractions from the BG296 extract were tested for activity, and no degradation was detected for either substrate in the presence of Mg2+ (Fig. 3). Thus, deletion of the yvaJ gene resulted in a strain that was missing the 3′-sensitive RNase activity.
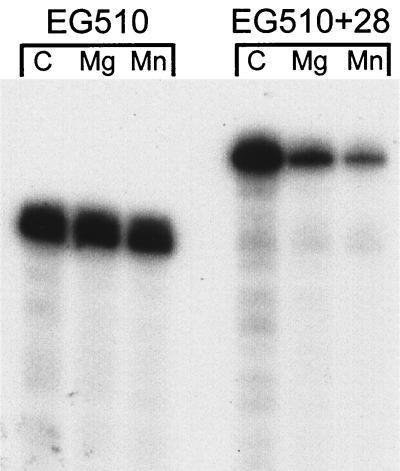
To further test the effect of 3′ structure on the RNase activity present in BG119, the EG510+28 RNA substrate was used (Fig. 2A). While the activity present in BG119 could not degrade EG510 RNA, it was able to degrade EG510+28 RNA (Fig. 4). The same fraction from the BG296 strain was unable to degrade either EG510 or EG510+28 RNA (data not shown). The sensitivity of EG510+28 RNA (but not EG510 RNA) to decay suggested strongly that the activity observed in the BG119 fraction, which was absent from BG296, was initiating at the 3′ end.
FIG. 4.
Decay of 32P-labeled RNAs in the presence of a purified fraction from BG119. EG510 RNA contains 12 unpaired nt at the 3′ end, and EG510+28 RNA contains 40 unpaired nt at the 3′ end. Incubation at 37°C for 20 min was in the absence of divalent cation (C) and in the presence of 1 mM Mg2+ or 1 mM Mn2+.
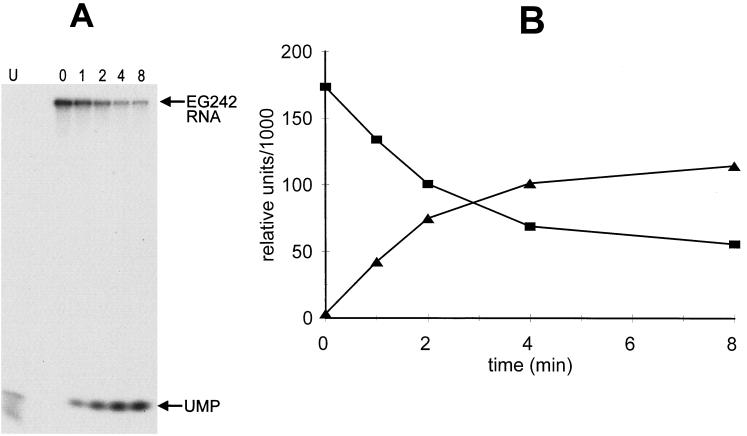
To test whether the activity was exonucleolytic, degradation products from EG242 RNA were run on a 20% polyacrylamide gel. The only product observed was a mononucleotide (Fig. 5A), and this accumulated inversely with the decay of the full-length RNA (Fig. 5B). Taken together, these results demonstrate that the yvaJ gene codes for a 3′-to-5′ exoribonuclease that is able to degrade rRNA. We propose that the B. subtilis yvaJ gene be renamed rnr.
FIG. 5.
Generation of monophosphate nucleosides from EG242 RNA. (A) Degradation products were separated on a 20% denaturing polyacrylamide gel. Time (in minutes) of incubation at 37°C are indicated at the top. The control lane (U) contained [32P]UTP. (B) Plot of EG242 decay (squares) and generation of mononucleotides (triangles) versus time. Relative units represent volumes as measured by a PhosphorImager scan (Molecular Dynamics).
We assume that 3′-to-5′ exoribonucleolytic activity that can degrade mRNA is an essential function in B. subtilis. We found that a B. subtilis strain lacking both RNase R and PNPase was viable, suggesting that at least one other 3′-to-5′ exoribonuclease is present that is involved in mRNA decay. The remaining ribonuclease activity in the BG296 extract is now a focus of our interest.
Acknowledgments
This work was supported by Public Health Service grant GM-48804 from the National Institutes of Health.
We thank David Hicks for advice on protein purification and George Mackie for suggesting the use of micrococcal nuclease.
REFERENCES
- 1.Bechhofer D H, Wang W. Decay of ermC messenger RNA in a polynucleotide phosphorylase mutant of Bacillus subtilis. J Bacteriol. 1998;180:5968–5977. doi: 10.1128/jb.180.22.5968-5977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng Z-F, Zuo Y, Li Z, Rudd K E, Deutscher M P. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J Biol Chem. 1998;273:14077–14080. doi: 10.1074/jbc.273.23.14077. [DOI] [PubMed] [Google Scholar]
- 3.Deutscher M P, Marlor C W, Zaniewski R. Ribonuclease T: new exoribonuclease possibly involved in end-turnover of tRNA. Proc Natl Acad Sci USA. 1984;81:4290–4293. doi: 10.1073/pnas.81.14.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deutscher M P, Reuven N B. Enzymatic basis for hydrolytic versus phosphorolytic mRNA degradation in Escherichia coli and Bacillus subtilis. Proc Natl Acad Sci USA. 1991;88:3277–3280. doi: 10.1073/pnas.88.8.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donovan W P, Kushner S R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci USA. 1986;83:120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farr G A, Oussenko I A, Bechhofer D H. Protection against 3′-to-5′ RNA decay in Bacillus subtilis. J Bacteriol. 1999;181:7323–7330. doi: 10.1128/jb.181.23.7323-7330.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasai T, Gupta R S, Schlessinger D. Exoribonucleases in wild type Escherichia coli and RNase II-deficient mutants. J Biol Chem. 1977;252:8590–8596. [PubMed] [Google Scholar]
- 8.Luttinger A, Hahn J, Dubnau D. Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis. Mol Microbiol. 1996;19:343–356. doi: 10.1046/j.1365-2958.1996.380907.x. [DOI] [PubMed] [Google Scholar]
- 9.Mitra S, Bechhofer D H. In vitro processing activity of Bacillus subtilis polynucleotide phosphorylase. Mol Microbiol. 1996;19:329–342. doi: 10.1046/j.1365-2958.1996.378906.x. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, Bechhofer D H. Properties of a Bacillus subtilis polynucleotide phosphorylase deletion strain. J Bacteriol. 1996;178:2375–2382. doi: 10.1128/jb.178.8.2375-2382.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]