Abstract
Asthma has striking disparities across ancestral groups, but the molecular underpinning of these differences is poorly understood and minimally studied. A goal of the Consortium on Asthma among African-ancestry Populations in the Americas (CAAPA) is to understand multi-omic signatures of asthma focusing on populations of African ancestry. RNASeq and DNA methylation data are generated from nasal epithelium including cases (current asthma, N = 253) and controls (never-asthma, N = 283) from 7 different geographic sites to identify differentially expressed genes (DEGs) and gene networks. We identify 389 DEGs; the top DEG, FN1, was downregulated in cases (q = 3.26 × 10−9) and encodes fibronectin which plays a role in wound healing. The top three gene expression modules implicate networks related to immune response (CEACAM5; p = 9.62 × 10−16 and CPA3; p = 2.39 × 10−14) and wound healing (FN1; p = 7.63 × 10−9). Multi-omic analysis identifies FKBP5, a co-chaperone of glucocorticoid receptor signaling known to be involved in drug response in asthma, where the association between nasal epithelium gene expression is likely regulated by methylation and is associated with increased use of inhaled corticosteroids. This work reveals molecular dysregulation on three axes – increased Th2 inflammation, decreased capacity for wound healing, and impaired drug response – that may play a critical role in asthma within the African Diaspora.
Subject terms: Gene expression, Epigenetics, Transcriptomics
Here, the authors suggest that molecular dysregulation on three axes may play a critical role in asthma within the African Diaspora. RNASeq and DNA methylation data are generated from nasal epithelium including cases and controls from seven different geographic sites.
Introduction
Asthma is the most common disease of childhood and its prevalence continues to increase in many parts of the world1. Despite advances in therapeutics and a better understanding of environmental risk factors, disparities between populations are profound, and cannot be entirely explained by non-genetic factors2–5. In the U.S., childhood asthma prevalence is 20% higher among African Americans compared to non-Hispanic children of European descent, and African American children are three times more likely to die from asthma6. Individuals of African ancestry have greater asthma morbidity and mortality both within2 and outside the U.S.7–9, and asthma prevalence is high in African countries and countries with populations of African ancestry9–11. There are notable differences in therapeutic response12; patients of African ancestry respond differently to inhaled corticosteroids (ICS) compared to patients of European ancestry13,14, and long acting beta agonizts (LABA) may have greater risk for adverse events in this group15. Genetic variants in the receptors for pharmaceutical agents used in asthma management may partially explain these differences in response12. To address gaps related to the genomic underpinnings of disparities in asthma, we established the Consortium on Asthma among African ancestry Populations in the Americas (CAAPA)16.
Genome-wide association studies (GWAS) have identified >170 loci for asthma17. CAAPA has contributed the single largest GWAS focused solely on the African Diaspora18, but populations of African ancestry remain woefully under-represented in large scale international studies19,20. In the recent Global Biobank Meta-analysis Initiative (GBMI), African ancestry individuals represented a mere 5% of the total number of asthma cases19. Nonetheless, the ancestry diversity in the GBMI facilitated identification of SNPs with different effects across ancestries and increased power to identify loci missed in European-only subsets. Importantly, the GBMI demonstrated that increased diversity and sample size of the discovery GWASs were primary drivers of improved polygenic risk score (PRS) accuracy in non-European populations.
While there is mounting evidence that asthma risk variants may play a role in the regulation of immune system pathways20,21, molecular mechanisms underlying asthma heterogeneity and their signals at GWAS loci are poorly described. With a few exceptions like the Chr17q locus, GWAS loci have offered limited insight into the biology of asthma, and even less into the complex heterogeneity and disparities in asthma. Transcriptomic approaches have demonstrated some success in profiling asthma endotypes22 and epigenetics can provide mechanistic understanding of the regulation of these transcriptomic signatures23, especially in the context of environment24,25. There are now multiple lines of evidence supporting the important role of the airway epithelium in the pathogenesis of asthma26. A meta-analysis of gene expression from airway epithelial cells has identified >400 differentially expressed genes, consistency between signatures from upper (nasal) and lower (bronchial) airway epithelial cells, and upregulation of Th2 pathway genes26 – pathways related to allergen specific T-helper 2 (Th2) cells that are centrally involved in allergic asthma. Transcriptomic signatures can distinguish “type 2 high” from “type 2 low” endotypes of asthma, and importantly, such signatures are now shown to be associated with response to ICS27, with the potential to also distinguish viral and non-viral asthma exacerbations28. Monoclonal antibodies including anti-IgE, anti-IL5, anti-IL5 receptor, and anti-IL4 receptor have advanced the management of severe asthma, with the choice of therapy predicated on the ability to define the endotype of asthma in the patient29.
We hypothesize that transcriptomic signatures from the nasal airway epithelium in asthma cases and controls representing the African Diaspora will allow us to validate previously identified gene expression signatures of asthma and, importantly, identify pathways of dysregulation that are relevant to the disparities observed with respect to asthma. We rely on nasal epithelium as a proxy for the airways given its ease of tissue collection on large numbers of individuals and the established correlation between signatures of asthma between nasal epithelium and bronchial tissue26,30. RNA sequencing and DNA methylation data from the same nasal epithelium samples in cases (individuals with current asthma) and controls (individuals never having asthma) representing 7 locations across the African Diaspora reveals dysregulation on three axes – increased Th2 inflammation, decreased capacity for wound healing in airway epithelium, and impaired drug response – that play a role in the development of asthma in individuals of African ancestry.
Results
Clinical characteristics
Cases (N = 253) with current asthma status and controls with never-asthma status (N = 283) were recruited from seven sites including 4 US-based locations and 3 international locations (Supplementary Data 1). The subjects were 70% adult and 38% male. Global genetic ancestry deconvolution revealed a wide range in African ancestry (YRI ancestry = 9–100%) representing diversity across the African Diaspora (Supplementary Data 1, Supplementary Fig 1). The highest African ancestry was observed in the subjects from Nigeria (average = 100% YRI), and the lowest was observed in the subjects from Brazil (average = 51% YRI). Cases had higher total serum IgE (p = 1.66 × 10−23), higher eosinophil counts (p = 3.42 × 10−05) and higher phadiotop (p = 2.27 × 10−21), reflecting the greater burden of the allergic phenotype in this group. Cases also had poorer lung function (FEV1; p = 7.30 × 10−16). Similar patterns were observed within each site comparing cases to controls, with differences also observed between sites. Asthma cases from all the three non-US recruitment locations had higher IgE compared to Chicago, but no difference in IgE levels was noted for the controls (Supplementary Data 1). Notably, cases from Salvador, Brazil had the most severe asthma (CASI score average = 8), and the accompanying highest rate of medication use (97.2%).
Single gene analysis
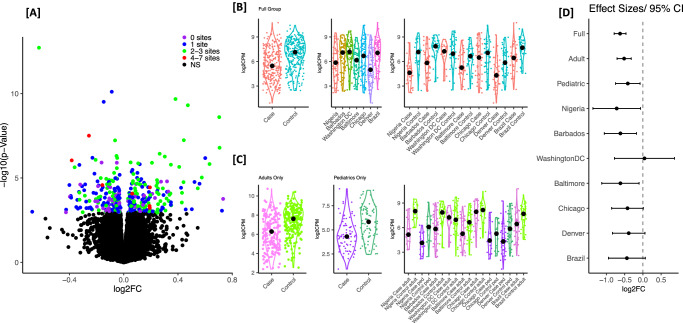
We identified 389 differentially expression genes (DEGs) between cases and controls (Supplementary Data 2); 41% of these DEGs had at least 2 sites with site-specific p < 0.05 (Fig. 1A). We evaluated replication of these signals in a published meta-analysis of 8 studies including nasal epithelium transcriptomics26. A total 16,269 genes were tested in both analyses, including 353 of the 389 DEGs we identified; 87 of the 353 genes were independently identified in the meta-analysis (q < 0.05, enrichment p-value = 9.97 × 10−27) and all but one showed the same direction of effect (Supplementary Data 2). An additional 64 genes were replicated at a nominal threshold (p < 0.05) in the meta-analysis with the same direction of effect (Supplementary Data 2). The 15 most significant DEGs (Supplementary Data 3, Supplementary Fig. 2) included genes known to play a role in wound healing (FN1, CDH11), immune response (VSIG4, HS3ST4), and asthma drug response (PTHHD4, SPTBN1, FKBP5). Additionally, SNTG2 is the target of multiple miRNAs related to asthma31, and PPP1R9A expression was previously determined to be influenced by IL-13 in mouse lung32. Effect sizes for the top 15 genes were generally consistent across the seven recruitment sites and, with the exception of RHEX2, and effect sizes were similar with overlapping confidence intervals between adult and pediatric subset analyses (Supplementary Fig. 2).
Fig. 1. Summary of the DEG analysis for active asthma in CAAPA.
Panel A Volcano plot of DEG analysis for asthma in the full combined group (N = 253 cases, N = 283 controls) from all 7 sites. Color represents the number of sites where the uncorrected significance for the DEG analysis within the site was p-value < 0.05, and genes that did not cross FDR of 0.05 in full combined analysis are retained as black. Panel B Combined gene expression for top gene FN1 by site. Panel C Gene expression for top gene FN1 stratified by adult vs. pediatrics. Panel D DEG effect sizes (log2 fold change and the 95% confidence interval) for top gene FN1 looking at the full combined analysis, analysis stratified by adults vs. pediatrics and the analysis within each site. CAAPA sites are ordered based on average African ancestry (%YRI) from highest (Nigeria) to lowest (Brazil). The test used in the DEG analysis was a moderated two-sided t-statistic. Source data are provided as a Source Data file.
The most significant DEG was FN1 (log2FC = −0.62, q = 3.26 × 10−9, Supplementary Data 3). Despite variability in FN1 expression across sites (Fig. 1B) there was consistently lower expression in cases (Fig. 1B), similar effect sizes in the adult and pediatric subsets, and consistent effects across all sites except Washington DC (Fig. 1C, D). FN1 was independently identified by Tsai et al.26 with a similar lower expression in asthma (z = −3.61, q = 1.25 × 10−2).
Pathway analysis on significant DEGs
Ingenuity Pathway Anlaysis (IPA) analysis on the N = 389 significant DEGs identified 1188 upstream regulators with p-value of overlap <0.05; the set of top 10 upstream regulators with notable roles in asthma are indicated in Supplementary Fig 3 and Supplementary Data 4. These include inflammatory cytokines IL4 (z = 0.096, p = 7.25 × 10−10, Supplementary Fig 3B) and TGFβ1 (z = −1.711, p = 5.47 × 10−8, Supplementary Fig 3C), both of which are known to play key roles in asthma. Interestingly, two asthma drugs – dexamethasone (z = 2.117, p = 4.31 × 10−10, Supplementary Fig 3D) and fluticasone propionate (z = 1.44, p = 9.42 × 10−08, Supplementary Fig 3E) – were also among the top 10 upstream regulators for the significant DEGs. The 188 genes identified as direct or indirect targets of these two drugs were not themselves related to medication use in CAAPA (Supplementary Data 5). Beyond its role in immune-related mechanisms for asthma, TGFβ1 has also been found to be a potent stimulus for FN1 expression in vascular and airway smooth muscle (ASM) cells, lung fibroblasts, and the alveolar epithelial cell lines33. In our study, it was identified as an upstream regulator for a network that includes three genes that are known to play a role together in airway remodeling and wound healing – FN1, COL3A1 and COL41 (Supplementary Fig 3C)34. All three genes have lower nasal epithelial expression among asthmatics (FN1 (log2FC = −0.62, q = 3.26 × 10−9); COL3A1 (log2FC = −0.32, q = 3.54 × 10−2); COL4A1 (log2FC = −0.24, q = 3.90 × 10−2), Supplementary Data 2).
Gene expression module analysis
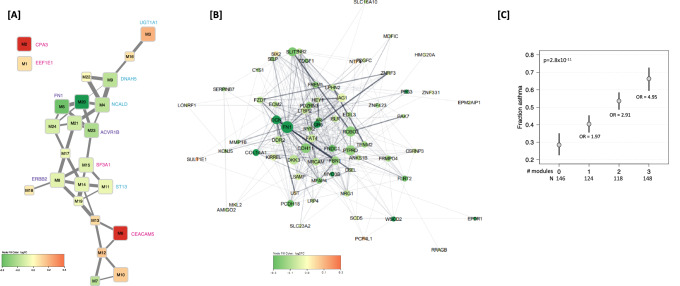
There were 24 weighted gene correlated network analysis (WGCNA) modules identified from analysis of N = 1326 genes (DEGs with FDR < 0.15). Of these, 16 modules ranging in size from N = 21–88 genes were significantly differentially expressed by asthma status (Table 1, Fig. 2); gene-module membership is shown in Supplementary Data 2. We note that none of these N = 1326 genes were differentially expressed based on nasal steroid (NS) usage 5 days prior to nasal epithelium sampling in cases (minimum q-value = 0.89 comparing 32 cases on NSs but not withholding compared to 36 cases on NS that withheld NS; Supplementary Data 6). Of the five modules upregulated in asthma cases, the strongest was a network (M6) with CEACAM5 as the hub gene (Fig. 2, Supplementary Fig 5A, log2FC = 0.32, q = 9.62 × 10−16). Also strongly upregulated in asthma cases, was the network (M2) with hub gene CPA3 (Fig. 2, Supplementary Fig 5B, log2FC = 0.54, q = 2.39 × 10−14). In pathways downregulated in asthma cases, hub genes were identified reflecting the importance of impaired wound healing: FN1 (Fig. 2B) as noted above, and ERBB2 35,36. Additionally, networks related to drug response are implicated with hub genes DNAH537(M9), NCALD38(M4), and ST1339(M11) from downregulated modules for asthma.
Table 1.
Significant WGCNA gene module differential expression analysis for asthma
| Module label | Hub gene | log2 FC | Adjusted p-value | # genes in module | # genes with DEG q < 0.05 | Most signifiicant | |
|---|---|---|---|---|---|---|---|
| WGCNA | STRING | DEG in module | |||||
| M6 | CEACAM5 | MET | 0.32 | 9.62E−16 | 71 | 26 | HS3ST4 |
| M2 | CPA3 | POSTN | 0.54 | 2.39E−14 | 81 | 45 | PTCHD4 |
| M5 | DKK3 | FN1 | −0.28 | 7.63E−09 | 72 | 34 | FN1 |
| M20 | C16orf89 | RIMS1,PPP1R9A,NR2F1 | −0.36 | 3.41E−08 | 33 | 14 | PPP1R9A |
| M23 | ACVR1B | CCND1 | −0.16 | 4.56E−08 | 23 | 5 | SUSD4 |
| M15 | SF3A1 | SMARCA2 | −0.06 | 1.61E−05 | 44 | 18 | SPTBN1 |
| M1 | PDCD10 | EEF1E1 | 0.07 | 3.74E−05 | 88 | 21 | ETAA1 |
| M4 | IFT172 | GLRB,NCALD,TP53BP1,PTPRT | −0.13 | 3.74E−05 | 77 | 17 | SLC13A3 |
| M3 | ENSG00000279476 | BCAN,PLAGL1,UGT1A1,PRRT2,GARNL3,CYP2A7 | 0.16 | 1.24E−04 | 78 | 15 | ENSG00000273599 |
| M21 | POLD2 | POLD2,UBB | −0.09 | 2.17E−04 | 31 | 11 | GNAS |
| M9 | DNAH10 | DNAH5 | −0.13 | 2.46E−04 | 58 | 15 | DNAH5 |
| M24 | RPL7A | RPL23A | −0.08 | 2.29E−03 | 21 | 5 | MT3 |
| M14 | SARS1 | VCP | −0.05 | 2.58E−03 | 44 | 11 | POFUT1 |
| M11 | ST13 | HSP90AB1 | −0.04 | 4.15E−03 | 48 | 9 | FAM169A |
| M8 | PRKCSH | ERBB2 | −0.05 | 6.25E−03 | 62 | 18 | VPS18 |
| M10 | ASPM | BIRC5,CCNA2 | 0.10 | 2.74E−02 | 48 | 11 | PARPBP |
Genes with q < 0.15 in full group DEG analysis for asthma (see Supplementary Table S1) were grouped into 24 modules using WGCNA. 24 modules were tested for association with asthma, and the 16 identified with q < 0.05 are shown in this table. Bold font indicates gene used to label the module in Fig. 2. Analysis was performed using a moderated t-statistic (two-sided).
Fig. 2. Differential module expression based on the N = 1326 DEGs with FDR < 0.15 for active asthma.
Panel A Module connectivity network for the 24 modules. Each node represents a module, and each edge represents a significant positive Pearson pairwise correlation of module expression (correlation >0.5). Node color intensity corresponds to log2FC in DE Module analysis for asthma (red upregulated in cases, green downregulated in cases). Differentially expressed modules are larger in size (q < 0.05). Edge weight indicates correlation (wider edges higher correlation of module expression). Panel B STRING network retrieved for genes assigned to module M5 with hub gene FN1. Each node represents a gene and each edge represents a protein-protein interaction with a stringdb score >0.15. Node color intensity corresponds to log2FC in DE analysis of asthma (red upregulated in cases, green downregulated in cases). Node size was made proportional to the number of interactions of the node divided by maximum number of interactions of a node in the gene module (dg/max dg of module). Unconnected nodes were not included. Edge weight and transparency indicate stringdb score (wider, darker edges indicate higher score). Panel C Fraction of asthma cases and ORs for asthma if an individual was in the upper median for any one, any two and all three modules (M4, M5, M6). Fitted probabilities (gray dots) and 95% confidence intervals (black lines) were derived from a logistic model with number of modules as an additive predictor. Source data are provided as a Source Data file.
The WGCNA modules reflected the same three axes of dysregulation in asthma cases as indicated from the top 15 DEGs and IPA upstream regulators on 389 DEGs; it should be noted that many of the associated modules and therefore the axes of dysregulation are correlated to varying degrees (Fig. 2A). The cumulative effect of these three axes is illustrated in a joint model examining dichotomized module expression focusing on the most significant module for each axis: Th2 inflammation (CEACAM5,M6), wound repair (FN1,M5), and drug response (NCALD,M4) (Fig. 2C). These modules are significantly correlated (M6-M4: R = −0.70, p < 0.001/M5-M6: R = −0.58, p < 0.001, and M5-M4: R = 0.29, p < 0.001). The ORs for asthma if an individual was in the upper median for any one, two and all three modules were 1.70 (95% CI 1.49–1.99), 2.91 (2.12–3.98), and 4.95 (3.09–7.93), respectively. This joint analysis provides evidence the greatest risk for asthma was when there is dysregulation along all three axes (Fig. 2D). The number of modules was significant in the additive logistic model (p = 2.8 × 10−11) and no departures from additivity were detected (p = 0.29 from a 2df likelihood ratio test).
Integration of gene expression with DNA methylation (DNAm)
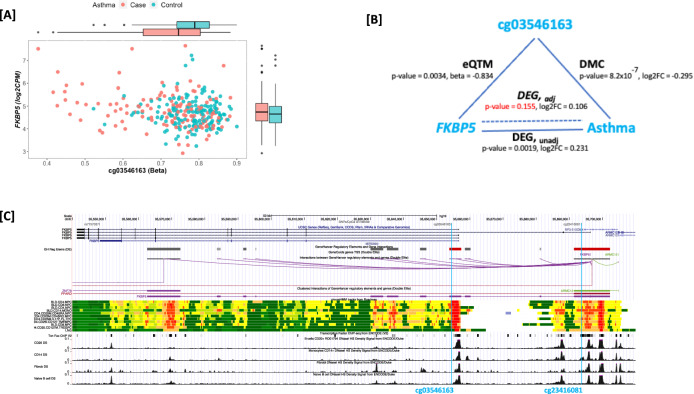
There were 8,418 eQTM tests performed for gene-CpG pairs comprising significant DEGs and CpGs mapping within 5 kb of the gene transcription start site or that were annotated by promoter-capture HiC in bronchial epithelial cells lying in putative enhancer regions for these genes. Of these, 918 gene-CpG pairs had uncorrected eQTM p < 0.05 (Supplementary Data 7); this included 288 unique genes and 915 unique CpGs. Testing for differential DNAm by asthma status, we found only five of these CpGs to be DMCs (i.e. significant for asthma at the Bonferroni level of p < 0.05/915, Supplementary Data 8): two CpGs for FKBP5 (cg03546163, cg23416081), two for TREML2 (cg26928682, cg18297196) and one for TMEM71 (cg27159719). We found cg03546163 to be the strongest predictor of FKBP5 expression and cg26928682 for TREML2 expression independent of other eQTMs for each gene. For FKBP5 (Fig. 3A,B), adjusting for methylation at cg03546163 strongly attenuated the association between gene expression and asthma, with a reduction in effect size (log2FC = 0.231 and 0.106 pre- and post- adjustment for methylation at cg03546163), and a loss in significance (p = 0.0019 and 0.155 pre- and post-adjustment for methylation at cg03546163). The same pattern was noted for TREML2 (Supplementary Fig 6) where the DEG lost significance after adjusting for methylation at cg26928682. However, the TMEM71 asthma DEG remained largely unchanged when adjusting for methylation at cg27159719 (Supplementary Fig 6). These results suggest an epigenetic mechanism of regulation of gene expression in asthma risk for both FKBP5 and TREML2.
Fig. 3. Epigenetic mechanism relating gene expression to asthma for FKBP5.
Panel A Scatter plot of methylation (beta) values at cg03546163 vs gene expression (log2 CPM) values for FKBP5 and box plots showing median, lower and upper quartiles, whiskers extending to the furthest data point no more than 1.5 times the distance between the lower and upper quartiles, and outliers, by asthma case and control status for N = 298 individuals. Panel B Effect sizes and unadjusted p-values from two-sided multivariate linear regression models for DMC analysis (cg03546163 and asthma, N = 331), eQTM analysis (cg03546163 and FKBP5 expression, N = 298) and DEG (FKBP5 expression and asthma, N = 298) analysis pre- and post-adjustment for methylation at the CpG (labeled DEG, unadj and DEG, adj). Panel C UCSC Genome Browser view of the FKBP5 locus, indicating locations of cg03546163 (pcHiC) and cg23416081 (5 kb of TSS) showing interaction between the GeneHancer regulatory elements at these two regions. Publicly available data from tracks displayed includes location of exonic and intronic gene regions from the UCSC gene annotation; regulatory elements, genes and their interactions from GeneHancer, in detailed and clustered views; chromHMM tracks from Roadmap; transcription factor CHIP-seq from ENCODE; and DNAse hypersensitivity density signal from ENCODE for CD20 + B-cells, CD14+ monocytes, fibroblasts and naïve B-cells. Source data are provided as a Source Data file.
The two CpGs recognized as eQTMs for FKBP5 map to regulatory features, and there is interaction between the two regulatory features possibly accounting for the correlation observed between the two CpGs and asthma risk (Fig. 3C). FKBP5 encodes FKBP prolyl isomerase 5, a cochaperone modulating glucocorticoid receptor (GR) activity associated with the inflammatory response40. While site-specific effects of gene expression at FKBP5 with asthma are consistent with overlapping confidence intervals, a markedly stronger effect was observed in the Brazil site (Supplementary Fig 2) which is also the site with the most severe asthmatics and highest inhaled corticosteroid (ICS) use in asthma cases (97.2%, Supplementary Data 1). FKBP5 is not differentially expressed by ICS use in the full CAAPA dataset (Supplementary Data 5, p = 0.615). However, given prior evidence that FKBP5 may be differentially expressed after dexamethasone treatment41, we performed analyses to tease apart the relationship between ICS use, methylation at cg03546163, asthma severity by CASI, and risk for asthma in the four US sites where both DNAm and RNAseq were available. Lower methylation at cg03546163 was strongly associated with asthma (β = −0.295, p = 8.2 × 10−7) in the full subset of asthma cases and controls (N = 331). When limited to those asthma cases not on moderate-high ICS use (N = 283), the association with asthma remained (β = −0.197, p = 1.04 × 10−3). In a subset case-only analysis of all asthmatics (N = 149), we observed a more significant association between methylation and asthma severity measured by CASI score (β = −0.393, p = 8.17 × 10−4), than between methylation and ICS usage (β = −0.314, p = 0.014).
Discussion
In this report, we focus on RNASeq data in nasal epithelium tissue from 536 subjects representing the African Diaspora to define transcriptomic profiles related to asthma. We then integrate these transcript data with DNA methylation also in the same tissue with the goal of identifying mechanisms of dysregulation that underlie molecular subtypes or endotypes of asthma. Limitations in our study include the inability to differentiate gene expression profiles of atopy from asthma given the high prevalence of atopy in our cases and controls, the restriction of methylation data to only the US-based recruitment sites, and our inability to investigate environmental risk factors and social determinants of health in asthma. Despite these limitations, we are uniquely positioned to investigate asthma multi-omics in populations that are historically under-represented in genomics research but bear a disproportionate burden of the disease and disease severity. Overall, we found N = 389 differentially expressed genes, and 16 differentially expressed modules that are associated with current asthma adjusting for differences by site and ancestry. We identify strong signatures related to wound healing and drug response at single-gene and network-based levels that may have identified additional endotypes for asthma with potential implications for targeted therapy in the future.
The most significantly differentially expressed gene in asthma cases from CAAPA was FN1 encoding fibronectin. Despite some between-site differences in overall expression of this gene, there was a consistent lower expression of FN1 in nasal epithelium in asthma cases compared to controls, and this differential expression of FN1 was replicated in data from Tsai et al.26. Fibronectin is an adhesion protein. Increased deposition of FN1, along with fibrillar collagen proteins (COL3A and COL4A) in the extracellular matrix (ECM) and subepithelial space of airways, results in airway wall thickening and airflow obstruction, thereby altering the structural properties of the airways and the functional properties of airway cells in asthma34. In airway epithelial cells (AEC) from children with asthma compared to healthy atopic and nonatopic controls, FN1 was the only ECM component whose expression was significantly lower in AECs from asthma cases42. Furthermore, wound healing models using AECs showed that a reduced capacity of AECs to secrete FN1 contributes to dysregulated AEC repair and impaired wound healing42.
There are several additional lines of support related to down regulation of genes resulting in impaired wound healing as a prominent feature of asthma risk in CAAPA. First, pathway analysis of the full set of significant DEGs identified TGFβ1 as an upstream regulator for a network of genes including FN1, COL3A and COL4A. Transforming growth factor, encoded by the TGFβ1 gene, increases the deposition of ECM proteins, including FN1, and stimulates FN1 expression in airway epithelial cells33. In our study, the expression of all three genes were lower in asthma cases, consistent with impaired wound healing. Second, using a systems biology approach, we identified three modules of co-expressed genes down-regulated in airway cells from the asthma cases. The hub genes from these modules, FN1 (M5), ERBB2 (M8), and ACVR1B (M23), are associated with wound healing or airway remodeling. ERBB2 encodes a member of the epidermal growth factor (EGF) receptor family of receptor tyrosine kinases that play a key role in epithelial differentiation, proliferation, and repair36. Wound models have shown lower ERBB2 activation in freshly brushed isolated human AECs from patients with asthma and diminished wound closure and cell proliferation compared to AECs from healthy controls36. ACVR1B encodes an activin A type IB receptor for the cytokine activin A which is closely related to TGFβ143. In the network of activin A and TGFβ1 regulation, activin A is a potential modulator of airway remodeling43.
The second axis of dysregulation in asthma cases was a network of upregulated genes that reflect the canonical Th2 pathway. The strongest evidence is seen in the two most differentially expressed modules M6 and M2, which have Th2-related hub genes (CEACAM5 and CPA3, respectively); both are upregulated in the asthma cases. CEACAM5 is an IL-13-regulated epithelial gene that is upregulated in severe asthma and associated with increased asthma exacerbations44–46. It has also been previously identified as a hub gene in a limited microarray-based analysis in 42 cases and 28 controls47. Carboxypeptidase A3 (CPA3) is a mast cell (MC) protease and there is a well-known switching of MCs expressing tryptase only (MCT) to MCs expressing tryptase, chymase and CPA3 (MCTC) with a dominant expansion of the latter in airways of asthma cases and more specifically in the context of severe asthma48. MCs are key effector cells in asthma that are involved in both early and late phase allergic responses49, and they are most notably involved in the Th2-high endotype of asthma49. Additionally, VSIG4 is the third strongest DEG in our study. It encodes V-set and Ig domain-containing 4 and is an inhibitory ligand on antigen presenting cells thereby regulating T cell responses: macrophages expressing VSIG4 play a role in inhibiting T cell proliferation and cytokine production50.
The third axis implicates dysregulated genes that could reflect drug responsiveness. The top 15 DEGs included PTCHD4, which was associated with airway disease and identified in a GWAS for oral corticosteroid use51; SPTBN1, which was identified in a GWAS for leukotriene modifier response in asthma52; and FKBP5, which plays a role in response to inhaled corticosteroid response as described below. The most significant upstream regulator identified by IPA was the corticosteroid dexamethasone – an oral corticosteroid used in the treatment of asthma, and the 6th strongest upstream regulator was fluticasone propionate – a commonly used steroid treatment for allergic rhinitis in nasal spray formulations. Importantly, we ruled out that the 188 gene targets of these two drugs are directly related to ICS medication use in the asthma cases; 89 genes were targets to both drugs.
The FKBP5 gene encodes FKBP prolyl isomerase 5 (FKBP5), a cochaperone modulating glucocorticoid receptor (GR) activity associated with an inflammatory response40. It was previously shown to be differentially expressed after dexamethasone treatment41. Furthermore, a prior study suggested tightly regulated epigenetic control of the expression of genes that modulate GR responsiveness40. Here, we identified FKBP5 in the 15 most differentially expressed genes for asthma and also show epigenetic variation at two interacting regulatory regions possibly accounting for the differential expression. We also observed the largest estimated effect size for this gene was within the Brazil site, which has the most severe asthma cases with greatest proportion of asthma cases on moderate to high ICS use. This observation may be reflective of high FKBP5 gene expression in these subjects resulting in a decreased responsiveness to ICS, and a consequent escalation of ICS dose. This hypothesis aligns with the requirement of higher doses of ICS in this group of moderate to severe asthma cases from Brazil. By the time the patients were enrolled in the cohort they had difficult-to-treat asthma, but in many cases the disease has been controlled on medium-high doses of ICS combined with formoterol, a long-acting beta 2 agonist bronchodilator (LABA). The severity of asthma could also be related to a long journey to proper treatment, by patients who have suffered from recurrent asthma attacks and symptoms before having access to proper management with free ICS-LABA. Overall, the expression of FKBP5 was higher in the asthma cases in all other sites, except for those recruited in Denver. In comparison with other CAAPA sites, Denver site participants had the lowest asthma severity indicated by having the lowest mean CASI score (2.81) and highest mean FEV1 (99.5%) predicted in asthma cases (Supplementary Table S1). The Denver site’s low mean CASI score is consistent with mild asthma (CASI ≤ 3)53,54; in comparison, the Brazil site’s mean CASI score of 8 was the highest for all sites, and has been associated with severe asthma53. The Denver site was also distinguished by being 100% pediatric enrollment, differing from the other CAAPA sites with 48.5–100% adult participant enrollment (Brazil site was 100% adult participants). These observations support the relevance of FKBP5 expression in more severe and persistent disease in CAAPA.
Our integrative multi-omics analyses revealed an important set of genes and co-expressed gene networks with relevant mechanistic roles in asthma that are differentially expressed in the nasal epithelium of asthma cases of African ancestry in CAAPA. This work reveals dysregulation of three axes – increased Th2 inflammation, decreased capacity for wound healing, and impaired drug response. Each is associated with risk for asthma and there is correlation between the axes themselves, but the impact of dysregulation on multiple axes bears a cumulative risk with an OR of 4.95 (95% CI = 3.09–7.93). Networks of genes implicating Th2 inflammation are now well documented, but our findings with respect to gene networks related to wound healing and drug response are not documented, and our findings may be implicating additional endotypes of asthma beyond the well-known type 2 high vs. type 2 low29. To date, effective choice of monoclonal antibodies in the management of severe asthma is dependent on the clinical and inflammatory profile of the patient that relates to known endotypes29. While future work will be needed to validate these additional axes of dysregulation for drug response and airway remodeling identified in CAAPA and determine if these transcriptomic signatures may be related to ancestry, our findings from this understudied ancestry group that bears significant health disparities in asthma offer the potential to expand our understanding of clinical heterogeneity in disease and treatment response.
Methods
Study subjects
Study subjects included African ancestry individuals with no history of COPD, emphysema or chronic bronchitis. CAAPA included adult (aged 18–89) and pediatric (children aged 8–12 and adolescents aged 13–17) individuals. Other exclusion criteria included: pregnancy, lung transplant, kyphoscoliosis, sarcoidosis, bronchiopulmonary dysplasia, cystic fibrosis, bronchiectasis, rheumatoid arthritis, Crohn’s disease, psoriasis, lung carcinoma, ciliary dyskinesia, lupus and active tuberculosis. Recruitment occurred at 3 non-US sites (Nigeria, Barbados and Salvador, Brazil) and four US sites (Denver, Baltimore, Washington DC and Chicago). Study subjects were asked to self-identify as African, African American, African Caribbean, African Brazilian or African-Other. Cases were first defined as subjects with ‘ever’ asthma confirmed by a physician (response = yes to the two questions: (1) Have you ever had asthma?; and (2) Was it confirmed by a doctor?). The final set of cases was further restricted to the subset of individuals with’current’ asthma described below. Controls were defined as subjects with no history of asthma (response = no to the question: (1) Have you ever had asthma?). All samples used for this study were obtained following written informed consent from participants. The University of Colorado (IRB#: 17-1807), Johns Hopkins University (IRB00179053), University of Chicago (IRB18-0466-CR001), National Institutes of Health (IRB#: P184385), University of West Indies (IRB#: 190604-A), University of Bahia (IRB#: 3.302.487) and University of Ibadan Institutional Review Boards approved the conduct of this study (IRB18-0840).
Sample collection
Nasal columnar epithelial cells from the posterior surface of the inferior turbinate were collected using cytology brushes and standardized protocols30,55. After collection, the brush was immediately submerged in Buffer RLT Plus (Qiagen Inc., Valencia, CA, USA) to lyse the cells and stored until DNA/RNA were extracted. Prior to any processing, a small sample of cells was smeared on a glass slide followed by fixation and H&E staining for quality control. Only samples with ≥80% ciliated epithelial cells visualized from slides were retained. DNA and RNA were extracted from the same nasal sample for multi-omics analysis.
Questionnaires & phenotype data
Study data were collected and managed using REDCap® electronic data capture tools hosted at Yale University56 to record health questionnaires from the subjects at all 7 sites. These included informed consent, recruitment forms, respiratory health questionnaires, an asthma severity questionnaire, pulmonary function tests data, complete blood count (CBC) with differentials, vitals collected at the time of visit, medications and physical examination information and date and time nasal and blood samples were collected from the patient. Whole blood collected in BD Vacutainer® EDTA tubes was sent to the clinical laboratory at each site to perform a complete blood count (CBC) with differentials. Serum samples were sent to the Johns Hopkins University School of Medicine Reference Laboratory for Dermatology, Allergy and Clinical Immunology (DACI) for total serum IgE (tIgE) and multi-allergen (phadiatop) measurements. Spirometry was conducted according to the ATS guidelines using a hand-held KoKo DigDoser (Louisville, CO).
Measurement of atopy, asthma severity, medication use and current asthma
Atopy was defined on the basis of the phadiotop and total serum IgE: if phadiotop was ≥0.36 PAU or IgE was >100KU/L the subject was defined to have atopy. The Composite Asthma Severity Index (CASI) questionnaire57 was administered to all asthma cases at each of the recruitment sites. CASI takes into consideration medication use and the corresponding treatment level, in determining asthma severity57. The scoring guidelines for a treatment component of the CASI test were adapted from the Expert Panel Report 3 (EPR3) asthma guidelines for determining the treatment categories associated with the different levels of medication and a list of medications used for systemic corticosteroid bursts during asthma exacerbations54,58. The CASI questionnaire was used to subset asthma cases to those with current asthma given the importance of current disease status for dynamic omics signatures captured in RNA and DNA methylation. Current asthma status was defined as CASI ≥ 1 in asthma cases. Finally, the CASI questionnaire was used to define two medication use groups: no treatment or albuterol as needed (score 0-1 on question 4b) and low to high dose inhaled corticosteroid use (score 2–5 on question 4b). Additionally, at the time of nasal epithelium sample collection, individuals were asked if there were on nasal steroids, and if they were able to withhold usage for 5 days prior to sampling: there were 68 cases on nasal steroids, of whom 32 did not withhold usage.
Clinical characteristics of study subjects
Clinical characteristics of study subjects used for generating multi-omic data sets are summarized in Supplementary Data 1. Adults ranged in age from 18 to 89, and pediatric subjects ranged in age from 8 to 17. Additional information on sample distributions by phenotypes and their associations is also shown in Supplementary Data 1. For each quantitative phenotype (CASI, tIgE, eosinophil count, phadiotop, FEV1, FEV1/FVC), we tested for differences between cases and controls in the full group using a linear model adjusting for age, sex, the first two PCs plus recruitment site site (Supplementary Data 1, Full Group). Additionally, because Chicago was the only site to recruit both adults and children, we used this as the reference group to compare phenotypes across sites. In those analyses, we used similar linear models for the quantitative phenotypes and a logistic model for medication use, stratifying by case and control status and adjusting for age, sex, and the first two PCs.
Omics data generation
Genotyping was performed on DNA extracted from blood clots from CPT tubes on samples from all 7 recruitment sites. Samples with DNA Integrity Number (DIN) ≥6 were run on Illumina’s Infinium® Multi-Ethnic Global BeadChip (MEGA). Following the Illumina Infinium protocol, idat files were generated and used to extract the genotype calls, perform data QC and downstream analysis. Genotype data was used to derive ancestry principal components (PCs). Genotyping was performed on samples from all 7 recruitment sites.
RNA samples from nasal airway epithelial cells collected across all 7 recruitment sites and that passed all the laboratory QC thresholds (at least 80% columnar cells per nasal slide assessment, nanodrop and Qubit concentration ≥30 ng/ul in 20 uL volume (a total amount of 600 ng), RIN ≥ 6 and 260/280 between 1.7 and 2.2) were sent for RNA sequencing to Psomagen. RNASeq batches were balanced with respect to site, asthma status, sex and age (adult vs. child) to minimize confounding. The Illumina TruSeq Stranded Total RNA with Ribo-Zero kit was used to prepare libraries, depleting the ribosomal RNA. RNA sequencing (RNAseq) was performed on the NovaSeq 6000 using 150 bp paired end reads and yielding at least a total of 80 million paired end reads per sample. RNASeq was performed on samples from all 7 recruitment sites.
DNA methylation (DNAm) quantification was performed using Illumina’s Infinium MethylationEPIC array® using genomic DNA from nasal airway epithelial cells collected across the 4 US recruitment sites that passed all the laboratory QC thresholds (at least 80% columnar cells per nasal slide assessment, nanodrop and Qubit concentration ≥15 ng/ul in 50 ul (a total amount of 750 ng), DIN ≥ 6 and 260/280 between 1.4 and 2.15). Bisulfite conversion was performed using the EZ DNA Methylation™ kit (Zymo Research) and DNAm quantification was performed using the Illumina Infinium protocol. Idat files generated from the Infinium protocol were used to perform the data QC and downstream analysis. Methylation chips were randomized with respect to site, asthma status, sex and age (adult vs. child) to minimize confounding.
Quality control (QC) and preprocessing
Quality control procedures were performed on genotype data to exclude any samples or variants that had missingness >3%; standard QC steps included: sex verification, heterozygosity checks and identity-by-descent (IBD) to look for any unexpected relatedness. Two individuals failed on call rate and four individuals were identified as sex mismatches in one or more omics data sets (dropped from all datasets). Thirteen individuals showed cryptic relatedness, as being part of a parent offspring pair, full sibs or half sibs pairs based on the IBD estimates; one independent subject was selected prioritizing case status from each relationship resulting in 7 individuals being dropped from all datasets. One sample was identified as a sample swap, duplicating another individual, and was dropped. A total of 14 samples were excluded and further ancestry analysis was limited to 673 individuals from all seven sites.
To perform RNAseq QC pre-alignment, QC, adapter trimming, and alignment of reads to GRCh38 were performed using FastQC [http://www.bioinformatics.babraham.ac.uk/projects/fastqc/], Picard tools [http://broadinstitute.github.io/picard], BBDUK [sourceforge.net/projects/bbmap/], and HISAT259, respectively. Raw counts were generated by CoCo60. Four samples were excluded due to sex mismatches, 3 samples were excluded due to unexpected relatedness, 4 samples were excluded due to failure of library preparation, and 7 samples were excluded due a high percentage of ribosomal RNA. After filtering asthma cases to include only current asthmatics, and available genotype data for ancestry PCs, analysis was limited to 536 individuals from all seven sites.
DNAm data QC was tested using the minfi R-package61. Samples were excluded based on the following metrics: 12 with mean of methylated and unmethylated signal <10.5; four with discordant sex. CpG sites were filtered as follows: N = 953 probes that had fewer calls detected (detection p > 0.01) in more than 20% of the samples; N = 30,247 probes that were in close proximity to SNPs (at the CpG site or in the single-base extension site for the array probe), yielding a total of 834,663 CpGs that were normalized using the quantile option for downstream analysis. After filtering asthma cases to include only current asthmatics, and available genotype data for ancestry PCs, analysis was limited to 331 individuals from four US-based recruitment sites: Baltimore, Chicago, Denver and Washington DC.
Quantification of principal components and global sample ancestry
We used KING62 to estimate relatedness and build a kinship matrix including all subjects from all the sites. SNPs with >5% missingness, <1% minor allele frequency and hardy Weinberg p < 1 × 10−6 were discarded. SNPs underwent linkage disequilibrium-pruning (removal of SNPs with an R2-value > 0.1 within every 50 SNP window) and PCA analyses were performed using a CAAPA-only dataset comprised of 673 samples and 512,925 SNPs. PCA was performed allowing for observed kinship using PC-AiR as implemented in the GENESIS R package63. Additionally, PCA was also performed including reference populations from the 1000 Genomes Project (85 Utah residents with Northern and Western European ancestry CEU (EUR), 88 Yoruba samples from Ibadan, Nigeria YRI (AFR) and 43 Native Americans selected from Mao et al.64 (AMR)) on 219,832 autosomal SNPs obtained after the merge with reference data. The elbow in the scree plot was used to identify the top two PCs (PC1 and PC2) as covariates for ancestry adjustment, Supplementary Fig 1.
To estimate global ancestry proportions, we first implemented cross-validation using ADMIXTURE65 to determine the number of reference populations (K) with K = 1–5, to infer the optimal number of ancestral reference groups needed. K = 3 accounted for the lowest cross-validation error. CAAPA samples were merged with 3 reference populations and with the set of 219,832 autosomal SNPs obtained after the merge, and using K = 3, we performed global ancestry estimation using ADMIXTURE and plotted the admixture estimates using the PONG visualization tool66 as shown in Supplementary Fig 1.
Differential gene expression analysis
Mean normalized counts were generated by DESeq267. Genes were first filtered across all sites combined to only include those with DESeq2 mean normalized count ≥ 20 and counts per million (CPM) > 0 in the percentage of samples corresponding to the proportion of asthma cases to never-asthma controls (e.g. in the full sample there were 47.20% cases, and we required CPM > 0 in 47.20% of the total sample); this was done to ensure expression variability was present in both cases and controls. Following filtering, 21,831, 21,789 and 21,887 genes were available for analysis in the full dataset, adult-only dataset and pediatric-only dataset, respectively. Differential gene expression analyses for case-control comparisons were performed on all subjects, and stratifying into adult and pediatric groups, plus stratifying subjects by site. All site analyses were performed on the same set of filtered genes generated for the analysis of all subjects above. Differential analysis in limma68 and edgeR69 was performed. Counts were transformed to log2 (CPM) using voom68 and a linear model was fit adjusting for relevant covariates: asthma status, age, sex, library preparation batch, site, RNA integrity number (RIN), GC content, and ancestry PCs 1 and 2. Sites were not included as a covariate in these site specific analyses. SVA70 was used to generate surrogate variables (SVs) for each stratified analysis and significant SVs were added to the model. Analysis was also performed in the subset of N = 253 asthma cases for medication use including the same covariates, and SVs derived on only these subjects.
Ingenuity pathway analysis
Differentially expressed genes with q < 0.05 in the analysis of all subjects were selected as input for Ingenuity pathway analysis (IPA)71. IPA upstream regulator analysis was utilized to identify molecules upstream of the selected DEGs that could potentially explain the observed expression differences between case and controls. Activation z-score (a measure of consistency between up/down gene regulation pattern and activation/inhibition pattern given by the IPA knowledgebase network) and p-value of overlap (a measure of significance of enrichment of regulated genes in the dataset, agnostic to direction) were generated for each upstream regulator, and gene targets of each regulator were obtained from the upstream regulator table. P-value of overlap measures of significance of enrichment of regulated genes in the dataset given a regulator, agnostic to direction; all upstream regulators have a generated p-value. IPA predicts the activation state of the regulator by assessing the consistency of direction of gene expression of gene targets in the dataset with activation/inhibition patterns given by the IPA knowledgebase relative to a random pattern. Activation z-score was calculated for regulators given that the direction of regulation is well defined based on literature findings in the IPA Knowledgebase and the underlying null model is appropriate; therefore, not all upstream regulators have an activation z-score.
Gene expression module analysis
Differentially expressed genes with q < 0.15 in the analysis of all subjects were selected for weighted gene coexpression network analysis (WGCNA)72. This more liberal significance threshold was used to cast a wide net for a systems biology analysis. WGCNA was performed on 1326 genes using mean normalized counts described above corrected for library preparation, batch, sex, RIN, and GC content with the following parameters: power was selected for which the scale-free topology fit index reached 0.9, network type = “signed”, TOM type = “signed”, deepSplit = 3, min module size = 15, max block size = 8000. Gene module differential expression analysis with average expression across genes in a module as the outcome was performed using limma according to the analysis pipeline described above for gene differential expression comparing current asthma cases to never-asthmatic controls without the use of SVs, but otherwise adjusting for the covariates age, sex, library preparation batch, site, RNA integrity number (RIN), GC content, and ancestry PCs 1 and 2. Multiple testing correction was performed using the Benjamini-Hochberg method and an FDR cutoff <0.05 was considered significant.
Significantly differentially expressed gene modules were visualized using cytoscape73 and STRINGdb74. Module expression was calculated as the mean expression of genes, using voom transformed counts corrected for sex, library preparation batch, RIN, and GC content using limma removeBatchEffect(), assigned to a module and STRING protein-protein interaction networks were retrieved for differentially expressed modules. Interactions with stringdb score >0.15 were considered significant and unconnected genes were omitted. Gene hubs for each module were determined as the gene with the highest connectivity in the module using the chooseTopHubInEachModule() WGCNA function or the gene with the highest connectivity in the module STRING network.
Multi-module analysis
To evaluate the cumulative risk for asthma across the three axes of dysregulation, we selected the most significant module based on q-value for Th2 inflammation (M6, CEACAM5), wound repair (M5, FN1), and drug response (M4, NCALD). Covariate effects (age, sex, site, RIN, GC content, library preparation batch, ancestry PC1 and PC2) were regressed out from module average expression. Each module was dichotomized with values of 0 and 1 at the median considering direction of effect of module association with asthma: for M6 where higher module expression is associated with asthma risk, the upper 50th percentile was labeled as = 1, and lower 50th percentile labeled as 0; for M4 and M5 where lower module expression is associated with asthma risk, lower 50th percentile was labeled as = 1 and upper 50th percentile was labeled as = 0. We then calculated the sum of these three binary covariates (the “number of modules” a sample is in the risk side of the median dichotomy) as a covariate of interest and used logistic regression models to investigate the association between this number and the fraction of asthma cases observed in our data. The number of modules was fit as a numeric term, but we also tested formally for departures from additivity by comparing this model in a likelihood ratio test to a logistic model with number of modules as a factor, allowing for an extra 2 degrees of freedom.
DNA Methylation and multi-omics analysis
For the set of N = 389 asthma DEGs, eQTM analysis was performed by fitting a linear model for each gene-CpG pair to test the association between gene expression and cis-CpG methylation using the Matrix eQTL R package75, testing all CpGs within 1 megabase of the nearest end of the gene. As we are modeling gene expression as the outcome, all the covariates used for differential expression analysis were included: case-control status, age, sex, library prep batch, site, RIN score, GC content, ancestry PC1 and PC2. As generation of SVs using SVA would need to be performed for each CpG, we instead generated PEER factors76 using the peer package in R with all covariates listed above and default parameters (Alpha_Prior_A = 0.001, Alpha_Prior_B = 0.01, Eps_Prior_A = 0.1, Eps_Prior_B = 10, Max_Iteration = 1000, Tol = 0.001). We included 60 PEER factors in our eQTM models. eQTM analysis was run on a total of 298 subjects where both RNASeq and DNAm data were available. CpGs were selected if they mapped within 5 kb of a gene transcription start site (upstream or downstream) or if they were annotated by promoter-capture HiC in bronchial epithelial cells77to lie in putative enhancer regions for these 389 genes. There were 8418 eQTM tests performed for gene-CpG pairs and significance for the eQTMs was defined as eQTM p < 0.05.
For the subset of CpGs identified as significant eQTMs, we tested for differential methylation (DMC analysis) using standard linear modeling approaches implemented through the limma R package68, including age, sex, site, plate, ancestry PC1 and PC2 and 12 latent factors estimated from CBCV-CorrConf R package78 included to adjust for additional unmeasured confounders such as cell composition differences. Differential methylation analysis was run between 149 cases and 182 controls for 915 CpGs. Significance was defined at the Bonferroni threshold of p < 0.05/915. For the subset of CpGs that were identified to be eQTMs and DMCs, and where there were multiple eQTMs per gene, a joint model was run to evaluate independence of the multiple CpGs.
Finally, in the 298 subjects that had both DNAm and RNAseq, conditional analysis was performed for the CpG’s identified as both eQTMs and DMCs to understand the effect of DNAm on the relationship between gene expression and asthma, i.e. was there a gene expression association with asthma after adjustment for the relevant DMC. The following models were run: DEG,unadj testing for differential gene expression including all the original covariates (asthma status, age, library preparation batch, site, RNA integrity number (RIN), GC content, ancestry PCs 1 and 2, 23 SVs), and DEG,adj testing for differential gene expression including all the original covariates and also including methylation at the CpG of relevance for the gene (i.e. peak eQTM for the gene). We examined the change in effect size and significance of the DEG between the DEG,unadj and DEG,adj models.
Replication
For the 21,831 genes tested in the DEG analysis of all subjects, we performed an exhaustive search for replication in a meta-analysis study of airway epithelium gene expression in asthma26. Briefly, Tsai et al. performed a meta-analysis of eight independent gene expression studies including both nasal and bronchial epithelium tissue. Full results from the meta-analysis were obtained from these authors and were compared to the CAAPA results by matching genes on Ensembl ID. Ensembl IDs were retrieved for the meta-analysis gene symbols from Ensembl Release 109 homo sapiens GRCh38 using pyensembl [https://github.com/openvax/pyensembl]. Each gene symbol in an observation was queried individually for matching Ensembl IDs and matched to Ensembl IDs in CAAPA. Where multiple observations in the meta-analysis matched an Ensembl ID in CAAPA, the observation with the highest number of studies (k) in the meta-analysis was selected. Enrichment of CAAPA DEGs in the meta-analysis DEGs was tested using a hypergeometric test. The total number of genes tested was determined as the number of unique Ensemble IDs retrieved in the meta-analysis full results that matched to Ensembl IDs included in the 21,831 genes tested in CAAPA.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank the numerous health care providers, and community clinics and co-investigators who assisted in the phenotyping and collection of samples, and the CAAPA participants for generously donating time and samples. We gratefully acknowledge the contributions of clinical recruiters and technicians from each recruitment site: Allison Schiltz, Alan Franklin, Elizabeth Davidson (Denver); Amy Bentley, Lin Lei, Carolyn Allen, Paule Joseph, Hermon Feron (Washington DC); Carly Jackson and Robert Stanaker (Chicago); Laura Grammer (Baltimore); Dr. Ayobami Bakare, Dayo Adepoju, Samuel Adekunle, Tope Ibigbami, Abayomi Odetunde, Sunkanmi Owolade, Joseph Eleyinmi (Nigeria); Pissamai Maul, Trevor Maul, Desiree Walcott, Andre Greenidge (Barbados); Luane Marques Mello, Gabriela Pimentel Pinheiro das Chagas, Cinthia Vila Nova Santana, Débora Inácio, Laila Trindade, Givaneide Lima, Tamires Carneiro, Candace Andrade, Helena Teixeira, Ryan Costa (Brazil). We are grateful for the support from the international state governments and universities from Barbados, Brazil and Nigeria who made this work possible. We acknowledge the support from James Kiley and Weiniu Gan at the National Heart, Lung and Blood Institute. Funding for this study was provided by National Institutes of Health (NIH) R01HL104608. We thank Christopher Arehart, Iain Konigsberg and Chris McKennan for their support and input on bioinformatic pipelines leveraged in this work.
Author contributions
Data analysis was performed by C.C., A.M. and W.L. Data analysis, and writing was carried out by B.S., M.P.B., S.C., M.C., R.K.J., K.K., E.E.T., G.S., M.D., S.N.P.K. and M.S.C. Scientific leadership and data acquisition were provided by H.W., E.T.N., B.L.G., G.A.A., O.S., A.G.F., N.N.H., C.N.R, R.C.L. and E.K. Data analysis, scientific leadership and writing were performed by I.R., A.H.L., C.O., M.A.T., K.C.B., R.A.M. and M.C.A. Scientific leadership and writing were provided by THB,APD,AAC,IVY,COO and CAF.
Peer review
Peer review information
Nature Communications thanks Ian Hall and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The RNASeq data generated in this study have been deposited in the GEO database under accession code GSE240567 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM7701971]. The Methylation data generated in this study have been deposited in the GEO database under accession code GSE250513. The genotype data generated in this study have been deposited in dbGAP database under accession code phs001123. Meta-analysis result from Tsai et al. are available in Supplementary Data 2. Source data are provided with this paper for all figures. Source data are provided with this paper.
Competing interests
K.C.B. declares Royalties from UpToDate. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Brooke Szczesny, Meher Preethi Boorgula.
These authors jointly supervised this work: Kathleen C. Barnes, Rasika A. Mathias.
Contributor Information
Kathleen C. Barnes, Email: kathleen.barnes@cuanschutz.edu
Rasika A. Mathias, Email: rmathias@jhmi.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-48507-7.
References
- 1.World Health Organization. Asthma Fact Sheets. https://www.who.int/news-room/fact-sheets/detail/asthma Accessed 5/10/2024.
- 2.Akinbami, L. J., Moorman, J. E. & Liu, X. Asthma prevalence, health care use, and mortality: United States, 2005-2009. 1–14 (National Health Statistics Report, 2011). [PubMed]
- 3.Barnes KC. Genomewide association studies in allergy and the influence of ethnicity. Curr. Opin. allergy Clin. Immunol. 2010;10:427–433. doi: 10.1097/ACI.0b013e32833de6ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes KC, Grant AV, Hansel NN, Gao P, Dunston GM. African Americans with asthma: genetic insights. Proc. Am. Thorac. Soc. 2007;4:58–68. doi: 10.1513/pats.200607-146JG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guilbert T, et al. Racial Disparities in Asthma-Related Health Outcomes in Children with Severe/Difficult-to-Treat Asthma. J. Allergy Clin. Immunol. Pr. 2019;7:568–577. doi: 10.1016/j.jaip.2018.07.050. [DOI] [PubMed] [Google Scholar]
- 6.U.S. Department of Health and Human Services, Office of Minority Health. Asthma and African Americans, https://minorityhealth.hhs.gov/asthma-and-african-americans.
- 7.Vergara C, et al. African ancestry is a risk factor for asthma and high total IgE levels in African admixed populations. Genet. Epidemiol. 2013;37:393–401. doi: 10.1002/gepi.21702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Obeng BB, Hartgers F, Boakye D, Yazdanbakhsh M. Out of Africa: what can be learned from the studies of allergic disorders in Africa and Africans? Curr. Opin. Allergy Clin. Immunol. 2008;8:391–397. doi: 10.1097/ACI.0b013e32830ebb70. [DOI] [PubMed] [Google Scholar]
- 9.Pearce N, et al. Worldwide trends in the prevalence of asthma symptoms: phase III of the International Study of Asthma and Allergies in Childhood (ISAAC) Thorax. 2007;62:758–766. doi: 10.1136/thx.2006.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adeloye D, Chan KY, Rudan I, Campbell H. An estimate of asthma prevalence in Africa: a systematic analysis. Croat. Med. J. 2013;54:519–531. doi: 10.3325/cmj.2013.54.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bateman ED, et al. Global strategy for asthma management and prevention: GINA executive summary. Eur. Respir. J. 2008;31:143–178. doi: 10.1183/09031936.00138707. [DOI] [PubMed] [Google Scholar]
- 12.Ortega VE, Meyers DA. Pharmacogenetics: implications of race and ethnicity on defining genetic profiles for personalized medicine. J. Allergy Clin. Immunol. 2014;133:16–26. doi: 10.1016/j.jaci.2013.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Federico MJ, Covar RA, Brown EE, Leung DY, Spahn JD. Racial differences in T-lymphocyte response to glucocorticoids. Chest. 2005;127:571–578. doi: 10.1378/chest.127.2.571. [DOI] [PubMed] [Google Scholar]
- 14.Wells KE, et al. Assessing differences in inhaled corticosteroid response by self-reported race-ethnicity and genetic ancestry among asthmatic subjects. J. Allergy Clin. Immunol. 2016;137:1364–1369 e1362. doi: 10.1016/j.jaci.2015.12.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ortega VE, et al. Effect of rare variants in ADRB2 on risk of severe exacerbations and symptom control during longacting beta agonist treatment in a multiethnic asthma population: a genetic study. Lancet Respir. Med. 2014;2:204–213. doi: 10.1016/S2213-2600(13)70289-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathias RA, et al. A continuum of admixture in the Western Hemisphere revealed by the African Diaspora genome. Nat. Commun. 2016;7:12522. doi: 10.1038/ncomms12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim KW, Ober C. Lessons Learned From GWAS of Asthma. Allergy Asthma Immunol. Res. 2019;11:170–187. doi: 10.4168/aair.2019.11.2.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daya M, et al. Association study in African-admixed populations across the Americas recapitulates asthma risk loci in non-African populations. Nat. Commun. 2019;10:880. doi: 10.1038/s41467-019-08469-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsuo K, et al. Multi-ancestry meta-analysis of asthma identifies novel associations and highlights the value of increased power and diversity. Cell Genom. 2022;2:100212. doi: 10.1016/j.xgen.2022.100212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han Y, et al. Genome-wide analysis highlights contribution of immune system pathways to the genetic architecture of asthma. Nat. Commun. 2020;11:1776. doi: 10.1038/s41467-020-15649-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demenais F, et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat. Genet. 2018;50:42–53. doi: 10.1038/s41588-017-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altman MC, et al. Endotype of allergic asthma with airway obstruction in urban children. J. Allergy Clin. Immunol. 2021;148:1198–1209. doi: 10.1016/j.jaci.2021.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang IV, et al. The Nasal Methylome: A Key to Understanding Allergic Asthma. Am. J. Respir. Crit. Care Med. 2017;195:829–831. doi: 10.1164/rccm.201608-1558LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morin A, et al. Epigenetic landscape links upper airway microbiota in infancy with allergic rhinitis at 6 years of age. J. Allergy Clin. Immunol. 2020;146:1358–1366. doi: 10.1016/j.jaci.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magnaye KM, et al. DNA methylation signatures in airway cells from adult children of asthmatic mothers reflect subtypes of severe asthma. Proc. Natl Acad. Sci. USA. 2022;119:e2116467119. doi: 10.1073/pnas.2116467119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai, Y. H., Parker, J. S., Yang, I. V. & Kelada, S. N. P. Meta-analysis of airway epithelium gene expression in asthma. Eur. Respir. J.51, 10.1183/13993003.01962-2017 (2018). [DOI] [PMC free article] [PubMed]
- 27.Woodruff PG, et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am. J. Respir. Crit. Care Med. 2009;180:388–395. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altman MC, et al. Transcriptome networks identify mechanisms of viral and nonviral asthma exacerbations in children. Nat. Immunol. 2019;20:637–651. doi: 10.1038/s41590-019-0347-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schleich, F., Bougard, N., Moermans, C., Sabbe, M. & Louis, R. Cytokine-targeted therapies for asthma and COPD. Eur. Respir. Rev. 32, 10.1183/16000617.0193-2022 (2023). [DOI] [PMC free article] [PubMed]
- 30.Poole A, et al. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J. Allergy Clin. Immunol. 2014;133:670–678 e612. doi: 10.1016/j.jaci.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi K, Ge MN, Chen XQ. Coordinated DNA Methylation and Gene Expression Data for Identification of the Critical Genes Associated with Childhood Atopic Asthma. J. Comput Biol. 2020;27:109–120. doi: 10.1089/cmb.2019.0194. [DOI] [PubMed] [Google Scholar]
- 32.Lewis CC, et al. Unique and overlapping gene expression patterns driven by IL-4 and IL-13 in the mouse lung. J. Allergy Clin. Immunol. 2009;123:795–804 e798. doi: 10.1016/j.jaci.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ge Q, et al. Differential deposition of fibronectin by asthmatic bronchial epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2015;309:L1093–L1102. doi: 10.1152/ajplung.00019.2015. [DOI] [PubMed] [Google Scholar]
- 34.Hocking DC. Fibronectin matrix deposition and cell contractility: implications for airway remodeling in asthma. Chest. 2002;122:275S–278S. doi: 10.1378/chest.122.6_suppl.275s. [DOI] [PubMed] [Google Scholar]
- 35.Inoue, H., Akimoto, K., Homma, T., Tanaka, A. & Sagara, H. Airway Epithelial Dysfunction in Asthma: Relevant to Epidermal Growth Factor Receptors and Airway Epithelial Cells. J. Clin. Med.9, 10.3390/jcm9113698 (2020). [DOI] [PMC free article] [PubMed]
- 36.Inoue H, et al. Dysfunctional ErbB2, an EGF receptor family member, hinders repair of airway epithelial cells from asthmatic patients. J. Allergy Clin. Immunol. 2019;143:2075–2085 e2010. doi: 10.1016/j.jaci.2018.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joo J, et al. Genome-wide association study in minority children with asthma implicates DNAH5 in bronchodilator responsiveness. Sci. Rep. 2022;12:12514. doi: 10.1038/s41598-022-16488-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sordillo, J. E. et al. Pharmacogenetic Polygenic Risk Score for Bronchodilator Response in Children and Adolescents with Asthma: Proof-of-Concept. J. Pers. Med.11. 10.3390/jpm11040319 (2021). [DOI] [PMC free article] [PubMed]
- 39.Vijverberg SJ, et al. ST13 polymorphisms and their effect on exacerbations in steroid-treated asthmatic children and young adults. Clin. Exp. Allergy. 2015;45:1051–1059. doi: 10.1111/cea.12492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zannas AS, Wiechmann T, Gassen NC, Binder EB. Gene-Stress-Epigenetic Regulation of FKBP5: Clinical and Translational Implications. Neuropsychopharmacology. 2016;41:261–274. doi: 10.1038/npp.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menke A, et al. Dexamethasone stimulated gene expression in peripheral blood is a sensitive marker for glucocorticoid receptor resistance in depressed patients. Neuropsychopharmacology. 2012;37:1455–1464. doi: 10.1038/npp.2011.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kicic A, et al. Decreased fibronectin production significantly contributes to dysregulated repair of asthmatic epithelium. Am. J. Respir. Crit. Care Med. 2010;181:889–898. doi: 10.1164/rccm.200907-1071OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karagiannidis C, et al. Activin A is an acute allergen-responsive cytokine and provides a link to TGF-beta-mediated airway remodeling in asthma. J. Allergy Clin. Immunol. 2006;117:111–118. doi: 10.1016/j.jaci.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 44.Hoda U, et al. Clinical and transcriptomic features of persistent exacerbation-prone severe asthma in U-BIOPRED cohort. Clin. Transl. Med. 2022;12:e816. doi: 10.1002/ctm2.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mumby S, et al. CEACAM5 is an IL-13-regulated epithelial gene that mediates transcription in type-2 (T2) high severe asthma. Allergy. 2022;77:3463–3466. doi: 10.1111/all.15465. [DOI] [PubMed] [Google Scholar]
- 46.Xu C, et al. Elevated CEACAM5 Levels in Patients with Asthma. Int Arch. Allergy Immunol. 2022;183:673–681. doi: 10.1159/000521754. [DOI] [PubMed] [Google Scholar]
- 47.Cao Y, et al. Identifying key genes and functionally enriched pathways in Th2-high asthma by weighted gene co-expression network analysis. BMC Med. Genomics. 2022;15:110. doi: 10.1186/s12920-022-01241-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pejler, G. The emerging role of mast cell proteases in asthma. Eur. Respir. J.54, 10.1183/13993003.00685-2019 (2019). [DOI] [PubMed]
- 49.Andersson C, Tufvesson E, Diamant Z, Bjermer L. Revisiting the role of the mast cell in asthma. Curr. Opin. Pulm. Med. 2016;22:10–17. doi: 10.1097/MCP.0000000000000228. [DOI] [PubMed] [Google Scholar]
- 50.Vogt L, et al. VSIG4, a B7 family-related protein, is a negative regulator of T cell activation. J. Clin. Investig. 2006;116:2817–2826. doi: 10.1172/JCI25673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, A. L. et al. Novel genetic variants associated with inhaled corticosteroid treatment response in older adults with asthma. Thorax,10.1136/thoraxjnl-2021-217674 (2022). [DOI] [PMC free article] [PubMed]
- 52.Dahlin A, et al. Genome-wide association study of leukotriene modifier response in asthma. Pharmacogenomics J. 2016;16:151–157. doi: 10.1038/tpj.2015.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mokhallati N, Schuler CL, Thomas S, Hossian MM, Guilbert TW. Use of the composite asthma severity index in a pediatric subspecialty clinic. Ann. Allergy Asthma Immunol. 2021;126:702–706. doi: 10.1016/j.anai.2021.01.036. [DOI] [PubMed] [Google Scholar]
- 54.Krouse RZ, et al. Minimally important differences and risk levels for the Composite Asthma Severity Index. J. Allergy Clin. Immunol. 2017;139:1052–1055. doi: 10.1016/j.jaci.2016.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang IV, et al. The nasal methylome and childhood atopic asthma. J. Allergy Clin. Immunol. 2017;139:1478–1488. doi: 10.1016/j.jaci.2016.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harris PA, et al. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inf. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wildfire JJ, et al. Development and validation of the Composite Asthma Severity Index–an outcome measure for use in children and adolescents. J. Allergy Clin. Immunol. 2012;129:694–701. doi: 10.1016/j.jaci.2011.12.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.National Asthma Education and Prevention Program. Expert Panel Report 3 (EPR-3): Guidelines for the Diagnosis and Management of Asthma-Summary Report 2007. J. Allergy Clin. Immunol.120, S94–S138 (2007). [DOI] [PubMed]
- 59.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deschamps-Francoeur G, Boivin V, Abou Elela S, Scott MS. CoCo: RNA-seq read assignment correction for nested genes and multimapped reads. Bioinformatics. 2019;35:5039–5047. doi: 10.1093/bioinformatics/btz433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fortin JP, Triche TJ, Jr., Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33:558–560. doi: 10.1093/bioinformatics/btw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manichaikul A, et al. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gogarten SM, et al. Genetic association testing using the GENESIS R/Bioconductor package. Bioinformatics. 2019;35:5346–5348. doi: 10.1093/bioinformatics/btz567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mao X, et al. A genomewide admixture mapping panel for Hispanic/Latino populations. Am. J. Hum. Genet. 2007;80:1171–1178. doi: 10.1086/518564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Behr AA, Liu KZ, Liu-Fang G, Nakka P, Ramachandran S. pong: fast analysis and visualization of latent clusters in population genetic data. Bioinformatics. 2016;32:2817–2823. doi: 10.1093/bioinformatics/btw327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robinson MD, McCarthy DJ, Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kramer A, Green J, Pollard J, Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, B. & Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol.4, 17 10.2202/1544-6115.1128 (2005). [DOI] [PubMed]
- 73.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019;18:623–632. doi: 10.1021/acs.jproteome.8b00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stegle O, Parts L, Durbin R, Winn J. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput. Biol. 2010;6:e1000770. doi: 10.1371/journal.pcbi.1000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Helling BA, et al. Altered transcriptional and chromatin responses to rhinovirus in bronchial epithelial cells from adults with asthma. Commun. Biol. 2020;3:678. doi: 10.1038/s42003-020-01411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McKennan C, Nicolae D. Estimating and accounting for unobserved covariates in high-dimensional correlated data. J. Am. Stat. Assoc. 2022;117:225–236. doi: 10.1080/01621459.2020.1769635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The RNASeq data generated in this study have been deposited in the GEO database under accession code GSE240567 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM7701971]. The Methylation data generated in this study have been deposited in the GEO database under accession code GSE250513. The genotype data generated in this study have been deposited in dbGAP database under accession code phs001123. Meta-analysis result from Tsai et al. are available in Supplementary Data 2. Source data are provided with this paper for all figures. Source data are provided with this paper.