Abstract
A novel sphingoglycolipid was isolated from Sphingomonas yanoikuyae, and its structure was identified as a galacturonosyl-β (1→1)-ceramide. This was a characteristic sphingoglycolipid present in S. yanoikuyae and certain other species of Sphingomonas, such as Sphingomonas mali, Sphingomonas terrae, and Sphingomonas macrogoltabidus, but not in the type species of Sphingomonas, Sphingomonas paucimobilis.
Sphingolipids are one of the most ubiquitous components of eukaryotic cell membranes. In contrast, the occurrence of sphingolipids in bacteria is rare. They have been reported as sphingomyelin in Mycoplasma, sphingophospholipid in Bacteroides and Bdellovibrio, aminoglycosphingolipid in Chlorobium, capnoid in Capnocytophaga, Cytophaga, Flavobacterium, Flexibacter, and Sporocytophaga, sphingoglycolipid (SGL) in Sphingomonas and Zymomonas, and ceramide in Bacteroides and Sphingobacterium (6–8, 10, 15, 19). SGL (2-N-2′-hydroxymyristoyl dihydrosphingosine-1-glucuronic acid) containing glucuronic acid was isolated first from cellular lipids of the type strain of Flavobacterium devorans ATCC 10829 (14, 18). A similar SGL was found in a group of deep-yellow pigmented organisms, including the type strain of Pseudomonas paucimobilis (5). We proposed previously a new genus Sphingomonas with the type species Sphingomonas paucimobilis, based on the existence of a unique SGL, the major type of ubiquinone (Q-10), 16S rRNA sequence, and phenotypic features. Furthermore, three new species, Sphingomonas yanoikuyae, Sphingomonas parapaucimobilis, and Sphingomonas adhaesiva, and one new combination, Sphingomonas capsulata, were described from the homology of DNA-DNA hybridization and phenotypic characterization (15). SGLs containing di-, tri-, and tetrasaccharides have been found in the type strain of S. paucimobilis (7, 13). The major ubiquitous SGL in Sphingomonas is a glucuronosyl ceramide which is called SGL-1.
During research leading to the proposal of a new genus, Sphingomonas, we have noticed the existence of a novel alkali-stable sphingoglycolipid (SGL-1′) in S. yanoikuyae migrating close to but distinctive from an SGL-1 spot on a thin-layer chromatogram (TLC) of silica gel G (Uniplate; Analtech, Newark, Del.) developed with an acidic solvent system, chloroform-methanol-acetic acid-water (100:20:12:5, by volume) (15). The objective of this study was to isolate SGL-1′ and to define its chemical structure. We have demonstrated here an alkali-stable lipid that is identical to SGL-1′ in S. yanoikuyae and other type strains of Sphingomonas mali, Sphingomonas terrae, and Sphingomonas macrogoltabidus.
Isolation of SGL-1′.
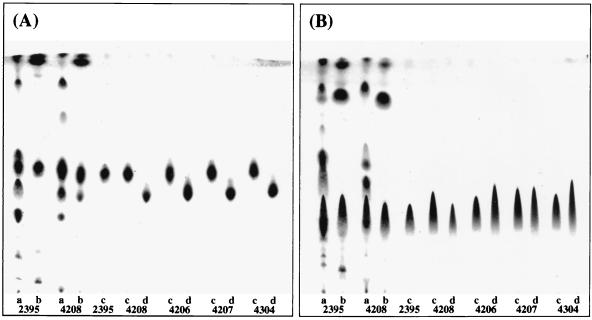
S. yanoikuyae EY 4208T, S. mali EY 4206T, S. terrae EY 4207T, and S. macrogoltabidus EY 4304T were used in this study. The detailed history, corresponding strain number, and culture conditions were described previously (11, 12, 15). The crude lipids were extracted by the method of Folch et al. (4). In brief, harvested bacteria were sonicated and lipids were extracted with a chloroform-methanol (2:1 and 1:3, by volume) mixture. After condensation of the organic phase, extractable lipids were hydrolyzed with 0.5 N KOH (30°C, 3 h), which was followed by neutralization. Alkali-stable lipids were again extracted with chloroform-methanol (2:1, vol/vol). Alkali-stable lipids in S. yanoikuyae EY 4208T revealed two major glycolipid spots on a TLC developed with an acidic solvent system (Fig. 1A). The upper spot, with an Rf value of 0.48, coincided with glucuronosyl ceramide (SGL-1) isolated from S. paucimobilis EY 2395T; however, the lower spot (SGL-1′) did not correspond to any sphingo- (or glycero-) glycolipid reported previously. When the plate was developed with the neutral solvent system, chloroform-methanol-water (65:25:4, by volume) (Fig. 1B), these two spots were united to form a long tailing spot resembling anionic phospholipid, suggesting that both compounds (SGL-1 and SGL-1′) had an anionic charge. SGL-1′ was purified by using TLC developed with an acidic solvent system until a single spot was obtained. Both SGL-1 and SGL-1′ were reactive with anthrone reagent to yield a brownish-purple color, but they did not react with Dittmer's and ninhydrin reagents. Similarly, two spots (SGL-1 and SGL-1′) were found ubiquitously in S. mali EY 4206T, S. terrae EY 4207T, and S. macrogoltabidus EY 4304T and were purified. They were analogous to those of S. yanoikuyae EY 4208T, although the spot of SGL-1′ was not seen in S. paucimobilis EY 2395T.
FIG. 1.
Thin-layer chromatograms of SGL-1 and SGL-1′. Shown are solvent systems chloroform-methanol-acetic acid-water (100:20:12:5, by volume) (A) and chloroform-methanol-water (65:25:4, by volume) (B). 2395, S. paucimobilis EY 2395T; 4208, S. yanoikuyae EY 4208T; 4206, S. mali EY 4206T; 4207, S. terrae EY 4207T; 4304, S. macrogoltabidus EY 4304T. Lipids used were crude lipids (a), alkali-stable lipids (b), SGL-1 (c), and SGL-1′ (d).
Molecular weight of SGL-1′.
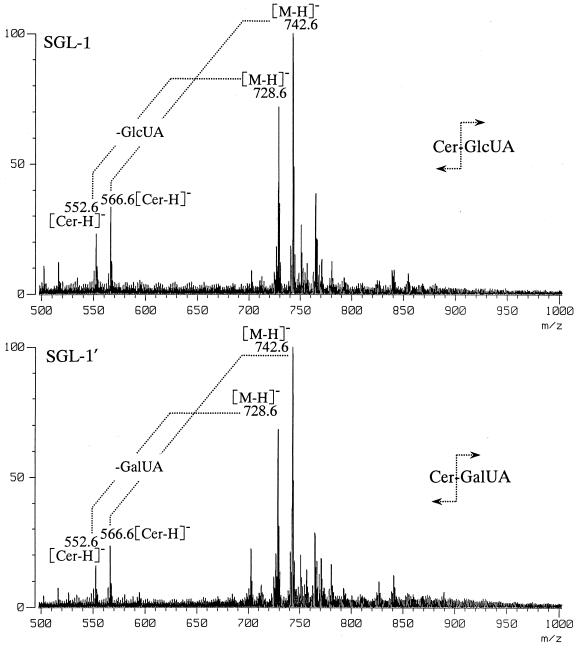
Molecular weight was determined by fast atom bombardment-mass spectrometry (FAB/MS) with triethanolamine as a matrix. FAB/MS analysis of SGL-1 and SGL-1′ showed the similar result of quasimolecular ions at m/z 742.6 and 728.6, respectively, due to [M-H]− as the major ions and fragment ions at m/z 566.6 and 552.6, respectively, due to ceramide moiety corresponding to two molecular species (Fig. 2). The molecular weight and fragmentation of SGL-1′ were essentially identical to those of SGL-1.
FIG. 2.
Negative FAB/MS spectra of SGL-1 and SGL-1′ from S. yanoikuyae EY 4208T. The major molecular ions were detected at m/z 742.6 and 728.6, and fragment ions of ceramide cleaved of carbohydrate moiety were detected at m/z 566.6 and 552.6.
Composition of fatty acids and long-chain bases in ceramide.
SGL-1′ was hydrolyzed with saturated Ba(OH)2 solution (100°C, 16 h). Long-chain bases were extracted with diethyl ether. After acidification of the residues, fatty acids were extracted with n-hexane. Trimethylsilyl (TMS) derivatives of long-chain bases and fatty acid methyl esters were identified by gas chromatography (GC) and GC-mass spectrometry (GC-MS). C20-sphingosine and C21-monocyclopropanoyl dihydrosphingosine were identified as the long-chain bases from fragment ions (C2-C3 cleavage of TMS derivative, m/z 132; [M-132]+, m/z 339; [M-103]+, m/z 368; [M-15]+, m/z 456 for C20-sphingosine; and C2-C3 cleavage of TMS derivative, m/z 132; [M-132]+, m/z 353; [M-103]+, m/z 382; [M-15]+, m/z 470 for C21-monocyclopropanoyl dihydrosphingosine) by GC-MS analysis (20) and TLC (Rf value and ninhydrin-positive spot). On the other hand, 2-hydroxymyristic acid was identified as the major fatty acid, based on the quasimolecular ion ([M]+, m/z 258) and characteristic fragment ions of 2-hydroxy fatty acid methyl ester ([M-59]+, m/z 199 and m/z 90 and 103).
Identification of carbohydrate moiety and linkage analysis.
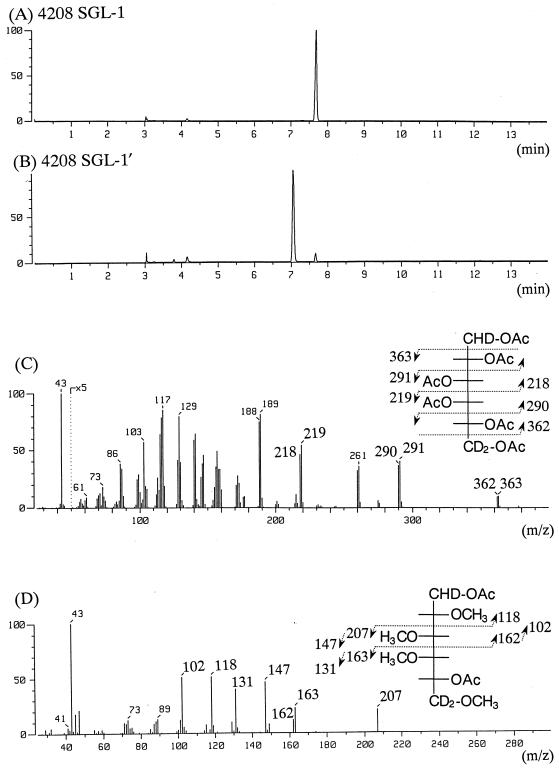
SGL-1′ was first reduced with LiAlD4 (70°C, 4 h) (18). After that, the alditol acetate derivative was obtained by hydrolysis with trifluoroacetic acid (120°C, 1 h), reduction with NaBD4 (room temperature, 2 h), and acetylation with acetic anhydride-pyridine (1:1, by volume). For partially methylated alditol acetate analysis, SGL-1′ was treated with dimethyl iodide after reduction with LiAlD4 (3). Alditol acetate and partially methylated alditol acetate derivatives were analyzed and characterized by GC and GC-MS (1, 2). In GC analysis, the alditol acetate derivative of SGL-1′ exhibited the same retention time as alditol acetate of galacturonic acid monohydrate (Sigma Chemical Co., St. Louis, Mo.) (the reference standard) and a different retention time than that of the alditol acetate derivative of SGL-1, which contained glucuronic acid (Fig. 3A and B). The mass spectrum showed characteristic fragment ions at m/z 218, 219, 290, 291, 362, and 363 (Fig. 3C). These fragment ions showed a signal of two mass units higher than those of the alditol acetate derivative of d-galactose, due to reduction with LiAlD4 at the C-6 position. These results suggest the presence of galacturonic acid as a carbohydrate moiety. Moreover, the GC-MS spectrum of the partially methylated alditol acetate derivative showed characteristic fragment ions of 2,3,4,6-tetra-O-methyl-1,5-di-O-acetyl galactitol at m/z 207, 147, 118, 163, 131, 162, and 102 (Fig. 3D). This result implies that the linkage of ceramide is at position C-1 of galacturonic acid. On the other hand, the coupling constant (J1.2) of galacturonic acid of SGL-1′ was 6.3 Hz (chemical shift, d = 5.693 ppm) from the nuclear magnetic resonance (NMR) analysis (Table 1). Heteronuclear coupling constants (1JH.C) were 162.1 Hz for SGL-1′. These data suggest that galacturonic acid of SGL-1′ is linked β-glycosidically to the long-chain base of lipid.
FIG. 3.
Total ion gas chromatograms and mass spectra of alditol acetate and partially methylated alditol acetate derivatives of SGL-1′ from S. yanoikuyae EY 4208T. Alditol acetate derivatives of SGL-1 (A), SGL-1′ (B and C), and a partially methylated alditol acetate derivative of SGL-1′ (D) are shown. The partially methylated alditol acetate derivative of SGL-1′ showed the fragment pattern of 2,3,4,6-tetra-O-methyl-1,5-di-O-acetyl galactitol. The fused silica capillary column SP-2380 was used, and the GC oven temperature was programmed to increase linearly at 5°C/min from 180 to 250°C.
TABLE 1.
NMR analysis of anomeric regions of SGL-1 and SGL-1′ derived from S. yanoikuyae EY 4208T
| Source of anomeric regions | Results of NMR analysis
|
|||
|---|---|---|---|---|
|
1H
|
13C
|
|||
| Chemical shift (ppm) | Coupling constant (J1.2 [Hz]) | Chemical shift (ppm) | Coupling constant (JC.H. [Hz]) | |
| SGL-1 | 4.853 | 1.0 | 98.975 | 172.2 |
| SGL-1′ | 5.693 | 6.3 | 90.443 | 162.1 |
Based on above findings, the difference between SGL-1 and SGL-1′ may be only in their carbohydrate moiety. Taken together, the molar ratio of galacturonic acid-fatty acid-long-chain base of SGL-1′ is 1:1:1 by the results of FAB/MS and component and quantitative analyses. The alkali-stable sphingoglycolipid SGL-1′ obtained from S. yanoikuyae appears to be most likely ceramide galacturonic acid (2-N-2′-hydroxymyristoyl C20-sphingosine-1-galacturonic acid and 2-N-2′-hydroxymyristoyl C21-monocyclopropanoyl dihydrosphingosine-1-galacturonic acid). In gram-negative bacteria, the presence of diacylglycerol-type glycolipids containing hexuronic acid and sphingoglycolipids containing glucuronic acid has been demonstrated (7, 10, 18), although the existence of sphingoglycolipid containing galacturonic acid has not yet been reported for any prokaryotic or eukaryotic cells. To our knowledge, this is the first study to demonstrate this type of acidic sphingoglycolipid. The relationship between SGL-1′ and the biologic activity of acidic sphingoglycolipid derived from bacteria and mammalian sources (9, 16, 17) is particularly interesting and will be discussed taxonomically and biochemically in the near future.
Acknowledgments
This work was supported in part by grants from Research on Environmental Health to E.Y. and Research on Emerging and Re-emerging Infectious Diseases (Ministry of Health and Welfare, Japan), and The United States-Japan Cooperative Medical Science Program against Tuberculosis and Leprosy.
REFERENCES
- 1.Bhat U R, Forsberg L S, Carlson R W. Structure of lipid A component of Rhizobium leguminosarum bv. phaseoli lipopolysaccharide. Unique nonphosphorylated lipid A containing 2-amino-2-deoxygluconate, galacturonate, and glucosamine. J Biol Chem. 1994;269:14402–14410. [PubMed] [Google Scholar]
- 2.Björndal H, Hellerqvist C G, Lindberg B, Svensson S. Gas-liquid chromatography and mass spectrometry in methylation analysis of polysaccharides. Angew Chem Int Ed Engl. 1970;9:610–619. [Google Scholar]
- 3.Ciucanu I, Kerek F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res. 1984;131:209–217. [Google Scholar]
- 4.Folch J, Lees M, Sloane Stanley G H. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1959;226:497–509. [PubMed] [Google Scholar]
- 5.Holmes B, Owen R J, Evans A, Malnick H, Willcox W R. Pseudomonas paucimobilis, a new species isolated from human clinical specimens, the hospital environment, and other sources. Int J Syst Bacteriol. 1977;27:133–146. [Google Scholar]
- 6.Jensen M T, Knudsen J, Olson J M. A novel aminoglycosphingolipid found in Chlorobium limicola f. thiosulfatophilum 6230. Arch Microbiol. 1991;156:248–254. [Google Scholar]
- 7.Kawahara K, Seydel U, Matsuura M, Danbara H, Rietschel E T, Zähringer U. Chemical structure of glycosphingolipids isolated from Sphingomonas paucimobilis. FEBS Lett. 1991;292:107–110. doi: 10.1016/0014-5793(91)80845-t. [DOI] [PubMed] [Google Scholar]
- 8.Miyagawa E, Azuma R, Suto T, Yano I. Occurrence of free ceramides in Bacteroides fragilis NCTC 9343. J Biochem. 1979;86:311–320. doi: 10.1093/oxfordjournals.jbchem.a132528. [DOI] [PubMed] [Google Scholar]
- 9.Miyazaki Y, Oka S, Yamaguchi S, Mizuno S, Yano I. Stimulation of phagocytosis and phagosome-lysosome fusion by glycosphingolipids from Sphingomonas paucimobilis. J Biochem. 1995;118:271–277. doi: 10.1093/oxfordjournals.jbchem.a124902. [DOI] [PubMed] [Google Scholar]
- 10.Tahara Y, Kawazu M. Isolation of glucuronic acid-containing glycosphingolipid from Zymomonas mobilis. Biosci Biotechnol Biochem. 1994;58:586–587. [Google Scholar]
- 11.Takeuchi M, Kawai F, Shimada Y, Yokota A. Taxonomic study of polyethylene glycol-utilizing bacteria: emended description of the genus Sphingomonas and new description of Sphingomonas macrogoltabidus sp. nov., Sphingomonas sanguis sp. nov. and Sphingomonas terrae sp. nov. Syst Appl Microbiol. 1993;16:227–238. [Google Scholar]
- 12.Takeuchi M, Sakane T, Yanagi M, Yamasato K, Hamana K, Yokota A. Taxonomic study of bacteria isolated from plants: proposal of Sphingomonas rosa sp. nov., Sphingomonas pruni sp. nov., Sphingomonas asaccharolytica sp. nov., and Sphingomonas mali sp. nov. Int J Syst Bacteriol. 1995;45:334–341. doi: 10.1099/00207713-45-2-334. [DOI] [PubMed] [Google Scholar]
- 13.Yabuuchi E, Kosako Y, Naka T, Suzuki S, Yano I. Proposal of Sphingomonas suberifaciens (van Bruggen, Jochimsen and Brown 1990) comb. nov., Sphingomonas natatoria (Sly 1985) comb. nov., Sphingomonas ursincola (Yurkov et al. 1997) comb. nov., and emendation of the genus Sphingomonas. Microbiol Immunol. 1999;43:339–349. doi: 10.1111/j.1348-0421.1999.tb02414.x. [DOI] [PubMed] [Google Scholar]
- 14.Yabuuchi E, Tanimura E, Ohyama A, Yano I, Yamamoto A. Flavobacterium devorans ATCC 10829: a strain of Pseudomonas paucimobilis. J Gen Appl Microbiol. 1979;25:95–107. [Google Scholar]
- 15.Yabuuchi E, Yano I, Oyaizu H, Hashimoto Y, Ezaki T, Yamamoto H. Proposals of Sphingomonas paucimobilis gen. nov. and comb. nov., Sphingomonas parapaucimobilis sp. nov., Sphingomonas yanoikuyae sp. nov., Sphingomonas adhaesiva sp. nov., Sphingomonas capsulata comb. nov., and two genospecies of the genus Sphingomonas. Microbiol Immunol. 1990;34:99–119. doi: 10.1111/j.1348-0421.1990.tb00996.x. [DOI] [PubMed] [Google Scholar]
- 16.Yamaguchi S, Miyazaki Y, Oka S, Yano I. Stimulation of phagocytosis and phagosome-lysosome (P-L) fusion of human polymorphonuclear leukocytes by sulfatide (galactosylceramide-3-sulfate) FEMS Immunol Med Microbiol. 1996;13:107–111. doi: 10.1016/0928-8244(95)00091-7. [DOI] [PubMed] [Google Scholar]
- 17.Yamaguchi S, Miyazaki Y, Oka S, Yano I. Stimulatory effect of gangliosides on phagocytosis, phagosome-lysosome fusion, and intracellular signal transduction system by human polymorphonuclear leukocytes. Glycoconj J. 1997;14:707–714. doi: 10.1023/a:1018517400380. [DOI] [PubMed] [Google Scholar]
- 18.Yamamoto A, Yano I, Masui M, Yabuuchi E. Isolation of a novel sphingoglycolipid containing glucuronic acid and 2-hydroxy fatty acid from Flavobacterium devorans ATCC 10829. J Biochem. 1978;83:1213–1216. doi: 10.1093/oxfordjournals.jbchem.a132015. [DOI] [PubMed] [Google Scholar]
- 19.Yano I, Imaizumi S, Tomiyasu I, Yabuuchi E. Separation and analysis of free ceramide containing 2-hydroxy fatty acids in Sphingobacterium species. FEMS Microbiol Lett. 1983;20:449–453. [Google Scholar]
- 20.Yano I, Tomiyasu I, Yabuuchi E. Long chain base composition of strains of three species of Sphingobacterium gen. nov. FEMS Microbiol Lett. 1982;15:303–307. [Google Scholar]