Abstract
The reaction of sulfamethoxazolehydrazonoyl chloride with thiosemicarbazones, bis-thiosemicarbazones, or 4-amino-3-mercapto-1,2,4-triazole in dioxane in the presence of triethylamine as a basic catalyst at reflux resulted in the regioselective synthesis of thiazoles and bis-thiazoles linked to azo-sulfamethoxazole as novel hybrid molecules. The structures of the new compounds were confirmed using a range of spectra. Each compound's antibacterial properties were evaluated using the agar well-diffusion technique, and most of them demonstrated significant potency. In silico investigations revealed that the described compounds had strong interactions with the binding sites of MurE ligase, tyrosyl-tRNA synthetase, and dihydropteroate synthase, demonstrating inhibitory activity.
Keywords: Sulfamethoxazole, Hydrazonoyl chloride, Thiazoles and bis-thiazoles, Docking, Antibacterial activity
Highlights
-
•
A range of new bis-thiazoles were synthesized and characterized using spectroscopic analyses.
-
•
All synthesized compounds were tested for antimicrobial activity.
-
•
Compounds 8b, 11a, and 16c showed the most promising activities among the three series.
-
•
The results were compared to that of the in silico molecular docking study.
1. Introduction
Infections due to bacteria are growing more frequent over the world, and they have the potential to have a significant impact on health and become the primary cause of disease [[1], [2], [3]]. Public health is severely threatened globally by the emergence of new microbial strains that are resistant to contemporary antibiotics [4,5]. One of the biggest shortcomings in the management of bacterial infections is that the antimicrobial treatments currently being developed are inadequate to address the growing threat of antibiotic resistance, according to the annual pipeline report by the World Health Organization. Several approaches have been proposed to address this issue, including the development and introduction of novel medications capable of eliminating multidrug resistance. Furthermore, decreasing the propagation of resistant infections and reducing the speed of resistance growth are key aims in this respect [6].
Various heterocyclic rings have piqued the interest of researchers as antimicrobial agents with novel modes of action in recent decades [7,8]. Sulfamethoxazole was one of the most used antimicrobial compounds, with good activity against a wide range of bacteria. It has been reported that it inhibits folic acid production in bacteria, which is necessary for nucleic acid production [[9], [10], [11]]. Nonetheless, the emergence of widespread resistance has severely compromised their once-critical role in managing bacterial infections [12]. Furthermore, thiazoles and their derivatives have been identified as important scaffolds in medicinal chemistry among the most diverse heterocyclic compounds. They showed significant antibacterial activity against a wide range of bacteria and pathogens, as well as a variety of biological activities [[13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25]]. Many powerful drugs contain a thiazole ring, including sulfathiazole (an antimicrobial) and abafungin (an antifungal drug). Moreover, compounds containing 1,2,4-triazole [[26], [27], [28], [29], [30], [31], [32], [33], [34], [35]] and/or 1,3,4-thiadiazines [32,36,37] are well-known for their broad biological activities and have attracted attention primarily as potent antibacterial agents. In addition, over the past few decades, the idea of molecular hybridization has generated a great deal of attention in the realm of drug creation. A new and effective synthetic technique for combining two or more different entities into a single molecule with unique biological features is the hybrid approach. The resulting scaffolds may be capable of overcoming drug resistance while also increasing activity and binding affinity [[38], [39], [40], [41]]. Based on the aforementioned hypotheses and our continued endeavor towards the synthesis of heterocycles [[42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55], [56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72]] we report herein the synthesis of some thiazoles, bis(thiazoles), and [1,2,4]triazolo[3,4-b] [1,3,4]thiadiazines linked to hydrazinyl-N-(5-methylisoxazol-3-yl)benzenesulfonamide moiety as novel hybrid molecules and evaluation their bioactivities against different bacterial strains and fungal strains; in an attempt to conquer sulfonamide resistance and find novel therapeutic options. Furthermore, the potential binding interactions of the novel compounds with the active sites of different target enzymes were investigated using molecular docking simulations.
2. Results and discussion
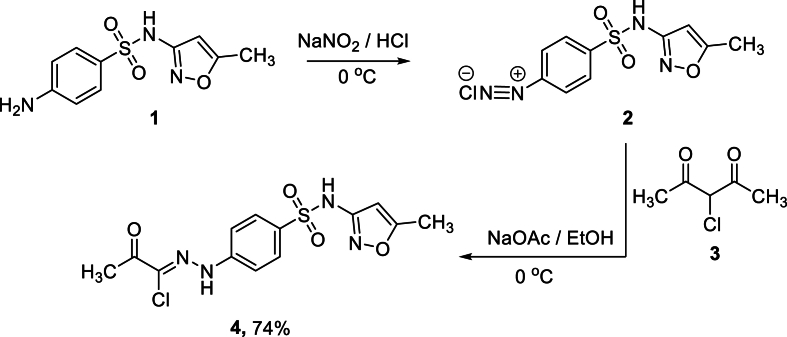
The N-(4-(N-(5-methylisoxazol-3-yl)sulfamoyl)phenyl)-2-oxopropanehydrazonoyl chloride 4 was chosen as an interesting synthon for a variety of valuable bioactive heterocyclic compounds.
In the first step, sulfamethoxazole diazonium chloride 2 is prepared from the corresponding sulfamethoxazole 1 and the appropriate quantities of both HCl and sodium nitrite. In the second step, the coupling reaction of 2 with 3-chloropentane-2,4-dione 3 in a basic KOH solution afforded the corresponding sulfamethoxazolehydrazonoyl chloride 4 (see Scheme 1). The constitution of compound 4 was confirmed based on the spectral analysis. Thus, the 1H NMR spectrum revealed three characteristic singlets at 2.29, 2.51, and 6.12 ppm for the two methyl groups and isoxazole-H, respectively. It also indicated the presence of characteristic two broad exchangeable signals at 11.01 and 11.28 for the two NH groups. The aromatic multiplet appeared at its expected position at 7.8–7.83 ppm.
Scheme 1.
Synthesis of N-(4-(N-(5-methylisoxazol-3-yl)sulfamoyl)phenyl)-2-oxopropanehydrazonoyl chloride 4.
The reactivity of the novel sulfamethoxazolehydrazonoyl chloride towards thiosemicarbazone was then investigated. High yields were obtained when the reaction was conducted at reflux in dioxane with triethylamine acting as a basic catalyst. The reaction was carried out in several solvents, including EtOH, CH3CN, H2O, dioxane, and DMF. While H2O may be used as a green solvent to carry out the reaction, other substrates' intrinsic insolubility has limited their use. The reactions continue in the absence of a solvent, but regretfully with extremely low yields. The use of chitosan in our reactions makes the isolation and purification of some derivatives time-consuming, even though it is an inexpensive, non-toxic base, eco-friendly, and highly reactive catalyst for building organic frameworks. Even after a considerable period, no residues of the products were found at room temperature, despite the reactions producing excellent yields of the products at the solvent's refluxing temperature. The findings demonstrated that, in terms of reaction yields, MW heating did not significantly outperform conventional heating.
Thus, the reaction of sulfamethoxazolehydrazonoyl chloride 4 with 1-(aryl)ethanone thiosemicarbazone 5a, 5b, and 1-(2-thienyl)ethanone thiosemicarbazone 5c in dioxane at reflux in the presence of few drops of TEA afforded (hydrazineyl)thiazol-5-yl)diazenyl)-N-(5-methylisoxazol-3-yl)benzenesulfonamide 8a-c in 70–80 % yields. The reaction occurs via an initial S-alkylation reaction of 5 with 4 to yield 6. Removal of water affords intermediate 7 that could tautomerize into the final isolable product 8 (Scheme 2).
Scheme 2.
Synthesis of (hydrazineyl)thiazol-5-yl)diazenyl)-N-(5-methylisoxazol-3-yl)benzenesulfonamide 8a-c.
Based on spectral data, the structures of compounds 8a-c were supported. Thus, the 1H NMR spectrum of 8a indicated four characteristic singlets at 2.29, 2.56, 2.61, and 6.11 ppm for the three methyl groups and isoxazole-4-H, respectively. It also indicated the presence of a characteristic broad exchangeable signal at 11.19 for the NH group. The aromatic multiplet appeared at their expected position at 7.45–8.34 ppm.
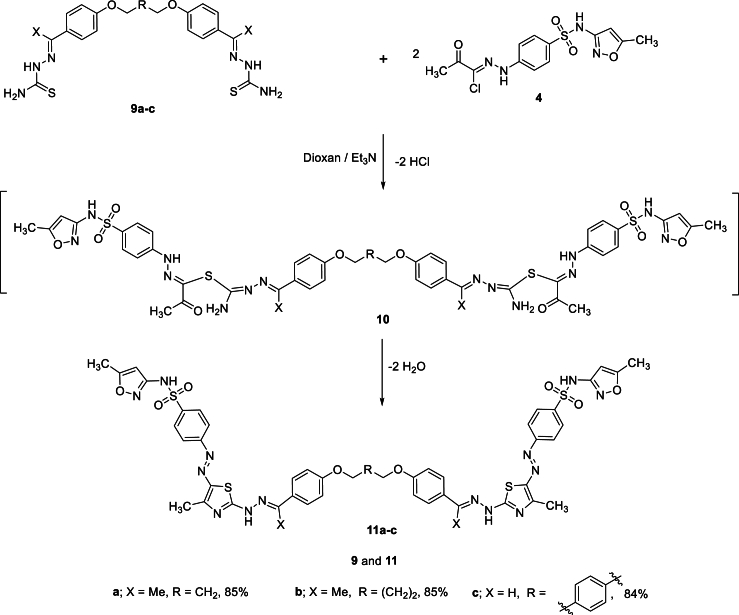
Stimulated by these results and in a trial to expand the scope of this reaction, we also investigated the cyclo-condensation reactions of bis-thiosemicarbazones with the sulfamethoxazolehydrazonoyl chloride (Scheme 3). Thus, a series of bis-(hydrazineyl)thiazol-5-yl)diazenyl)-N-(5-methylisoxazol-3-yl)benzenesulfonamides that are linked to propane 11a, butane 11b, or benzene 11c core via phenoxymethyl linkage were prepared in good yields via the reaction of the corresponding bis-thiosemicarbazones 9a-c with sulfamethoxazolehydrazonoyl chloride 4 in ethanolic – triethylamine solution. The reaction occurs via an initial bis-S-alkylation reaction of 9 with 4 to yield intermediate 10 that loses two water molecules to give 11 (Scheme 2).
Scheme 3.
Synthesis of bis-(hydrazineyl)thiazol-5-yl)diazenyl)-N-(5-methylisoxazol-3-yl)benzenesulfonamides 11.
It is worth noting that bis-thiosemicarbazones 9a-c were prepared via the condensation of acetophenones 12a and 12b or bis-aldehydes 12c with thiosemicarbazide 13 (Scheme 4) [[73], [74], [75]].
Scheme 4.
Synthesis of bis-thiosemicarbazones 9a-c.
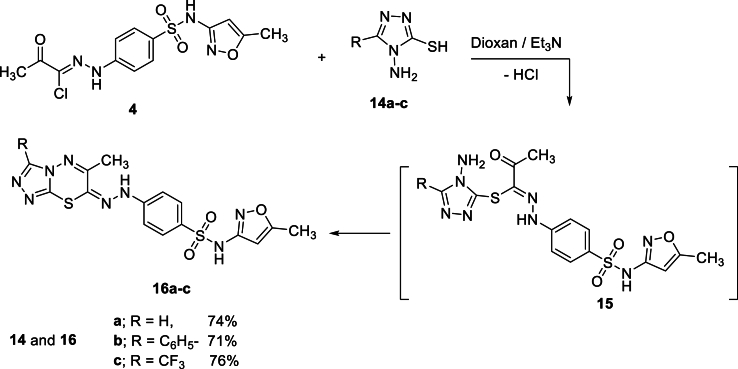
Furthermore, the reaction of sulfamethoxazolehydrazonoyl chloride 4 with 4-amino-3-mercapto-1,2,4-triazole derivatives 14a-c in ethanol in the presence of few drops of triethylamine as a catalyst at reflux produced the novel [1,2,4]triazolo[3,4-b][1,3,4]thiadiazin-7-ylidene)hydrazineyl)-N-(5-methylisoxazol-3-yl)benzenesulfonamide 16a-c (Scheme 5). The reaction pathway may involve S-alkylation to yield S-alkyl-aminotriazole intermediates 15 that undergo intramolecular cyclocondensation to afford the target products 16a-c in good yields (Scheme 5).
Scheme 5.
Synthesis of [1,2,4]triazolo[3,4-b] [1,3,4]thiadiazin-7-ylidene)hydrazineyl)-N-(5-methylisoxazol-3-yl)benzenesulfonamide 16a-c.
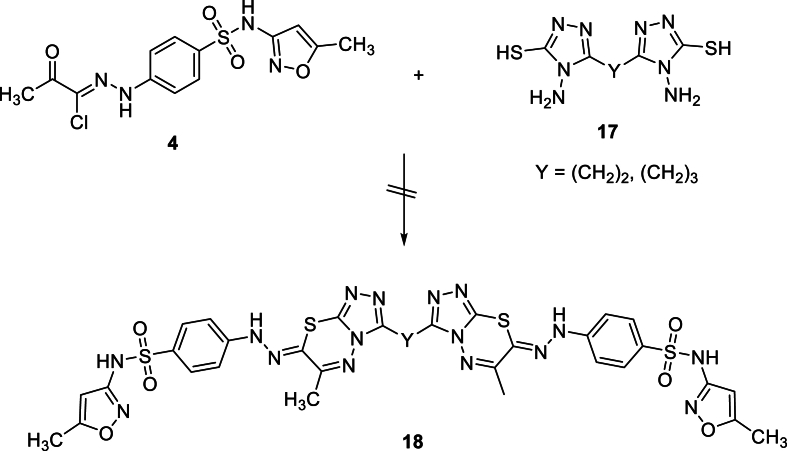
On the other hand, repeated attempts to prepare bis-thiadiazoles 18, by the reaction of 1 mol of bis(4-amino-4H-1,2,4-triazole-3-thiols) 17 with 2 mol of sulfamethoxazolehydrazonoyl chloride 4 were unsuccessful. Instead, the reaction yielded a combination of non-isolable compounds that could not be purified at this time (see Scheme 6).
Scheme 6.
Attempted synthesis of bis-thiadiazoles 18.
2.1. In vitro antimicrobial activity
In this study, the synthesized compounds 8a-c, 11a–c, and 16a-c have been evaluated in vitro for their inhibitory action on the growth of five bacterial strains “(Gram‐positive bacterial species (Staphylococcus aureus and Bacillus subtilis) and Gram-negative bacterial species (Escherichia coli, Proteus vulgaris, and Pseudomonas aeruginosa)) in addition to two fungal strains (one filamentous fungus (Aspergillus fumigatus) and one yeast species (Candida albicans))”. Table 1 lists the antibacterial activity results, that were acquired through the measurement of the inhibition zone (mm) with the classical agar well diffusion method [76]. Gentamycin and ketoconazole have been utilized as the standard antibacterial and antifungal drugs, respectively, with remarkable inhibition zone diameters against the examined microorganisms. Overall, the investigated derivatives exhibited remarkable antibacterial activities with variable potencies. The antibacterial properties of the (hydrazineyl)thiazol-5-yl)diazenyl)-N-(5-methylisoxazol-3-yl)benzenesulfonamide derivatives 8a-c (ranged from 10 to 17 mm) was greater than their corresponding bis-(hydrazineyl)thiazol-5-yl)diazenyl)-N-(5-methylisoxazol-3-yl)benzenesulfonamide derivatives 11a-c (from no activity to 14 mm) and [1,2,4]triazolo[3,4-b][1,3,4]thiadiazin-7-ylidene)hydrazineyl)-N-(5-methylisoxazol-3 yl)benzenesulfonamide derivatives 16a-c (from no activity to 15 mm). In addition, the synthesized derivatives were non-toxic against the fungus C. albicans.
Table 1.
Antibacterial and antifungal activities of the newly synthesized compounds.
| Tested microorganisms |
Sample No. |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 8a | 8b | 8c | 11a | 11b | 11c | 16a | 16b | 16c | Control | |
| Fungi | Ketoconazole | |||||||||
| Aspergillus fumigatus | NA | NA | NA | NA | NA | NA | NA | NA | NA | 17 |
| Candida albicans | NA | NA | NA | NA | NA | NA | NA | NA | NA | 20 |
| Gram-positive Bacteria | Gentamycin | |||||||||
| Staphylococcus aureus | 14 | 16 | 15 | 10 | 9 | NA | 13 | NA | 15 | 24 |
| Bacillus subtilis | 12 | 17 | 16 | NA | NA | NA | NA | 8 | 10 | 26 |
| Gram-negative Bacteria | Gentamycin | |||||||||
| Escherichia coli | 10 | 15 | 14 | 11 | 10 | NA | 10 | NA | 12 | 30 |
| Proteus vulgaris | 13 | 17 | 15 | 14 | 8 | NA | 15 | NA | 13 | 25 |
| Pseudomonas aeruginosa | 12 | 13 | 13 | 10 | 11 | NA | 11 | 10 | 11 | 32 |
“Mean zone of inhibition in mm beyond well diameter (6 mm) produced on a range of microorganisms”.
“The test was done using the diffusion agar technique, well diameter: 6.0 mm (100 μL was tested).
Positive control for fungi (Ketoconazole, 100 μg/mL) and Positive control for bacteria (Gentamycin, 4 μg/mL)”.
NA: No activity. The sample was tested at 10 mg/ml concentration.
2.2. Structure-activity relationship
The structure-activity relationship (SAR) investigations could serve as a valuable tool for the subsequent rational design of the studied compounds and contribute to assessing numerous areas of drug discovery, from primary screening to lead optimization. Sulphonamide, sulfamethazine, and sulfadiazine drugs are a significant family of antibiotics with a broad spectrum of activity that is particularly potent against several bacterial and fungal infections, such as Staphylococcus aureus, Escherichia coli, Pneumocystis carinii, Klebsiella, Salmonella, and Enterobacter species [77]. As illustrated in the current work (Scheme 7 and Table 1), sulfonamide derivatives 8a-c have been determined to be the most potent of the three investigated series of synthesized derivatives. It was noted that compound 8b bearing 4-methoxyphenyl group, with electron-donating methoxy group attached to phenyl ring, exhibited higher antibacterial properties than thienyl-containing derivative 8c and 4-nitrophenyl group bearing derivative 8a, with electron-withdrawing nitro group attached to phenyl ring, with the inhibition zone in the range of 17 to 13 mm, 16 to 13 mm, and 14 to 10 mm, respectively, against the different tested bacterial strains. On the other side, investigating the bis-sulfonamide derivatives 11a-c antibacterial activities, compound 11a (inhibition zone: no activity-14 mm) with a propyl-linker exhibited good antimicrobial activity, whereas compound 11b (inhibition zone: no activity-11 mm) having a butyl-linker exhibited weaker activity. Replacing the aforementioned aliphatic linkers with the 1,4-dimethylphenyl linker in derivative 11c remarkably abolished the compound 11c activity. In the SAR analysis of the third sulfonamide derivatives, 16a-c, 16c with strong electron-withdrawing trifluoromethyl moiety revealed good antibacterial potency (inhibition zone: 10–15 mm), nevertheless, 16a with H-atom had a reduced activity (inhibition zone: no activity-15 mm). While 16b bearing phenyl moiety displayed almost no activity except for B. subtilis and P. aeruginosa with 8 and 10 mm inhibition zones, respectively.
Scheme 7.
Design strategy of compounds 8, 11, and 16.
2.3. Molecular docking study
Chemotherapeutic drugs such as antibiotics are used to either suppress or eradicate pathogens. According to Zessel et al., sulfonamides function as structural mimics and competitive antagonists of p-aminobenzoic acid (PABA) in the formation of folic acid, which is necessary for the bacteria to continue replicating DNA and survive (10.1016/j.chemosphere.2013.11.038). Thus, molecular docking has been utilized to give additional insight into the potential molecular mechanism of the synthesized compounds. Molecular docking has greatly contributed to the pursuit of potentially new compounds of therapeutic interest as an innovative approach for rationalizing, forecasting the affinities of compounds at a molecular basis, and assessing the suitable orientations and binding of compounds under study at pockets of receptor proteins [[78], [79], [80]]. The primary objective is to investigate how our novel sulfonamides will interact with different bacterial proteins, namely MurE ligase (PDB ID: 4C13), tyrosyl-tRNA synthetase (PDB ID: 1JIJ), and dihydropteroate synthase (PDB ID: 1AJ0), that are considered as crucial targets for finding broad-spectrum antibiotics [[81], [82], [83], [84], [85], [86], [87]].
Docking results were first validated by self-docking of the reference co-crystallized ligand (2-amino-6-hydroxymethyl-7,8-dihydro-3H-pteridin-4-one) with its corresponding receptor (PDB ID: 1AJ0). RMSD value was found to be less than 1 Å and a remarkable superimposition of the re-docked and co-crystallized ligand within the active site was exhibited. In addition, the docking protocol was successful and reproduced the majority of the key interactions between the ligand (re-docked (cyan) and co-crystalized (green)) and the amino acids of its corresponding protein active site, as illustrated in Fig. 1.
Fig. 1.
Superimposition of the re-docked (carbon atoms with cyan color) and the co-crystallized ligand (carbon atoms with green color) within the active site of its protein receptor (dihydropteroate synthase, PDB ID: 1AJ0). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
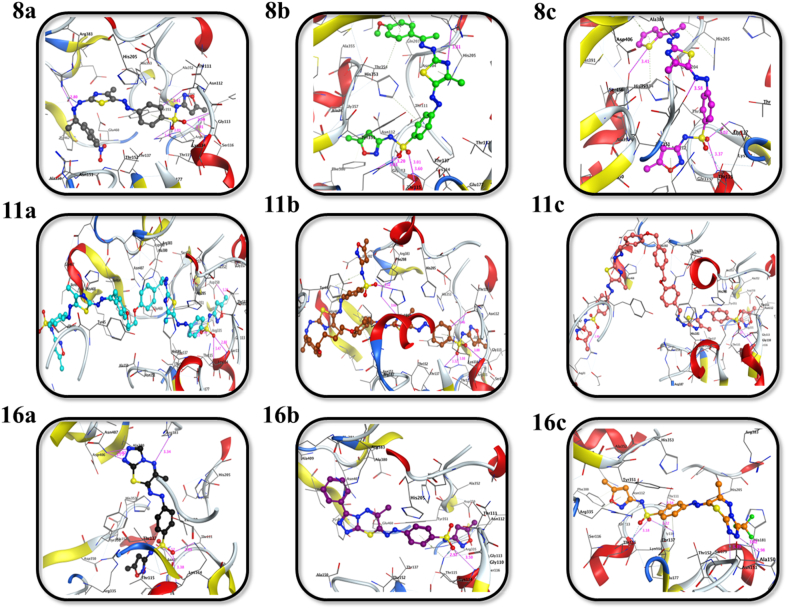
MurE ligase is involved in the peptidoglycan biosynthesis of the bacterial cell wall through the addition of m-DAP to UDP-MurNAc-L-Ala-D-Glu [81,82], and it is a highly attractive drug target due to it being unique in bacteria and absent in human cells [88,89]. Fig. 2, Fig. 3, and Table 2 revealed significant interaction profiles (binding affinities) of the sulfonamide derivatives (8a-c, 11a–c, and 16a-c) with the binding cavity of MurE ligase (Pdb ID: 4C13) with significant docking scores ranging from −8.07 to −12.12 kcal mol−1. Sulfonamides 8b, 11a, and 16c were found to be the most potent among the synthesized derivatives 8a-c, 11a–c, and 16a-c, as evidenced by their binding scores of −10.15, −12.12, and −8.31 kcal mol−1, respectively. Analogue 8b formed nine non-covalent interactions with Lys114, Thr115, NE and NH2 of Arg335, Arg383, His205, His353 Thr354 and Met379 residues. Seven non-covalent interactions were observed for 11a with Asp350, Lys114, Thr115, NE, CD, and NH2 of Arg335, Arg383, and His468 residues. Furthermore, 16c established nine non-covalent interactions with Thr111, N, and NZ of Lys114, Thr115, ND2, and CA of Asn151, Arg187, and Arg335 residues.
Fig. 2.
2D representations of the putative intermolecular interaction of the synthesized sulfonamide derivatives (8a-c, 11a-c, and 16a-c) against MurE ligase (PDB ID: 4C13) active site residues.
Fig. 3.
3D representations of the putative intermolecular interaction of the synthesized sulfonamide derivatives (8a-c, 11a-c, and 16a-c) against MurE ligase (PDB ID: 4C13) active site residues.
Table 2.
Molecular docking reports of the synthesized sulfonamide derivatives (8a-c, 11a-c, and 16a-c) with bacterial protein MurE ligase (PDB ID: 4C13).
| Protein (PDB ID) | Compd. No. | Docking score (kcal/mol) | Ligand | Receptor | Type of interaction | Distance | E (kcal/mol) |
|---|---|---|---|---|---|---|---|
| 4C13 | 8a | −9.46 | C 2 | O THR 111 | H-donor | 3.41 | −0.7 |
| O 12 | N GLY 113 | H-acceptor | 2.99 | −0.5 | |||
| O 12 | N LYS 114 | H-acceptor | 2.91 | −1.0 | |||
| O 13 | NE ARG 335 | H-acceptor | 3.01 | −3.9 | |||
| O 13 | NH2 ARG 335 | H-acceptor | 3.35 | −0.9 | |||
| N 31 | NH2 ARG 383 | H-acceptor | 2.80 | −1.6 | |||
| 6-ring | NZ LYS 114 | pi-cation | 4.04 | −0.9 | |||
| 8b | −10.15 | O 12 | N LYS 114 | H-acceptor | 3.01 | −0.7 | |
| O 12 | N THR 115 | H-acceptor | 3.60 | −0.5 | |||
| O 13 | NE ARG 335 | H-acceptor | 3.03 | −3.7 | |||
| O 13 | NH2 ARG 335 | H-acceptor | 3.20 | −2.0 | |||
| N 26 | NE ARG 383 | H-acceptor | 3.31 | −1.5 | |||
| 5-ring | CE1 HIS 205 | pi-H | 3.93 | −0.6 | |||
| 6-ring | CE1 HIS 353 | pi-H | 4.18 | −0.5 | |||
| 6-ring | CG2 THR 354 | pi-H | 4.53 | −0.5 | |||
| 6-ring | CE MET 379 | pi-H | 3.78 | −0.7 | |||
| 8c | −9.89 | S 40 | O HIS 353 | H-donor | 3.41 | −1.3 | |
| O 12 | N LYS 114 | H-acceptor | 3.37 | −1.0 | |||
| O 12 | NZ LYS 114 | H-acceptor | 3.02 | −5.3 | |||
| O 13 | N THR 115 | H-acceptor | 3.43 | −1.0 | |||
| N 23 | OH TYR 351 | H-acceptor | 3.58 | −0.5 | |||
| C 45 | 5-ring HIS 205 | H-pi | 4.68 | −0.5 | |||
| 5-ring | CD ARG 335 | pi-H | 4.36 | −0.8 | |||
| 5-ring | CG2 THR 354 | pi-H | 4.45 | −0.5 | |||
| 5-ring | CE MET 379 | pi-H | 3.74 | −0.9 | |||
| 5-ring | 5-ring HIS 353 | pi-pi | 3.98 | −0.0 | |||
| 11a | −12.12 | C 96 | OD2 ASP 350 | H-donor | 3.28 | −0.5 | |
| O 12 | N LYS 114 | H-acceptor | 2.88 | −0.7 | |||
| O 12 | N THR 115 | H-acceptor | 3.47 | −1.0 | |||
| O 13 | NE ARG 335 | H-acceptor | 3.20 | −2.5 | |||
| O 13 | NH2 ARG 335 | H-acceptor | 3.21 | −2.0 | |||
| N 26 | NE ARG 383 | H-acceptor | 3.65 | −1.5 | |||
| 5-ring | CD ARG 335 | pi-H | 4.34 | −1.0 | |||
| 5-ring | 5-ring HIS 468 | pi-pi | 3.94 | −0.0 | |||
| 11b | −11.98 | C 2 | O THR 111 | H-donor | 3.46 | −0.7 | |
| S 24 | OG1 THR 152 | H-donor | 4.06 | −0.7 | |||
| O 12 | NE ARG 335 | H-acceptor | 3.08 | −3.4 | |||
| O 12 | NH2 ARG 335 | H-acceptor | 3.32 | −1.3 | |||
| O 13 | N LYS 114 | H-acceptor | 3.00 | −0.9 | |||
| N 26 | NH2 ARG 383 | H-acceptor | 3.60 | −1.5 | |||
| O 89 | NE2 HIS 181 | H-acceptor | 3.12 | −0.8 | |||
| C 81 | 6-ring TYR 45 | H-pi | 4.04 | −1.0 | |||
| N 91 | 6-ring TYR 45 | H-pi | 4.37 | −1.1 | |||
| 6-ring | NZ LYS 114 | pi-cation | 4.02 | −0.9 | |||
| 6-ring | ND2 ASN 151 | pi-H | 3.73 | −0.5 | |||
| 11c | −11.53 | O 13 | NH1 ARG 31 | H-acceptor | 2.99 | −4.5 | |
| O 95 | N LYS 114 | H-acceptor | 3.36 | −0.9 | |||
| O 95 | NZ LYS 114 | H-acceptor | 3.12 | −3.8 | |||
| O 96 | CA ASN 112 | H-acceptor | 3.45 | −0.7 | |||
| 5-ring | CG2 VAL 47 | pi-H | 4.08 | −0.5 | |||
| 5-ring | CD ARG 335 | pi-H | 3.78 | −1.1 | |||
| 16a | −8.28 | O 12 | CA ASN 112 | H-acceptor | 3.52 | −0.6 | |
| O 13 | N LYS 114 | H-acceptor | 3.38 | −1.1 | |||
| O 13 | NZ LYS 114 | H-acceptor | 2.89 | −6.9 | |||
| N 28 | NE ARG 383 | H-acceptor | 3.34 | −1.3 | |||
| N 31 | ND2 ASN 407 | H-acceptor | 2.92 | −0.5 | |||
| N 32 | ND2 ASN 407 | H-acceptor | 2.75 | −0.7 | |||
| 16b | −8.07 | O 13 | N LYS 114 | H-acceptor | 3.50 | −0.5 | |
| O 13 | NZ LYS 114 | H-acceptor | 2.92 | −5.4 | |||
| 6-ring | NZ LYS 114 | pi-cation | 4.23 | −0.6 | |||
| 5-ring | CD ARG 335 | pi-H | 3.88 | −0.7 | |||
| 16c | −8.31 | C 2 | O THR 111 | H-donor | 3.54 | −0.6 | |
| O 12 | N LYS 114 | H-acceptor | 3.18 | −2.1 | |||
| O 12 | NZ LYS 114 | H-acceptor | 3.22 | −4.0 | |||
| O 13 | N THR 115 | H-acceptor | 3.25 | −2.0 | |||
| N 31 | ND2 ASN 151 | H-acceptor | 3.02 | −0.6 | |||
| N 32 | CA ASN 151 | H-acceptor | 3.39 | −0.9 | |||
| F 35 | NH1 ARG 187 | H-acceptor | 2.98 | −0.7 | |||
| 6-ring | NZ LYS 114 | pi-cation | 4.05 | −0.5 | |||
| 5-ring | CD ARG 335 | pi-H | 4.37 | −0.9 |
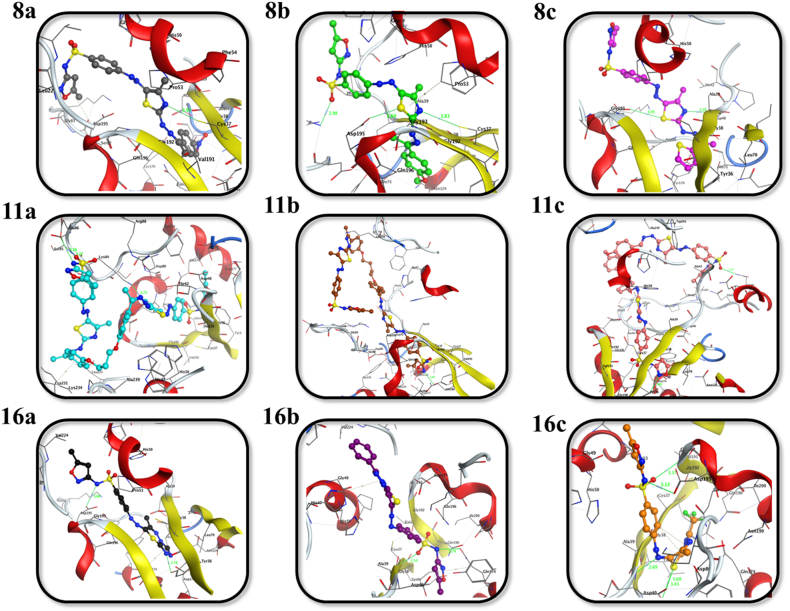
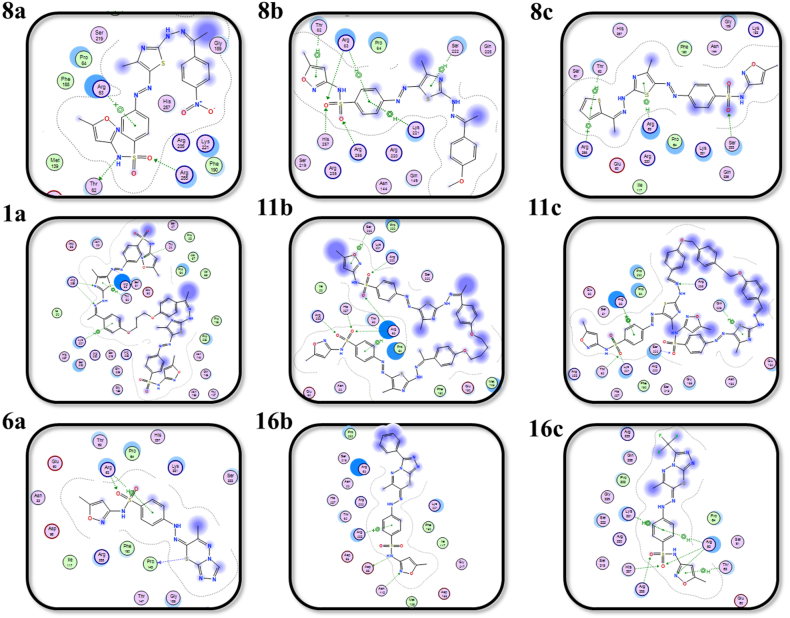
Furthermore, the bacterial enzyme tyrosyl-tRNA synthetase (TyrRS) contributes to the process of amino acids binding to their corresponding tRNAs that are required for the production of bacterial proteins [83,84]. TyrRS is selected as an emerging druggable target because of its essentiality for bacterial survival. As illustrated in Fig. 4, Fig. 5, and Table 3, all of the synthesized sulfonamides were efficiently docked into the TyrRS pocket, with binding energies in the range of −6.96 to −10.65 kcal mol−1. With binding scores of −8.72, −10.65, and −7.19 kcal mol−1, the investigation of the docking data revealed that 8b, 11a, and 16c had significant binding affinities among the synthesized derivatives. Notably, the sulfonamide compounds 8b, 11a, and 16c interacted with TyrRS pocket amino acid residues through five non-covalent interactions. Compound 8b associated with Asp195, Cys37, Lys84, Pro53 and His47 residues, 11a had its interaction with Glu86, Ser85, NZ and CA of Lys84, and Lys231 residues, whereas 16c bound to OD1, OD2 and N of Asp40, Asp195 and Gly193 residues.
Fig. 4.
2D representations of the putative intermolecular interaction of the synthesized sulfonamide derivatives (8a-c, 11a-c, and 16a-c) against tyrosyl-tRNA synthetase (PDB ID: 1JIJ) active site residues.
Fig. 5.
3D representations of the putative intermolecular interaction of the synthesized sulfonamide derivatives (8a-c, 11a-c, and 16a-c) against tyrosyl-tRNA synthetase (PDB ID: 1JIJ) active site residues.
Table 3.
Molecular docking reports of the synthesized sulfonamide derivatives (8a-c, 11a-c, and 16a-c) with bacterial protein tyrosyl-tRNA synthetase (PDB ID: 1JIJ).
| Protein (PDB ID) | Compound | Docking score (kcal/mol) | Ligand | Receptor | Type of interaction | Distance | E (kcal/mol) |
|---|---|---|---|---|---|---|---|
| 1JIJ | 8a | −8.15 | N 26 | SG CYS 37 | H-donor | 3.93 | −1.4 |
| 5-ring | CB ASP 195 | pi-H | 4.54 | −0.5 | |||
| 8b | −8.72 | S 24 | OD2 ASP 195 | H-donor | 3.64 | −1.0 | |
| N 26 | SG CYS 37 | H-donor | 3.83 | −0.9 | |||
| O 12 | NZ LYS 84 | H-acceptor | 2.98 | −6.9 | |||
| 5-ring | CG PRO 53 | pi-H | 4.33 | −0.9 | |||
| 5-ring | 5-ring HIS 47 | pi-pi | 3.91 | −0.0 | |||
| 8c | −8.30 | S 24 | OD2 ASP 80 | H-donor | 4.00 | −0.6 | |
| S 24 | OD1 ASP 195 | H-donor | 3.95 | −3.0 | |||
| N 26 | N ASP 40 | H-acceptor | 3.18 | −4.1 | |||
| 11a | −10.65 | N 14 | OE2 GLU 86 | H-donor | 3.16 | −1.1 | |
| O 13 | N SER 85 | H-acceptor | 3.22 | −1.7 | |||
| N 70 | NZ LYS 84 | H-acceptor | 2.75 | −8.2 | |||
| 6-ring | CA LYS 84 | pi-H | 4.46 | −0.7 | |||
| 6-ring | CB LYS 231 | pi-H | 3.95 | −1.0 | |||
| 11b | −10.44 | S 24 | OD1 ASP 195 | H-donor | 4.06 | −0.8 | |
| N 17 | OH TYR 36 | H-acceptor | 2.83 | −1.4 | |||
| 5-ring | CG GLN 174 | pi-H | 4.25 | −1.1 | |||
| 11c | −10.25 | O 13 | NE2 GLN 95 | H-acceptor | 3.27 | −1.7 | |
| N 100 | OH TYR 36 | H-acceptor | 2.82 | −1.6 | |||
| 6-ring | CE LYS 84 | pi-H | 4.64 | −0.7 | |||
| 5-ring | CG GLN 174 | pi-H | 4.25 | −1.1 | |||
| 16a | −6.96 | N 14 | OD2 ASP 195 | H-donor | 3.06 | −6.4 | |
| N 32 | OH TYR 36 | H-acceptor | 2.78 | −1.6 | |||
| 5-ring | CG GLN 174 | pi-H | 3.86 | −0.7 | |||
| 16b | −7.19 | N 14 | OE1 GLN 174 | H-donor | 2.74 | −0.5 | |
| O 13 | N GLY 38 | H-acceptor | 2.54 | −1.7 | |||
| 5-ring | CE1 HIS 47 | pi-H | 4.10 | −0.8 | |||
| 5-ring | CG GLN 174 | pi-H | 3.71 | −0.7 | |||
| 16c | −7.19 | N 14 | OD1 ASP 195 | H-donor | 3.13 | −4.2 | |
| S 25 | OD1 ASP 40 | H-donor | 3.69 | −0.5 | |||
| S 25 | OD2 ASP 40 | H-donor | 3.43 | −1.5 | |||
| O 12 | N GLY 193 | H-acceptor | 3.17 | −1.3 | |||
| N 24 | N ASP 40 | H-acceptor | 2.69 | −2.5 |
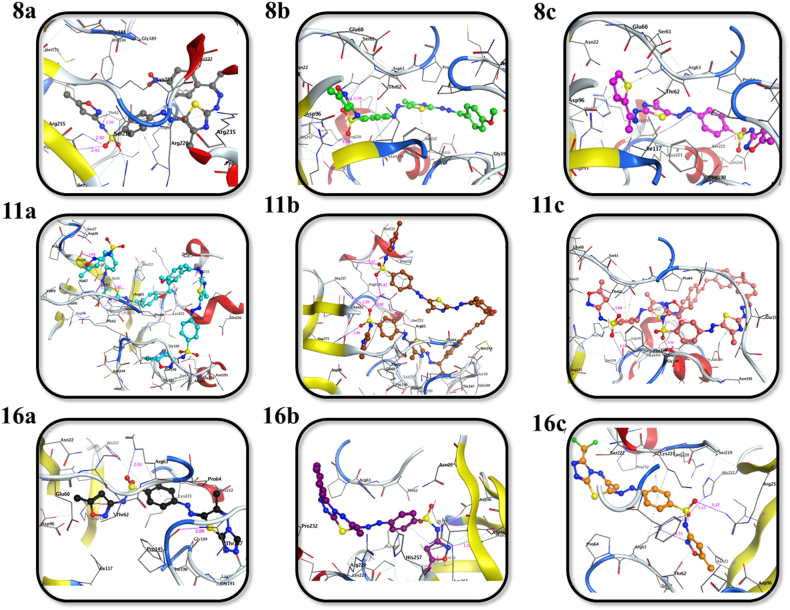
Eventually, dihydropteroate synthase (DHPS) has a prominent role in bacterial folate biosynthesis required for amino acids and nucleic acids production through the formation of dihydropteroate from para-aminobenzoic acid and dihydropterin pyrophosphate [[85], [86], [87]]. The most frequently utilized DHPS inhibitors are a family of synthetic compounds known as sulfonamides, which function as competitive inhibitors [10.1006/bbrc.1999.0695]. Sulfonamide derivatives (8a-c, 11a–c, and 16a-c) exhibited good binding scores of −6.45 to −10.75 kcal mol−1 against DHPS (PDB ID: 1AJ0) active site, as depicted in Fig. 6, Fig. 7 and Table 4. Compound 8b had good binding affinity among sulfonamides 8a-c with a binding score of −8.05 kcal mol−1 through nine non-covalent interactions with NE, NH2, and CG of Arg63, His257, Arg255, Thr62, CA, and CG of Lys221 and Ser222 residues. Compounds 11a and 16c displayed binding scores of −10.75 and −6.92 kcal mol−1 compared to their groups 11a-c and 16a-c. Compound 11a established seven interactions with OG and CA of Ser61, NH1, and NH2 of Arg255, Thr24, Thr62, and Lys221 residues, and compound 16c had eight interactions with NH2, NE and CG of Arg63, His257, Arg255, Thr62, and CA and CG of Lys221 residues.
Fig. 6.
2D representations of the putative intermolecular interaction of the synthesized sulfonamide derivatives (8a-c, 11a-c, and 16a-c) against dihydropteroate synthase (PDB ID: 1AJ0) active site residues.
Fig. 7.
3D representations of the putative intermolecular interaction of the synthesized sulfonamide derivatives (8a-c, 11a-c, and 16a-c) against dihydropteroate synthase (PDB ID: 1AJ0) active site residues.
Table 4.
Molecular docking reports of the synthesized sulfonamide derivatives (8a-c, 11a-c, and 16a-c) with bacterial protein dihydropteroate synthase (PDB ID: 1AJ0).
| Protein (PDB ID) | Compound | Docking score (kcal/mol) | Ligand | Receptor | Type of interaction | Distance | E (kcal/mol) |
|---|---|---|---|---|---|---|---|
| 1AJ0 | 8a | −7.80 | N 14 | OG1 THR 62 | H-donor | 2.94 | −1.2 |
| O 13 | NH1 ARG 255 | H-acceptor | 2.92 | −4.0 | |||
| O 13 | NH2 ARG 255 | H-acceptor | 2.93 | −3.9 | |||
| 6-ring | NE ARG 63 | pi-cation | 3.74 | −0.7 | |||
| 8b | −8.05 | O 12 | NE ARG 63 | H-acceptor | 3.39 | −0.7 | |
| O 12 | NH2 ARG 63 | H-acceptor | 3.08 | −3.6 | |||
| O 12 | NE2 HIS 257 | H-acceptor | 3.33 | −0.8 | |||
| O 13 | NH1 ARG 255 | H-acceptor | 3.06 | −3.0 | |||
| 5-ring | N THR 62 | pi-H | 4.12 | −0.6 | |||
| 6-ring | CG ARG 63 | pi-H | 4.39 | −0.7 | |||
| 6-ring | CA LYS 221 | pi-H | 4.79 | −0.5 | |||
| 6-ring | CG LYS 221 | pi-H | 3.93 | −0.7 | |||
| 5-ring | CB SER 222 | pi-H | 3.97 | −0.6 | |||
| 8c | −7.89 | O 13 | OG SER 222 | H-acceptor | 3.17 | −1.0 | |
| 5-ring | N THR 62 | pi-H | 4.06 | −1.2 | |||
| 5-ring | CG ARG 63 | pi-H | 4.06 | −1.0 | |||
| 5-ring | NH2 ARG 255 | pi-cation | 3.66 | −1.8 | |||
| 11a | −10.76 | S 68 | OG SER 61 | H-donor | 3.88 | −0.8 | |
| N 65 | NH1 ARG 255 | H-acceptor | 2.96 | −6.2 | |||
| N 70 | NH2 ARG 255 | H-acceptor | 3.19 | −0.7 | |||
| N 73 | CA SER 61 | H-acceptor | 3.22 | −1.0 | |||
| N 91 | OG1 THR 24 | H-acceptor | 3.01 | −1.7 | |||
| 5-ring | N THR 62 | pi-H | 4.10 | −0.5 | |||
| 6-ring | CG LYS 221 | pi-H | 3.66 | −0.5 | |||
| 11b | −10.55 | O 12 | NH1 ARG 255 | H-acceptor | 2.88 | −4.8 | |
| O 13 | NH2 ARG 63 | H-acceptor | 2.82 | −4.7 | |||
| O 13 | NE2 HIS 257 | H-acceptor | 2.99 | −0.8 | |||
| O 89 | NH2 ARG 63 | H-acceptor | 3.02 | −3.4 | |||
| O 90 | NH1 ARG 220 | H-acceptor | 3.07 | −1.6 | |||
| 6-ring | CG ARG 63 | pi-H | 3.77 | −0.8 | |||
| 5-ring | CB SER 233 | pi-H | 4.66 | −0.6 | |||
| 11c | −8.93 | O 12 | N SER 222 | H-acceptor | 3.46 | −0.9 | |
| N 74 | NH2 ARG 235 | H-acceptor | 3.04 | −4.5 | |||
| O 95 | OG1 THR 62 | H-acceptor | 2.66 | −1.7 | |||
| O 96 | NZ LYS 221 | H-acceptor | 3.12 | −1.2 | |||
| 6-ring | CG ARG 63 | pi-H | 4.37 | −0.6 | |||
| 6-ring | NE ARG 63 | pi-cation | 3.81 | −0.6 | |||
| 5-ring | NE2 GLN 226 | pi-H | 3.89 | −0.6 | |||
| 16a | −6.83 | S 25 | O PRO 145 | H-donor | 3.99 | −0.5 | |
| O 13 | NE ARG 63 | H-acceptor | 2.91 | −4.4 | |||
| 6-ring | CG ARG 63 | pi-H | 4.36 | −0.5 | |||
| 16b | −6.46 | N 14 | OD2 ASP 96 | H-donor | 2.89 | −10.3 | |
| N 17 | ND2 ASN 115 | H-acceptor | 3.22 | −3.0 | |||
| 6-ring | NH1 ARG 255 | pi-cation | 3.66 | −0.5 | |||
| 16c | −6.92 | O 12 | NH2 ARG 63 | H-acceptor | 3.15 | −3.0 | |
| O 12 | NE2 HIS 257 | H-acceptor | 3.25 | −0.6 | |||
| O 13 | NH1 ARG 255 | H-acceptor | 3.37 | −0.8 | |||
| N 17 | NE ARG 63 | H-acceptor | 3.51 | −0.6 | |||
| 5-ring | N THR 62 | pi-H | 4.13 | −1.4 | |||
| 6-ring | CG ARG 63 | pi-H | 4.51 | −0.5 | |||
| 6-ring | CA LYS 221 | pi-H | 4.58 | −0.8 | |||
| 6-ring | CG LYS 221 | pi-H | 3.95 | −0.5 |
Collectively, the aforementioned molecular docking findings for the synthesized derivatives demonstrated that 8b, 11a, and 16c were the most potent against the pocket of the investigated druggable proteins, including MurE ligase, tyrosyl-tRNA synthetase, and dihydropteroate synthase, with lower binding energy among their corresponding synthesized derivatives. This led to increased compatibility with the experimental antibacterial activities.
2.4. Toxicological properties
The potential safety/toxicity parameters of the most potent antibacterial derivative 8b were evaluated using the ProTox-II online server (https://tox-new.charite.de/protox_II). Based on ProTox-II prediction, 8b was found to belong to class V with a predicted half-lethal dose (LD50) of 3471 mg/kg. As illustrated in Fig. 8 and Table 5, compound 8b was found to be non-neurotoxin, non-nephrotoxic, non-cardiotoxic, non-immunotoxin, non-mutagenic, non-cytotoxic, and non-ecotoxic with probability ranging from 0.54 to 0.99, nonetheless, it exhibited hepatotoxicity, respiratory toxicity, carcinogenicity, and nutritional toxicity with probability ranged from 0.5 to 0.61. Additionally, 8b exhibited no effect against numerous proteins involved in nuclear receptor signaling pathways, including aryl hydrocarbon receptor, androgen receptor, androgen receptor ligand binding domain, aromatase, oestrogen receptor alpha, oestrogen receptor ligand binding domain, and peroxisome proliferator-activated receptor gamma. Additionally, 8b was inactive against several proteins involved in the stress response pathway, including nuclear factor (erythroid-derived 2)-like 2/antioxidant responsive element, heat shock factor response element, mitochondrial membrane potential, phosphoprotein (tumour suppressor) p53, and ATPase family AAA domain-containing protein 5. Compound 8b exhibited inactivity towards several molecular initiating events, including Thyroid hormone receptor alpha, Thyroid hormone receptor beta, Transtyretrin, Ryanodine receptor, GABA receptor, Glutamate N-methyl-d-aspartate receptor, alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionate receptor, Kainate receptor, Acetylcholinesterase, Constitutive androstane receptor, Pregnane X receptor, NADH-quinone oxidoreductase, Voltage-gated sodium channel, and Na+/I- symporter. In addition, 8b was found to be inactive against different cytochrome P450 proteins that are implicated in drug metabolism, such as CYP1A2, CYP2C19, CYP2D6, CYP3A4, and CYP2E1, except activity against CYP2C9.
Fig. 8.
Toxicity radar for derivative 8b.
Table 5.
The predicted toxicity for compound 8b using ProTox-II.
| Classification | Target | Prediction | Probability |
|---|---|---|---|
| Organ toxicity | Hepatotoxicity | Active | 0.50 |
| Neurotoxicity | Inactive | 0.82 | |
| Nephrotoxicity | Inactive | 0.54 | |
| Respiratory toxicity | Active | 0.57 | |
| Cardiotoxicity | Inactive | 0.70 | |
| Toxicity endpoints | Carcinogenicity | Active | 0.58 |
| Immunotoxicity | Inactive | 0.99 | |
| Mutagenicity | Inactive | 0.61 | |
| Cytotoxicity | Inactive | 0.92 | |
| BBB-barrier | Inactive | 0.56 | |
| Ecotoxicity | Inactive | 0.75 | |
| Clinical toxicity | Inactive | 0.58 | |
| Nutritional toxicity | Active | 0.61 | |
| Tox21-Nuclear receptor signaling pathways | Aryl hydrocarbon Receptor | Inactive | 0.95 |
| Androgen Receptor | Inactive | 0.95 | |
| Androgen Receptor Ligand Binding Domain | Inactive | 0.99 | |
| Aromatase | Inactive | 0.98 | |
| Estrogen Receptor Alpha | Inactive | 0.96 | |
| Estrogen Receptor Ligand Binding Domain | Inactive | 0.98 | |
| Peroxisome Proliferator-Activated Receptor Gamma | Inactive | 0.96 | |
| Nuclear factor (erythroid-derived 2)-like 2/antioxidant responsive element | Inactive | 0.98 | |
| Heat shock factor response element | Inactive | 0.98 | |
| Mitochondrial Membrane Potential | Inactive | 0.77 | |
| Phosphoprotein (Tumor Suppressor) p53 | Inactive | 0.96 | |
| ATPase family AAA domain-containing protein 5 | Inactive | 0.97 | |
| Molecular Initiating Events | Thyroid hormone receptor alpha | Inactive | 0.90 |
| Thyroid hormone receptor beta | Inactive | 0.78 | |
| Transtyretrin | Inactive | 0.97 | |
| Ryanodine receptor | Inactive | 0.98 | |
| GABA receptor | Inactive | 0.96 | |
| Glutamate N-methyl-d-aspartate receptor | Inactive | 0.92 | |
| alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate receptor | Inactive | 0.97 | |
| Kainate receptor | Inactive | 0.99 | |
| Acetylcholinesterase | Active | 0.50 | |
| Constitutive androstane receptor | Inactive | 0.98 | |
| Pregnane X receptor | Inactive | 0.92 | |
| NADH-quinone oxidoreductase | Inactive | 0.97 | |
| Voltage-gated sodium channel | Inactive | 0.95 | |
| Na+/I- symporter (NIS) | Inactive | 0.98 | |
| Metabolism | Cytochrome CYP1A2 | Inactive | 0.87 |
| Cytochrome CYP2C19 | Inactive | 0.63 | |
| Cytochrome CYP2C9 | Active | 0.52 | |
| Cytochrome CYP2D6 | Inactive | 0.82 | |
| Cytochrome CYP3A4 | Inactive | 0.65 | |
| Cytochrome CYP2E1 | Inactive | 0.99 |
3. Conclusion
We devised a new method for producing novel hybrid compounds including thiazoles or bis-thiazoles coupled to azo-sulfamethoxazole. We attempted to apply green chemistry to the design of synthetic protocols while minimizing environmental impact. The synthetic process in use has gentle reaction conditions, is straightforward to operate, has a large structural diversity, and tolerates functional groups well. We believe that combining these heterocyclic systems with adaptable structural motifs in a single molecule will improve the biological activities of the resultant heterocyclic systems. The antibacterial screening of the produced compounds revealed that several of the new derivatives displayed exceptional activity, with compounds 8b, 11a, and 16c remaining the most powerful among the three series. The molecular docking data were inconsistent with the antibacterial experimental results, but they could provide useful information about a potential mechanism of action by illustrating the thermodynamic associations that formed when these compounds bound to the crucial proteins' active sites. Our future research will focus on improving the structure of our compounds by including solubilizing groups into the primary core of compounds, to improve their pharmacokinetic characteristics.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
CRediT authorship contribution statement
Mostafa E. Salem: Writing – original draft, Methodology, Data curation. Ismail A. Abdelhamid: Writing – review & editing, Conceptualization. Ahmed H.M. Elwahy: Writing – review & editing, Conceptualization. Mohamed A. Ragheb: Writing – original draft, Software, Formal analysis, Data curation. Arwa sultan Alqahtani: Methodology, Formal analysis. Magdi E.A. Zaki: Methodology, Formal analysis. Faisal K. Algethami: Methodology, Data curation. Huda Kamel Mahmoud: Writing – original draft, Supervision, Methodology.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e31082.
Contributor Information
Ismail A. Abdelhamid, Email: ismail_shafy@yahoo.com, ismail_shafy@cu.edu.eg.
Ahmed H.M. Elwahy, Email: aelwahy@hotmail.com, aelwhy@cu.edu.eg.
Appendix A. Supplementary data
The following is/are the supplementary data to this article.
References
- 1.Geweely N.S.I. Non-toxic fumigation and alternative control techniques against fungal colonization for preserving archaeological oil painting. Int. J. Bot. 2006;2:353–362. [Google Scholar]
- 2.Geweely N.S. Novel inhibition of some pathogenic fungal and bacterial species nby new synthetic phytochemical coumarin derivatives. Ann. Microbiol. 2009;59:359–368. [Google Scholar]
- 3.Geweely N.S. Anticandidal cytotoxicity, antitumor activities, and purified cell wall modulation by novel Schiff base ligand and its metal (II) complexes against some pathogenic yeasts. Arch. Microbiol. 2009;191:687–695. doi: 10.1007/s00203-009-0497-4. [DOI] [PubMed] [Google Scholar]
- 4.Prestinaci F., Pezzotti P., Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog. Glob. Health. 2015;109:309–318. doi: 10.1179/2047773215Y.0000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aslam B., Wang W., Arshad M.I., Khurshid M., Muzammil S., Rasool M.H., Nisar M.A., Alvi R.F., Aslam M.A., Qamar M.U., Salamat M.K.F., Baloch Z. Antibiotic resistance: a rundown of a global crisis. Infect. Drug Resist. 2018;11:1645–1658. doi: 10.2147/IDR.S173867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moretta A., Scieuzo C., Salvia R., Popović Ž.D., Sgambato A., Falabella P. Tools in the era of multidrug resistance in bacteria: applications for new antimicrobial peptides discovery. Curr. Pharm. Des. 2022;28:2856–2866. doi: 10.2174/1381612828666220817163339. [DOI] [PubMed] [Google Scholar]
- 7.Turan-Zitouni G., Çavuşoğlu B.K., Sağlık B.N., Çevik U.A. Synthesis and antimicrobial activities of some novel thiazole compounds. Turkish J. Biochem. 2018;43:220–227. doi: 10.1515/TJB-2017-0093/MACHINEREADABLECITATION/RIS. [DOI] [Google Scholar]
- 8.O'Connell K.M.G., Hodgkinson J.T., Sore H.F., Welch M., Salmond G.P.C., Spring D.R. Combating multidrug-resistant bacteria: current strategies for the discovery of novel antibacterials. Angew. Chem. Int. Ed. 2013;52:10706–10733. doi: 10.1002/ANIE.201209979. [DOI] [PubMed] [Google Scholar]
- 9.Raz R., Chazan B., Kennes Y., Colodner R., Rottensterich E., Dan M., Lavi I., Stamm W. Empiric use of trimethoprim‐sulfamethoxazole (TMP‐SMX) in the treatment of women with uncomplicated urinary tract infections, in a geographical area with a high prevalence of TMP‐SMX–resistant uropathogens. Clin. Infect. Dis. 2002;34:1165–1169. doi: 10.1086/339812. [DOI] [PubMed] [Google Scholar]
- 10.McCarty J.M., Richard G., Huck W., Tucker R.M., Tosiello R.L., Shan M., Heyd A., Echols R.M. A randomized trial of short-course ciprofloxacin, ofloxacin, or trimethoprim/sulfamethoxazole for the treatment of acute urinary tract infection in women. Am. J. Med. 1999;106:292–299. doi: 10.1016/S0002-9343(99)00026-1. [DOI] [PubMed] [Google Scholar]
- 11.Zhanel G.G., Karlowsky J.A., Harding G.K., Carrie A., Mazzulli T., Low D.E., Hoban D.J., Hoban D.J. A Canadian national surveillance study of urinary tract isolates from outpatients: comparison of the activities of trimethoprim-sulfamethoxazole, ampicillin, mecillinam, nitrofurantoin, and ciprofloxacin. The Canadian Urinary Isolate Study Group. Antimicrob. Agents Chemother. 2000;44:1089–1092. doi: 10.1128/AAC.44.4.1089-1092.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eliopoulos G.M., Huovinen P. Resistance to trimethoprim-sulfamethoxazole. Clin. Infect. Dis. 2001;32:1608–1614. doi: 10.1086/320532. [DOI] [PubMed] [Google Scholar]
- 13.Niu Z.-X., Wang Y.-T., Zhang S.-N., Li Y., Chen X.-B., Wang S.-Q., Liu H.-M. Application and synthesis of thiazole ring in clinically approved drugs. Eur. J. Med. Chem. 2023;250 doi: 10.1016/j.ejmech.2023.115172. [DOI] [PubMed] [Google Scholar]
- 14.Guo J., Xie Z., Ruan W., Tang Q., Qiao D., Zhu W. Thiazole-based analogs as potential antibacterial agents against methicillin-resistant Staphylococcus aureus (MRSA) and their SAR elucidation. Eur. J. Med. Chem. 2023;259 doi: 10.1016/j.ejmech.2023.115689. [DOI] [PubMed] [Google Scholar]
- 15.Gaikwad N.D., Patil S.V., Bobade V.D. Synthesis and antimicrobial activity of novel thiazole substituted pyrazole derivatives. J. Heterocycl. Chem. 2013;50:519–527. doi: 10.1002/jhet.1513. [DOI] [Google Scholar]
- 16.El-Hag Ali G.A.M., Helal M.H., Mohamed Y.A., Ali A.A., Ammar Y.A. Synthesis, characterization and antimicrobial activity of thiazole, bis-thiazole, pyridone and bispyridone derivatives. J. Chem. Res. 2010;34:459–464. doi: 10.3184/030823410X12812857779516. [DOI] [Google Scholar]
- 17.Liaras K., Geronikaki A., Glamočlija J., Ćirić A., Soković M. Thiazole-based chalcones as potent antimicrobial agents. Synthesis and biological evaluation. Bioorg. Med. Chem. 2011;19:3135–3140. doi: 10.1016/j.bmc.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Wang L.L., Battini N., Bheemanaboina R.R.Y., Zhang S.L., Zhou C.H. Design and synthesis of aminothiazolyl norfloxacin analogs as potential antimicrobial agents and their biological evaluation. Eur. J. Med. Chem. 2019;167:105–123. doi: 10.1016/J.EJMECH.2019.01.072. [DOI] [PubMed] [Google Scholar]
- 19.Nastasă C., Vodnar D.C., Ionuţ I., Stana A., Benedec D., Tamaian R., Oniga O., Tiperciuc B. Antibacterial evaluation and virtual screening of new thiazolyl-triazole schiff bases as potential DNA-gyrase inhibitors. Int. J. Mol. Sci. 2018;19:222. doi: 10.3390/IJMS19010222. 19 (2018) 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dilek Alt|ntop M., Özdemir A., Atl| Ö., Cantürk Z., Baysal M., As|m Kaplanc|kl| Z. Synthesis and evaluation of new thiazole derivatives as potential antimicrobial agents. Lett. Drug Des. Discov. 2016;13:903–911. doi: 10.2174/1570180813666160226001021. [DOI] [Google Scholar]
- 21.Altintop M.D., Ödemir A., Turan-Zitouni G., Ilgin S., Atli Ö., Demirel R., Kaplancikli Z.A. A novel series of thiazolyl–pyrazoline derivatives: synthesis and evaluation of antifungal activity, cytotoxicity and genotoxicity. Eur. J. Med. Chem. 2015;92:342–352. doi: 10.1016/J.EJMECH.2014.12.055. [DOI] [PubMed] [Google Scholar]
- 22.Kheder N.A., Mabkhot Y.N. Synthesis and antimicrobial studies of some novel bis-[1,3,4]thiadiazole and bis-thiazole pendant to thieno[2,3-b]thiophene moiety. Int. J. Mol. Sci. 2012;13:3661–3670. doi: 10.3390/ijms13033661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahmoodi N.O., Khalili B., Rezaeianzade O., Ghavidast A. One-pot multicomponent synthesis of indol-3-yl-hydrazinyl thiazoles as antimicrobial agents. Res. Chem. Intermed. 2016;42:6531–6542. doi: 10.1007/s11164-016-2478-y. [DOI] [Google Scholar]
- 24.Chhabria M.T., Patel S., Modi P., Brahmkshatriya P.S. Thiazole: a review on chemistry, synthesis and therapeutic importance of its derivatives. Curr. Top. Med. Chem. 2016;16:2841–2862. doi: 10.2174/1568026616666160506130731. [DOI] [PubMed] [Google Scholar]
- 25.Desai N.C., Bhatt N., Somani H., Trivedi A. Synthesis, antimicrobial and cytotoxic activities of some novel thiazole clubbed 1,3,4-oxadiazoles. Eur. J. Med. Chem. 2013;67:54–59. doi: 10.1016/j.ejmech.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gümrükçüoǧlu N., Uǧraş S., Uǧraş H.I., Çakir Ü. Synthesis, extraction and antibacterial studies of some new bis-1,2,4-triazole derivatives part II. J. Incl. Phenom. Macrocycl. Chem. 2012;73:359–367. doi: 10.1007/S10847-011-0072-X/FIGURES/3. [DOI] [Google Scholar]
- 27.Pervaram S., Ashok D., Rao B.A., Sarasija M., Reddy C.V.R. Design and synthesis of new 1,2,3-triazole-pyrazole hybrids as antimicrobial agents. Russ. J. Gen. Chem. 2017;87:2454–2461. doi: 10.1134/S1070363217100280. [DOI] [Google Scholar]
- 28.Ulusoy N., Gürsoy A., Ötük G. Synthesis and antimicrobial activity of some 1,2,4-triazole-3-mercaptoacetic acid derivatives. Farm. 2001;56:947–952. doi: 10.1016/S0014-827X(01)01128-4. [DOI] [PubMed] [Google Scholar]
- 29.El Ashry E.S.H., El Tamany E.S.H., El Fattah M.E.D.A., Boraei A.T.A., Abd El-Nabi H.M. Regioselective synthesis, characterization and antimicrobial evaluation of S-glycosides and S, N-diglycosides of 1,2-Dihydro-5-(1H-indol-2-yl)-1,2,4-triazole-3-thione. Eur. J. Med. Chem. 2013;66:106–113. doi: 10.1016/j.ejmech.2013.04.047. [DOI] [PubMed] [Google Scholar]
- 30.Zoumpoulakis P., Camoutsis C., Pairas G., Soković M., Glamočlija J., Potamitis C., Pitsas A. Synthesis of novel sulfonamide-1,2,4-triazoles, 1,3,4-thiadiazoles and 1,3,4-oxadiazoles, as potential antibacterial and antifungal agents. Biological evaluation and conformational analysis studies. Bioorg. Med. Chem. 2012;20:1569–1583. doi: 10.1016/j.bmc.2011.12.031. [DOI] [PubMed] [Google Scholar]
- 31.Alam S. Synthesis, antibacterial and antifungal activity of some new [1,2,4]-triazole-3-thiones. J. Chem. Sci. 2004;116:325–331. [Google Scholar]
- 32.Prakash O., Aneja D.K., Hussain K., Lohan P., Ranjan P., Arora S., Sharma C., Aneja K.R. Synthesis and biological evaluation of dihydroindeno and indeno [1,2-e] [1,2,4]triazolo [3,4-b] [1,3,4]thiadiazines as antimicrobial agents. Eur. J. Med. Chem. 2011;46:5065–5073. doi: 10.1016/j.ejmech.2011.08.019. [DOI] [PubMed] [Google Scholar]
- 33.V Dabholkar V., Ansari F.Y. Synthesis and biological studies of bis (thiadiazole/triazole) by sonication, acta pol. Pharm. ñ Drug Res. 2008;65:521–526. [PubMed] [Google Scholar]
- 34.Mallemula V.R., Sanghai N.N., Himabindu V., Chakravarthy A.K. Synthesis and characterization of antibacterial 2-(pyridin-3-yl)-1H-benzo[d]imidazoles and 2-(pyridin-3-yl)-3H-imidazo[4,5-b]pyridine derivatives. Res. Chem. Intermed. 2015;41:2125–2138. doi: 10.1007/s11164-013-1335-5. [DOI] [Google Scholar]
- 35.Sumangala V., Poojary B., Chidananda N., Arulmoli T., Shenoy S. Facile synthesis, cytotoxic and antimicrobial activity studies of a new group of 6-aryl-3-[4-(methylsulfonyl)benzyl]-7H-[1,2,4]triazolo[3,4-b][1,3,4] thiadiazines. Eur. J. Med. Chem. 2012;54:59–64. doi: 10.1016/j.ejmech.2012.04.024. [DOI] [PubMed] [Google Scholar]
- 36.Kendre B.V., Landge M.G., Bhusare S.R. Synthesis and biological evaluation of some novel pyrazole, isoxazole, benzoxazepine, benzothiazepine, and benzodiazepine derivatives bearing an aryl sulfonate moiety as antimicrobial and anti-inflammatory agents. Arab. J. Chem. 2015;12:2091–2097. doi: 10.1016/j.arabjc.2015.01.007. [DOI] [Google Scholar]
- 37.Li Z., Zhu A., Yang J. One-Pot three-component mild synthesis of 2-Aryl-3-(9-alkylcarbazol-3-yl)thiazolin-4-ones. J. Heterocycl. Chem. 2012;49:1458–1461. doi: 10.1002/jhet. [DOI] [Google Scholar]
- 38.Gontijo V.S., Viegas F.P.D., Ortiz C.J.C., de Freitas Silva M., Damasio C.M., Rosa M.C., Campos T.G., Couto D.S., Tranches Dias K.S., Viegas C. Molecular hybridization as a tool in the design of multi-target directed drug candidates for neurodegenerative diseases. Curr. Neuropharmacol. 2019;18:348–407. doi: 10.2174/1385272823666191021124443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Viegas-Junior Claudio, Barreiro Eliezer J., Fraga Carlos Alberto Manssour, Hybridization Molecular. A useful tool in the design of new drug prototypes. Curr. Med. Chem. 2007;14:1829–1852. doi: 10.2174/092986707781058805. [DOI] [PubMed] [Google Scholar]
- 40.Elshemy H.A.H., Zaki M.A. 2017. Design and Synthesis of New Coumarin Hybrids and Insight into Their Mode of Antiproliferative Action. [DOI] [PubMed] [Google Scholar]
- 41.Alkhzem A.H., Woodman T.J., Blagbrough I.S. Design and synthesis of hybrid compounds as novel drugs and medicines. RSC Adv. 2022;12:19470–19484. doi: 10.1039/D2RA03281C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ibrahim N.S., Mohamed M.F., Elwahy A.H.M., Abdelhamid I.A. Biological activities and docking studies on novel bis 1,4-DHPS linked to arene core via ether or ester linkage. Lett. Drug Des. Discov. 2018;15:1036–1045. doi: 10.2174/1570180815666180105162323. [DOI] [Google Scholar]
- 43.Ghozlan S.A.S., Abdelmoniem A.M., Butenschön H., Abdelhamid I.A. Discrepancies in the reactivity pattern of azaenamines towards cinnamonitriles: synthesis of novel aza-steroid analogs. Tetrahedron. 2015;71:1413–1418. doi: 10.1016/j.tet.2015.01.026. [DOI] [Google Scholar]
- 44.Fathi E.M., Sroor F.M., Mahrous K.F., Mohamed M.F., Mahmoud K., Emara M., Elwahy A.H.M., Abdelhamid I.A. Design, synthesis, in silico and in vitro anticancer activity of novel bis-furanyl-chalcone derivatives linked through alkyl spacers. ChemistrySelect. 2021;6:6202–6211. doi: 10.1002/slct.202100884. [DOI] [Google Scholar]
- 45.Al-Awadi N.A., Abdelhamid I.A., Al-Etaibi A.M., Elnagdi M.H. Gas-phase pyrolysis in organic synthesis: rapid green synthesis of 4-quinolinones. Synlett. 2007:2205–2208. doi: 10.1055/s-2007-985573. [DOI] [Google Scholar]
- 46.Sroor F.M., Abdelmoniem A.M., Abdelhamid I.A. Facile synthesis, structural activity relationship, molecular modeling and in vitro biological evaluation of new urea derivatives with incorporated isoxazole and thiazole moieties as anticancer agents. ChemistrySelect. 2019;4:10113–10121. doi: 10.1002/slct.201901415. [DOI] [Google Scholar]
- 47.Mohamed M.F., Sroor F.M., Ibrahim N.S., Salem G.S., El-Sayed H.H., Mahmoud M.M., Wagdy M.A.M., Ahmed A.M., Mahmoud A.A.T., Ibrahim S.S., Ismail M.M., Eldin S.M., Saleh F.M., Hassaneen H.M., Abdelhamid I.A. Novel [l,2,4]triazolo[3,4-a]isoquinoline chalcones as new chemotherapeutic agents: block IAP tyrosine kinase domain and induce both intrinsic and extrinsic pathways of apoptosis. Invest. New Drugs. 2021;39:98–110. doi: 10.1007/s10637-020-00987-2. [DOI] [PubMed] [Google Scholar]
- 48.Ghozlan S.A.S., Ahmed A.G., Abdelhamid I.A. Regioorientation in the addition reaction of α-substituted cinnamonitrile to enamines utilizing chitosan as a green catalyst: unambiguous structural characterization using 2D-HMBC NMR spectroscopy. J. Heterocycl. Chem. 2016;53:817–823. doi: 10.1002/jhet.2341. [DOI] [Google Scholar]
- 49.Helmy M.T., Sroor F.M., Mahrous K.F., Mahmoud K., Hassaneen H.M., Saleh F.M., Abdelhamid I.A., Mohamed Teleb M.A. Anticancer activity of novel 3-(furan-2-yl)pyrazolyl and 3-(thiophen-2-yl)pyrazolyl hybrid chalcones: synthesis and in vitro studies. Arch. Pharm. (Weinheim) 2022;355 doi: 10.1002/ardp.202100381. [DOI] [PubMed] [Google Scholar]
- 50.Mohamed M.F., Ibrahim N.S., Elwahy A.H.M., Abdelhamid I.A. Molecular studies on novel antitumor bis 1,4-dihydropyridine derivatives against lung carcinoma and their limited side effects on normal melanocytes. Anti Cancer Agents Med. Chem. 2018;18:2156–2168. doi: 10.2174/1871520618666181019095007. [DOI] [PubMed] [Google Scholar]
- 51.WalyEldeen A.A., Sabet S., El-Shorbagy H.M., Abdelhamid I.A., Ibrahim S.A. Chalcones: promising therapeutic agents targeting key players and signaling pathways regulating the hallmarks of cancer. Chem. Biol. Interact. 2023;369 doi: 10.1016/j.cbi.2022.110297. [DOI] [PubMed] [Google Scholar]
- 52.Darwish E.S., Abdelhamid I.A., Nasra M.A., Abdel-Gallil F.M., Fleita D.H. A one-pot Biginelli synthesis of 6-unsubstituted 5-aroylpyrimidin-2(1H)- ones and 6-acetyl-1,2,4-triazin-3(2H)-ones. Helv. Chim. Acta. 2010;93:1204–1208. doi: 10.1002/hlca.200900355. [DOI] [Google Scholar]
- 53.Kamel M.G., Sroor F.M., Othman A.M., Mahrous K.F., Saleh F.M., Hassaneen H.M., Abdallah T.A., Abdelhamid I.A., Teleb M.A.M. Structure-based design of novel pyrazolyl–chalcones as anti-cancer and antimicrobial agents: synthesis and in vitro studies. Monatsh. Chem. 2022;153:211–221. doi: 10.1007/s00706-021-02886-5. [DOI] [Google Scholar]
- 54.Ghozlan S.A.S., Abdelhamida I.A., Ibrahim H.M., Elnagdia M.H. Studies with 2-arylhydrazononitriles: a new convenient synthesis of 2, 4-disubstituted- 1,2,3-triazole-5-amines. ARKIVOC (Gainesville, FL, U. S.) 2006;2006:53–60. doi: 10.3998/ark.5550190.0007.f07. [DOI] [Google Scholar]
- 55.Al-Awadi N.A., Abdelkhalik M.M., Abdelhamid I.A., Elnagdi M.H. Pyrolytic methods in organic synthesis: novel routes for the synthesis of 3-oxoalkanenitriles, 2-acyl anilines, and 2-aroyl anilines. Synlett. 2007:2979–2982. doi: 10.1055/S-2007-992355. [DOI] [Google Scholar]
- 56.Ghozlan S.A.S., Mohamed M.H., Abdelmoniem A.M., Abdelhamid I.A. Synthesis of pyridazines and fused pyridazines via [3+3] atom combination using chitosan as a green catalyst. ARKIVOC (Gainesville, FL, U. S.) 2009:302–311. doi: 10.3998/ark.5550190.0010.a27. [DOI] [Google Scholar]
- 57.Sayed O.M., Mekky A.E.M., Farag A.M., Elwahy A.H.M. 3,4-Bis(bromomethyl)thieno[2,3-b]thiophene: versatile precursors for novel bis(triazolothiadiazines), bis(quinoxalines), bis(dihydrooxadiazoles), and bis(dihydrothiadiazoles) J. Heterocycl. Chem. 2016;53:1113–1120. doi: 10.1002/jhet.2373. [DOI] [Google Scholar]
- 58.Ibrahim Y.A., Abbas A.A., Elwahy A.H.M. New trends in the chemistry of condensed heteromacrocycles Part B: macrocyclic formazans. J. Heterocycl. Chem. 2004;41:135–149. doi: 10.1002/JHET.5570410202. [DOI] [Google Scholar]
- 59.Ibrahim Y.A., Elwahy A.H.M., Abbas A.A. New synthesis of macrocyclic crown-formazans from pyruvic acid derivatives. Tetrahedron. 1994;50:11489–11498. doi: 10.1016/S0040-4020(01)89286-3. [DOI] [Google Scholar]
- 60.Elwahy A.H.M., Abbas A.A. Synthetic communications : an international journal for rapid communication of synthetic organic chemistry bis (β -difunctional) compounds : versatile starting materials for novel bis (heterocycles) Synth. Commun. 2000;30:2903–2921. doi: 10.1080/00397910008087441. [DOI] [Google Scholar]
- 61.Mekky A.E.M., Elwahy A.H.M. Synthesis of novel benzo-substituted macrocyclic ligands containing thienothiophene subunits. J. Heterocycl. Chem. 2014;51:E34–E41. doi: 10.1002/jhet.2012. [DOI] [Google Scholar]
- 62.Elwahy A.H., Shaaban M.R. Synthesis of pyrido- and pyrimido-fused heterocycles by multi-component reactions (Part 3) Curr. Org. Synth. 2014;11:835–873. doi: 10.2174/157017941106141023114039. [DOI] [Google Scholar]
- 63.Barsoum B.N., Khella S.K., Elwaby A.H.M., Abbas A.A., Ibrahim Y.A. Evaluation of some new 14- and 15-crown-formazans as carriers in cesium ion selective electrodes. Talanta. 1998;47:1215–1222. doi: 10.1016/S0039-9140(98)00204-5. [DOI] [PubMed] [Google Scholar]
- 64.Salem M.E., Darweesh A.F., Farag A.M., Elwahy A.H.M. 2-Bromo-1-(1H-pyrazol-4-yl)ethanone: versatile precursors for novel mono-, bis- and poly{6-(1H-pyrazol-4-yl)-[1,2,4]triazolo[3,4-b][1,3,4]thiadiazines. Tetrahedron. 2016;72:712–719. doi: 10.1016/j.tet.2015.12.024. [DOI] [Google Scholar]
- 65.Ibrahim Y.A., Abbas A.A., Elwahy A.H.M. Selective synthesis and structure of 2-N-and 3-S-glucosyl-1, 2, 4-triazoles of potential biological interest. Carbohydr. Lett. 1999;3:331–338. [Google Scholar]
- 66.Elwahy A.H.M., Abbas A.A. Synthesis of N -pivot lariat ethers. J. Heterocycl. Chem. 2008;45:1–65. doi: 10.1002/jhet.5570450101. [DOI] [Google Scholar]
- 67.Elwahy A.H.M., Abbas A.A., Kassab R.M. Unexpected synthesis of novel condensed heteromacrocycles. Synthesis. 2002:260–264. [Google Scholar]
- 68.Darweesh A.F., Abd El-Fatah N.A., Abdel-Latif S.A., Abdelhamid I.A., Elwahy A.H.M., Salem M.E. Synthesis and DFT studies of novel aminoimidazodipyridines using 2-(3H-imidazo[4,5-b]pyrid-2-yl)acetonitrile as an efficient key precursor. ARKIVOC (Gainesville, FL, U. S.) 2021;2021:23–37. doi: 10.24820/ARK.5550190.P011.415. [DOI] [Google Scholar]
- 69.Radwan I.T., Elwahy A.H.M., Darweesh A.F., Sharaky M., Bagato N., Khater H.F., Salem M.E. Design, synthesis, docking study, and anticancer evaluation of novel bis-thiazole derivatives linked to benzofuran or benzothiazole moieties as PI3k inhibitors and apoptosis inducers. J. Mol. Struct. 2022;1265 doi: 10.1016/j.molstruc.2022.133454. [DOI] [Google Scholar]
- 70.Diab H.M., Salem M.E., Abdelhamid I.A., Elwahy A.H.M. Synthesis of novel star-shaped molecules based on a 1,3,5-triazine core linked to different heterocyclic systems as novel hybrid molecules. RSC Adv. 2020;10:44066–44078. doi: 10.1039/d0ra09025e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salem M.E., Ahmed A.A., Shaaban M.R., Shibl M.F., Farag A.M. Regioselective synthesis and ab initio calculations of fused heterocycles thermally and under microwave irradiation. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015;148:175–183. doi: 10.1016/j.saa.2015.03.102. [DOI] [PubMed] [Google Scholar]
- 72.Elnagdi M.H., Al-Awadi N.A., Abdelhamid I.A. Chapter 1 recent developments in pyridazine and condensed pyridazine synthesis. Adv. Heterocycl. Chem. 2009;97:1–43. doi: 10.1016/S0065-2725(08)00201-8. [DOI] [Google Scholar]
- 73.Hosny M., Salem M.E., Darweesh A.F., Elwahy A.H.M. Synthesis of novel bis(thiazolylchromen-2-one) derivatives linked to alkyl spacer via phenoxy group. J. Heterocycl. Chem. 2018;55:2342–2348. doi: 10.1002/jhet.3296. [DOI] [Google Scholar]
- 74.Salem M.E., Darweesh A.F., Mekky A.E.M., Farag A.M., Elwahy A.H.M. 2-Bromo-1-(1 H -pyrazol-4-yl)ethanone: versatile precursor for novel mono- and bis[pyrazolylthiazoles] J. Heterocycl. Chem. 2017;54:226–234. doi: 10.1002/jhet.2571. [DOI] [Google Scholar]
- 75.Salem M.E., Darweesh A.F., Shaaban M.R., Elwahy A.H.M. Synthesis of novel bis-and poly(hydrazinylthiazole) linked to benzofuran or benzothiazole as new hybrid molecules. ARKIVOC (Gainesville, FL, U. S.) part v. 2019:73–88. doi: 10.24820/ark.5550190.p010.810. [DOI] [Google Scholar]
- 76.Balouiri M., Sadiki M., Ibnsouda S.K. Methods for in vitro evaluating antimicrobial activity: a review. J. Pharm. Anal. 2016;6:71–79. doi: 10.1016/J.JPHA.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ovung A., Bhattacharyya J. Sulfonamide drugs: structure, antibacterial property, toxicity, and biophysical interactions. Biophys. Rev. 2021;132(13):259–272. doi: 10.1007/S12551-021-00795-9. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Diab H.M., Elwahy A.H.M., Ragheb M.A., Abdelhamid I.A., Mahmoud H.K. Facile one-pot, three-component synthesis and antimicrobial screening of novel hexahydropyrimido[4,5-b]quinolinediones incorporating phenoxylacetamide core as novel hybrid molecules via Hantzsch reaction. J. Mol. Struct. 2023;1287 doi: 10.1016/j.molstruc.2023.135721. [DOI] [Google Scholar]
- 79.Ragab M.S., Shehata M.R., Shoukry M.M., Haukka M., Ragheb M.A. Oxidative DNA cleavage mediated by a new unexpected [Pd(BAPP)][PdCl4] complex (BAPP = 1,4-bis(3-aminopropyl)piperazine): crystal structure, DNA binding and cytotoxic behavior. RSC Adv. 2022;12:1871–1884. doi: 10.1039/D1RA07793G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ragab M.S., Soliman M.H., Shehata M.R., Shoukry M.M., Ragheb M.A. Design, synthesis, spectral characterization, photo-cleavage, and in vitro evaluation of anticancer activities of new transition metal complexes of piperazine based Schiff base-oxime ligand. Appl. Organomet. Chem. 2022;36 doi: 10.1002/AOC.6802. [DOI] [Google Scholar]
- 81.Saha N., Azam M.A. MurE inhibitors as antibacterial agents: a review. J. Incl. Phenom. Macrocycl. Chem. 2020;98:127–136. doi: 10.1007/s10847-020-01018-6. [DOI] [Google Scholar]
- 82.Zoeiby E., Sanschagrin F., Levesque R.C., El Zoeiby A., Sanschagrin F., Levesque R.C. Structure and function of the Mur enzymes: development of novel inhibitors. Mol. Microbiol. 2003;47:1–12. doi: 10.1046/J.1365-2958.2003.03289.X. [DOI] [PubMed] [Google Scholar]
- 83.Xiao Z.P., Ma T.W., Liao M.L., Feng Y.T., Peng X.C., Li J.L., Li Z.P., Wu Y., Luo Q., Deng Y., Liang X., Zhu H.L. Tyrosyl-tRNA synthetase inhibitors as antibacterial agents: synthesis, molecular docking and structure-activity relationship analysis of 3-aryl-4-arylaminofuran-2(5H)-ones. Eur. J. Med. Chem. 2011;46:4904–4914. doi: 10.1016/J.EJMECH.2011.07.047. [DOI] [PubMed] [Google Scholar]
- 84.Sun J., Lv P.C., Zhu H.L. Tyrosyl-tRNA synthetase inhibitors: a patent review. Expert Opin. Ther. Pat. 2017;27:557–564. doi: 10.1080/13543776.2017.1273350. [DOI] [PubMed] [Google Scholar]
- 85.Capasso C., Supuran C.T. Sulfa and trimethoprim-like drugs-antimetabolites acting as carbonic anhydrase, dihydropteroate synthase and dihydrofolate reductase inhibitors. J. Enzyme Inhib. Med. Chem. 2014;29:379–387. doi: 10.3109/14756366.2013.787422. [DOI] [PubMed] [Google Scholar]
- 86.Griffith E.C., Wallace M.J., Wu Y., Kumar G., Gajewski S., Jackson P., Phelps G.A., Zheng Z., Rock C.O., Lee R.E., White S.W. The structural and functional basis for recurring sulfa drug resistance mutations in Staphylococcus aureus dihydropteroate synthase. Front. Microbiol. 2018;9:1369. doi: 10.3389/FMICB.2018.01369/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Capasso C., Supuran C.T. Bact. Resist. To Antibiot. From Mol. to Man. John Wiley & Sons, Ltd; 2019. Dihydropteroate synthase (sulfonamides) and dihydrofolate reductase inhibitors; pp. 163–172. [DOI] [Google Scholar]
- 88.Billones J.B., Bangalan M.A.T. Structure-based discovery of inhibitors against MurE in methicillin-resistant Staphylococcus aureus. Orient. J. Chem. 2019;35:618–625. doi: 10.13005/ojc/350216. [DOI] [Google Scholar]
- 89.Zouhir A., Jemli S., Omrani R., kthiri A., Jridi T., sebei K. In silico molecular analysis and docking of potent antimicrobial peptides against MurE enzyme of methicillin resistant Staphylococcus aureus. Int. J. Pept. Res. Ther. 2021;27:1253–1263. doi: 10.1007/S10989-021-10165-4/TABLES/7. [DOI] [Google Scholar]
Further reading
- 90.Group C.C. Chemical computing group (CCG) | computer-aided molecular design. Chem. Comput. Gr. 2020 https://www.chemcomp.com/ [Google Scholar]
- 91.Ragheb M.A., Omar R.S., Soliman M.H., Elwahy A.H.M., Abdelhamid I.A. Synthesis, characterization, DNA photocleavage, in silico and in vitro DNA/BSA binding properties of novel hexahydroquinolines. J. Mol. Struct. 2022;1267 doi: 10.1016/j.molstruc.2022.133628. [DOI] [Google Scholar]
- 92.Ragheb M.A., Abdelwahab R.E., Darweesh A.F., Soliman M.H., Elwahy A.H.M., Abdelhamid I.A. Hantzsch‐like synthesis, DNA photocleavage, DNA/BSA binding, and molecular docking studies of bis(sulfanediyl)bis(tetrahydro‐5‐deazaflavin) analogs linked to naphthalene core. Chem. Biodivers. 2022;19 doi: 10.1002/cbdv.202100958. [DOI] [PubMed] [Google Scholar]
- 93.Bank P.D. Rcsb PDB: homepage, rcsb pdb. 2019. https://www.rcsb.org/
- 94.Banerjee P., Eckert A.O., Schrey A.K., Preissner R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018;46:W257–W263. doi: 10.1093/NAR/GKY318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.