ABSTRACT
Background
The salivary microbiome may interact with chemoradiotherapy through dynamic changes in microbial composition and systemic immunity. We aimed to explore the association between the salivary microbiome and response to chemoradiotherapy in initially inoperable patients with local advanced esophageal squamous cell carcinoma (LAESCC).
Methods
Salivary and peripheral blood samples were collected before and after chemoradiotherapy. The microbiome and metabolic pathways were analyzed by 16S ribosomal RNA sequencing and liquid chromatography tandem mass spectrometry/Mass spectrometry analyses.
Results
The salivary microbiome exhibited characteristic variations between patients and healthy controls. A significant correlation was found between Prevotella_salivae, Saccharibacteria_TM7_G3_bacterium_HMT_351, and Veillonellaceae_G1_bacterium_HMT_129 and pathological complete response (pCR) in initially inoperable patients who underwent surgery. The PICRUSt suggested that immune diseases and cell motility were different in tumor compared to normal groups. KEGG enrichment analysis showed enriched lipid metabolism, signal transduction, and membrane transport in the tumor group. CD3+CD8 T cells, IL6, IL10, and IFNγ exhibited an increasing trend during the treatment process of chemoradiotherapy.
Conclusions
Our study demonstrated that variations in specific saliva taxa associated with host immunomodulatory cells and cytokines could be promising for early efficacy prediction of chemoradiotherapy in initially inoperable patients with LAESCC.
KEYWORDS: Salivary, microbiome, local advanced esophageal squamous cell carcinoma, chemoradiotherapy, initially inoperable
Introduction
The human oral cavity is the initial gateway to the digestive tract, and contains more than 700 bacterial species, protozoa, fungi, and viruses. Saliva is the most common fluid covering the oral cavity [1]. It is involved in the physiological processes with proteins and enzymes, such as mastication, swallowing, and speech [2]. Recent studies have shown that disorders of the salivary microbiome are associated with multiple diseases, including periodontitis, tooth reduction, dental caries [3], chronic obstructive pulmonary diseases [4], cardiovascular diseases, and oral cancer [5,6]. Previous studies investigated the microbial characteristics of esophageal diseases, such as reflux-related esophagitis, Barrett’s esophagus, and esophageal carcinoma [7]. A relationship has been observed between patients with cancer with different gut microbial compositions and various treatment outcomes. Metagenomic studies have shown that intratumoral and gut microbiomes are associated with the response to neoadjuvant chemoradiotherapy in cancer [8,9]. Another study reported that transitions in the oral and gut microbiomes are associated with HPV+ oropharyngeal squamous cell carcinoma following definitive chemoradiotherapy [10]. Salivary microbiome changes can distinguish the responses to chemoradiotherapy in patients with oral cancer [11]. An increasing number of studies have shown that microbial communities induce immune and inflammatory responses, which in turn promote cancer progression [12]. With advances in gene sequencing technology, more information on the microbiota has been obtained to compare species composition, diversity, and complexity of different cancers [13]. However, few studies have focused on the relationship between microorganisms and initially inoperable patients with local advanced esophageal squamous cell carcinoma (LAESCC).
Carcinoma of the esophagus is one of the most common malignant tumors worldwide [14]. Squamous cell carcinoma is a more prevalent pathological subtype of esophageal cancer in East Asia than adenocarcinoma [15]. New paradigms including chemoradiotherapy and immunotherapy, have replaced traditional approaches for cancer in the last few decades [16]. However, chemoradiotherapy remains a critical conversion treatment for local regional cancer patients with unsatisfactory survival benefits especially for clinical T4b or lymph node metastatic cancer [17]. How to select those patients who respond well to chemoradiotherapy sequential operation deserves further investigation. In addition to the pathological complete response (pCR), novel markers are essential for monitoring the treatment response. Increasing studies have proved that the microorganisms in saliva play a key role in the diagnosis, prognosis, and survival of cancer, which warrants further study.
Chemoradiotherapy is a compulsory therapeutic regimen for local advanced esophageal cancer [18]. The microbiota has been demonstrated to boost treatment interactions among the microbiome, microbial metabolites and immune microenvironment [19,20]. However, little is known about the relationship between the salivary microbiome and treatment response in initially inoperable patients with LAESCC. In the present study, we hypothesized that the salivary microbiome alters the post-treatment effects of similar chemoradiotherapies. Therefore, we aimed to characterize the salivary microbiome and response prediction factors during chemoradiotherapy in initially inoperable patients with LAESCC.
Methods
Participants and sample collection
The participants included 79 initially inoperable patients with LAESCC before chemoradiotherapy, 8 patients after chemoradiotherapy, and 10 healthy controls recruited from XXX. All patients were verified by pathological examination of hematoxylin-eosin staining and immunohistochemistry. Healthy controls were enrolled from 10 volunteers with no digestive diseases, as confirmed by electronic gastroscopy. Informed consent was obtained from all participants. This study was approved by the Institutional Review Board of the XXX.
The exclusion criteria were as follows: any infectious diseases, oral or gastrointestinal tumors, history of gastrointestinal surgery, medication history of antibiotics and microecological preparations (probiotics, prebiotics, or symbiotics), and use of adrenocortical hormones or other immunosuppressive drugs in the past 1 month.
Five milliliters (mL) of saliva were collected immediately in a quiet state between 6.00–7.00am. The participants were restricted from eating or drinking anything before saliva collection. Salivary samples were stored in specific collection tubes at −80°C before further analysis. Fresh peripheral blood samples were centrifuged at 1200 rpm for 5 min. Meanwhile, the upper layer and lymphocytes were stored at −80°C for testing, respectively.
DNA extraction, amplification, and 16s rRNA sequencing
After centrifugation, the salivary genomic DNA of the supernatant was collected from 2 mL of saliva by sodium dodecyl sulfate. The total microbial genomic DNA was extracted from patients’ and healthy controls’ specimens using a DNA Kit (Qiagen, Shanghai, China). The quantity and purity of DNA were testified by NanoDrop 2000 UV-Vis Spectrophotometer (Thermo Scientific, Wilmington, MA, USA). After genomic DNA was extracted from the sample, the V3+V4 region of the 16S rDNA was amplified using a specific primer with barcode (Primer sequence: 341F: CCTACGGGNGGCWGCAG; 806 R: GGACTACHVGGGTATCTAAT). Purified amplification products (amplicons) were connected to the sequencing joints. The 16S rDNA target region of the ribosomal RNA gene was amplified by polymerase chain reaction (PCR) (95°C for 5 min, followed by 30 cycles at 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min and a final extension at 72°C for 7 min) using primers. A 50 µL mixture contained 10 µL of 5 × Q5@ Reaction Buffer, 10 µL of 5 × Q5@ High GC Enhancer, 1.5 µL of 2.5 mM dNTPs, 1.5 µL of each primer (10 µM), 0.2 µL of Q5@ High-Fidelity DNA Polymerase, and 50 ng of template DNA. A sequencing library was constructed, and sequenced using Illumina (San Diego, CA, USA). Amplicons were evaluated on 2% agarose gels and purified using AMPure XP Beads (Beckman, CA, USA) according to the manufacturer’s instructions. Sequencing libraries were generated using the Illumina DNA Prep Kit (Illumina, CA, USA) following the manufacturer’s recommendations. The library quality was assessed with the ABI StepOnePlus Real-Time PCR System (Life Technologies, Foster City, USA). At the end, 2 × 250 bp paired-end reads were generated by sequencing on the Novaseq 6000 platform. Studies on microbial diversity have included alpha and beta diversity, species analyses, functional studies and environmental relationships.
16s rRNA data processing and analysis
Raw Fastq files were merged using FLASH software (version 1.2.11) and quality-filtered with Trimmomatic using the following criteria: (i) reads were truncated at any site receiving an average quality score of < 20 over a 50 base pair (bp) sliding window, which retained sequences with overlaps > 10 bp and mismatches of no more than 2 bp, and (ii) sequence data were demultiplexed and assigned to samples based on barcodes (exact matches) and primers (two nucleotide mismatches were allowed). The reads containing ambiguous bases were removed. Paired reads were overlapped as raw tags using FLASH with a minimum overlap of 10 bp and a mismatch error rate of 2%. Operational taxonomic units (OTUs) were clustered with a 97% sequence similarity cutoff using UPARSE (version 7.1, https://drive5.com/uparse/) and a novel ‘greedy’ algorithm that simultaneously performed chimera filtering and OTU clustering. Taxonomic assignment of each sequence was carried out using the Ribosomal Database Project (RDP) Classifier algorithm (http://rdp.cme.msu.edu/) against the Silva (SSU123) 16S rRNA database with a confidence threshold of 70%.
Metabolic analysis
The 100 μL sample was transferred to the 400 μL extraction solution (methanol: acetonitrile = 1:1 (V/V), containing the internal standard mixture of isotope labels. The mixture was ultrasonicated for 10 min in an ice water bath. Then the sample was centrifuged at 12,000 rpm at 4°C for 15 min. Supernatants were collected in the sample bottles for machine testing. For polar metabolites, an ultra-high performance liquid chromatographic instrument of Vanquish (Thermo Fisher Scientific) was used in this project. The target compounds were separated by Waters ACQUITY UPLC BEH Amide (2.1 mm × 50 mm, 1.7 μm) liquid chromatography column. Phase A was aqueous, containing 25 mmol/L ammonium acetate and 25 mmol/L ammonia, and phase B was acetonitrile. The Orbitrap Exploris 120 mass spectrometer is capable of primary and secondary mass spectrometry data acquisition under the control software (Xcalibur, version 4.4, Thermo). Detailed parameters were as follows: sheath gas flow rate, 50 Arb, Aux gas flow rate, 15 Arb, Capillary temperature, 320°C, Full ms resolution 60,000, MS/MS resolution 15,000, Collision energy, SNCE 20/30/40, Spray Voltage, 3.8kV (positive) or −3.4kV (negative).
Positive and negative ion modes were used to detect metabolites, which could make the metabolite coverage higher and the detection effect better. Quality control samples are typically used for quality control when metabolomic studies are based on mass spectrometry. For preliminary visualization of differences between different groups of samples, the unsupervised dimensionality reduction method of principal component analysis (PCA) was applied to all samples using the R package models (http://www.r-project.org/). The variable importance in the projection (VIP) score of the (O)PLS model was applied to rank the metabolites that best distinguished the two groups. Receiver operating characteristic (ROC) curve analysis was performed to evaluate the predictive power of each discriminant metabolite using the R pROC package. Area under the curve (AUC) was computed by the numerical integration of the ROC curves. The metabolite signature with the largest area under the ROC curve was identified as having the strongest predictive power for discriminating between the two groups. The metabolites were mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathways for annotation and enrichment analysis. Through the hypothesis test of the p-value calculation and FDR correction, the pathways with Q value ≤ 0.05 were defined as significantly enriched pathways.
Flow cytometry and enzyme-linked immunosorbent assay
A total of 100 μL whole blood were added with FITC anti-mouse CD3 antibody (Beijing tongsheng, China), APC anti-mouse CD4 antibody (Beijing tongsheng, China), and PerCP/Cyanine5.5 anti-mouse CD8 antibody (Beijing tongsheng, China). The mixture was then incubated for 30 minutes in the dark at room temperature. Next, 2 mL of the red cell lysate was added at room temperature in the dark for 10 minutes. Besides, it was centrifuged at 1500 rpm for 5 minutes and cell precipitate was added to 500 μL phosphate buffer and detected by flow analyzer.
A total of 1000 μL serum was centrifugated and collected to further testing. All anti-human antibodies (Raisecare, China) of cytokines (IL4, IL6, IL10, IL12p70, IL17, TNFα, IFNγ) were pre-packaged on a high-affinity enzyme-labeled plate. Samples for testing, standard substances, and biotinylated detection antibodies were mixed in the holes of enzyme label plates. After incubation at 37°C for 45 minutes, the samples were combined with the detection antibody. The immune complex was then washed to remove unbound substances and labeled with horseradish peroxidase and Streptavidin-horseradish peroxidase. The color reactions were terminated using a termination solution and the absorbance was measured at 450 nm.
Statistical analysis
Statistical analyses were carried out by GraphPad Prism (version 9) and the SPSS 25. The paired t-test and Kruskal-Wallis test were used to compare demographic characteristics. Alpha diversity indexes were calculated using MOTHUR software (version 1.30.1). Principal coordinates analysis (PCoA) based on the weighted and unweighted UniFrac distance and analysis of similarities (ANOSIM) were performed to compare the global microbial composition at the operational taxonomic unit (OTU) level. Welch’s t-test and the Wilcoxon Rank sum test were used to identify the species. Discriminant microbiome species were performed using the linear discriminant analysis effect size (LEfSe). The linear discriminant analysis score (LDA score) indicated the effect size of each OTU, and OTUs with an LDA score > 3.0 were defined as differentially abundant OTUs. Phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) was used to identify KEGG biochemical pathways, and the results were visualized as a heatmap using the Multiple Experiment Viewer (version 4.9.0). p values of less than 0.05 were considered statistically significant.
Results
Demographic characteristics of all individuals
The demographic characteristics of all patients (n = 87) and healthy controls (n = 10) are depicted in Table 1. The median age was 72 years in the pre-treatment chemoradiotherapy group (T1), 75 years in the post-treatment chemoradiotherapy group (T2), and 70 years in the normal healthy controls (NC). 59(74.6%) patients were male and 20 (25.4%) were female in the T1 group. All patients who were initially inoperable were diagnosed with local advanced cancer without distant metastasis. The number of primary tumors of grades 1, 2, 3, and 4 in the T1 group were 0 (0.0%), 7 (8.8%), 63 (79.7%), and 9 (11.5%), respectively. The number of cases of lymph node metastasis of grade 0–1 and 2–3 in the T1 group were 45 (56.9%), and 34 (43.1%), respectively.
Table 1.
Basic characteristics of the overall population.
| Variables | Patients before chemoradiotherapy (n = 79) | Patients post chemoradiotherapy (n = 8) | Healthy control (n = 10) |
|---|---|---|---|
| Age(median) | 72 years | 75 years | 70 years |
| Gender | |||
| Male | 59(74.6%) | 4(50.0%) | 5(50.0%) |
| Female | 20(25.4%) | 4(50.0%) | 5(50.0%) |
| ECOG | |||
| 0–1 | 73(92.4%) | 8(100.0%) | 10(100.0%) |
| 2 | 5(7.6%) | 0(0.0%) | / |
| Tumor grade | |||
| 1 | 0(0.0%) | 0(0.0%) | / |
| 2 | 7(8.8%) | 0(0.0%) | / |
| 3 | 63(79.7%) | 7(87.5%) | |
| 4 | 9(11.5%) | 1(12.5%) | |
| Lymph node metastasis | |||
| 0–1 | 45(56.9%) | 7(87.5%) | / |
| 2–3 | 34(43.1%) | 1(12.5%) | / |
| Disease location | |||
| Cervical | 5(7.6%) | 1(12.5%) | / |
| Upper thoracic | 17(21.5%) | 3(37.5%) | / |
| Middle thoracic | 31(39.2%) | 2(25.0%) | / |
| Lower thoracic | 26(31.7%) | 2(25.0%) | / |
| Pathological complete response | |||
| Yes | 8(30.7%) | / | / |
| No | 18(69.3%) | / | / |
Abbreviations: Eastern Cooperative Oncology Group (ECOG) Performance Status.
Variations in microbiome profiles between cancer patients and normal healthy controls
To explore the multiple roles of the salivary microbiome in cancer patients, 79 salivary samples from patients with LAESCC before, and 8 after chemoradiotherapy were recruited. The total taxonomy fill and stack was shown in Figure 1a,b. The NC group showed a significantly higher alpha diversity (Chao index) than the T1 and T2 groups. The same higher Chao index was observed in the T1 group compared to that in the T2 group (Figure 1c-f). Meanwhile, no alpha diversity (assessed with the Shannon indexes) changed throughout the treatment among patients with T1 versus T2. Significant difference in beta diversity (NMDA, and PCoA index) was found between the T1/T2 versus NC groups (Figure 1g-j). The relative abundance of the microbiome composition was shown at seven levels (Domain, Phylum, Class, Order, Family, Genus, Species) among all groups (Figure 2a,b).
Figure 1.
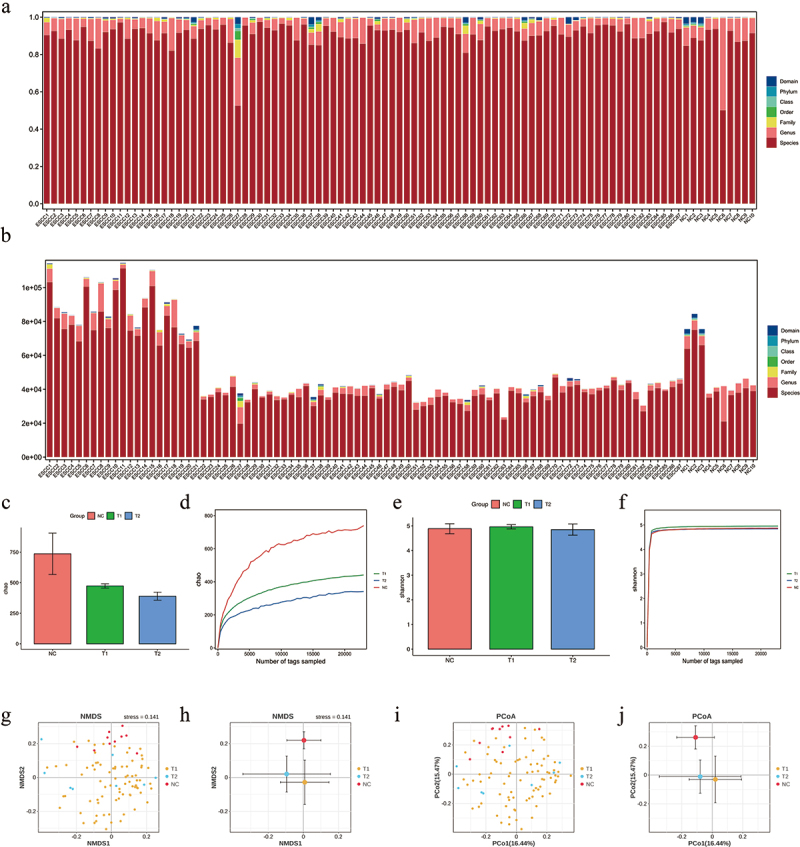
The results of taxonomy, α and β diversity analysis for initially inoperable patients with esophageal squamous cell carcinoma (LAESCC) before chemoradiotherapy (T1), post chemoradiotherapy group (T2), and 10 healthy subjects without any general medical history (NC). (a-b) Taxonomy fill and stack analysis among patients’ groups and control. (c-d) α diversity analysis of Chao1 and Shannon index among patients’ groups and control. (e-f) β diversity analysis. (g-j) Analysis indices were NMDS and PCOA analysis.
Figure 2.
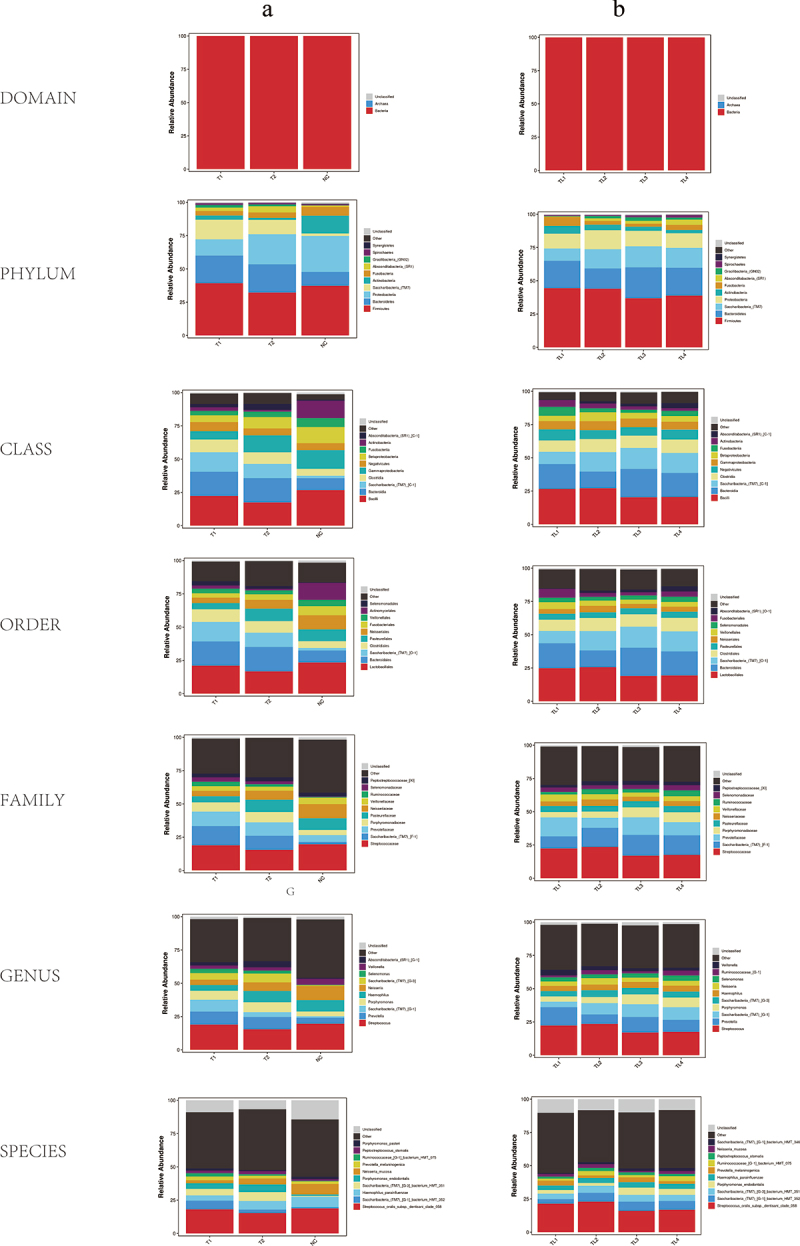
Relative abundance of microbiome composition in seven levels (Domain, Phylum, Class, Order, Family, Genus, Species). (a) Relative abundance of microbiome composition among patients’ groups and control. (b) Relative abundance of microbiome composition among patients’ groups of different tumor locations (TL1: Cervical, TL2: Upper thoracic, TL3: Middle thoracic, TL4: Lower thoracic).
Variant microbial communities lead to differences in the metabolic pathways in cancer and NC groups
To further evaluate the diagnostic role of the salivary microbiome, LEfSe analysis was used to identify the potential biomarkers (Figure 3a,b). The cladogram, and bubble plot indicated that 19 species (e.g. Saccharibacteria_TM7, and Prevotella) were significantly enriched in the T1 group compared to those in the NC group (Figure 3c). The indicative biomarkers between the patient and normal control groups were plotted by Venn and ROC curves (Figure 3d,e). PICRUSt was used to predict the metagenomes from the 16S data of the total cohort with Welch’s T test. This implies that immune diseases and cell motility were significantly different between T1 and NC groups (Figure 3f). No relationship was found between the microbiome and age, gender, ECOG, tumor grade, lymph node metastasis, or disease location (p < 0.05, Table 2).
Figure 3.
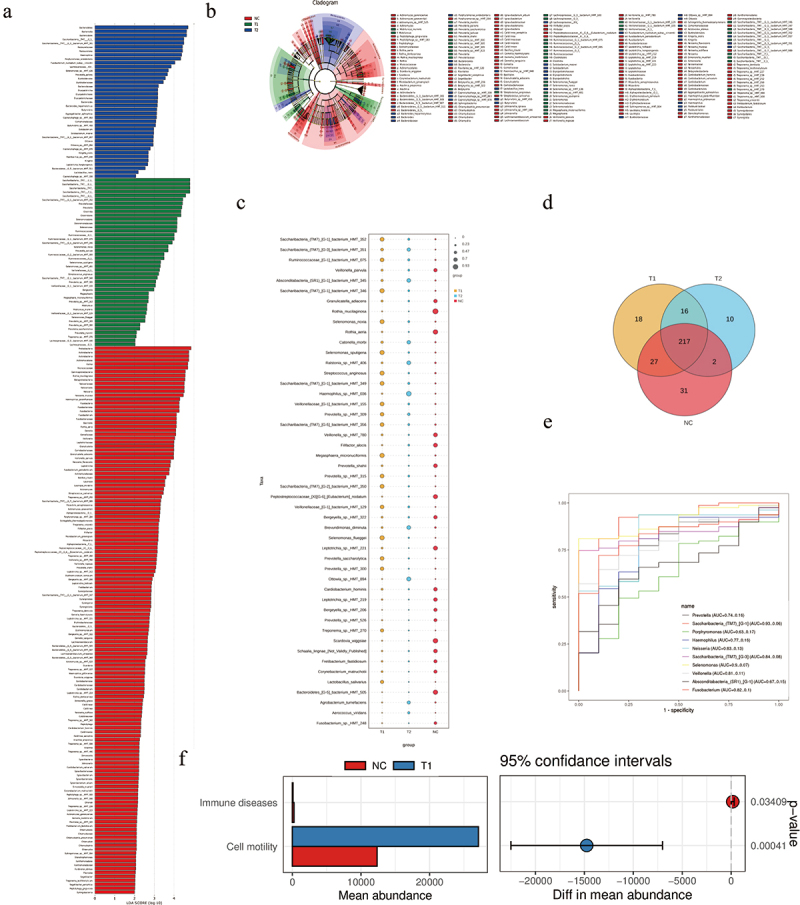
(a-b-c) LEfSe comparison, cladogram, and bubble plot between initially inoperable patients with LAESCC before chemoradiotherapy (T1), post chemoradiotherapy group (T2), and healthy controls (NC). (d)The Venn plot of indicative biomarkers in patients’ groups and control. (e)The result of ROC curve analysis showing a significant high AUC value for cancer diagnosis. (f) The microbiome functions of patients and control were predicted by PICRUSt2 analysis with Welch’s T test.
Table 2.
Association between representative microbiome and clinical characteristics in patients before chemoradiotherapy (n = 79).
| Variables | Prevotella_salivae |
Saccharibacteria_TM7_G3_bacterium_HMT_351 |
Veillonellaceae_G1_bacterium_HMT_129 |
|---|---|---|---|
| p value | |||
| Age (median, years) | |||
| ≥72 | >0.05 | >0.05 | >0.05 |
| <72 | |||
| Gender | |||
| Male | >0.05 | >0.05 | >0.05 |
| Female | |||
| ECOG | |||
| 0–1 | >0.05 | >0.05 | >0.05 |
| 2 | |||
| Tumor grade | |||
| 1 | >0.05 | >0.05 | >0.05 |
| 2 | |||
| 3 | |||
| 4 | |||
| Lymph node metastasis | |||
| 0–1 | >0.05 | >0.05 | >0.05 |
| 2–3 | |||
| Disease location | |||
| Cervical | >0.05 | >0.05 | >0.05 |
| Upper thoracic | |||
| Middle thoracic | |||
| Lower thoracic | |||
Abbreviations: Eastern Cooperative Oncology Group (ECOG) Performance Status.
To seek the metabolic differences in the salivary microbiome, non-targeted metabolic analysis was used to identify valuable biomarkers. The numbers of significant metabolites in the patients’ and controls’ groups are presented in Figure 4a. KEGG enrichment analysis revealed that the significant pathways are lipid metabolism, signal transduction, and membrane transport (Figure 4b). Besides, it was also involved in the development of digestive system. The relationship between the metabolome and valuable microbiomes were plotted in Figure 4c among initially inoperable patients (T1). Pathways in the T2 group were similar to those in the T1 group.
Figure 4.
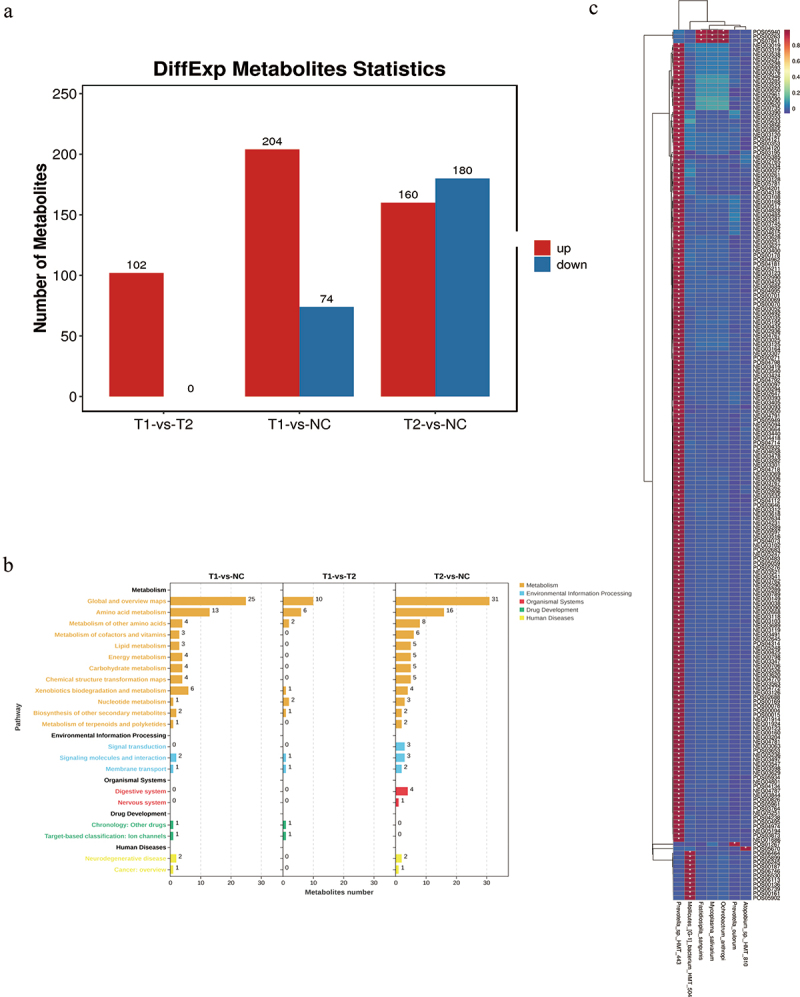
(a) Number of significant metabolites in patients’ groups and control. (b) KEGG enrichment analysis of potential pathways in initially inoperable patients with LAESCC group. (c) Heatmap of metabolome and microbiome in initially inoperable patients with LAESCC group.
Integrated correlation analysis of pCR and candidate microbes, immune cells and immunomodulatory cytokines in initially inoperable patients with LAESCC during chemoradiotherapy
A significant association was found between potential biomarkers (Prevotella_salivae, Saccharibacteria_TM7_G3_bacterium_HMT_351, and Veillonellaceae_G1_bacterium_HMT_129) and pCR in initially inoperable patients who underwent surgery (Figure 5a-c). To further investigate the dynamic changes among the candidate microbes, immune cells and immunomodulatory cytokines were selected for comparison at baseline and after the chemoradiotherapy. There was an increasing trend of CD3+CD8 T cells, IL6, IL10, and IFNγ during the treatment process of chemoradiotherapy (Figure 5d-l, *<0.05, **<0.01, ***<0.001). It suggested that the potential microbes may be associated with CD3+CD8 T cells, IL6, IL10, and IFNγ in the pCR response of chemoradiotherapy.
Figure 5.
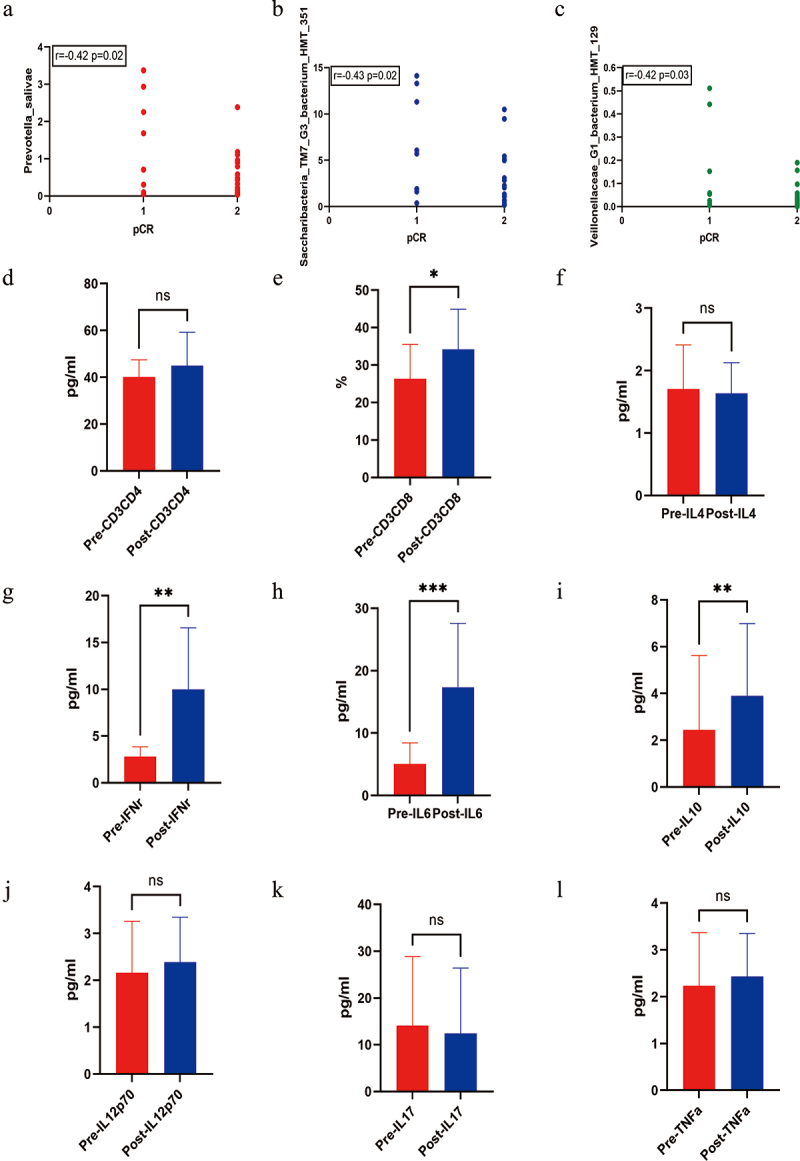
(a-b-c) Correlation of microbiome and pathological complete response (pCR) in initially inoperable patients with LAESCC. (d-l) Comparison analysis of immune cells and cytokines before and after chemoradiotherapy in patients with pCR (*<0.05, **<0.01, ***<0.001).
Discussion
Mutual interactions between the microbiome and chemoradiotherapy have been demonstrated to affect the complex mechanism between the microbiome and the host response [13]. The prediction of the efficacy of neoadjuvant chemoradiotherapy was misty, which deserves further investigation for initially inoperable LAESCC. This present study of the salivary microbiome and immunomodulators in blood samples revealed the characteristics of cancer versus normal controls, and pCR versus non-pCR groups. Microbiome signatures, combined with host immunomodulatory cells and cytokines, are correlated with chemoradiotherapy outcomes. The predictive functions of immune diseases, cell motility, lipid metabolism, signal transduction, and membrane transport are involved in cancer progression.
Numerous studies have suggested that the microbiome of the inner tumor or oral cavity is associated with cancer diagnosis, progression, survival benefits, and adverse effects. Some reported that the microbiome detection patterns were associated with the presence and severity of Barrett’s esophagus [21]. The Lachnospira, Bacteroides, Streptococcus, and Bifidobacterium achieved a high accuracy in esophageal cancer diagnosis [7]. An abundance of Porphyromonas (P.) gingivalis in the oral cavity is associated with an increased risk of esophageal cancer [22]. The oral and esophageal microbiomes are useful for distinguishing precancerous esophageal lesions from squamous cell carcinoma [23]. Oral microorganisms can cause esophageal cancer by inducing mild chronic inflammation and distal esophageal cancer [24]. The characteristics of the gut microbiome affect the severity of acute radiation-induced esophagitis [25]. Interestingly, the microbiome was more abundant in patients with leukemia undergoing radiotherapy with milder gastrointestinal dysfunction. The administration of propionate and tryptophan to mice caused long-term radioprotection, and reduced in pro-inflammatory responses [26]. The number of amplicons, but not alpha or beta diversity, in post-radiation samples with cervical cancer was significantly lower compared with the baseline samples [27]. Additionally, the differentially abundant microbiome inside tumor tissues has been identified as a critical marker of tumorigenesis in spatial and cellular heterogeneity in cancer [28]. Eubacterium_coprostanoligenes_group and Prevotella were independent predictors of disease-free survival. The predicting classifier for recurrence and metastasis risk yielded an area under the curve of 0.83 at 3 years and 0.86 at 5 years [29]. Consistent with previous studies, our study focused on patients with a large tumor burden. A general notion exists that some novel microbiome biomarkers are useful for initially inoperable patients with LAESCC. In addition, we assessed microbiome biomarkers in patients with pCR versus non-pCR who underwent surgery. The effects of pCR are similar to those of rectal and non-small cell lung cancers [8,9]. The abundance of the genera Fusobacterium, Parvimonas, and Dialister were enhanced in higher T classifications (T3–4) and advanced stages (IV) [30]. No similar trend was found in our study, which may be attributed to the various tumors and heterogeneity.
Chemotherapy influences the diversity, abundance, and treatment response of the oral microbiome [31]. Accumulating evidence supported not only the functional role of gut microbiome in cancer development and progression but also its role in defining the efficacy and toxicity of chemotherapeutic agents (5-fluorouracil [32], cyclophosphamide [33], irinotecan [34], gemcitabine [35], methotrexate [33]). The TIMER mechanism consists of Translocation, Immunomodulation, Metabolism, Enzymatic degradation, and Reduced diversity, which represent the strategies of interaction and toxicity during therapy, and how changes in the microbiome itself could influence the therapy response [36]. Several platinum-based treatment regimens can alter the dynamic composition of the microbiome [37]. Cisplatin inhibited the growth of 7 gram-negative, 8 gram-positive bacterial strains, 7 yeast strains, and 7 mold strains [38]. Cisplatin combined with cepharanthine hydrochloride increased the effect of chemotherapy and reduced the side effects on microbes and intestinal mucosal immunity in mice by activating TLR4/MYD88 immune signaling and TNFR death receptors [39]. Similar 16S rRNA gene sequencing and metabolomic analysis were found in a mouse model of C57BL/6 [40]. Carboplatin was also found to reduce the abundance of several taxa in patients with ovarian cancer [37]. The Fusobacterium nucleatum in cancer tissues promoted ESCC progression and chemoresistance via secretion of a chemotherapy-induced senescence-associated secretory phenotype [41]. Moreover, bacterial enzymes alleviated the drastic effects of irinotecan on cancer cells [42].
As for radiotherapy, it disrupts the microbiome and those products in turn influence the effectiveness of anticancer therapies [43]. Radiotherapy reshaped the tumor microenvironment via the microbiome, which consists of anti-inflammatory and pro-inflammatory cells and cytokines. Oral probiotics, prebiotics, and drug interventions could repair functions and reshape the tumor microenvironment [19]. Radiotherapy changed the salivary proteome before, during, and after treatment of head and neck cancer by 35-fold alpha-enolase. Compared to healthy volunteers, salivary proteins before radiotherapy showed elevated levels of cystatin-C, lysozyme C, histatin-1, and proline-rich proteins [44]. The relative abundance of Lactobacillaceae in fecal samples showed a partial or complete response in patients with ESCC receiving chemoradiotherapy [45]. In mouse models of breast cancer and melanoma, intestinal fungi exhibit antitumor immune responses following radiation, whereas, bacteria exhibit the opposite responses. Furthermore, elevated intratumoral Dectin-1 is negatively associated with survival in patients with breast cancer, and is required for the radiation effects of commensal fungi in mouse models. Depletion of bacteria with antibiotics reduces the efficacy of oxaliplatin and cyclophosphamide by reducing myeloid and T-helper 17 responses. The Akkermansia and Bifidobacterium species are also critical for the response to immunotherapies targeting programmed cell death protein 1 (PD-1)/programmed death-ligand 1 and cytotoxic T lymphocyte associated protein 4 [46]. In our study, chemoradiotherapy did not affect the dynamic changes in microbiomes during treatment, but some metabolites changed obviously. This finding implies that a detailed functional mechanism maybe involved in the metabolites, which needs to be further verified.
Generally, the inflammation, immune responses, microbial components, and toxic products are the main mechanisms promoting the malignant transformation of the esophagus by the microbiome [47]. This present study found that baseline levels of Prevotella_salivae, Saccharibacteria_TM7_G3_bacterium_HMT_351, and Veillonellaceae_G1_bacterium_HMT_129 were positively associated with pCR. Meanwhile, the CD3+CD8 T cells, IL6, IL10, and IFNγ were obviously changed in patients with pCR. It suggested that some particular microbial species with some immunoregulatory cells and cytokines (CD3+CD8 T cells, IL6, IL10, and IFNγ) could be useful for chemoradiotherapy in patients with LAESCC. Changes in the gut microbiome led to changes in glycerophospholipid metabolism level, which may affect the expression of immune-related cytokines IFNγ and IL2 but not CD4/8 in the tumor microenvironment, resulting in a different therapeutic effect of PD-1 antibody in MSS-type colorectal tumor-bearing mice [48,49]. The activation of the LPS-TLR4-NF-κB pathway of gastrointestinal microbiota may contribute to inflammation and malignant transformation [50]. Dendritic cell antigen presentation was modulated by the gut microbiota via a radiotherapy-induced antitumor immune response (CD8+). Vancomycin potentiated the radiotherapy-induced antitumor immune response and tumor growth inhibition dependent on cytolytic CD8+ T cell/IFNγ elicitation and the abscopal effect [25,51].
Several limitations should be further analyzed and discussed. First, the number of salivary and serum samples was small without sufficient dynamic monitoring, which deserves further verification before, during and after treatment. A better model should be verified in a larger number of patients. Second, long-term survival outcomes including event-free and overall survival need to be further explored. Third, the number of immune cell markers in the patients’ blood samples was insufficient to evaluate the total expression of specific immune subsets. Besides, the taxonomic scope of fungi was not included in the present analysis. Finally, some animal experiments and cell cultures are warranted to determine the detailed mechanisms of certain special microbiomes. Despite these limitations, this study provided some useful information for future research.
Conclusions
Our study demonstrated that variations in specific saliva taxa associated with host immunomodulatory cells and cytokines could be promising for the early efficacy prediction of chemoradiotherapy in initially inoperable patients with LAESCC.
Acknowledgments
We are grateful to thank those patients who are suffering from cancer for providing the kind help.
Funding Statement
This study was supported by the National Natural Science Foundation of China [82102821], WU JIEPING Medical Foundation [320.6750.2023-05-100], and Health Research Project of Anhui Province [AHWJ2023BAc20038].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Authors’ contributions
The authors take responsibility for the accuracy and completeness of the data and analyses. HY, LXY and QD had full access to all of the data in the study and took responsibility for the integrity of the data and the accuracy of the data analysis. Study design: HY, LXY and QD; Statistical analysis: HY, LXY; Manuscript draft: HY, LXY. Other authors have participated in data collection. All authors have seen and approved the manuscript.
Availability of data and materials
The datasets analyzed in the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was in accordance with the Declaration of Helsinki. The study protocol was approved by the Ethics Committee of the first affiliated hospital affiliated to University of Science and Technology of China(2023-ky202). Informed consent was obtained from all subjects or their legal guardian.
References
- [1].Gao L, Xu T, Huang G, et al. Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell. 2018;9(5):488–13. doi: 10.1007/s13238-018-0548-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Song X, Greiner-Tollersrud OK, Zhou H.. Oral microbiota variation: a risk factor for development and poor prognosis of esophageal cancer. Dig Dis Sci. 2022;67(8):3543–3556. doi: 10.1007/s10620-021-07245-2 [DOI] [PubMed] [Google Scholar]
- [3].Kahharova D, Pappalardo V, Buijs MJ, et al. Microbial indicators of dental health, dysbiosis, and early childhood caries. J Dent Res. 2023;102(7):759–766. doi: 10.1177/00220345231160756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhang X, Li X, Xu H, et al. Changes in the oral and nasal microbiota in pediatric obstructive sleep apnea. J Oral Microbiol. 2023;15(1):2182571. doi: 10.1080/20002297.2023.2182571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kageyama S, Takeshita T, Takeuchi K, et al. Characteristics of the salivary microbiota in patients with various digestive tract cancers. Front Microbiol. 2019;10:1780. doi: 10.3389/fmicb.2019.01780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dan W, Peng L, Yan B, et al. Human microbiota in esophageal adenocarcinoma: pathogenesis, diagnosis, prognosis and therapeutic implications. Front Microbiol. 2021;12:791274. doi: 10.3389/fmicb.2021.791274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Deng Y, Tang D, Hou P, et al. Dysbiosis of gut microbiota in patients with esophageal cancer. Microb Pathog. 2021;150:104709. doi: 10.1016/j.micpath.2020.104709 [DOI] [PubMed] [Google Scholar]
- [8].Yi Y, Shen L, Shi W, et al. Gut microbiome components predict response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer patients: a prospective, longitudinal study. Clin Cancer Res. 2021;27(5):1329–1340. doi: 10.1158/1078-0432.CCR-20-3445 [DOI] [PubMed] [Google Scholar]
- [9].Qiu B, Xi Y, Liu F, et al. Gut microbiome is associated with the response to chemoradiotherapy in patients with non-small cell lung cancer. Int J Radiat Oncol Biol Phys. 2023;115(2):407–418. doi: 10.1016/j.ijrobp.2022.07.032 [DOI] [PubMed] [Google Scholar]
- [10].Oliva M, Schneeberger P, Rey V, et al. Transitions in oral and gut microbiome of HPV+ oropharyngeal squamous cell carcinoma following definitive chemoradiotherapy (ROMA LA-OPSCC study). Br j cancer. 2021;124(9):1543–1551. doi: 10.1038/s41416-020-01253-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Medeiros M, The S, Bellile E, et al. Salivary microbiome changes distinguish response to chemoradiotherapy in patients with oral cancer. Microbiome. 2023;11(1):11. doi: 10.1186/s40168-023-01677-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Matson V, Chervin C, Gajewski TF. Cancer and the microbiome—influence of the commensal microbiota on cancer, immune responses, and immunotherapy. Gastroenterology. 2021;160(2):600–613. doi: 10.1053/j.gastro.2020.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Belstrom D. The salivary microbiota in health and disease. J Oral Microbiol. 2020;12(1):1723975. doi: 10.1080/20002297.2020.1723975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhu H, Ma X, Ye T, et al. Esophageal cancer in China: practice and research in the new era. Int J Cancer. 2023;152(9):1741–1751. doi: 10.1002/ijc.34301 [DOI] [PubMed] [Google Scholar]
- [15].Moody S, Senkin S, Islam SMA, et al. Mutational signatures in esophageal squamous cell carcinoma from eight countries with varying incidence. Nat Genet. 2021;53(11):1553–1563. doi: 10.1038/s41588-021-00928-6 [DOI] [PubMed] [Google Scholar]
- [16].An L, Li M, Jia Q. Mechanisms of radiotherapy resistance and radiosensitization strategies for esophageal squamous cell carcinoma. Mol Cancer. 2023;22(1):22. doi: 10.1186/s12943-023-01839-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sugimura K, Miyata H, Tanaka K, et al. Multicenter randomized phase 2 trial comparing chemoradiotherapy and docetaxel plus 5-fluorouracil and cisplatin chemotherapy as initial induction therapy for subsequent conversion surgery in patients with clinical T4b esophageal cancer. Ann Surg. 2021;274(6):e465–e472. doi: 10.1097/SLA.0000000000004564 [DOI] [PubMed] [Google Scholar]
- [18].Buckstein MH, Anker CJ, Chuong MD, et al. CROSSing into new therapies for esophageal cancer. Int J Radiat Oncol Biol Phys. 2022;113(1):5–10. doi: 10.1016/j.ijrobp.2021.12.177 [DOI] [PubMed] [Google Scholar]
- [19].Liu J, Liu C, Yue J. Radiotherapy and the gut microbiome: facts and fiction. Radiat Oncol. 2021;16(1):9. doi: 10.1186/s13014-020-01735-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhou CB, Zhou Y, Fang JY. Gut microbiota in cancer immune response and immunotherapy. Trends Cancer. 2021;7(7):647–660. doi: 10.1016/j.trecan.2021.01.010 [DOI] [PubMed] [Google Scholar]
- [21].Okereke IC, Miller AL, Jupiter DC, et al. Microbiota detection patterns correlate with presence and severity of Barrett’s esophagus. Front Cell Infect Microbiol. 2021;11:555072. doi: 10.3389/fcimb.2021.555072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Peters BA, Wu J, Pei Z, et al. Oral microbiome composition reflects prospective risk for esophageal cancers. Cancer Res. 2017;77:6777–6787. doi: 10.1158/0008-5472.CAN-17-1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li Z, Dou L, Zhang Y, et al. Characterization of the oral and esophageal microbiota in esophageal precancerous lesions and squamous cell carcinoma. Front Cell Infect Microbiol. 2021;11:714162. doi: 10.3389/fcimb.2021.714162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li X, Zhu S, Zhang T, et al. Association between oral microflora and gastrointestinal tumors (Review). Oncol Rep. 2021;46(2). doi: 10.3892/or.2021.8111 [DOI] [PubMed] [Google Scholar]
- [25].Lin MQ, Wu YH, Yang J, et al. Gut microbiota characteristics are associated with severity of acute radiation-induced esophagitis. Front Microbiol. 2022;13:883650. doi: 10.3389/fmicb.2022.883650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Guo H, Chou WC, Lai Y, et al. Multi-omics analyses of radiation survivors identify radioprotective microbes and metabolites. Science. 2020;370(6516):370. doi: 10.1126/science.aay9097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tsakmaklis A, Vehreschild M, Farowski F, et al. Changes in the cervical microbiota of cervical cancer patients after primary radio-chemotherapy. Int J Gynecol Cancer. 2020;30(9):1326–1330. doi: 10.1136/ijgc-2019-000801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Galeano Nino JL, Wu H, LaCourse KD, et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature. 2022;611(7937):810–817. doi: 10.1038/s41586-022-05435-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yuan X, Lau HC, Shen Y, et al. Tumour microbiota structure predicts hypopharyngeal carcinoma recurrence and metastasis. J Oral Microbiol. 2023;15(1):2146378. doi: 10.1080/20002297.2022.2146378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lau HC, Shen Y, Huang H, et al. Cross-comparison of microbiota in the oropharynx, hypopharyngeal squamous cell carcinoma and their adjacent tissues through quantitative microbiome profiling. J Oral Microbiol. 2022;14(1):2073860. doi: 10.1080/20002297.2022.2073860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hong BY, Sobue T, Choquette L, et al. Chemotherapy-induced oral mucositis is associated with detrimental bacterial dysbiosis. Microbiome. 2019;7(1):66. doi: 10.1186/s40168-019-0679-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].LaCourse KD, Zepeda-Rivera M, Kempchinsky AG, et al. The cancer chemotherapeutic 5-fluorouracil is a potent Fusobacterium nucleatum inhibitor and its activity is modified by intratumoral microbiota. Cell Reports. 2022;41(7):111625. doi: 10.1016/j.celrep.2022.111625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Corley C, McElroy T, Sridharan B, et al. Physiological and cognitive changes after treatments of cyclophosphamide, methotrexate, and fluorouracil: implications of the gut microbiome and depressive-like behavior. Front Neurosci. 2023;17:1212791. doi: 10.3389/fnins.2023.1212791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Parvez MM, Basit A, Jariwala PB, et al. Quantitative investigation of irinotecan metabolism, transport, and gut microbiome activation. Drug Metab Dispos. 2021;49(8):683–693. doi: 10.1124/dmd.121.000476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Miyake M, Oda Y, Owari T, et al. Probiotics enhances anti‐tumor immune response induced by gemcitabine plus cisplatin chemotherapy for urothelial cancer. Cancer Sci. 2023;114(3):1118–1130. doi: 10.1111/cas.15666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chrysostomou D, Roberts L, Marchesi JR, et al. Gut microbiota modulation of efficacy and toxicity of cancer chemotherapy and immunotherapy. Gastroenterology. 2023;164(2):198–213. doi: 10.1053/j.gastro.2022.10.018 [DOI] [PubMed] [Google Scholar]
- [37].Chambers LM, Esakov Rhoades E, Bharti R, et al. Disruption of the gut microbiota confers cisplatin resistance in epithelial ovarian cancer. Cancer Research. 2022;82(24):4654–4669. doi: 10.1158/0008-5472.CAN-22-0455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gong S, Feng Y, Zeng Y, et al. Gut microbiota accelerates cisplatin-induced acute liver injury associated with robust inflammation and oxidative stress in mice. J Transl Med. 2021;19(1):147. doi: 10.1186/s12967-021-02814-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhou P, Li Z, Xu D, et al. Cepharanthine hydrochloride improves cisplatin chemotherapy and enhances immunity by regulating intestinal microbes in mice. Front Cell Infect Microbiol. 2019;9:225. doi: 10.3389/fcimb.2019.00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhenya Zhai JL, Niu K-M, Lin C, et al. Integrated metagenomics and metabolomics to reveal the effects of policosanol on modulating the gut microbiota and lipid metabolism in hyperlipidemic C57BL/6 mice. Front Endocrinol. 2021;12:722055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhang JW, Zhang D, Yin HS, et al. Fusobacterium nucleatum promotes esophageal squamous cell carcinoma progression and chemoresistance by enhancing the secretion of chemotherapy-induced senescence-associated secretory phenotype via activation of DNA damage response pathway. Gut Microbes. 2023;15(1):2197836. doi: 10.1080/19490976.2023.2197836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wallace BD, Wang H, Lane KT, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330(6005):831–835. doi: 10.1126/science.1191175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Danckaert W, Spaas M, Sundahl N, et al. Microbiome and metabolome dynamics during radiotherapy for prostate cancer. Radiotherapy And Oncology. 2023;189:109950. doi: 10.1016/j.radonc.2023.109950 [DOI] [PubMed] [Google Scholar]
- [44].Ventura TMO, Ribeiro NR, Taira EA, et al. Radiotherapy changes the salivary proteome in head and neck cancer patients: evaluation before, during, and after treatment. Clin Oral Investig. 2022;26(1):225–258. doi: 10.1007/s00784-021-03995-5 [DOI] [PubMed] [Google Scholar]
- [45].Sasaki T, Matsumoto Y, Murakami K, et al. Gut microbiome can predict chemoradiotherapy efficacy in patients with esophageal squamous cell carcinoma. Esophagus. 2023;20(4):691–703. doi: 10.1007/s10388-023-01004-0 [DOI] [PubMed] [Google Scholar]
- [46].Shiao SL, Kershaw KM, Limon JJ, et al. Commensal bacteria and fungi differentially regulate tumor responses to radiation therapy. Cancer Cell. 2021;39(9):1202–1213.e6. doi: 10.1016/j.ccell.2021.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zhou J, Sun S, Luan S, et al. Gut microbiota for esophageal cancer: role in carcinogenesis and clinical implications. Front Oncol. 2021;11:717242. doi: 10.3389/fonc.2021.717242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Xu X, Lv J, Guo F, et al. Gut microbiome influences the efficacy of pd-1 antibody immunotherapy on MSS-Type colorectal cancer via metabolic pathway. Front Microbiol. 2020;11:814. doi: 10.3389/fmicb.2020.00814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Roviello G, Iannone L, Bersanelli M, et al. The gut microbiome and efficacy of cancer immunotherapy. Pharmacology & Therapeutics. 2022;231:107973. doi: 10.1016/j.pharmthera.2021.107973 [DOI] [PubMed] [Google Scholar]
- [50].Lv J, Guo L, Liu JJ, et al. Alteration of the esophageal microbiota in Barrett’s esophagus and esophageal adenocarcinoma. World J Gastroenterol. 2019;25(18):2149–2161. doi: 10.3748/wjg.v25.i18.2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Uribe-Herranz M, Rafail S, Beghi S, et al. Gut microbiota modulate dendritic cell antigen presentation and radiotherapy-induced antitumor immune response. J Clin Invest. 2020;130(1):466–479. doi: 10.1172/JCI124332 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed in the current study are available from the corresponding author on reasonable request.


