Abstract
The gene region coding for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17 is located on a 13-kb insert of plasmid pEG12. Upstream of the previously described six open reading frames (ORFs) soxABCDEF with a partial sequence of soxA and soxF (C. Wodara, F. Bardischewsky, and C. G. Friedrich, J. Bacteriol. 179:5014–5023, 1997), 4,350 bp were sequenced. The sequence completed soxA, and uncovered six new ORFs upstream of soxA, designated ORF1, ORF2, and ORF3, and soxXYZ. ORF1 could encode a 275-amino-acid polypeptide of 29,332 Da with a 61 to 63% similarity to LysR transcriptional regulators. ORF2 could encode a 245-amino-acid polypeptide of 26,022 Da with the potential to form six transmembrane helices and with a 48 to 51% similarity to proteins involved in redox transport in cytochrome c biogenesis. ORF3 could encode a periplasmic polypeptide of 186 amino acids of 20,638 Da with a similarity to thioredoxin-like proteins and with a putative signal peptide of 21 amino acids. Purified SoxXA, SoxYZ, and SoxB are essential for thiosulfate or sulfite-dependent cytochrome c reduction in vitro. N-terminal and internal amino acid sequences identified SoxX, SoxY, SoxZ, and SoxA to be coded by the respective genes. The molecular masses of the mature proteins determined by electrospray ionization spectroscopy (SoxX, 14,834 Da; SoxY, 11,094 Da; SoxZ, 11,717 Da; and SoxA, 30,452 Da) were identical or close to those deduced from the nucleotide sequence with differences for the covalent heme moieties. SoxXA represents a novel type of periplasmic c-type cytochromes, with SoxX as a monoheme and SoxA as a hybrid diheme cytochrome c. SoxYZ is an as-yet-unprecedented soluble protein. SoxY has a putative signal peptide with a twin arginine motif and possibly cotransports SoxZ to the periplasm. SoxYZ neither contains a metal nor a complex redox center, as proposed for proteins likely to be transported via the Tat system.
The mechanism of the oxidation of sulfur or hydrogen sulfide to sulfuric acid, a major reaction of the global sulfur cycle, is unknown. To uncover the mechanism of this reaction and the proteins involved, we selected a genetically accessible strain of a thiobacterium. Strains of the genus Paracoccus are facultatively lithoautotrophic, neutrophilic bacteria able to grow with various organic compounds and inorganic electron donors such as molecular hydrogen or thiosulfate for autotrophic carbon dioxide fixation (reviewed in reference 16). Paracoccus pantotrophus isolated as Thiosphaera pantotropha (38) is accessible to gene transfer via conjugation (10). The strain was reclassified as Paracoccus denitrificans (30), and recently it was suggested to divide the species of Paracoccus denitrificans beyond the criteria of Wayne et al. (50) into P. denitrificans and P. pantotrophus. The former type strain P. denitrificansT was transferred to P. pantotrophus with the former Thiosphaera pantotropha as type strain P. pantotrophusT (37).
The only gene region known so far coding for lithotrophic sulfur oxidation was localized by transposon Tn5 mutagenesis in P. pantotrophus GB17 (10). The wild-type gene region has been cloned on a 13-kb EcoRI/SalI fragment, sequenced, and further analyzed. Six open reading frames (ORFs), designated soxABCDEF, were identified, of which soxA and soxF were incomplete (31, 54, 56). The soxB gene product predicts a protein of a molecular mass of 61,897 Da which is 20.7% identical to 5′-nucleotidase of Escherichia coli (56). The adjacent soxCD genes code for a new type of heterotetrameric sulfite dehydrogenase, consisting of two SoxC and two diheme-carrying SoxD subunits (36, 56). soxE predicts a diheme cytochrome with a molecular mass of 25,926 Da. The partial soxF predicts a polypeptide of 247 amino acid residues with a high identity of 47.4% to a flavoprotein of Chromatium vinosum, which is considered to be involved in hydrogen sulfide metabolism (17) but not to be essential for phototrophic growth with hydrogen sulfide (11).
The mechanism of oxidation of sulfur or hydrogen sulfide to sulfite is not known. However, various enzymes have been characterized from phototrophic or lithotrophic bacteria that are able to oxidize, for example, hydrogen sulfide to sulfur or thiosulfate to tetrathionate or to hydrolyze polythionates (16, 22). From the neutrophilic thiobacterium Paracoccus versutus (formerly Thiobacillus versutus [49]), a close relative of P. pantotrophus (20), the thiosulfate-oxidizing enzyme system has been characterized biochemically as reviewed by Kelly et al. (21, 22). It is located in the periplasm (27) and is composed of four periplasmic proteins: enzyme A (16,000 Da), enzyme B (63,000 Da), cytochrome c552.5 (56,000 Da, composed of identical subunits of 29,000 Da), and cytochrome c551 (260,000 Da), composed of six subunits (43,000 Da). P. versutus sulfite dehydrogenase (44,000 Da) is intimately associated with cytochrome c551 and has been described as not being required for thiosulfate oxidation in vitro, though it stabilizes the reconstituted thiosulfate-oxidizing system.
Biochemical studies with cell extracts of P. pantotrophus GB17 demonstrated that four fractions of proteins eluted from Q Sepharose are required to reconstitute thiosulfate and sulfite-oxidizing activity in vitro using horse heart cytochrome c as the electron acceptor. SoxC (43,442 Da) was identified as a novel type of molybdenum cofactor containing sulfite dehydrogenase which did not carry a b5-type heme on the same polypeptide but which was associated with SoxD (37,637 Da), a diheme c-type cytochrome. SoxCD was isolated as an α2β2 heterotetramer present as one component required for thiosulfate-oxidizing activity (36).
In this study we complete the nucleotide sequence of soxA, identify other genes of the sox gene region, and identify the function of the gene products involved in sulfur-oxidizing ability in vitro. We here describe six new ORFs designated ORF1, ORF2, and ORF3 and the soxXYZ 5′ of soxA. Of the sox gene products, we purified three proteins SoxXA, SoxYZ, and SoxB which are, in addition to SoxCD, essential for thiosulfate-dependent cytochrome c reduction. From N-terminal and internal amino acid sequences we have verified that SoxXA and SoxYZ are encoded by soxX, soxA, soxY, and soxZ, and we have obtained evidence that these proteins are located in the periplasm of P. pantotrophus GB17.
MATERIALS AND METHODS
Medium and growth conditions.
P. pantotrophus strain GB17T, which is identical to strain LMD82.5 (38), was used throughout this study. P. pantotrophus was cultivated at 30°C. Seed cultures were cultivated mixotrophically in mineral medium, pH 8.0 (10), with 40 mM sodium thiosulfate and 20 mM disodium succinate. Lithotrophic mass cultivation was performed in a 300-liter fermentor (Bioengineering, Wald, Switzerland) with a 220-liter working volume. Cells were harvested by cross-flow filtration and stored at −20°C as described previously (36).
DNA techniques.
Standard DNA techniques (40) were used. Plasmid DNA was isolated using the high pure plasmid isolation kit (Boehringer Mannheim) according to the manufacturer's protocol. DNA sequencing was performed by primer walking with the thermostable DNA polymerase of Thermus aquaticus and 7-deaza-dGTP (Amersham-Buchler, Braunschweig, Germany) by the dideoxy-chain termination method (41) using fluorescent primers and an automated DNA sequencing system (Li-Cor; MWG-Biotech, Munich, Germany).
The nucleotide sequence was analyzed with the BLAST search program package (1). Deduced polypeptides were analyzed by PC/GENE (Intelligenetics, Inc., Mountainview, Calif.) and PROSIS (Hitachi Software Engineering, San Bruno, Calif.). Transmembrane regions and protein orientations were predicted by TMpred (18). Signal peptides and protein localization was predicted by the PSORT program package (32). DNA-binding helix-turn-helix motifs of deduced proteins were predicted according to the Pfam multiple sequence alignments (3, 43, 44).
Preparation of cell extracts.
Thawed cells were resuspended in 55 mM potassium phosphate buffer, pH 7.5, at a ratio of 1 g (wet weight) plus 1.5 ml of buffer. About 0.2 mg of DNase I was added to 60 ml of cell suspension. Cells were disrupted by using a French press, the resulting cell extract was subjected to differential centrifugation as described earlier, and the 200,000 × g supernatant was referred to as crude extract (36). Proteins were purified at 0 to 4°C. Cell extracts were brought to different degrees of ammonium sulfate saturation by dropwise addition of a saturated solution at 0°C. Crude extract (38 ml) was brought to 44% saturation of ammonium sulfate. The supernatant of the precipitate was brought to 65% saturation, and the pellet was resuspended, dialyzed as described, yielded a final volume of 25 ml, and designated the A65% fraction (36). This fraction was applied to a Q Sepharose Fast Flow column (diameter, 2.6 cm; length, 4.0 cm). The column was equilibrated with 25 mM sodium potassium phosphate buffer, pH 6.5, containing 2 mM sodium thiosulfate, 1 mM magnesium sulfate, and 1 μM phenylmethylsulfonyl fluoride, which was designated the stabilizing buffer. Protein was eluted from the column with a step gradient of 400 ml for each step at 0.05, 0.10, 0.15, 0.20, 0.25, 0.30, 0.35, or 0.40 M sodium chloride in the same buffer at a flow rate of 5.0 ml/min. Fractions of 20 ml were collected. Eluted protein was monitored at 280 nm. Peak fractions were collected from each sodium chloride step gradient.
SoxYZ purification.
SoxYZ was eluted mainly at 0 M and partly at 0.05 M sodium chloride. The 0 M sodium chloride eluate (100 ml) was concentrated to 20 ml by ultrafiltration using an Amicon PM10 membrane. The extract was brought to 30% saturation of ammonium sulfate. Precipitated proteins were removed by centrifugation. The supernatant (28.5 ml) was applied to a Phenylsepharose HP-column (diameter, 1.6 cm; length, 4 cm) equilibrated against stabilizing buffer saturated with ammonium sulfate to 30%. Protein was eluted with 240 ml of a linear gradient of 30 to 0% ammonium sulfate at a flow rate of 3 ml/min. Fractions of 4 ml were collected and examined for thiosulfate-reconstituting activity and by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). SoxYZ containing fractions were pooled, dialyzed against 2 liters of stabilizing buffer for 4 h, and concentrated by Amicon ultrafiltration. The final preparation (16 ml) contained 0.90 mg of homogeneous SoxYZ/ml, as judged by SDS-PAGE.
SoxB purification.
SoxB was eluted from the Q Sepharose column at 0.25 M sodium chloride. The pool fractions (100 ml) were concentrated to 4 ml by Amicon ultrafiltration, subjected to gel filtration using Sephacryl S-200 HR (column diameter, 1.6 cm; length, 60 cm), and equilibrated with stabilizing buffer, and the protein was eluted with the same buffer at a flow rate of 45 ml/h. The SoxB-containing fractions were pooled (10 ml) and brought to 30% saturation of ammonium sulfate, and the protein was applied on a Phenylsepharose column (diameter, 1.6 cm; length, 4 cm) equilibrated against stabilizing buffer being 30% saturated with ammonium sulfate. Protein was eluted with 240 ml of a linear gradient of 30 to 0% ammonium sulfate at a flow rate of 3 ml/min. Fractions of 4 ml were collected and examined for the thiosulfate-reconstituting activity and by SDS-PAGE. SoxB-containing fractions were pooled (18 ml) and contained 0.35 mg of SoxB/ml with a purity of 95%, as determined by densitometric analysis from SDS-polyacrylamide gels.
SoxXA purification.
Cytochrome SoxXA was eluted from Q Sepharose at 0.30 M sodium chloride. The eluate (100 ml) was concentrated to 4 ml by Amicon ultrafiltration using a PM10 membrane and was subjected to gel filtration by Sephacryl S-200 HR (column diameter, 1.6 cm; length, 60 cm) at a flow rate of 45 ml/h. The column was equilibrated, and the protein was eluted with stabilizing buffer. Fractions of 2.3 ml were collected and examined for SoxXA activity as described above. Moreover, the absorption of the heme chromophore was monitored at 416 nm. Fractions 33 to 35 were pooled, and 7 ml was applied to a Resource Q column (total volume, 6 ml), equilibrated against bis(2-hydroxy-ethyl)iminotris(hydroxmethyl)methane (Bis-Tris buffer; pH 6.0) instead of phosphate buffer but containing the additions described above for the stabilizing buffer. Protein was eluted from the Resource Q column by using 400 ml of a linear 0 to 0.50 M sodium chloride gradient at a flow rate of 5 ml/min, and fractions of 4.5 ml were collected. The SoxXA-containing fractions were pooled (13.5 ml) and dialyzed against 2 liters of stabilizing buffer for 4 h.
SoxCD purification.
Sulfite dehydrogenase was purified from the 0.35 M sodium chloride eluate to homogeneity as described previously (36), which included the same steps as for purification of SoxXA.
Enzyme assays.
The assay (1.0 ml) for determination of the thiosulfate-dependent cytochrome c reduction rate contained 2.0 μmol of disodium thiosulfate, 50 μmol of sodium-potassium phosphate buffer (pH 7.5), 35 nmol of horse heart cytochrome c, and extract. When the A65% fraction was assayed for enzyme activity, 25 μl of extract (32.2 mg of protein/ml) was used. After chromatography on Q Sepharose, 0.10 ml of the pools of the 0, 0.05, 0.25, 0.30, and 0.35 M sodium chloride pool fractions were added to the assay. When pure proteins were assayed for enzyme activity, 0.50 nmol (each) of SoxB, SoxCD, SoxXA, and SoxYZ was added to the assay mixture unless otherwise stated.
The assay (1.0 ml) for determination of the hydrogen sulfide-dependent cytochrome c reduction rate contained 10 nmol of sodium sulfide, 50 mM sodium-potassium phosphate buffer (pH 6.0), and 35 nmol of horse heart cytochrome c. The pure proteins were washed twice by concentrating each preparation 20-fold, diluting them with stabilizing buffer (but without sodium thiosulfate), concentrating them, and diluting them again. The proteins were added to the assay at concentrations described for the determination of the thiosulfate-oxidizing activity.
For the determination of the sulfite-dependent cytochrome c reduction rate, the assay was performed with 100 μmol of Tris-HCl (pH 7.5) and otherwise contained the same components as described above except that hydrogen sulfide was omitted and 100 nmol of sodium sulfite was added.
One unit of enzyme activity was defined as the reduction of one μmol of horse heart cytochrome c 550 per min at 30°C, as determined by using a molar extinction coefficient of 27.8 cm2/mol. The protein content was determined according to the method of Bradford (8).
Protein techniques.
Immunoblot (Western) analysis (48) was performed according to the “semidry” procedure using the multiphor electrophoretic system (Pharmacia, Freiburg, Germany) with antibodies raised from rabbits. SoxB antigens were detected with antibodies raised against SoxB heterologously expressed in E. coli and purified under denaturing conditions with 6 M guanidine hydrochloride by metal chelate affinity chromatography as described previously (54, 55). SoxZ was detected with antibodies raised against the oligopeptide HKMESGQRDADGKLI (OP-Z) comprising a highly immunogenic epitope deduced from the SoxZ nucleotide sequence. For detection of sulfite dehydrogenase, antibodies against the denatured SoxCD protein were obtained as described previously (36).
The molecular masses of native proteins were determined by nondenaturing PAGE using a linear gradient of 5 to 27.5% polyacrylamide according to the method of Andersson et al. (2).
The molecular masses of denatured proteins were determined by SDS-PAGE according to the method of Laemmli (26, 51). Nondenatured or denatured proteins were stained with Coomassie blue as described earlier (48). Proteins were quantified from polyacrylamide gels by laser densitometry (Ultroscan; Pharmacia, Freiburg, Germany).
N-terminal and internal amino acid sequences were determined from pure proteins by automated Edman degradation using a protein sequencer system model 494A/190A (Applied Biosystems, Foster City, Calif.) as detailed previously (13).
Analytical procedures.
Densitometric analysis of Coomassie blue-stained proteins separated by denaturing SDS-PAGE or nondenaturing gradient PAGE was done as described earlier (13). Determination of molecular masses of Sox proteins was performed by nano-electrospray mass spectrometry with a Finnigan LCQ mass spectrometer equipped with a micromanipulator for the correct positioning of the nanospray needle. The needles and the ion source were made according to the original design of the EMBL group (35). Peptides of tryptic digested SoxB and SoxYZ were determined by matrix-assisted laser desorption ionization (MALDI) at the facilities of the Max-Delbrück-Centrum, Berlin, Germany.
Heme moieties were quantified from pure SoxXA according to the pyridine hemochrome method of Berry and Trumpower (6). Heme staining of proteins from SDS-PAGE included reduction of the sample by mercaptoethanol but not heating. Heme staining was performed according to the protocol of Francis and Becker (14).
The metal content of homogeneous proteins was determined by total reflection X-ray fluorescence analysis as described previously (24). Proteins were diluted 1:1 with nitric acid and contained an internal standard of 5 μg of selenium/ml. The sample (10 μl) was applied to a quartz glass plate and analyzed for 400 s.
Fluorescence excitation and emission spectra were determined according to the principal procedure described elsewhere (19). Purified SoxYZ (10 μM; 0.45 ml) was treated with 50 μl of potassium iodide-iodine (2% [wt/vol]–1% [wt/vol]) acidified to pH 2.5 with 10 mM phosphoric acid and incubated at 95°C for 40 min. Precipitated protein was separated by centrifugation. To the clear supernatant, 2 M ammonium hydroxide was added in a ratio of 1:1 (vol/vol).
Nucleotide sequence accession number.
The nucleotide sequence was submitted to the EMBL data library under accession no. X79242 PDSOX.
RESULTS
Sequence analysis.
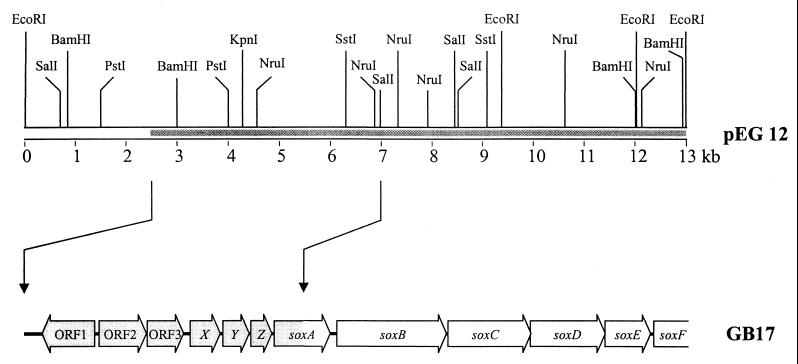
Previously, 6.2 kb of the 13-kb insert of pEG12 harboring the sox gene region of P. pantotrophus were sequenced. Sequence analysis uncovered the six ORFs soxABCDEF up to the end of the cloned DNA with a partial sequence of soxA and soxF (54, 56). The nucleotide sequence of 4,350 bp was determined from both strands 5′ of the SalI restriction site which had cleaved soxA (Fig. 1). Sequence analysis revealed coding characteristics according to codon preference analysis (45). The sequence completed soxA and uncovered six new ORFs upstream of soxA designated ORF1, ORF2, and ORF3 and soxXYZ (Fig. 2). Nucleotides 0 to 319 revealed no coding characteristics. Also, the nucleotide sequence of soxB was partially reexamined.
FIG. 1.
Physical map of plasmid pEG12 and sox-relevant ORFs.
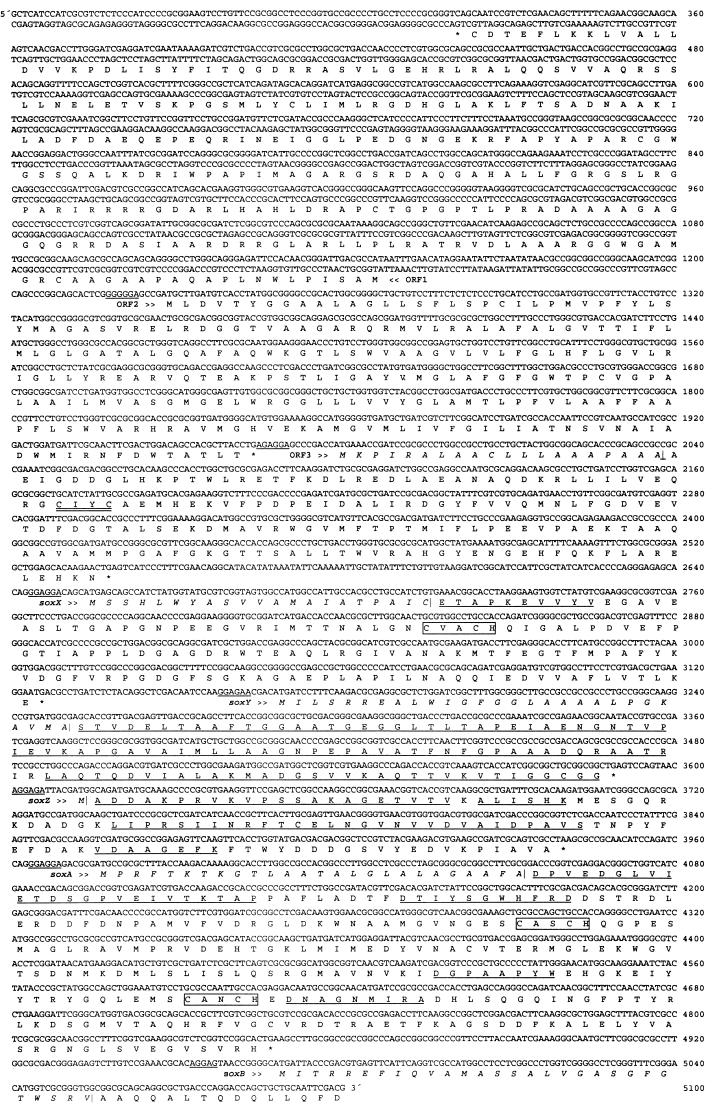
FIG. 2.
Nucleotide sequence and deduced amino acid sequence of the sox gene region of P. pantotrophus GB17. Underlined nucleotide sequences indicate proposed ribosome-binding sites; underlined amino acid sequences indicate sequences determined by Edman degradation; double-underlined amino acid sequence indicates the thioredoxin motif; and boxes indicate heme-binding motifs.
ORF1.
ORF1 was located from nucleotide 1147 to 323 on the 3′ strand and was oriented in the opposite direction to the other ORFs. ORF1 had the potential to encode a polypeptide of 275 amino acids (29,332 Da) with an alkaline pI of 10.59. Analysis of the deduced amino acid sequence did not indicate the presence of a leader peptide. The primary sequence of the putative ORF1 gene product showed identities of 42 to 43% in overlaps of 68 to 72 amino acids with members of the LysR family of transcriptional regulators such as HlyU of Vibrio cholerae and NolR of Sinorhizobium meliloti (Table 1). The identities related exclusively to the C-terminal ends of HlyU and NolR, which are significantly smaller than the putative ORF1 gene product with polypeptides of 100 to 130 amino acids. This observation may indicate the common characteristic for the DNA- and effector-binding domain or may point to different start codons at nucleotides 1081 and 769, resulting in putative polypeptides of 253 and 149 amino acid residues, respectively. Helix-turn-helix motifs are characteristic for DNA-binding proteins, and such a motif is predicted from nucleotides 571 to 339 according to the Pfam-A Prosite search and GenBank (3, 43, 44).
TABLE 1.
Amino acid sequence identities and similarities of Sox proteins to other proteins
| ORF and protein (no. of amino acids) | Organism | Function | % (aa overlap)a
|
Reference | |
|---|---|---|---|---|---|
| Identity | Similarity | ||||
| ORF1 (275) | |||||
| HLYU | Vibrio cholerae | Transcriptional activator | 43 (72) | 63 (72) | 53 |
| NOLR | Rhizobium leguminosarum | Repressor of nodulation | 42 (68) | 61 (68) | 25 |
| NOLR | Sinorhizobium meliloti | Transcriptional repressor | 42 (69) | 62 (69) | 25 |
| ORF2 (245) | |||||
| CCDA | Bacillus subtilis | Cytochrome c, biogenesis protein | 29 (230) | 48 (230) | 42 |
| CCDA | Treponema pallidum | dtob | 29 (224) | 48 (224) | 15 |
| CCDA | Helicobacter pylori | dto | 25 (222) | 51 (222) | 47 |
| DsbD | Escherichia coli | dto | 21 (211) | 34 (211) | 46 |
| ORF3 (186) | |||||
| aq1669 | Aquifex aeolicus | Putative protein | 34 (92) | 57 (92) | 12 |
| aq1343 | Aquifex aeolicus | dto | 29 (87) | 50 (87) | 12 |
| TRXA2 | Aquifex aeolicus | Thioredoxin | 27 (77) | 50 (77) | 12 |
| SoxX (157), PETJ | Heliobacillus mobilis | Cytochrome c553 | 30 (86) | 45 (86) | 57 |
| SoxY (140), aq1810 | Aquifex aeolicus | Putative protein | 40 (90) | 55 (90) | 12 |
| SoxZ (109), aq1809 | Aquifex aeolicus | Putative protein | 36 (110) | 47 (110) | 12 |
| SoxA (290) | |||||
| aq1807 | Aquifex aeolicus | Putative protein | 28 (224) | 43 (224) | 12 |
| NIRA | Pseudomonas aeruginosa | Nitrate reductase, chain A | 22 (102) | 43 (102) | 33 |
| Cytochrome c551 | Chlorobium limicola | Cytochrome c551 | 27 (257) | 57 (290) | 23 |
| SoxB (564), SoxB | Aquifex aeolicus | P. pantotrophus SoxB | 39 (584) | 59 (584) | 12 |
Numbers in parentheses indicate the amino acid (aa) residue overlap, i.e., the number of amino acid residues that overlapped.
dto, same function as above.
ORF2.
ORF2 was located from nucleotides 1228 to 1962 on the 5′ strand and was separated from ORF1 by 80 nucleotides with a G+C content of 58.8% ORF2 had the potential to encode a protein of 245 amino acids of 26,022 Da with a pI of 9.96. A putative ribosome-binding site was located five nucleotides 5′ of the start codon. The deduced amino acid sequence did not indicate the presence of a signal peptide (32, 49). The potential ORF2 gene product revealed highly hydrophobic regions with the potential to form six transmembraneous α-helices. The deduced primary structure revealed identities of 25 to 29% in an overlap of 222 to 230 amino acids to predicted transmembraneous proteins involved in cytochrome c biogenesis of Bacillus subtilis, Treponema pallidum, Helicobacter pylori, and E. coli (Table 1). Such involvement was suggested in the transport of reductant through the cytoplasmic membrane with a thioredoxin as redox mediator. For DsbD of E. coli the thioredoxin-like moiety is fused to a polypeptide predicted to form a transmembraneous channel (46).
ORF3.
ORF3 was located from nucleotides 1979 to 2536 and was separated from ORF2 by 16 nucleotides. ORF3 had the potential to encode a polypeptide of 186 amino acids with a molecular mass of 20,638 Da. A putative well-conserved ribosome-binding site was located nine nucleotides 5′ of the start codon. A closer analysis revealed a possible signal peptide of 21 amino acids (see Fig. 7). The putative cleavage site for the signal peptidase was in accordance with the “−1, −3 rule” (49) and suggested a periplasmic location of the mature ORF3 gene product (18,719 Da; pI 4.64). Within the deduced polypeptide a CxxC motif was located diagnostic for thioredoxin-like proteins. In fact, an amino acid sequence comparison revealed a 27 to 34% identity to a thioredoxin-like protein of Aquifex aeolicus and other putative gene products of unknown function of A. aeolicus (Table 1 [12]). ORF2, together with the putative periplasmic mature ORF3 gene product, may bring about the transport of reductant from the periplasm, a result comparable to the suggested function for DsbD of E. coli (46).
FIG. 7.
Predicted signal peptides of sox gene products. Superscript a, N terminus of the mature protein was verified by Edman degradation; superscript b, N terminus was blocked.
The stop codon of ORF3 was followed by a stretch of 117 nucleotides to the start codon of the subsequent soxX gene. The G+C content of this intervening sequence was 43.6% and differed significantly of the genomic content of P. pantotrophus GB17 of 68%. This sequence contained a short inverted repeat with a potential to form a hairpin structure with a free energy of formation of −8.6 kJ/mol.
soxX.
The soxX gene was located from nucleotides 2654 to 3124. soxX had the potential to encode a polypeptide of 157 amino acids with a molecular mass of 16,421 Da. A putative ribosome-binding site was located five nucleotides 5′ of the start codon. Analysis of the deduced amino acid sequence (32, 49) suggested the presence of a leader peptide of 21 amino acids (see Fig. 7) with a well-conserved cleavage site for the signal peptidase (Fig. 2). The predicted cleavage site of SoxX was verified from its N-terminal amino acid sequence from the purified mature protein (14,216 Da) as determined by Edman degradation (Table 2). A conserved putative CxxCH motif for covalent linkage of a heme moiety suggested this peptide to be a monoheme c-type cytochrome. A comparison of the deduced primary structure of SoxX with entries in the SWISSPROT and GenBank databases revealed only one significant relationship with an identity of 30% in an 86-amino-acid overlap to cytochrome c553 of Heliobacillus mobilis (Table 1).
TABLE 2.
Determination of amino acid sequences of N termini and internal peptides of Sox proteins of P. pantotrophus GB17a
| Organism and protein | Peptide or N terminusb | Amino acid sequence |
|---|---|---|
| P. pantotrophus GB17 | ||
| SoxX | N | ETAPKEVVYV |
| SoxY | N | STVDELTAAFTG |
| T1 | AATR | |
| T4 | AQTTVK | |
| T7 | MADGSVVK | |
| T19 | VTIGGCGG | |
| T39 | LAQTQDVIALAK | |
| T64 | STVDELTAAFTGGAATGEGGLTLTAPEIAENGNTVPIEVK | |
| T76 | APGAVAIMLLAAGNPEPAVATFNFGPAAADQR | |
| SoxZ | N | ADDAKPRVKVPSSAKA |
| T17 | AGETVTV | |
| T17 | VKVPSSA | |
| T18 | ALISHK | |
| T22 | LIPR | |
| T22 | SIINR | |
| T24 | VDAAGEFK | |
| T69 | FTXELNGVNVVXVAIXPAVS | |
| SoxA | N | DPVEDGLVIETDSGPVEIVTKTAP |
| F17 | DNAGNMIRA | |
| F35 | DGPAAPYW | |
| F42 | DTIYSGWHPR | |
| P. versutusSoxZ | N | ADDAKPPVVV |
The amino acid sequences of internal peptides of the Sox proteins were analyzed after tryptic digestion, except for SoxA, which was digested by aspartate–N-endopeptidase.
N, N terminus.
soxY.
The soxY gene was located from nucleotides 3168 to 3587 and was separated by 40 nucleotides from soxX. soxY had the potential to encode a polypeptide of 140 amino acids (molecular mass of 13,830 Da). A putative ribosome-binding site was located five nucleotides 5′ of the start codon (Fig. 2). Analysis of the deduced amino acid sequence (32, 49) suggested a leader peptide of 28 amino acids (see Fig. 7) with a twin arginine motif diagnostic for proteins with complex redox centers (5, 7, 52). Amino acid sequence analysis revealed no indication for a prosthetic group linked to SoxY (data not shown). Comparison of the deduced SoxY primary sequence revealed only one significant identity of 40% to the putative aq1810 gene product of A. aeolicus, whose function is unknown (Table 1 [12]).
soxZ.
The soxZ gene was located from nucleotides 3612 to 3938 and was separated by 24 nucleotides 3′ of soxY. soxZ had the potential to encode a polypeptide of 109 amino acids (molecular mass of 11,850 Da). A putative ribosome-binding site was located six nucleotides 5′ of the start codon. Sequence analysis of the deduced amino acid sequence did not indicate a signal peptide. Comparison of the deduced SoxZ amino acid sequence with entries in the SWISSPROT and GenBank databases revealed only a single significant identity of 36% in a 110-amino-acid overlap to the putative aq1809 gene product of A. aeolicus, whose function is unknown (12).
soxA.
The soxA gene was separated from soxZ by 37 nucleotides and extended from nucleotide 3976 to nucleotide 4845. soxA had the potential to encode a protein of 290 amino acids (molecular mass of 31,884 Da). A putative well-conserved ribosome-binding site was located seven nucleotides 5′ of the start codon. Analysis of the deduced amino acid sequence (49) suggested a leader peptide of 26 amino acids (Fig. 2; see also Fig. 7) to give a mature polypeptide of molecular mass of 29,353 Da. Two putative conserved CxxCH heme-binding motifs at positions 80 to 84 and 180 to 184 of the mature polypeptide suggested SoxA to be a diheme c-type cytochrome. A comparison of the deduced primary structure of SoxA with entries in the SWISSPROT and GenBank databases revealed an identity of 27.2% in a 257-amino-acid overlap to the 258 residues containing cytochrome c551 from the phototrophic green sulfur bacterium Chlorobium limicola strain Tassajara (23) (Table 1). The c-type cytochrome of C. limicola exhibits only one heme-binding site. The deduced primary structure of SoxA suggested a hybrid protein and a new class of c-type cytochromes. Other similarities related to partial amino acid sequences of the putative aq1807 protein of A. aeolicus and to nitrate reductase of Pseudomonas aeruginosa (Table 1).
soxB.
The previously reported poor ribosome-binding site and the deduced signal peptide (54) were not in accordance with the general rules (45, 49). Therefore, the nucleotide sequence was reexamined and corrected for one nucleotide at position 4998 (Fig. 2). This correction caused a significant change: the start codon of the soxB gene was located 123 nucleotides 3′ of soxA with a well-conserved putative ribosome-binding site 11 nucleotides 5′ of the start codon. soxB had the potential to encode a protein of 564 amino acids (molecular mass of 61,897 Da). Further analysis uncovered a putative signal peptide of 29 amino acids to give a predicted mature protein with a molecular mass of 58,786 Da. The putative signal peptide contained a twin arginine motif (see Fig. 7) characteristic of periplasmic proteins transported through the cytoplasmic membrane via the twin-arginine-transport (Tat) system (5, 7, 52).
Purification of proteins essential for sulfur oxidation.
Of the proteins required for thiosulfate oxidation, SoxB was identified to be coded for by the soxB gene (54). The molybdoenzyme sulfite dehydrogenase, being intimately associated with cytochrome c551, was purified and characterized previously and was identified to be coded for by the soxCD genes (36, 56). To examine if the products of the new genes characterized above were required in thiosulfate oxidation the proteins essential for thiosulfate-dependent cytochrome c reduction were purified.
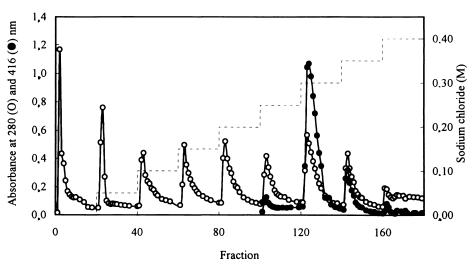
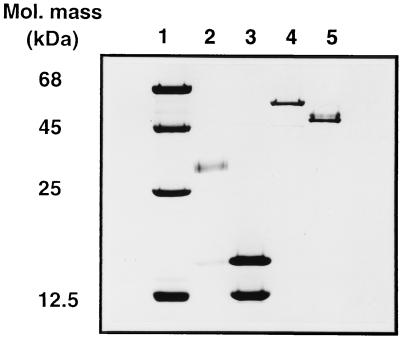
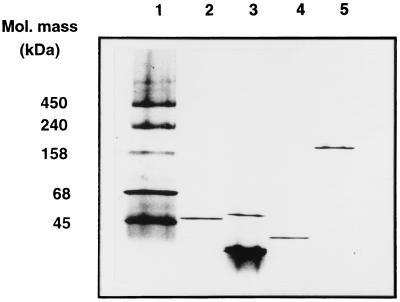
Of the different fractions eluted by step gradients from Q Sepharose (Fig. 3), those eluted at 0, 0.25, 0.30, and 0.35 M sodium chloride reconstituted the thiosulfate-dependent cytochrome c reducing activity (data not shown). From each of these fractions one protein was purified as described in Materials and Methods in order to reconstitute the maximum thiosulfate-oxidizing activity: SoxYZ, SoxB, SoxXA, and SoxCD (Table 3). The chromogenic fractions forming the 0.30, 0.35, and 0.40 M NaCl pools contained cytochromes, as evident from the absorption at 416 nm (Fig. 3). From the fractions specified above SoxYZ, SoxXA, and SoxCD were purified to homogeneity, while the SoxB preparation exhibited a minor contaminating protein (Fig. 4).
FIG. 3.
Chromatography of proteins of the A65% fraction on Q Sepharose. Symbols: ○, protein absorption at 280 nm; ●, heme absorption at 416 nm; −, sodium chloride step gradient.
TABLE 3.
Proteins required for thiosulfate- and sulfite-dependent cytochrome c reduction
| Sox protein added to the assaya
|
Cytochrome c reduction rate (nmol/min)
|
||||
|---|---|---|---|---|---|
| XA | YZ | B | CD | Thiosulfate dependent | Sulfite dependent |
| + | + | + | + | 6.51 | 4.50 |
| + | − | + | + | 0 | 0 |
| + | + | − | + | 0 | 0 |
| + | + | + | − | 2.77 | 2.84 |
| − | + | + | + | 0.29 | 0 |
A total of 0.50 nmol of each protein was used per assay. For sulfite-dependent activity, thiosulfate was eliminated from the buffer as described in Materials and Methods.
FIG. 4.
SDS-PAGE analysis of purified Sox proteins. Lane 1, 5.0 μg of marker proteins (1.5 μg of purified proteins was applied to each well). Lane 2, SoxXA; lane 3, SoxYZ; lane 4, SoxB; lane 5, SoxCD.
SoxB.
The SoxB protein was purified to near homogeneity as judged from denaturing SDS-PAGE (Fig. 4), and contained 95.6% SoxB, as determined from densitometric analysis (data not shown). The molecular mass of SoxB as determined by SDS-PAGE was 59,000 Da (Fig. 4) and as determined by nondenaturing gradient PAGE was 47,000 Da (Fig. 5). The preparation (6.0 mg of protein/ml) was colorless. Quantitative determination of the metal content of this preparation revealed 9.8 μg of manganese/ml, an amount equivalent to 1.75 atoms of Mn/mol of SoxB, using the molecular mass of the deduced mature protein (58,786 Da). The amount of iron was 0.12 and the amount of copper was 0.16 atom per mol of SoxB; other metals were below the detection limit.
FIG. 5.
Nondenaturing gradient PAGE analysis of Sox proteins. Lane 1, 10 μg of marker proteins (1.5 μg of purified proteins was applied to each well). Lane 2, SoxB; lane 3, SoxYZ; lane 4, SoxXA; lane 5, SoxCD.
The N-terminal amino acid sequence of SoxB could not be determined since the N terminus was blocked. The putative twin-arginine motif within the signal peptide (see Fig. 7) diagnostic for proteins with complex redox centers (5, 52) prompted us to search for a possible cofactor of SoxB. UV-VIS absorption spectra of SoxB (1.8 mg of protein/ml) and difference spectra against an equivalent concentration of bovine serum albumin revealed no indication of a chromophore (data not shown). Therefore, the molecular mass of SoxB was determined by electrospray ionization mass spectroscopy. Two major masses of 58,630 and 58,813 Da were observed, and the latter was similar to that deduced from the nucleotide sequence (molecular mass of 58,786 Da) with the predicted signal peptide cleavage site (Fig. 2; see also Fig. 7).
SoxYZ.
The protein isolated from the 0 M sodium chloride eluate of Q Sepharose essential to reconstitute thiosulfate and sulfite oxidizing activity was designated SoxYZ. Under denaturing SDS-PAGE, the protein split into subunits of 12,000 and 16,000 Da (Fig. 4). Nondenaturing gradient PAGE revealed bands at 29,000, 31,000, and 50,000 Da (Fig. 5). The split bands might have indicated an equilibrium of a heterodimeric and heterotetrameric structure of SoxYZ under the experimental conditions applied. Using electrospray ionization mass spectroscopy, two masses of 11,094 and 11,717 Da were determined. The N-terminal amino acid sequence of the 12,000-Da subunit (Table 2) was identical to that deduced from the nucleotide sequence of soxY, identifying this subunit as SoxY (deduced molecular mass of the mature peptide of 10,977 Da), and confirmed the predicted cleavage site for the signal peptidase.
The putative signal peptide with the twin-arginine motif of SoxY indicated the presence of a complex redox center. However, the molecular mass determined by electrospray ionization mass spectroscopy (11,094 Da) was slightly different from that predicted from the nucleotide sequence (10,977 Da) and that determined by SDS-PAGE (12,000 Da). The mass difference was not resolved from the determination of 110 amino acid residues (of 112 of the mature protein) of the N terminus and from internal peptides after tryptic digestion. This sequence confirmed the deduced amino acid sequence (Table 2, Fig. 2). Also, the molecular masses of the peptides T19, T39, T64, and T76 determined by MALDI-TOF were identical to those predicted from the amino acid sequences (data not shown). These data did not support the presence of a prosthetic group or covalently bound moiety of SoxY. Analysis of SoxYZ by X-ray fluorescence did not indicate the presence of a metal in this protein (data not shown). Therefore, the mass difference of 117 Da is presently unexplained.
The N-terminal amino acid sequence of the 16,000-Da subunit (Table 2) identified this protein as SoxZ (deduced molecular mass of 11,849 Da) and confirmed the absence of a leader peptide, as predicted from the respective analysis (32, 49). However, the methionine residue was cleaved off, resulting in a mature polypeptide of 11,718 Da. The molecular mass of the mature SoxZ as determined by electrospray ionization spectroscopy (17,717 Da) confirmed the predicted mass. In addition, determination of 70 of 108 amino acid residues of the N terminus and internal peptides after tryptic digestion of SoxZ confirmed 64.8% of the amino acid sequence of the mature SoxZ as deduced from the nucleotide sequence. Also, the molecular masses of the SoxZ internal peptides deduced from the nucleotide sequence were identical with the MALDI-TOF mass determinations. The discrepancy in the mass determination of SoxZ by SDS-PAGE and by electrospray ionization may have resulted from amino acid composition and/or structural characteristics that may have disturbed the ideal micelle structure required for the size-dependent mobility of the protein.
From an enzyme A preparation of P. versutus (molecular mass of 16,000 Da) essential for thiosulfate oxidation, the N terminus was determined, which appeared to be highly similar to the SoxZ of P. pantotrophus (Table 2).
SoxXA.
The 0.30 M sodium chloride eluate of Q Sepharose was highly chromogenic (Fig. 3). The protein purified from this fraction required to reconstitute thiosulfate and sulfite oxidizing activity was designated SoxXA (Table 3). Under denaturing SDS-PAGE the protein split into two subunits of 16,000 and 29,000 Da (Fig. 4). The molecular mass as determined by size exclusion chromatography from nondenaturing gradient PAGE was 38,000 Da (Fig. 5). Masses of 14,834 and 30,452 Da were determined from the identical preparation by electrospray ionization spectroscopy. The N-terminal amino acid sequence of the 12,000-Da subunit (Table 2) was identical to that deduced from the nucleotide sequence and identified this polypeptide as SoxX. The N terminus and internal amino acid sequences of the 29,000-Da subunit identified this polypeptide as SoxA and confirmed 19.3% of its amino acid sequence deduced from the nucleotide sequence (Table 2).
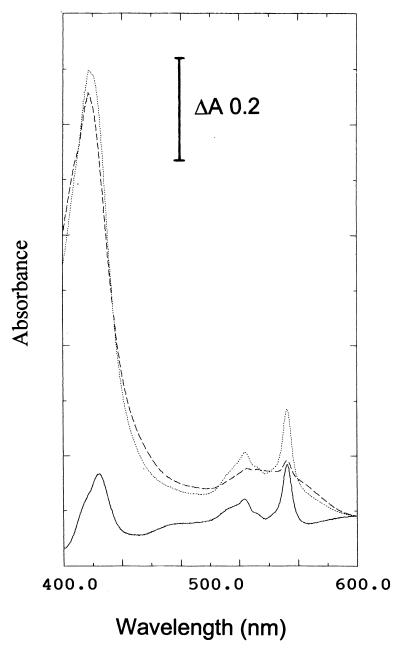
The dithionite-reduced minus air-oxidized difference spectrum of homogeneous SoxXA revealed peaks at 552.5 and 520 nm and identified this protein as c-type cytochrome (Fig. 6). The molar extinction coefficient of SoxXA at 552.5 nm calculated from the data of Fig. 6 was 38.5 cm2/mol. Inverse matrix of extinction coefficients as detailed by Berry and Trumpower (6) were used to calculate the concentrations of cytochromes and to differentiate these for the a, b, and c types. According to this method the heme c content of SoxXA was quantified by the pyridine hemochrome spectroscopic analysis to be 2.73 μmol of heme c and no heme b or heme a per 0.17 mg of pure SoxXA/ml, and these data suggested there was 2.53 mol of heme c per mol of SoxXA. Quantification of the metal content of purified SoxXA with protein concentrations ranging from 5.0 and 50.6 mg of SoxXA/ml by total reflection X-ray fluorescence analysis revealed 10.3 and 100 μg of iron/ml, a value corresponding to 1.63 mol of Fe/mol of SoxXA.
FIG. 6.
Absorption and dithionite-reduced minus air-oxidized difference spectra of SoxXA of P. pantotrophus GB17. The spectra were recorded at 20°C; the SoxXA concentration was 0.5 mg/ml. Lines: ..., dithionite reduced; ---, air oxidized; —, reduced minus air-oxidized difference spectrum.
Reconstitution of the Sox system.
To achieve an optimal thiosulfate-dependent cytochrome c-reducing rate, SoxXA, SoxYZ, SoxB, and SoxCD were applied in the assay at concentrations of 0.25 μM each (Table 3). However, the reconstitution of the multienzyme system followed Michaelis-Menten-type kinetics for each of the components examined. Using 0.25 μM SoxCD, SoxB, and SoxYZ in the assay for thiosulfate-dependent cytochrome c reduction the concentrations of SoxXA were altered from 0.05 to 1.0 μM. Under these conditions a Michaelis constant, Km, of 0.034 μM for SoxXA was determined from the linear double-reciprocal plot (data not shown). Alteration of the concentration of SoxCD at fixed concentrations of the other proteins of 0.25 μM revealed a Km of 0.025 μM SoxCD (data not shown).
Catalytic properties of the reconstituted Sox system.
Using purified proteins the reaction of the reconstituted Sox system with thiosulfate as electron donor was linear with time and continued to be constant almost to the complete reduction of horse heart cytochrome c. Without SoxCD a significant activity of 42.5% was observed after a lag phase of about 5 min. Without SoxXA a rate of 4.4% was observed compared with an assay containing the four Sox proteins (Table 3).
The maximum sulfite-dependent cytochrome c reduction rate required SoxXA, SoxYZ, and SoxB in addition to SoxCD. Surprisingly, without SoxCD a significant rate of 63.2% was observed (Table 3). This implied that, using pure proteins, SoxXA, SoxYZ, and SoxB exhibited sulfite-oxidizing activity. When the system was reconstituted from the pools after Q Sepharose chromatography containing the SoxXA, SoxYZ, and SoxB proteins, only 2.9% of the sulfite-oxidizing activity was obtained compared to that obtained with SoxCD (36). This may have indicated the presence of a factor within these pools inhibiting sulfite oxidation in the absence of SoxCD.
The number of electrons released per mole of thiosulfate by the multienzyme system of P. pantotrophus GB17 was determined from the reconstituted system using purified proteins. The amount of thiosulfate in the assay was lowered to 5 nmol, and the amount of horse heart cytochrome c was increased to 70 nmol. Under these conditions 37.7 nmol of cytochrome c was reduced within 10 min (Table 4). The molar ratio of thiosulfate over reduced cytochrome c of 1:7.58 suggested that eight electrons were released per mol of thiosulfate. Upon omission of SoxCD, from the assay only 11.3 nmol of cytochrome c were reduced, suggesting that only 2 electrons were released per mol of thiosulfate under these conditions (Table 4).
TABLE 4.
Thiosulfate- and sulfite-dependent degree of cytochrome c reduction mediated by the sulfur-oxidizing enzyme systema
| Electron donor | Concn (μM) | Sox protein
|
Cytochrome c reduced (nmol) | Ratio (Mol of cyt c reduced/mol of electron donor | |||
|---|---|---|---|---|---|---|---|
| XA | YZ | B | CD | ||||
| Thiosulfate | 5 | + | + | + | + | 37.7 | 7.54 |
| 5 | + | + | + | − | 11.3 | 2.26 | |
| Sulfite | 20 | + | + | + | + | 42.3 | 2.15 |
| 20 | + | + | + | − | 36.9 | 1.85 | |
For thiosulfate-dependent cytochrome c reduction, the assays contained 50 μmol of potassium phosphate buffer (pH 7.5), 0.5 nmol of the Sox proteins as indicated, and 70 nmol of horse heart cytochrome c. With sodium sulfite as the electron donor, the assay contained 100 μmol of Tris-HCl (pH 7.5).
DISCUSSION
Seven new genes, ORF1, ORF2, and ORF3 and soxXYZA, located within the gene cluster coding for lithotrophic thiosulfate oxidation, have been described, and the function of four of these in cytochrome c-dependent thiosulfate oxidation has been demonstrated. (i) Purification and determination of partial amino acid sequences of the soxXYZA gene products have shown that these genes encode the respective peptides. (ii) Biochemical studies have shown that SoxXA and SoxYZ form heterodimeric proteins and are essential for the biochemical reaction. (iii) SoxYZ represents a novel protein, and SoxXA represents a novel cytochrome. (iv) Sequence analysis suggested that the soxXA and soxYZ gene products were located in the periplasm and that SoxXA and SoxYZ were isolated from the soluble fraction.
Analysis of the nucleotide sequence suggested that the ORF1 gene product was related to the LysR type of transcriptional regulators. The transcription start of ORF1 is uncertain. The putative gene products lack characteristics for a signal peptide or a membrane anchor and are, therefore, considered cytoplasmic. The direction of transcription was in the opposite direction and distantly located from the ORF2 and the subsequent ORFs described here. The ORF2 gene product had the potential to form six transmembrane helices and is therefore predicted to be located within the cytoplasmic membrane. Opposite helices carry two cysteine residues which may be redox active. Such a characteristic is found for similar proteins predicted to be involved in redox transport required for cytochrome c biogenesis (34, 42, 46). The predicted structure of the ORF2 gene product was analogous to that of the DsbD (DipZ) protein of E. coli, whose function was suggested to be in the transport of reductant (34, 46). Such transport may be mediated by a thioredoxin that is possibly coded for by ORF3. Its deduced amino acid sequence exhibits the CxxC motif characteristic of thioredoxins, it exhibits a significant identity to TRXA2 (a thioredoxin of A. aeolicus), and evidence for the periplasmic location of the mature ORF3 product is given from the characteristics for a signal peptide. Evidence has been obtained that the sulfur-oxidizing enzyme system of P. pantotrophus GB17 is located in the periplasm (36, 56). Therefore, the function of the products of ORF2 and ORF3 may be in the transport of reductant to the cytoplasm of the cell for energy transformation via oxidative phosphorylation or for biosynthesis, and site-directed mutagenesis will elucidate their role in sulfur oxidation.
SoxXA is a new type of periplasmic c-type cytochrome with respect to its primary as well as its quaternary structure. SoxXA is suggested to contain three heme moieties, as evidenced by sequence analysis and hemochrome quantification. The content of 1.6 mol of iron per mol of SoxXA does not match this suggestion, and further analysis will elucidate this discrepancy. SoxA appears to be a new type of cytochrome c with the only significant identities to the putative cytochrome aq1807 of A. aeolicus and cytochrome c551 of C. limicola. From the latter strain amino acid sequence identity applies to the first heme-binding motif. From the genomic sequence of A. aeolicus, polypeptides such as aq1807, TRXA2, aq1810, and aq1809 were deduced (12) that are strikingly similar to ORF3 and SoxY, SoxZ, SoxA, and SoxB. No similarity of the deduced A. aeolicus proteins was detected with regard to SoxX, SoxC, or SoxD. These proteins are essential for the growth of P. pantotrophus GB17. The absence of homologues of these essential proteins in A. aeolicus suggests that this strain, in contrast to A. pyrophilus, cannot grow with reduced inorganic sulfur compounds, as has been determined (K. O. Stetter, personal communication).
SoxYZ is an as-yet-unprecedented protein with respect to its primary sequences and quaternary structure. Although deduced gene products have been reported from A. aeolicus (12) with significant similarities to SoxY and SoxZ, neither its quaternary structure nor its function has been described previously. SoxYZ is essential for cytochrome c-dependent sulfur-oxidizing activity in vitro. The deduced soxY gene product exhibits a twin-arginine motif characteristic of proteins transported through the cytoplasmic membrane via the Tat system (7, 52). Neither SoxY nor SoxZ contains a metal, a chromophore, a prosthetic group, or a posttranslational modification, as is evident from X-ray fluorescence metal analysis, UV-VIS spectroscopy, and molecular mass determination by electrospray ionization spectroscopy. No signal peptide can be deduced from soxZ. Size exclusion chromatography by denaturing gradient PAGE demonstrates that SoxYZ appears as a heterodimer and as a heterotetramer. Therefore, the twin-arginine motif of the SoxY leader peptide may be essential for the cotransport of SoxZ through the cytoplasmic membrane. Cotranslocation in a hitchhiker fashion has been suggested for the two subunits of the membrane-bound hydrogenase-2 of E. coli (39), which is located at the periplasmic side of the cytoplasmic membrane. The small hydrogenase subunit containing a twin-arginine motif cotransports the large subunit carrying the [NiFe] cofactor whose N-terminal sequence is colinear with the sequence predicted from the nucleotide sequence (39). This proposal was supported by a recent report of the membrane-bound hydrogenase of R. eutropha, also located at the periplasmic side of the cytoplasmic membrane (4).
SoxY and SoxZ each contain a single cysteine. The role of these cysteines is presently unresolved with respect to (i) the quaternary structure, covalent linkage of the two polypeptides for cotranslocation; (ii) the mechanism of inorganic sulfur oxidation; or (iii) its function as a redox mediator. It is noteworthy that the SoxY homolog of A. aeolicus, the predicted aq1810 gene product lacks a cysteine, while aq1809 does not. Future studies will examine the role of the cysteines of SoxYZ.
SoxZ of P. pantotrophus GB17 is very likely equivalent to enzyme A of P. versutus. The N terminus of enzyme A is almost identical to that of SoxZ. The molecular mass of enzyme A of P. versutus is 16,200 Da (28). It is accompanied by a polypeptide of 14,600 Da, as determined by SDS-PAGE, which might be equivalent to SoxY. Enzyme A of P. versutus binds thiosulfate with sulfite as a competitive inhibitor, and the protein was suggested to function in thiosulfate cleavage (29). Since SoxYZ is essential for thiosulfate and for sulfite oxidation of P. pantotrophus GB17, the function of SoxYZ in P. pantotrophus is not restricted to thiosulfate cleavage and, together with the SoxB protein, suggests its central role in sulfur oxidation.
The deduced primary structure of SoxB contains a signal peptide with a twin-arginine motif. The chemical nature of the moiety blocking the N terminus has not been identified so far, and its impact on the molecular mass is not known. The mass difference of 27 Da appears to be too small to account for a covalently bound redox center. However, SoxB contains two atoms of manganese per mol of SoxB. Thus, this protein is again strikingly similar to enzyme B of P. versutus, for which a binuclear manganese cluster has been reported (9). A manganese cluster may account for the protein translocation via the Tat system presumably also present in P. pantotrophus GB17. Of the deduced gene products of the sox gene region, twin-arginine motifs within the leader peptide have been found for SoxY, SoxB, SoxC, and SoxF (56). It should be noted, however, that the motifs vary with respect to the consensus sequence (S/T)RRxFLK proposed earlier (5, 7, 52).
The stoichiometry of thiosulfate oxidation of 8 mol of cytochrome c reduced per mol of thiosulfate oxidized eliminates the possibility of the involvement of oxygen. This conclusion is in accordance with an earlier finding of the thiosulfate-oxidizing enzyme system of P. versutus (29).
ACKNOWLEDGMENTS
We thank A. V. Bohlen for metal analysis, J. Ringk for excellent technical assistance, L. Naumann for assistance in purification of SoxYZ, and D. P. Kelly for a sample of enzyme A of P. versutus.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson L O, Borg H, Mikaelsson M. Molecular weight estimations of proteins by electrophoresis in polyacrylamide gels of graded porosity. FEBS Lett. 1972;20:199–202. doi: 10.1016/0014-5793(72)80793-2. [DOI] [PubMed] [Google Scholar]
- 3.Bateman A, Birney E, Durbin R, Eddy S R, Finn R D, Sonnhammer E L L. Pfam 3.1: 1313 multiple alignments match the majority of proteins. Nucleic Acids Res. 1999;27:260–262. doi: 10.1093/nar/27.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhard M, Friedrich B, Siddiqui R A. Ralstonia eutropha TF93 is blocked in Tat-mediated protein export. J Bacteriol. 2000;182:581–588. doi: 10.1128/jb.182.3.581-588.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berks B C. A common export pathway for proteins binding complex redox factors. Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 6.Berry E A, Trumpower B L. Simultaneous determinnation of hemes a, b, and c from pyridine hemochrome spectra. Anal Biochem. 1986;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 7.Bogsch E G, Sargent F, Stanley N R, Berks B C, Robinson C, Palmer T. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J Biol Chem. 1998;273:18003–18006. doi: 10.1074/jbc.273.29.18003. [DOI] [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Cammack R, Chapman A, Lu W-P, Karagouni A, Kelly D P. Evidence that protein B of the thiosulfate-oxidizing system of Thiobacillus versutus contains a binuclear manganese cluster. FEBS Lett. 1989;253:239–243. [Google Scholar]
- 10.Chandra T S, Friedrich C G. Tn5-induced mutations affecting sulfur-oxidizing ability (Sox) of Thiosphaera pantotropha. J Bacteriol. 1986;166:446–452. doi: 10.1128/jb.166.2.446-452.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahl C. Insertional gene inactivation in a phototrophic sulphur bacterium: APS-reductase-deficient mutants of Chromatium vinosum. Microbiology. 1996;142:3363–3372. doi: 10.1099/13500872-142-12-3363. [DOI] [PubMed] [Google Scholar]
- 12.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olson G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 13.Fischer J, Quentmeier A, Kostka S, Kraft R, Friedrich C G. Purification and characterization of the hydrogenase from Thiobacillus ferrooxidans. Arch Microbiol. 1996;165:289–296. doi: 10.1007/s002030050329. [DOI] [PubMed] [Google Scholar]
- 14.Francis R T, Becker R R. Specific indication of hemoproteins in polyacrylamide gels using a double-staining process. Anal Biochem. 1984;136:509–514. doi: 10.1016/0003-2697(84)90253-7. [DOI] [PubMed] [Google Scholar]
- 15.Fraser C M, Norris S J, Weinstock G M, White O, Sutton G G, Dodson R, Gwinn M, Hickey E K, Clayton R, Ketchum K A, Sodergren E, Hardham J M, McLeod M P, Salzberg S, Peterson J, Khalak H, Richardson D, Howell J K, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton M D, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Watthey L, Weidman J, Smith H O, Venter J C. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 16.Friedrich C. Physiology and genetics of sulfur-oxidizing bacteria. Adv Microb Physiol. 1998;39:235–289. doi: 10.1016/s0065-2911(08)60018-1. [DOI] [PubMed] [Google Scholar]
- 17.Fukumori Y, Yamanaka T. Flavocytochrome c of Chromatium vinosum: some enzymatic properties and subunit structure. J Biochem. 1979;85:1405–1414. doi: 10.1093/oxfordjournals.jbchem.a132467. [DOI] [PubMed] [Google Scholar]
- 18.Hofmann K, Stoffel W. TMbase - a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;347:166–176. [Google Scholar]
- 19.Johnson J L, Rajagopalan K V. Structural and metabolic relationship between the molybdenum cofactor and urothione. Proc Natl Acad Sci USA. 1982;79:6856–6860. doi: 10.1073/pnas.79.22.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katayama Y, Hiraishi A, Kuraishi H. Paracoccus thiocyanatus sp. nov., a new species of thiocyanate-utilizing facultative chemolithotroph, and transfer of Thiobacillus versutus to the genus Paracoccus as Paracoccus versutus comb. nov. with emendation of the genus. Microbiology. 1995;141:1469–1477. doi: 10.1099/13500872-141-6-1469. [DOI] [PubMed] [Google Scholar]
- 21.Kelly D P. Physiology and biochemistry of unicellular sulfur bacteria. In: Schlegel H G, Bowien B, editors. Autotrophic bacteria. 1989. pp. 193–217. Brock/Springer, Berlin, Germany. [Google Scholar]
- 22.Kelly D P, Shergill J, Lu W-P, Wood A P. Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Leeuwenhoek. 1997;71:95–107. doi: 10.1023/a:1000135707181. [DOI] [PubMed] [Google Scholar]
- 23.Klarskov K, Verté F, Van Driessche G, Meyer T E, Cusanovich M A, Van Beeumen J. The primary structure of soluble cytochrome c551 from the phototrophic green sulfur bacterium Chlorobium limicola, strain Tassajara, reveals a novel c-type cytochrome. Biochemistry. 1998;37:10555–10562. doi: 10.1021/bi9806706. [DOI] [PubMed] [Google Scholar]
- 24.Klockenkämper R, von Bohlen A. Elemental analysis of environmental samples by TXRF: a review. X-Ray Spectrom. 1996;25:156–162. [Google Scholar]
- 25.Kondorosi E, Pierre M, Cren M, Haumann U, Buire M, Hoffmann B, Schell J, Kondorosi A. Identification of NolR, a negative transacting factor controlling the nod regulon in Rhizobium meliloti. J Mol Biol. 1991;222:885–896. doi: 10.1016/0022-2836(91)90583-r. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Lu W-P. A periplasmic location for the thiosulfate-oxidizing multi-enzyme system from Thiobacillus versutus. FEMS Microbiol Lett. 1986;34:313–317. [Google Scholar]
- 28.Lu W-P, Kelly D P. Purification and some properties of two principal enzymes of the thiosulphate-oxidizing multi-enzyme system from Thiobacillus A2. J Gen Microbiol. 1983;129:3549–3564. [Google Scholar]
- 29.Lu W-P, Swoboda B E P, Kelly D P. Properties of the thiosulfate-oxidizing multi-enzyme system from Thiobacillus versutus. Biochim Biophys Acta. 1985;828:116–122. [Google Scholar]
- 30.Ludwig W, Mittenhuber G, Friedrich C G. Transfer of Thiosphaera pantotropha to Paracoccus denitrificans. Int J Syst Bacteriol. 1993;43:363–367. doi: 10.1099/00207713-43-2-363. [DOI] [PubMed] [Google Scholar]
- 31.Mittenhuber G, Sonomoto K, Egert M, Friedrich C G. Identification of the DNA region responsible for sulfur-oxidizing ability of Thiosphaera pantotropha. J Bacteriol. 1991;173:7340–7344. doi: 10.1128/jb.173.22.7340-7344.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakai K, Kaneshisa M. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 33.Nurizzo D, Silvestrini M C, Mathieu M, Cutruzzola F, Bourgeois D, Fulop V, Hajdu J, Brunori M, Tegoni M, Cambillau C. N-terminal arm exchange is observed in the 2.15 Å crystal structure of oxidized nitrite reductase from Pseudomonas aeruginosa. Structure. 1997;5:1157–1171. doi: 10.1016/s0969-2126(97)00267-0. [DOI] [PubMed] [Google Scholar]
- 34.Page M D, Saunders N F, Ferguson S J. Disruption of the Pseudomonas aeruginosa dipZ gene, encoding a putative protein-disulfide reductase, leads to partial pleiotropic deficiency. Microbiology. 1997;143:3111–3122. doi: 10.1099/00221287-143-10-3111. [DOI] [PubMed] [Google Scholar]
- 35.Prinz, H., A. Lavie, A. Scheidig, O. Spangenberg, and M. Konrad. Binding of nucleotides to guanylate kinase, ras p21 and to nucleoside diphosphate kinase studied by nano-electrospray mass spectrometry. J. Biol. Chem., in press. [DOI] [PubMed]
- 36.Quentmeier A, Kraft R, Kostka S, Klockenkämper R, Friedrich C G. Characterization of a new type of sulfite dehydrogenase from Paracoccus pantotrophus GB17. Arch Microbiol. 2000;173:117–125. doi: 10.1007/s002039900118. [DOI] [PubMed] [Google Scholar]
- 37.Rainey F A, Kelly D P, Stackebrandt E, Burghardt J, Hiraishi A, Katayama Y, Wood A P. A re-evaluation of the taxonomy of Paracoccus denitrificans and a proposal for the combination Paracoccus pantotrophus comb. nov. Int J Syst Bacteriol. 1999;49:645–651. doi: 10.1099/00207713-49-2-645. [DOI] [PubMed] [Google Scholar]
- 38.Robertson L A, Kuenen J G. Thiosphaera pantotropha gen. nov. sp. nov., a facultatively anaerobic, facultative autotrophic sulphur bacterium. J Gen Microbiol. 1983;129:2847–2855. [Google Scholar]
- 39.Rodrigue A, Chanal A, Beck K, Müller M, Wu L-F. Cotranslocation of a periplasmic enzyme complex by a hitchhiker mechanism through the bacterial Tat pathway. J Biol Chem. 1999;274:13223–13228. doi: 10.1074/jbc.274.19.13223. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Maniatis T, Fritsch E F. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 41.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schiött T, von Wachenfeldt C, Hederstedt L. Identification and characterization of the ccdA gene, required for cytochrome c synthesis in Bacillus subtilis. J Bacteriol. 1997;179:1962–1973. doi: 10.1128/jb.179.6.1962-1973.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sonnhammer E L L, Eddy S R, Birney E, Bateman A, Durbin R. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 1998;26:320–322. doi: 10.1093/nar/26.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sonnhammer E L L, Eddy S R, Durbin R. Pfam: a comprehensive database of protein families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 45.Steinrücke P, Ludwig B. Genetics of Paracoccus denitrificans. FEMS Microbiol Rev. 1993;104:83–118. doi: 10.1016/0378-1097(93)90505-v. [DOI] [PubMed] [Google Scholar]
- 46.Stewart E J, Katzen F, Beckwith J. Six conserved cysteines of the membrane protein DsbD are required for the transfer of electrons from the cytoplasm to the periplasm of E. coli. EMBO J. 1999;18:5963–5971. doi: 10.1093/emboj/18.21.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 48.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 50.Wayne L G, Brenner D J, Colwell R R, Grimont P A D, Kandler O, Krichevsky M I, Moore L H, Moore W E C, Murray R G E, Stackebrandt E, Starr M P, Trüper H G. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int J Syst Bacteriol. 1987;37:463–464. [Google Scholar]
- 51.Weber K, Pringle J R, Osborn M. Measurements of molecular weights by electrophoresis on SDS-polyacrylamide gels. Methods Enzymol. 1972;26:3–27. doi: 10.1016/s0076-6879(72)26003-7. [DOI] [PubMed] [Google Scholar]
- 52.Weiner, Bilous J H P T, Shaw G M, Lubitz S P, Frost L, Thomas G H, Cole J A, Turner R J. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell. 1998;93:93–101. doi: 10.1016/s0092-8674(00)81149-6. [DOI] [PubMed] [Google Scholar]
- 53.Williams S G, Attridge S R, Manning P A. The transcriptional activator HlyU of Vibrio cholerae: nucleotide sequence and role in virulence gene expression. Mol Microbiol. 1993;9:751–760. doi: 10.1111/j.1365-2958.1993.tb01735.x. [DOI] [PubMed] [Google Scholar]
- 54.Wodara C, Kostka S, Egert M, Kelly D P, Friedrich C G. Identification and sequence analysis of the soxB gene essential for sulfur oxidation of Paracoccus denitrificans GB17. J Bacteriol. 1994;176:6188–6191. doi: 10.1128/jb.176.20.6188-6191.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wodara C. Molekulare Analyse der Schwefel-Oxidation kodierenden DNA-Region von Paracoccus denitrificans GB17. Ph.D. thesis. Dortmund, Germany: University of Dortmund; 1995. [Google Scholar]
- 56.Wodara C, Bardischewsky F, Friedrich C G. Cloning and characterization of sulfite dehydrogenase, two c-type cytochromes, and a flavoprotwein of Paracoccus denitrificans GB17: essential role of sulfite dehydrogenase in lithotrophic sulfur oxidation. J Bacteriol. 1997;179:5014–5023. doi: 10.1128/jb.179.16.5014-5023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiong J, Inoue K, Bauer C E. Tracking molecular evolution of photosynthesis by characterization of a major photosynthesis gene cluster from Heliobacillus mobilis. Proc Natl Acad Sci USA. 1998;95:14851–14856. doi: 10.1073/pnas.95.25.14851. [DOI] [PMC free article] [PubMed] [Google Scholar]