Abstract
We have identified in the Streptococcus pneumoniae genome sequence a two-component system (TCS13, Blp [bacteriocin-like peptide]) which is closely related to quorum-sensing systems regulating cell density-dependent phenotypes such as the development of genetic competence or the production of antimicrobial peptides in lactic acid bacteria. In this study we present evidence that TCS13 is a peptide-sensing system that controls a regulon including genes encoding Blps. Downstream of the Blp TCS (BlpH R) we identified open reading frames (blpAB) that have the potential to encode an ABC transporter that is homologous to the ComA/B export system for the competence-stimulating peptide ComC. The putative translation product of blpC, a small gene located downstream of blpAB, has a leader peptide with a Gly-Gly motif. This leader peptide is typical of precursors processed by this family of transporters. Microarray-based expression profiling showed that a synthetic oligopeptide corresponding to the processed form of BlpC (BlpC*) induces a distinct set of 16 genes. The changes in the expression profile elicited by synthetic BlpC* depend on BlpH since insertional inactivation of its corresponding gene abolishes differential gene induction. Comparison of the promoter regions of the blp genes disclosed a conserved sequence element formed by two imperfect direct repeats upstream of extended −10 promoter elements. We propose that BlpH is the sensor for BlpC* and the conserved sequence element is a recognition sequence for the BlpR response regulator.
Signaling mechanisms controlling multicellular behavior of bacteria have attracted much attention in current research. In gram-negative bacteria, homoserine-lactone-based communication systems are prominent. Research in this area led to the term “quorum sensing” for phenomena that are controlled by cell density (12). In gram-positive bacteria, quorum sensing is accomplished by signaling systems that depend on the secretion and sensing of small peptides (11, 19). At least two different mechanisms for sensing the presence of pheromone-like peptides are known (21). The first involves import of the peptide and interaction with an intracellular factor (22); the second involves binding to the extracellular portion of a membrane-bound histidine kinase. This leads to the autophosphorylation of the kinase and subsequent activation, e.g., phosphorylation of a cognate response regulator that mediates changes in gene expression. Quorum-sensing systems regulate a plethora of cellular functions. In Staphylococcus aureus, the AgrC-AgrA-system is involved in the density-dependent regulation of virulence (18). In Lactobacillus strains, the production of bacteriocins is dependent on peptide-regulated two-component systems (TCS) (4, 10). In Streptococcus pneumoniae, the development of genetic competence (the natural ability to take up DNA) has been shown to be regulated by the comC-DE system (29).
The com system of S. pneumoniae was the first quorum-sensing system for which a biological function was defined. After a long search for a pheromone-like substance, biochemical purification led to the identification of a small cationic peptide (17 amino acids [aa] long) termed CSP (competence-stimulating peptide) (14). Genomic sequence analysis revealed that CSP is encoded as a larger precursor protein by the comC gene. The CSP precursor is exported by ComA, an ABC-type transporter specialized for the export of peptides, and ComB, an associated transmembrane (TM) protein. Concomitant with the export, the N-terminal leader sequence is cleaved at a characteristic Gly-Gly motif.
ComD and the response regulator ComE form a functional TCS which is involved in the regulation of genes important for competence. A comparison of the CSP and ComD sequences of different pneumococcal strains revealed significant sequence variations within both the CSP peptide sequence and the N-terminal region of ComD. It was suggested that such a variation would allow for regulation of competence in a strain-specific manner (15). Based on these comparisons of closely related ComD sequences representing different pherotypes, it has been suggested that CSP peptide can bind to the N-terminal, extracellular part of the histidine kinase ComD (16).
Recently, we have identified and compared 13 different TCSs encoded by the genome of S. pneumoniae (20). In addition, eight of these TCS were also shown to be involved in virulence in a respiratory tract mouse infection model including the Blp (bacteriocin-like peptide) TCS (486hk/rr pair) (36). In this study we provide evidence that the Blp TCS is part of a quorum-sensing regulon, termed blp, that shares many features with the CSP-controlled com regulon, e.g., density-dependent gene regulation and strain variability of the peptide pheromone and its proposed binding region. Microarray-based analysis was used to reveal genes regulated by this peptide pheromone, and comparative genomic studies propose that most of the regulated genes may have bacteriocin-related functions.
MATERIALS AND METHODS
Bacterial strains, cell growth, and handling.
S. pneumoniae strains R6, KNR.7/87, and LSP22 (R6 hk13′::pAS1::′hk13 (Table 1) were grown at 37°C in Todd-Hewitt medium in a 10% CO2 atmosphere. Additional strains used for sequencing of the blpC gene and the sensing domain of blpH are described in Table 1. For BlpC* induction, a 100-ml culture was split in two aliquots; to half of the culture, BlpC* peptide was added to 250 ng/ml. Cell pellets from 40 ml of culture were collected at various times after addition of the peptide and snap-frozen in liquid nitrogen prior to RNA extraction. The BlpH-deficient mutant LSP22 was obtained by insertional mutagenesis. A PCR product generated using primers HK13EF and HK13RB corresponding to an internal blpH gene segment was cut with restriction enzymes EcoRI and BamHI and cloned into pAS1, yielding the integrational plasmid pRPL54 (pAS1::′blpH′::pAS1). Insertional S. pneumoniae mutants were isolated and characterized as previously described (20).
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Relevant features | Reference |
|---|---|---|
| S. pneumoniae | ||
| R6 | Subclone of R36A, derivative of D39 | 34 |
| KNR.7/87 | Serotype 4 clinical isolate; TIGR sequence | 1 |
| D39 | Serotype 2; NCTC 7466 | 3 |
| 1 | Clinical isolate; Roche strain collection | 20 |
| 1711 | Clinical isolate; Roche strain collection | 20 |
| 4241 | ATCC reference strain | P. Geslin (Paris, France) |
| LSP22 | R6 hk13′::pAS1::′hk13 | This study |
| Plasmids | ||
| pJDC9 | Vector (Ery); nonreplicative in S. pneumoniae | 7 |
| pAS1 | Ery; pJDC9 derivative | 27 |
| pRPL54 | pAS1 derivative carrying a 401-bp fragment targeting hk13 (nt 456–857) | This study |
| Oligonucleotides | ||
| HK13EF | GAGAAAGGGAATTCTCGATAAAGC | This study |
| HK13RB | CTGACTGGGATCCTATAAGACGGAGa | This study |
Underlined nucleotides were modified to create an in-frame stop codon and an EcoRI restriction site.
Synthetic peptides.
The synthetic peptides corresponding to the expected processed forms of BlpC from strain R6 (BlpCR6) and of BlpCKNR.7/87 were synthesized using the continuous-flow, solid-phase synthesis method on a Pioneer peptide synthesis system, starting from Tenta Gel S RAM resin (3.0 g, 0.25 mmol/g; Rapp Polymere GmbH, Tübingen, Germany), using standard protocols (2).
DNA sequence analysis.
Potential coding sequences were derived from the genomic sequence of S. pneumoniae KNR.7/87 as described previously (20). Additional nucleotide sequencing was performed with the ABI Prism dRhodamine terminator cycle sequencing Ready Reaction kit and with the AmpliTaq DNA polymerase (Perkin-Elmer/ABI).
Probe selection, open reading frame (ORF) coverage, and array design.
An antisense oligonucleotide array (ROEZ06a) covering both genomes of S. pneumoniae and Haemophilus influenzae was custom designed by Affymetrix (Santa Clara, Calif.). In this report, we focus on the S. pneumoniae genome sequence, for which over 130,000 oligonucleotide probes complementary to S. pneumoniae strain KNR.7/87 were selected. Antisense refers to the target nucleic acid; i.e., the oligonucleotide probes on microarray have the sequence of the coding strand. This microarray is of the latest generation of Affymetrix chips with feature size reduced to 24 μm. A total of 1,973 potential S. pneumoniae gene sequences as predicted by GeneMark software and 323 intergenic regions larger than 200 bp were selected. The oligonucleotide probe selection (25-mers) and array fabrication were performed by Affymetrix according to published procedures (24, 38). Each gene represented on the ROEZ06a microarray has in general 25 probe pairs and at least 20 probe pairs for very short genes. A probe pair consists of a perfect match probe and a mismatch probe that is identical except for a single base change in the central position (24). The position of the oligonucleotide on each gene is determined by sequence uniqueness criteria and based on empirical rules for the selection of oligonucleotides likely to hybridize with high specificity and sensitivity (24).
RNA extraction and cDNA labeling.
The ability to isolate pure, intact RNA is critical to the success of genomewide expression analysis. Cell pellets from 40-ml cultures were snap-frozen in liquid nitrogen, and RNA was prepared as previously described (9). An additional purification step was performed through Qiagen RNeasy columns as instructed by the manufacturer in order to remove abundant small RNA molecules (tRNAs and 5S rRNA). The hybridization target was prepared by an optimized reverse transcription reaction from total RNA in the presence of random hexanucleotides and biotin-labeled dATP. Total RNA was not heat denatured prior to reverse transcription to avoid extensive priming to the rRNA. Random hexamers (7.5 μg) and total RNA (25 μg) were added to a reaction mixture containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 0.5 mM dCTP, 0.5 mM dGTP, 0.5 mM dTTP, 0.175 mM dATP, 0.3 mM biotin-labeled dATP (NEN), and 5,000 U of Superscript II reverse transcriptase (Life Technologies). Target cDNA synthesis was performed at 37°C overnight in a final volume of 100 μl. Following cDNA reactions, the RNA template was degraded by incubation for 30 min at 65°C in 0.25 M NaOH followed by neutralization with HCl and ethanol precipitation. The resulting cDNA was then fragmented by a partial DNase treatment. Fragmentation was performed in RQI buffer (Promega) in the presence of 0.1 U of DNase I for 5 min at 37°C in a 100-μl reaction volume. Fragmented cDNA was then ethanol precipitated and quantified by spectrophotometric measurement.
Hybridization and staining procedures.
Hybridization solutions contained 100 mM MES (N-morpholinoethanesulfonic acid), 1 M Na+, 20 mM EDTA, and 0.01% Tween 20. In addition, the solutions contained unlabeled fragmented yeast RNA (3 mg/ml; Roche Molecular Biochemicals) and acetylated bovine serum albumin (1.5 mg/ml; Sigma). Prior to hybridization, microarrays were prewarmed at room temperature, rinsed twice with hybridization buffer, and prehybridized for 10 min at 40°C. The hybridization mixture containing 10 μg of biotin-labeled cDNA was denatured at 95°C during 5 min, cooled to hybridization temperature, and centrifuged quickly to pellet all nonsolubilized material. Finally, 230 μl of this hybridization mixture was loaded on the chip for overnight hybridization at 40°C with mixing on a rotator at 60 rpm. After hybridization, the hybridization mixture was removed from the array and stored frozen. The arrays were rinsed twice with 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])–0.01% Tween 20 and washed in the same buffer at 40°C for 20 min. A stringent wash was then performed with 0.5× SSPE–0.01% Tween 20 for 15 min at 45°C. Following washing, the hybridized cDNA was labeled with streptavidin-phycoerythrin conjugate (3 μg/ml; Molecular Probes) and acetylated bovine serum albumin (2 mg/ml) in 6× SSPE–0.01% Tween 20 for 10 min at 40°C. Further signal amplification was also performed with a biotinylated antistreptavidin antibody and subsequent staining with streptavidin-phycoerythrin conjugate as described previously (6). A final washing step was performed in 6× SSPE–0.01% Tween 20 for 10 min at 40°C prior to scanning.
Microarray data analysis.
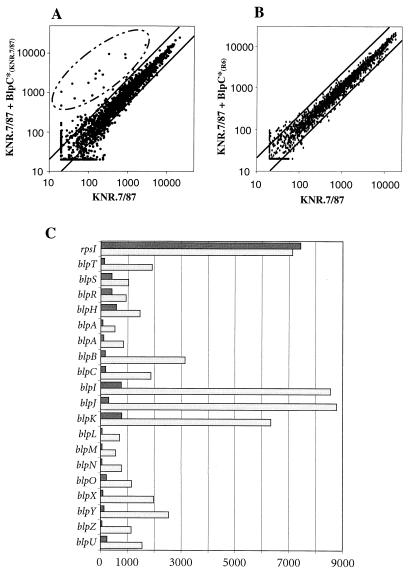
Microarrays were scanned at 570-nm, 3-μm resolution with a gene chip scanner (Affymetrix) and analyzed as previously described (24). The signal intensity for each gene is calculated as the average intensity difference, represented by [Σ(PM − MM)/(number of probe pairs)], where PM and MM denote perfect-match and mismatch probes. The average intensity difference value was then averaged for all experiments performed in duplicate to increase reproducibility. Intensity ratios are defined as the value of the average intensity difference in the condition where the gene is expressed at the highest level divided by the average intensity difference in the condition where the gene is expressed at the lowest level. A plus or minus sign is then applied to discriminate between gene induction and repression in comparison to the control condition. To reduce extreme intensity ratios for genes below the detection limit, we also arbitrarily set the minimum average intensity difference for each gene to the value of 20, which corresponds to the noise (see Fig. 2A and B).
FIG. 2.
Genes specifically induced by BlpC*. (A) Strain KNR.7/87 was grown to an optical density at 600 nm of 0.3, and the synthetic peptide BlpC*KNR.7/87 was added to a final concentration of 250 ng/ml to half of the culture. Cell pellets were collected after 30 min, RNA was extracted, and cDNA was labeled and hybridized to the microarray as described in Materials and Methods. The scatter graph shows the correlation for the intensities of all transcripts obtained for KNR.7/87 (x axis) versus KNR.7/87 30 min after addition of BlpC*KNR.7/87 (y axis). These values are the average of two independent experiments. The two solid lines flanking the diagonal indicate a difference of a factor of 2. Relevant changes involved 16 genes (circled). Due to the frameshift in blpA sequenced from strain KNR.7/87, this gene is represented by two independent probe sets on the microarray. (B) Same intensity scatter graph with KNR.7/87 after addition of BlpC*R6. KNR.7/87 (x axis) versus KNR.7/87 30 min after addition of BlpC*R6 (y axis). No genes were found to be differentially expressed. (C) Bar graph showing the normalized average intensity differences for a selection of genes in KNR.7/87 (dark bars) and KNR.7/87 30 min after addition of BlpC*KNR.7/87 (light bars). Genes are sorted as they appear on the contigs as depicted in Fig. 1. All 16 genes of the blp regulon plus the TCS operon (blpSRH) and a control ribosomal protein gene (rpsI) are shown (blpA again represented twice as two independent probe sets). The y axis shows average intensity differences calculated as described in Materials and Methods and is representative of the relative transcript abundance.
Northern blot analysis.
Total RNA (15 μg per lane) was electrophoresed on a formaldehyde gel and transferred to a nylon filter (GeneScreen). The RNA size standard was purchased from Gibco. [α32P]UTP-labeled RNA antisense probes were produced by in vitro transcription reaction from PCR-generated DNA fragments using a Lig'nScribe kit (Ambion). Probe blpT covers nucleotides 81 to 541, probe blpR covers nucleotides 1054 to 1528, probe blpA covers nucleotides 4850 to 5403, probe blpJ covers nucleotides 7861 to 8126, probe blpL covers nucleotides 8858 to 9391, probe blpN covers nucleotides 10655 to 11038, and probe blpY covers nucleotides 12929 to 13553. All sequence numbering is based on EMBL entry AJ276401.
Nucleotide sequence accession number.
The blp gene cluster sequence is available at GenBank as accession no. AJ276401.
RESULTS
Genomic characterization of a putative quorum-sensing TCS.
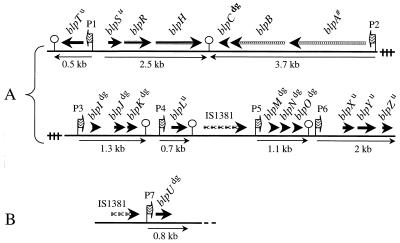
The sequences of the response regulator (RR13, BlpR) and the histidine kinase (HK13, BlpH) encoded by TCS13 display characteristic features of proteins constituting TCSs known to be regulated by short peptides and belonging to the Agr family (20). Immediately upstream of blpR, an ORF (blpS) was identified. The expected translation product shares significant sequence homology to the potential DNA binding domain of BlpR, and its structural gene is probably cotranscribed with blpRH (Fig. 1A). Downstream of blpH but transcribed in the opposite direction (Fig. 1A) we found a short ORF, blpC, encoding a peptide similar to the competence signaling peptide ComC (14, 15). In particular, the BlpC peptide contains a characteristic Gly-Gly motif that is believed to be important for its processing to the shorter mature signaling peptide (see below and Fig. 3A). Further downstream of blpC, two genes, blpA and blpB, were identified (Fig. 1A). These two genes are highly related to ComA and ComB (ABC transporter), which have been shown to be important for the export and processing of the ComC peptide. BlpA and BlpB share 65% and 31% identity with ComA and ComB, respectively. In strain KNR.7/87, the coding sequence of blpA contains a 4-bp insertion (compared to comA) that results in a change of the reading frame and in premature termination of the coding sequence. To determine whether this change is unique to strain KNR.7/87, the equivalent regions of 13 other clinical isolates and laboratory strains were sequenced. In total, six strains had no insertion and seven strains (including R6 and D39) displayed the same 4-bp (AAGC) insertion (data not shown). Further experiments would be required to determine if this frameshift affects functionality of the transporter in these strains.
FIG. 1.
Genomic organization of the blp regulon. Genes induced by BlpC* are clustered and were found on two contigs (A and B). Physical locations of these two contigs on the S. pneumoniae chromosome remain unknown. Genes as predicted by the GeneMark software are shown as thick arrows above the bold line. Patterned arrows were used for clarity. Genes encoding peptides with a leader of the double-glycine type are indicated with dg (superscript), and blp genes of unknown function are marked with u (superscript). Potential promoter regions with a conserved regulatory element comprised of two direct repeats upstream of extended −10 elements are shown as waved arrows and numbered P1 to P7. Promoter numbering is the same as in Fig. 5. The hairpin-like symbols indicate positions of inverted repeats that have the potential to form transcriptional terminators. Light arrows below the bold line indicate the transcriptional map with transcript sizes as measured by Northern blotting. blpA# indicates that the blpA gene exists in at least two allelic forms.
FIG. 3.
Sequence alignments for BlpC and BlpH. (A) Amino acid sequences of the various forms of the ComC (Csp) and BlpC signal peptide precursors. Amino acid residues in the leader peptides that match a reported consensus sequence (10) are shown in bold. Shading indicates amino acid similarity within the predicted mature signal peptide. (B) Comparison of two regions of the sensor domain of the BlpH histidine kinase from six S. pneumoniae strains. Significant substitutions are indicated in bold. Additional significant nonconservative amino acid substitutions are shaded. Boxes enclose long stretches of significant amino acid substitutions of the BlpH kinase from S. pneumoniae D39 and R6. A line above the sequence indicates stretches predicted to form TM segments II and V.
BlpC* peptide induces transcription of 16 genes.
We hypothesized that the Blp TCS together with the ABC transporter and the putative signaling peptide form a cell-cell signaling system with BlpC* as the signal. To test this hypothesis, the peptide corresponding to the predicted mature pheromone BlpC*KNR.7/87 was synthesized. To investigate the potential signaling effects of the synthetic BlpC*KNR.7/87 putative pheromone, the peptide was added to a concentration of 250 ng/ml (0.11 μM) to exponentially growing strains KNR.7/87 and R6 (optical density at 600 nm of 0.3). Subsequent changes in gene expression were monitored using a custom-designed genomic Affymetrix microarray (9; A. de Saizieu, N. Balmelle, B. Weber, C. Gardès, W. Keck, and R. Hakenbeck, unpublished data). This microarray allows expression probing of 1,973 predicted ORFs and 323 intergenic regions that are longer than 200 bp. Time course analysis indicated that BlpC*KNR.7/87 dramatically induces expression of a number of genes in strain KNR.7/87 but not in strain R6. Maximum gene induction was reached 10 min after the addition of the pheromone peptide and remained high until stationary phase (data not shown). The observed changes are visualized on the intensity scatter graph in Fig. 2A. These data were generated from two independent experiments and averaged. Based on standard deviation criteria, we consider as significant average intensity difference ratios greater than 2.5 when the average intensity difference is greater than 200 arbitrary units. Altogether, 16 genes are significantly up-regulated more than eightfold. In cases where the basal expression level was below the detection limit on the microarray, fold induction refers to fold above the noise level (set to 20 arbitrary units). An intensity bar graph is also shown in Fig. 2C. The gene coding for the small subunit ribosomal protein I (rpsI) is shown as an internal control. The genes blpS, blpR, and blpH were also found to be induced but not more than by a factor of 2, which probably reflects a bona fide induction, as it is consistent for the three genes, and the standard deviations were around 25% of the mean value. Most of the highly induced genes are located near the blpHR TCS, suggesting they all form a functional unit (Fig. 1A). Interestingly, the same set of genes (among others) is up-regulated in culture reaching stationary phase, suggesting a cell density-dependent quorum-sensing system (data not shown).
Allelic variation of the peptide pheromone BlpC*.
The fact that the BlpC*KNR.7/87 peptide induced changes in gene expression in strain KNR.7/87 and not in strain R6 suggests that there may be strain-specific peptide pheromones (pherotypes). It is well documented that the related Com system displays sequence variation within the signaling peptide ComC and its proposed target, the N-terminal domain of the histidine kinase ComD (15). Here we analyzed the sequence of DNA fragments amplified from genomic regions encoding BlpC and the N-terminal sequence of BlpH of six S. pneumoniae strains. Figure 3A shows the alignment of BlpC sequences together with peptide sequences derived from the related ComC signaling peptide. All of these sequences, including the six BlpC sequences, contain a highly conserved N-terminal half that ends in a Gly-Gly motif. The C-terminal portions of the signaling peptides vary significantly. For the six S. pneumoniae strains analyzed, three forms of BlpC* peptides were found. The N-terminal part of BlpH (proposed binding region for BlpC*) was also determined within the same six strains. Similarly, strain-dependent sequence variations were observed (Fig. 3B). However, strains that code for identical BlpC* peptides code for histidine kinases that are almost identical in their predicted extracellular domains, supporting the view that the kinase binds the BlpC* signaling peptide. Therefore, we postulate that at least three different blp pherotypes exist in S. pneumoniae, and we demonstrated that there is no blp regulon cross-induction between KNR.7/87 and R6.
Characterization of BlpC*-regulated operons.
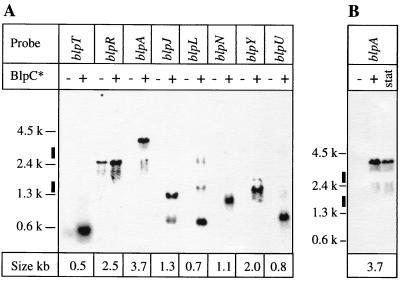
Detailed analysis of the expression data, including overall signal intensities of individual genes, induction factor upon peptide stimulation, and inspection of the genomic sequence allowed us to identify at least eight different operons regulated by BlpC* (Fig. 1). To validate this genomic prediction, systematic Northern blot analysis was performed with probes from each predicted operon. As shown in Fig. 4A, blpT, blpL, and blpU probes revealed main transcripts of 500, 700, and 800 bp, respectively. The blpR probe produced a tricistronic transcript of 2.5 kb covering blpSRH. About a twofold induction was also measured for the blpSRH transcript after induction by BlpC*, confirming the data obtained from the microarray. The blpA probe detected a 3.7-kb transcript covering blpABC. Three additional tricistronic operons were also identified with blpJ, blpN, and blpY probes of 1.3, 0.9, and 2 kb, respectively. With the exception of blpSRH, none of the transcripts were detected in the absence of external synthetic BlpC* peptide. All sizes for the main transcripts were in agreement with genomic prediction. The result in Fig. 4B confirms the natural induction of the blp gene cluster, exemplified by the blpABC probe, when cultures reach stationary phase.
FIG. 4.
Northern blot transcript analysis of KNR.7/87. Total RNA was isolated as described for microarray analysis. RNA probes were prepared and labeled with [32P]UTP using Lig'nScribe (Ambion). Probes correspond to the coding sequences of the genes indicated above the lanes as described in Materials and Methods. (A) Each probe was hybridized against two samples, KNR.7/87 and the same strain 30 min after addition, or in the absence, of synthetic BlpC*, indicated as + or − BlpC* above each lane. The size of the main transcript measured with each probe is indicated below each pair of lanes. The thick vertical lines indicate positions of 23S and 16S rRNAs. (B) An additional lane (stat) containing the same amount of total RNA isolated from stationary-phase KNR.7/87 culture was added. A blpA probe was used in the hybridization. The faint bands revealed by the blpA, blpJ, and blpL probes might correspond to transcript forms resulting from additional promoters, transcriptional readthrough, regulatory mRNA processing, cross-hybridization, or interference with rRNA.
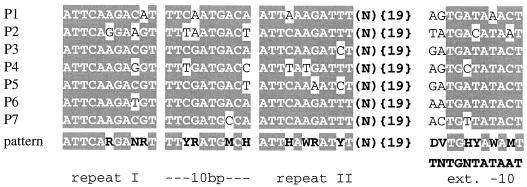
To search for conserved promoter elements, the regions immediately upstream of the potential start sites were aligned. An extended −10 region could be identified upstream of each operon. Furthermore, two direct 11-bp-long repeats spaced by exactly 10 bp were present in each promoter sequence with the exception of blpSRH (Fig. 5). Moreover, these repeats were all spaced by 23 bp from the extended −10 sites. The good conservation of these sequence elements as well as their precise positioning with regard to the extended −10 region suggested that they might constitute a binding site for a specific regulatory protein. Moreover, the blp consensus promoter element is in some features similar to the consensus sequence of the binding sites for the PlnC response regulator, whose DNA binding domain is related to that of BlpR (31). Taken together, the data suggest that BlpC* gene induction is mediated by the BlpHR TCS. We generated from the alignment a potential consensus binding site and used the program FINDPATTERN to search the genomic sequence of S. pneumoniae for occurrence of additional binding sites. The search failed to identify any additional promoter, indicating that the microarray identified all of the Blp-regulated genes represented in the available genome sequence.
FIG. 5.
Alignment of potential regulatory sequence elements identified upstream of operons induced upon entry into stationary phase or by external application of the synthetic pheromone peptide BlpC*. Two direct imperfect repeats (11 bp) separated by an extremely conserved spacer element of 10 bp are located 19 bp upstream of an extended −10 hexamer. The most conserved nucleotide positions are shaded. A pattern deduced from these elements is indicated using the GCG code for the degenerated nucleotide positions.
BlpC*-mediated gene activation depends on BlpH.
The genomic organization of the blpHR locus strongly suggests that the protein encoded by blpH constitutes the receptor and signal transducer for the pheromone BlpC*. To provide further evidence for such a mechanism, we analyzed gene induction mediated by BlpC*R6 in the wild-type strain R6 and in an R6 mutant lacking a functional blpH gene (R6 blpH′::pAS1::′blpH). We first established that BlpC*R6 also induced the blp cluster when the peptide was added to exponentially growing R6 cells but not KNR.7/87 cells (Fig. 2B). BlpC*R6 induced blpAB, blpK, blpU, blpT, and blpZ in R6 cells, whereas when BlpC*R6 was added to the BlpH-deficient R6 mutant, none of the blp genes were induced (not shown). Our data clearly indicate that BlpC* can induce gene expression only in the presence of a functional BlpH, confirming that it probably represents the BlpC* pheromone sensor.
In previously described genomic DNA hybridization experiments, the blpC, blpIJ, blpL, blpMNO, and blpXY genes of R6 were not detected with the KNR.7/87 oligonucleotide array (de Saizieu et al., unpublished), suggesting deletions or important sequence variation for the above-mentioned genes. As expected, expression analysis of R6 treated with its cognate signal BlpC*R6 did not reveal induction of the corresponding genes. This finding shows that there is significant allelic variation within some blp genes in addition to blpC as described above.
The blp cluster encodes potential bacteriocin peptides and immunity proteins.
Most of the BlpRH-regulated genes code for relatively small peptides. Detailed in silico sequence analysis revealed that the potential translation products of seven blp ORFs distributed over three operons (blpIJK, blpMNO, and blpU [Fig. 1]) contain a Gly-Gly motif characteristic of peptides that are secreted and processed by ComA/B-like proteins (Fig. 6). A common feature of the corresponding processed forms is a high alanine/glycine content. Homology searches and overall amino acid composition suggest that the individual predicted mature products of the blpIJK and blpMNO operons function synergistically such as described for the Streptococcus thermophilus two-peptide bacteriocin thermophilin 13 (25). Indeed, the predicted mature forms of BlpI and BlpJ are very similar to the ThmA and ThmB components of the two-peptide bacteriocin produced by S. thermophilus (Fig. 6). BlpU and BlpK are almost identical, differing only in one amino acid residue within the exported portion.
FIG. 6.
Amino acid comparison for the predicted Blp precursors with a typical Gly-Gly motif. Identical amino acid residues are shown in bold, while less conserved residues are shaded within the leader peptide. Note the high content of glycine and alanine residues of the expected processed forms. Below BlpJ and BlpI are compared with the components of the two-component thermophilin 13, a pore-forming bacteriocin produced by S. thermophilus (25). Conserved residues are shaded in gray.
Bacteriocin producers often show a specific immunity to their bacteriocin through the production of immunity determinants. Immunity proteins that protect the producer against the activity of two-peptide bacteriocins usually have a high pI, are larger than 110 aa, and contain several predicted TM segments (10). According to our computer analyses, BlpY (pI 10.69; 229 aa; five TM segments), BlpX (pI 10.78; 132 aa; two TM segments), and BlpL (pI 10.51; 134 aa; four TM segments) display these features. The product of blpT is probably a cytosolic protein.
DISCUSSION
Using oligonucleotide microarray technology for whole genome expression profiling, we have identified an S. pneumoniae regulon (blp) composed of genes for regulation, synthesis, export, and processing of Blps as well as their inducing factors. Treatment of S. pneumoniae log-phase cells with a synthetic form of the signal peptide BlpC* elicits a specific genetic response. Differential expression of the genes responding to BlpC* has also been observed at high-density/stationary phase. The cell density-dependent induction of the blp genes indicates that the blp regulon is a functional quorum-regulated system. We infer that BlpC is synthesized, exported, and processed in S. pneumoniae KNR.7/87 and R6.
A R6 mutant deficient in the BlpH histidine kinase does not respond to the synthetic form of BlpC*R6, corroborating our view that BlpH is the sensor for BlpC*. BlpH is related to a distinct group of histidine kinases including ComD of S. pneumoniae and AgrC of Staphylococcus aureus. Several (five to seven) potential TM segments characterize members of this group. Biochemical experiments have been reported that indicate a direct interaction of an octapeptide derived from AgrD with the AgrC sensor domain. It has been shown in vitro that autophosphorylation of AgrC is stimulated by a synthetic form of the cognate ligand (23). We postulate a similar mechanism for the regulation of the BlpH autokinase activity by BlpC*.
Our sequence analysis revealed a strain-dependent sequence covariation of BlpC* and its potential cognate receptor BlpH. Most of the significant amino acid substitutions are located in regions around the potential TM segments II and V (Fig. 3B). It is tempting to speculate that these parts of BlpH form a structure that might interact with BlpC*. The covariation of BlpH and its postulated ligand BlpC* is reminiscent of the covariance described for the AgrC octapeptide and the ComC* (CSP)-ComD receptor-ligand pairs (15, 17).
blpC is clearly autoinduced since it belongs to the genes induced upon BlpC* stimulation. Autoinduction allows signal amplification and is well known for regulatory systems controlling fast switches to distinct developmental programs. The activity of components controlling these switches is typically limited by feedback loops that prevent an overshoot of the response (35). Overshoot of the blp response might be avoided via a negative feedback through increased bacteriocin production. Stimulated synthesis of bacteriocin-like peptides could lead to a decrease in signal peptide processing and transport.
A common feature of BlpR-related response regulators (ComE, AgrA, PlnC, and PlnD) is a high content of basic residues within the effector output domains, suggesting a common DNA binding mode (10). Recently, the target DNA binding site for ComE has been identified. It consists of two imperfect direct repeats (9 bp) spaced by a segment of 12 nucleotides (37). Similar regulatory elements were shown to be the target DNA binding sites for PlnC and PlnD two-response regulators controlling bacteriocin production in Lactobacillus plantarum (10, 28, 31). We postulate that the direct repeats found in the blp promoters are the target DNA binding sites for BlpR. Compared to known regulatory elements targeted by Agr-like response regulators, the direct repeats upstream of the blp operons are separated by a highly conserved spacer element. Conservation of this spacer element might indicate an important regulatory role.
blpS has the potential to code for a protein homologous to the proposed DNA binding domain of the BlpR and AgrA/ComE type of response regulators but lacks the typical receiver domain. The pairwise arrangement of blpR and blpS is reminiscent of the tandem organization of plnC and plnD, the genes for full-length agrA-like response regulators of L. plantarum (10). The role of BlpS remains to be determined.
The DNA sequence upstream of the blpSRH operon contains an element with good similarity to a canonical extended −10 promoter element (32). The presence of a promoter at this site is in agreement with the blpSRH transcript size obtained by Northern analysis. The −10 promoter-like element is located close to the conserved regulatory element of blpT, which is transcribed in the divergent orientation (Fig. 1). Expression analysis using the microarray and Northern technique revealed a slight but significant increase in blpSRH mRNA levels upon BlpC* stimulation, despite the nonconforming orientation and position of the inverted repeats with respect to the blpSHR promoter. The apparent responsiveness of the blpSRH operon expression to BlpC* treatment might be due to an interference with activation of blpT expression.
Strain-dependent sequence variation is not restricted to blpC and blpH but extends to the genes for the Blps and their associated ORFs for potential immunity determinants (de Saizieu et al., unpublished). Diversity is typical for bacteriocins, and the genetic mechanisms leading to this diversity are often based on horizontal gene transfer and other variability-generating events (30). Bacteriocin diversity has been proposed to be the outcome of intense microbial competition and emergence of resistance in the target organisms. It is plausible to speculate that certain parts of the blp locus are subject to variability-generating phenomena similar to those described for the evolution of bacteriocins. In this context, it is interesting that two copies of the insertion sequence (IS) element IS1381 are located close to blp genes (33). IS elements can participate in homology-dependent reactions by providing homology for insertions/deletions or for additive recombination events.
Bacteriocins are commonly defined as compounds produced by bacteria that selectively inhibit or kill closely related species. It has been reported that several S. pneumoniae strains produce bacteriocins inhibiting the growth of some S. pneumoniae strains whereas certain strains exhibit immunity (26). It is possible that the genetic determinants for pneumocin production and immunity correspond to some of the blp genes.
Recently, an S. pneumoniae mutant deficient in BlpHR (486hh/rr pair) was tested in a respiratory tract infection model. The mutants were significantly attenuated, and bacterial cell counts in mice lungs decreased within a 48-h time period compared to a wild-type strain (36). Taken together, the Blp TCS and therefore blp gene products appear to be important for S. pneumoniae survival in the lung. Lungs are usually sterile and thus no microbial competitors for S. pneumoniae should be present, raising the question of the biological role of a bacteriocin regulon in the lung. BlpI and BlpJ are homologous to components of the broad-host-range pore-forming thermophilin 13. In vitro measurements have shown that thermophilin 13 does not need a specific membrane composition or proteinaceous receptor for its activity. In this regard, it is worth mentioning that S. pneumoniae displays beta-hemolytic activity, and a second hemolysin distinct from pneumolysin has been postulated (5). Hemolytic activity has also been described for a two-component lantibiotic encoded by the conjugative plasmid pAD1 of Enterococcus faecalis that contributes to animal virulence (8, 13). In this context, it is tempting to speculate that some of the blp gene products might compromise host cells, thus contributing to the proliferation of S. pneumoniae in the lung. Alternatively, additional strain-specific genes not present in the available DNA genome sequence data and therefore not represented on the S. pneumoniae chip might belong to the blp regulon, and corresponding gene products might contribute to proliferation in the lung.
The present study clearly demonstrates that the microarray technology is a valuable tool to analyze bacterial expression in response to a defined stimulus. We believe that bacterial expression profiling using the microarray technology will also help to elucidate the complex regulatory networks operating in pathogenic bacteria during the course of an infection. These new insights will allow development of new concepts for the prevention, diagnosis, or treatment of bacterial infections.
ACKNOWLEDGMENTS
We gratefully acknowledge Detlef Wolf, Clemens Broger, and Martin Neeb for help with bioinformatics. We thank Eric Kitas for chemical synthesis of the BlpC* peptide. We also thank Karin Kuratli, Nadège Balmelle, and Katharina Rupp for excellent technical assistance.
REFERENCES
- 1.Aaberge L S, Eng J, Lermark G, Lovik M. Virulence of Streptococcus pneumoniae in mice: a standardized method for preparation and frozen storage of the experimental bacterial inoculum. Microb Pathog. 1995;18:141–152. doi: 10.1016/s0882-4010(95)90125-6. [DOI] [PubMed] [Google Scholar]
- 2.Atherton E, Sheppard R. Solid phase peptide synthesis: a practical approach. Oxford, United Kingdom: IRL Press; 1989. [Google Scholar]
- 3.Avery O T, MacLeod C M, MacCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. J Exp Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brurberg M B, Nes I F, Eijsink V G. Pheromone-induced production of antimicrobial peptides in Lactobacillus. Mol Microbiol. 1997;26:347–360. doi: 10.1046/j.1365-2958.1997.5821951.x. [DOI] [PubMed] [Google Scholar]
- 5.Canvin J R, Paton J C, Boulnois G J, Andrew P W, Mitchell T J. Streptococcus pneumoniae produces a second haemolysin that is distinct from pneumolysin. Microb Pathog. 1997;22:129–132. doi: 10.1006/mpat.1996.0098. [DOI] [PubMed] [Google Scholar]
- 6.Certa U, de Saizieu A, Mous J. Hybridization analysis of labeled RNA by oligonucleotide arrays. 2000. Methods Mol. Biol., in press. [DOI] [PubMed] [Google Scholar]
- 7.Chen J D, Morrison D. Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning DNA from Streptococcus pneumoniae. Gene. 1988;64:155–164. doi: 10.1016/0378-1119(88)90489-1. [DOI] [PubMed] [Google Scholar]
- 8.Chow J W, Thal L A, Perri M B, Vazquez J A, Donabedian S M, Clewell D B, Zervos M J. Plasmid-associated haemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob Agents Chemother. 1993;37:2474–2477. doi: 10.1128/aac.37.11.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Saizieu A, Certa U, Warrington J, Gray C, Keck W, Mous J. Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nat Biotechnol. 1998;16:45–48. doi: 10.1038/nbt0198-45. [DOI] [PubMed] [Google Scholar]
- 10.Diep D B, Håvarstein L S, Nes I F. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J Bacteriol. 1996;178:4472–4483. doi: 10.1128/jb.178.15.4472-4483.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunny G M, Leonard B A. Cell-cell communication in gram-positive bacteria. Annu Rev Microbiol. 1997;51:527–564. doi: 10.1146/annurev.micro.51.1.527. [DOI] [PubMed] [Google Scholar]
- 12.Fuqua W C, Winans S C, Greenberg E P. Quorum-sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilmore M S, Segarra R A, Booth M C, Bogie C P, Hall L R, Clewell D B. Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to lantibiotic determinants. J Bacteriol. 1994;176:7335–7344. doi: 10.1128/jb.176.23.7335-7344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Håvarstein L S, Coomaraswamy G, Morrison D A. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–1144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Håvarstein L S, Hakenbeck R, Gaustad P. Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J Bacteriol. 1997;179:6589–6594. doi: 10.1128/jb.179.21.6589-6594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Håvarstein L S, Morrison D A. Quorum sensing and peptide pheromones in streptococcal competence for genetic transformation. In: Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C.: American Society for Microbiology; 1999. pp. 9–192. [Google Scholar]
- 17.Ji G, Beavis R, Novick R P. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 18.Ji G, Beavis R C, Novick R P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleerebezem M, Quadri L E, Kuipers O P, de Vos W M. Quorum-sensing by peptide pheromones and two-component signal transduction systems in Gram-positive bacteria. Mol Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 20.Lange R, Wagner C, de Saizieu A, Flint N, Molnos J, Stieger M, Caspers P, Kamber M, Keck W, Amrein K E. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene. 1999;237:223–234. doi: 10.1016/s0378-1119(99)00266-8. [DOI] [PubMed] [Google Scholar]
- 21.Lazazzera B A, Grossman A D. The ins and outs of peptide signaling. Trends Microbiol. 1998;6:288–294. doi: 10.1016/s0966-842x(98)01313-4. [DOI] [PubMed] [Google Scholar]
- 22.Lazazzera B A, Solomon J M, Grossman A D. An exported peptide functions intracellularly to contribute to cell density signaling in Bacillus subtilis. Cell. 1997;89:917–925. doi: 10.1016/s0092-8674(00)80277-9. [DOI] [PubMed] [Google Scholar]
- 23.Lina G, Jarraud S, Ji G, Greenland T, Pedraza A, Etienne J, Novick R P, Vandenesch F. Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol Microbiol. 1998;28:655–662. doi: 10.1046/j.1365-2958.1998.00830.x. [DOI] [PubMed] [Google Scholar]
- 24.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, Brown E L. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 25.Marciset O, Jeronimus-Stratingh M C, Mollet B, Poolman B. Thermophilin 13, a nontypical antilisterial poration complex bacteriocin, that functions without a receptor. J Biol Chem. 1997;272:14277–14284. doi: 10.1074/jbc.272.22.14277. [DOI] [PubMed] [Google Scholar]
- 26.Mindich L. Bacteriocins of Diplococcus pneumoniae. I. Antagonistic relationships and genetic transformations. J Bacteriol. 1966;92:1090–1098. doi: 10.1128/jb.92.4.1090-1098.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mortier-Barrière I, de Saizieu A, Claverys J-P, Martin B. Competence-specific induction of recA is required for full recombination proficiency during transformation in Streptococcus pneumoniae. Mol Microbiol. 1998;27:159–170. doi: 10.1046/j.1365-2958.1998.00668.x. [DOI] [PubMed] [Google Scholar]
- 28.Nes I F, Eijsink V G H. Regulation of group II peptide bacteriocin synthesis by quorum-sensing mechanisms. In: Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C.: American Society for Microbiology; 1999. pp. 175–192. [Google Scholar]
- 29.Pestova E V, Håvarstein L S, Morrison D A. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol. 1996;21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- 30.Riley M A. Molecular mechanisms of bacteriocin evolution. Annu Rev Genet. 1998;32:255–278. doi: 10.1146/annurev.genet.32.1.255. [DOI] [PubMed] [Google Scholar]
- 31.Risøen P A, Håvarstein L S, Diep D B, Nes I F. Identification of the DNA-binding sites for two response regulators involved in control of bacteriocin synthesis in Lactobacillus plantarum C11. Mol Gen Genet. 1998;259:224–232. doi: 10.1007/pl00008627. [DOI] [PubMed] [Google Scholar]
- 32.Sabelnikov A G, Greenberg B, Lacks S A. An extended −10 promoter alone directs transcription of the DpnII operon of Streptococcus pneumoniae. J Mol Biol. 1995;250:144–155. doi: 10.1006/jmbi.1995.0366. [DOI] [PubMed] [Google Scholar]
- 33.Sanchez-Beato A R, Garcia E, Lopez R, Garcia J L. Identification and characterization of IS1381, a new insertion sequence in Streptococcus pneumoniae. J Bacteriol. 1997;179:2459–2463. doi: 10.1128/jb.179.7.2459-2463.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith M, Guild W. A plasmid in Streptococcus pneumoniae. J Bacteriol. 1979;137:735–739. doi: 10.1128/jb.137.2.735-739.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thieffry D, Huerta A M, Perez-Rueda E, Collado-Vides J. From specific gene regulation to genomic networks: a global analysis of transcriptional regulation in Escherichia coli. Bioessays. 1998;20:433–440. doi: 10.1002/(SICI)1521-1878(199805)20:5<433::AID-BIES10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 36.Throup J P, Koretke K K, Bryant A P, Ingraham K A, Chalker A F, Ge Y, Marra A, Wallis N G, Brown J R, Holmes D J, Rosenberg M, Burnham M K. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol Microbiol. 2000;35:566–576. doi: 10.1046/j.1365-2958.2000.01725.x. [DOI] [PubMed] [Google Scholar]
- 37.Ween O, Gaustad P, Håvarstein L S. Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol Microbiol. 1999;33:817–827. doi: 10.1046/j.1365-2958.1999.01528.x. [DOI] [PubMed] [Google Scholar]
- 38.Wodicka L, Dong H, Mittmann M, Ho M-H, Lockhart D J. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]