Abstract
Most of the proteins in the circulation are N-glycosylated, shaping together the total blood N-glycome (TBNG). Glycosylation is known to affect protein function, stability, and clearance. The TBNG is influenced by genetic, environmental, and metabolic factors, in part epigenetically imprinted, and responds to a variety of bioactive signals including cytokines and hormones. Accordingly, physiological and pathological events are reflected in distinct TBNG signatures. Here, we assess the specificity of the emerging disease-associated TBNG signatures with respect to a number of key glycosylation motifs including antennarity, linkage-specific sialylation, fucosylation, as well as expression of complex, hybrid-type and oligomannosidic N-glycans, and show perplexing complexity of the glycomic dimension of the studied diseases. Perspectives are given regarding the protein- and site-specific analysis of N-glycosylation, and the dissection of underlying regulatory layers and functional roles of blood protein N-glycosylation.
Keywords: glycosylation, glycan, glycomics, glycome, protein glycosylation, mass spectrometry, blood, biomarker
1. Introduction
Glycoconjugates are glycan-modified proteins and lipids that coat the surface of all human cells but also of most microorganisms, impacting a myriad of biological processes across all life domains.1 They play pivotal roles in shaping cell–cell interactions, are entry points for pathogens, influence protein secretion, function, stability and half-life, and build up the extracellular matrix.2 Glycoconjugates are overall less abundant at the intracellular space, yet are important mediators of cellular homeostasis.2 Next to being ubiquitous, glycans exhibit remarkable structural diversity.3 Complex networks of enzymes tailor the repertoire of glycans in a nontemplate driven, co- and post-translational biosynthetic process called glycosylation. During this process, serial action of glycosidases and glycosyltransferases gives rise to glycosylated proteins, lipids, and even ribonucleic acids.4,5
The majority of secreted proteins in the human body are glycosylated, and glycans on secretory proteins gradually mature as they transit the compartments of the endoplasmic reticulum and the Golgi apparatus, under the regulation of genetic, cellular, metabolic, and environmental influences.6−12 The resulting diversity brings about an estimated ≥7000 glycan structures a selection of which will be found attached to any specific glycosylation sites (microheterogeneity) with varying degree of site-occupancy (macroheterogeneity), collectively defining the protein glycoform profile and forming the human glycome and glycoproteome.13
Glycoproteins are arguably the most studied type of glycoconjugates. Protein glycosylation is one of the most prevalent post-translational modification adding a layer of complexity that goes beyond genetic coding, allowing for subtle alterations in protein activity, stability, and interactions.14,15N- and O-glycosylation are the most frequent types, distinguished by the specific site on the protein backbone they extend from–asparagine residues at Asn-X-Ser/Thr motifs and serine or threonine residues, respectively.2 Changes in protein glycosylation have been described for many cell- and tissue (de)differentiation processes and appear to coincide with normal physiological processes as well as pathologies.16,17 Numerous well-established examples highlight the role of glycosylation in inherited and acquired human diseases.16,18,19 However, for many examples, the causative or contributory role of altered protein glycosylation remains elusive and is therefore an ongoing area of investigation.20 Such gaps arise due to a lack of mature tools to study and manipulate glycosylation, the utilization of diverse and low-throughput analytical methods on small sample sizes, unsuitable cohorts, flawed or inconsistent data analysis workflows, lack of replication, a restricted set of glycan features analyzed, or a combination thereof.21 Consequently, crucial information regarding key glycan structural features, and the role of glycosylation on many proteins (e.g., of major blood proteins) and in disease development is frequently missing or poorly understood.
Complementary techniques for analyzing protein glycosylation include lectin binding assays as well as hydrophilic interaction liquid chromatography (HILIC) and capillary electrophoresis with fluorescence detection of reducing end-tagged glycans.22,23 Mass spectrometry (MS) has become the key technology for assessing glycans from body fluids, cells and tissues of interest.24−26 MS allows for the concomitant detection and relative quantification of a large number of glycoforms at high sensitivity. Some of these approaches offer the possibility to discern changes in key structural features (glycosylation traits), including antennary and core fucosylation, bisection, galactosylation and (linkage-specific) sialylation, as well as antennarity and glycan type in a rapid single measurement23,27 (Figure 1). Deriving information on the relative contribution of the above-mentioned glycosylation traits can be particularly valuable in case-control settings, because they may be indicative of altered biosynthetic pathways in a compressed, easily interpretable way.28,29
Figure 1.
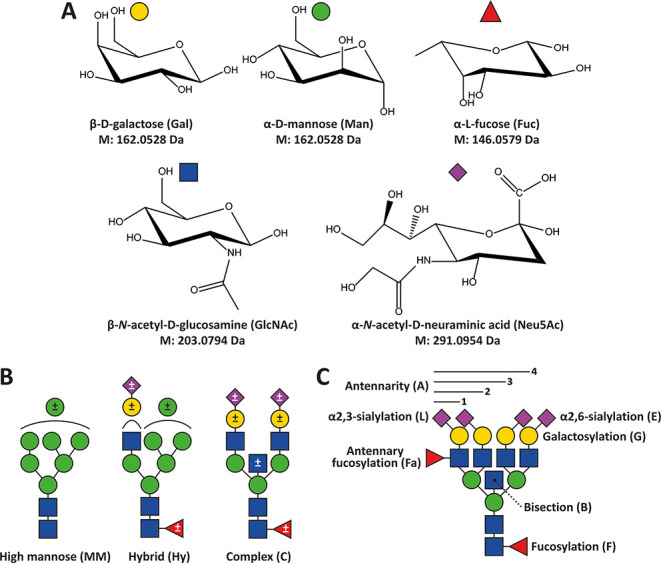
Monosaccharide diversity and glycan types characterizing human N-glycans with illustrative examples of glycosylation traits calculated therefrom. (A) Common monosaccharide constituents of human blood protein-associated N-glycans and their monoisotopic residue mass. (B) General types of N-glycans in humans. Optional modifications are indicated by ± in the symbols. Neuraminic (sialic) acids can be either α2,3- or α2,6-linked. Other potential modifications include phosphorylation, sulfation, sialic acid acetylation, or LacdiNAc repeats. (C) Glycosylation traits used to describe N-glycans in this study. Sialic acids with undefined linkages are denoted with the abbreviation S throughout.
Of all human tissues and body fluids, blood and its components are the most extensively studied regarding their glycosylation. The total blood N-glycome (TBNG) appears to be dominated by circulating proteins, i.e. serum and plasma that are devoid of cellular components, with the whole blood N-glycome, including cellular components, hardly differing from total plasma N-glycome (TPNG).30 Serum and plasma N-glycomes do show some minor, albeit consistent, differences, reflecting their slightly different glycoproteomic composition caused in part by fibrinogen consumption with blood clotting.31 In the context of this paper, TBNG will be used as an umbrella term capturing TPNG and the total serum N-glycome (TSNG).
Blood proteins undergo substantial glycosylation in hepatocytes and plasma cells, making blood, serum and plasma readily accessible sources for glycan biomarker discovery.17 Like any phenotypic trait, the TBNG reflects interindividual differences. These variations are influenced by factors such as age and sex, as well as by environmental and dietary conditions.6,32 In diseases, TBNG demonstrates distinct glycosylation patterns that are associated with a wide range of pathologies: this includes everything from congenital disorders of glycosylation (CDG) to cancer, neurological disorders, as well as systemic inflammation, infectious diseases, and metabolic conditions.33−41
In this Perspective, the spotlight will be on the TBNG, exploring an area of the human glycome which is relatively well mapped due to unified analytical approaches such as matrix-assisted laser desorption/ionization–mass spectrometry (MALDI-MS)-based workflows with sufficient throughput for the measurement of hundreds-to-thousands of samples in clinical cohorts and the ability to resolve functionally important sialic acid linkage isomers.27,36,42 The TBNG glycomic workflow established in our laboratory builds on the initial enzymatic release of N-glycans from circulatory proteins, linkage-specific sialic acid derivatization, cotton HILIC-based enrichment, MALDI-MS measurement, followed by a largely automated data extraction and analysis workflow.27,36,43,44
After providing brief historical context on protein N-glycosylation in genetic diseases and polymorphisms, we re-explore the blood N-glycomic signatures of major diseases obtained in our laboratory over the past decade and the influence of key demographic confounders thereon. TBNGs of different disease types and diseases, including 4 oncological,35,41,45,46 1 metabolic,37 1 infectious33 and 2 inflammatory gastrointestinal diseases,38,47 will be integrated, compared and discussed. We conclude by contextualizing the acquired findings, delve into current and future challenges, and highlight translational examples within the field of glyco(proteo)mics.
2. Impact of Genetic Variation on Blood Protein Glycosylation
A classic example of the use of glycan biomarkers is in the detection of rare monogenic diseases called congenital disorders of glycosylation (CDG), causing developmental and/or neurological symptoms. These diseases arise due to the defect of glycan biosynthetic or metabolic pathways, commonly embodied in systemic alterations in the glycoproteome.48−50 Historically, transferrin isoelectric focusing has served as a benchmark method for indicating the presence of CDGs, but MS-based techniques have additionally been implemented in both the clinical and research laboratories, allowing for the targeted investigation of glycoproteins such as transferrin or immunoglobulin G, but also the global analysis of the TBNG and the site- and protein-specific glycoproteome within the CDG spectrum.50−54
Beyond CDGs, aspects of normal genetic diversity, particularly polymorphisms, have been shown to impact glycosylation, with the ABO blood group system serving as a prime example of such polymorphism.55 As for the TBNG, an extensive population study described specific TBNG glycosylation phenotypes displaying high heritability, contributing to both interindividual variation within populations and variations between populations.56 Furthermore, genome-wide association studies (GWAS) revealed potential regulators of glycosylation events.57 A remarkable example is a GWAS study identifying transcription factor hepatocyte nuclear factor alpha (HNF1α) as a master regulator of antennary fucosylation. Next to loss-of-function mutations in this gene upon normal genetic diversity, also its epigenetic silencing resulted in the disappearance of the corresponding glycans of plasma protein origin in HILIC-ultrahigh-performance liquid chromatography measurements.7,58HNF1α is a risk factor of monogenic diabetes (type 3 maturity-onset diabetes of the young (MODY)), and such patients’ circulatory glycoproteins are characterized by a lowered ratio of antennary fucosylated tri-antennary to nonfucosylated tri-antennary glycans, which turns out to be of differential diagnostic value when discriminating MODY from other types of diabetic conditions.59 Another gene that was found to associate with variation within the blood N-glycome is SLC9A9, which codes for an endosomal proton pump.60 Polymorphisms in this gene have been proposed to be implicated in attention-deficit hyperactive disorder (ADHD), albeit further evidence and validation cohorts are necessary in order to establish a causative link.60,61 Intriguingly, the genetic associations of the glycome of immunoglobulin G and transferrin appeared to be divergent, reflecting the fact that those proteins are mainly B cell (plasma cell) and hepatocyte-derived, respectively.12 This study exemplifies the power of large-scale, protein specific insights. It also shows the power and relevance of studying protein glycosylation in tissue- and cell-type-resolved contexts, revealing the existence of cell-type-specific regulatory programs. Additional genetic variants directly or indirectly involved in glycosylation biosynthetic pathways are discussed elsewhere.12,57
3. A Cross-Study Overview of the Measured Glyco-Traits
To showcase the commonalities and differences of TPNG signatures of prevalent human diseases, we undertook a cross-disease study meta-analysis approach (Supplementary Figures and Methods). To begin, we initiated a preliminary, integrated analysis of the TPNGs from eight studies,33,35,37,38,41,45−47 each concentrating on distinct diseases or disease groups, yet unified through the use of a standardized methodology.27,36,43,44 The human blood N-glycome, characterized by its diversity and phenotypic variability, presents numerous challenges. While immunoglobulins and certain low-abundance regulatory proteins have extrahepatic origin, a significant portion of blood proteins, including albumin, clotting factors, acute phase proteins like α1-acid-glycoprotein, and transport proteins, are synthesized in the liver. The clinical conditions examined in this report, while distinct, each have the potential to trigger remodeling of liver protein production. This, in turn, suggests a significant hepatic influence on the blood N-glycome. Despite the use of standardized methods across the studies, questions remain about the consistency of glycans measured in various studies under differing clinical conditions. This aspect has not been systematically explored to date.
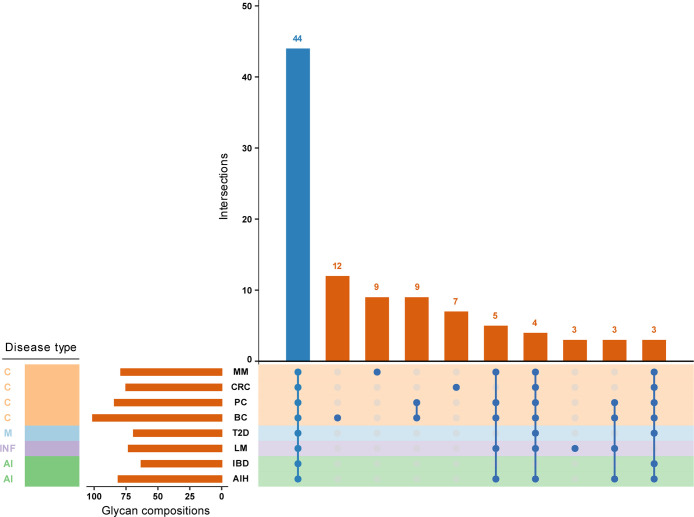
Figure 2 shows an UpSet plot, a tool for visualizing overlaps and unique elements across multiple sets. This figure reveals that, while the number of measured glycan compositions per data set varies, ranging from 63 to 101 features, nonetheless, a core set of 44 glycan compositions is consistently measured across all data sets (Table S1). Similarly to blood proteins, glycans linked to them likewise show large interindividual comparability, resulting in a large overlap of the glycome constituents between individuals and cohorts. Furthermore, given the outlined sources of variability, these findings underscore a significant degree of analytical consistency. It is also important to mention the presence of unique single-set intersections. For example, these intersections are evident in almost every oncological condition examined in the overview (Figure 2). While one might be inclined to interpret them as “disease-specific compositions”, the commonalities and differences in glycome coverage may rather reflect technical differences of those studies with regard to sensitivity, resolution, and data curation settings.
Figure 2.
UpSet plot displaying the overlaps and unique elements among the measured glycan compositions across eight studies included in the report. The size of each intersection is indicated at the top of the corresponding vertical bar, while the number of measured glycan compositions per data set is shown on the horizontal bars. Disease types are categorized as follows: C – cancer, AI – autoimmune and inflammatory conditions, INF – infectious-, and M – metabolic disease. Abbreviations of diseases (studies): MM – multiple myeloma,41 CRC – colorectal cancer,45 PC – pancreatic cancer,35 BC – breast cancer,46 T2D – type 2 diabetes,37 LM – leishmaniasis,33 IBD – inflammatory bowel disease (composed of ulcerative colitis and Crohn’s disease),38 AIH – autoimmune hepatitis.47
4. Influences of Demographic Factors on Blood Protein Glycosylation
The consistent exploration of the association between blood protein glycosylation and major demographic factors such as age and sex began in the last decades of the previous century. Boosted by the technological advances of recent years, this has led to the description of age- and sex-related differences in the human blood N-glycome.6,9,32,56,62−64
In the past few decades, various groups of investigators consistently revealed that glycosylation of the single blood protein, immunoglobulin G (IgG), characteristically changes upon aging. The observation was the accumulation of undergalactosylated IgG with progressing age, a phenomenon that is proposed to particularly well reflect chronic systemic inflammation (inflammaging) in both health and disease.63,65−68 Inspired by these findings, later large-scale studies additionally extended their investigation to the TBNG, characterized by more diversity in their N-glycosylation, confirming and finding new associations of the N-glycome with age, sex, their interaction, body mass index, metabolic health or smoking.6,9,32,64
We previously explored the associations of N-glycome with major demographic factors, specifically age and sex.36 This exploration was conducted by using individual linear regression models for assessing age-related variations and logistic regression models for sex-related differences in each measured trait. Here, we decided to introduce an additional exploration layer by employing linear models that include interaction terms (Supplementary Figures and Methods). This way, sex-specific and age-associated glycosylation differences become apparent together with insights into the intricate differences of age-associations between the sexes.
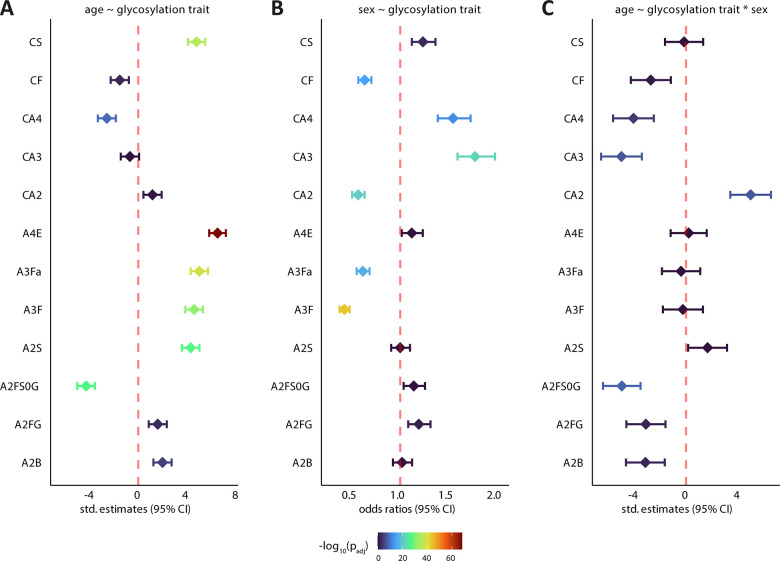
For our analysis, we followed the approach outlined in this previous report,36 pooling all control samples from the referenced studies. To ensure comparability across cohorts, we recalculated the glycosylation traits uniformly (Table S2). The total pool of control samples comprised 1776 observations. Figure S1 illustrates a density plot of the age distribution across each sex stratum. Notably, while the median age skews toward the higher end (62 years), the distribution across sex strata is well-balanced. Here we constructed two linear models for each glycosylation trait. The first linear model examined simple associations with age, while the second incorporated sex as an interaction term (Figure 3A and C). Standardized coefficients in linear regression (also known as beta coefficients or standardized regression coefficients) shown on the x-axis are used to compare the relative strength and direction of relationships between variables on a uniform scale, indicating the average change in standard deviations of the dependent variable (age) for each one standard deviation increase in a predictor variable (glycosylation trait). We also incorporated logistic regression models using sex as an outcome (Figure 3B). Figure 3 presents a summary of the most significant glyco-traits identified in each type of model. Table S3 summarizes the outcomes of all of the models.
Figure 3.
Summary of the most significant associations between glycosylation traits and demographic factors in the pooled control group across eight studies. (A) Linear models for age ∼ glycosylation trait, where the standardized estimates on the x-axis indicate the change in age (in standard deviations) for a one standard deviation increase/decrease in the glycosylation trait. (B) Logistic regression models for sex ∼ glycosylation trait, with odds ratios above 1 suggesting the trait is more prevalent in females than in males. (C) Linear models with interaction terms (age ∼ glycosylation trait × sex) demonstrate how the effect of glycosylation traits on age varies by sex; a positive coefficient suggests a stronger association in females, while a negative one indicates a stronger association in males. An example glycosylation trait, A2B, represents the proportion of glycan species with a bisecting GlcNAc (B) among all di-antennary glycans (A2). A2FS0G denotes the levels of galactosylated species (G) within nonsialylated (S0) and fucosylated (F) di-antennary species (A2). Other glycosylation traits follow similar analogies (see Figure 1C, Table S2 for details).
The trait A2B (level of bisection within di-antennary glycans) may serve as an example of the information conveyed by Figure 3. Figure 3A shows a significant relationship between age and this glyco-trait: as age increases, as do A2B levels, in a predictable manner. Figure 3B shows that the odds ratio of the trait is around zero; this indicates that variations in this trait do not significantly affect whether someone is likely to be male or female, meaning that A2B are overall similar for males and females. For example, an odds ratio above 1 would indicate that this glycosylation trait is more likely associated with female sex. However, Figure 3C, showing the models with an interaction term, indicates that the relationship between age and the A2B glyco-trait differs across sexes. In other words, sex influences the relationship between age and this glyco-trait. Specifically, the negative std estimates of A2B suggest that the increase in A2B is more explicit in males. For a visual representation of this interaction, Figure S2 displays a plot of the predicted values. This plot clearly illustrates that the increase in A2B with age is less pronounced in females than in males.
The major sex associations are represented in Figure 3B, with the most significant ones suggesting sex-dependent changes in fucosylation and N-glycan antennarity. The odds ratios significantly lower than 1 for both core- and antennary fucosylation (CF, A3Fa, A3F) suggest that fucosylation is lower in females compared to males. This pattern is also observed for di-antennary glycans (CA2). Conversely, the presence of multiantennary structures (CA3, CA4) is associated with odds ratios higher than 1, indicating a relatively higher likelihood of these structures being more abundant in females.
In accordance with literature reports and expectations, galactosylation of fucosylated, nonsialylated di-antennary glycans, hallmarking IgG Fc galactosylation, (A2FS0G) displayed the most remarkable negative association with age (std estimate: −4.28; p-value = 3 × 10–28) (Figure 3A).6,65,69−71 Additionally, we confirm previous observations on significant intersex differences in this glyco-trait, showing a stronger effect for males than females (Figure 3C).32 On the other hand, galactosylation of fucosylated di-antennary glycans (A2FG) showed an opposite trend, presumably due to the contribution of high abundant blood proteins enriched in sialylation in this glyco-trait, unlike IgG.
Indeed, di-antennary sialylation (A2S) exhibited a positive age association, whereas tri- (A3S) and tetra-antennary sialylation (A4S) displayed no or weak age associations (Table S3). Our methodological approach additionally allowed the dissection of changes related to sialylation in a linkage-specific fashion. Interestingly, directionally uniform alterations were observed for di-antennary sialyl-linkage variants (A2E, A2L), opposing linkage-specific effects were observed in case of tri- (A3E, A3L) and tetra-antennary (A4E, A4L) structures (Table S3), as described before.36 The strongest and most significant positive association with age was attributed to α2,6-sialylation of tetra-antennary glycans (A4E, std estimate: 6.5, p-value = 6.1 × 10–69), accompanied by a substantial decrease in its α2,3-sialylated counterpart (A4L; std estimate: −3.0, p-value = 6.7 × 10–4) (Figure 3A, Table S3). Based on the findings on decreased tetra-antennary structures (CA4) which are known carriers of highly sialylated glycans, one may speculate a parallel, substantial remodeling of the linkage-specific sialylation status upon aging, with potential implications on glycoprotein clearance and/or function. No sizable sex-specific associations were found for these glycosylation traits.
The change in di-antennary structures (CA2) was directionally opposing but in magnitude similar to tri- antennary structures. On the other hand, CA4, as aforementioned, decreased more pronouncedly over the lifespan of healthy individuals (Figure 3A). We therefore suggest that the observed phenomenon may be a trade-off, resulting in a shift toward di-antennary (CA2) glycans, due to either the relative decrease of tri- and tetra-antennary counterparts, the relative increase of di-antennary ones, or the combination thereof. Another related observation was the increase in sialylation of A2S, which largely outweighed the increase in associated di-antennary structures (CA2) (Figure 3A). Also, overall sialylation within complex-type glycans (CS), regardless of antenna number, profoundly increased with age (Figure 3A). This raises the challenging question of what are the key drivers of these changes, which may include differences in glycoprotein abundance as well as differences in site-specific glycosylation. The compiled data reveal an abundance increase in di-antennary structures (CA2), but an even more pronounced increase in sialylation levels on these (A2S, A2E, A2L), reflecting an alteration in protein makeup and/or glycosylation status (Figure 3A, Table S3). All this at the expense of decreasing abundance of proteins carrying tri- and tetra-antennary glycans (CA3, CA4), of which overall sialylation hardly changes, as afore described. The additive nature of this effect may be additionally conveyed by an increased overall sialylation (CS). When we studied the influence of sex on age for these effects, we found the observed phenomena to be less pronounced in females than in males for tri- and tetra-antennary sialylation and structures (CA3, CA4), while the reverse effect was seen for di-antennary ones (CA2) (Figure 3C).
Tri-antennary fucosylated ones (A3F) exhibited an increase as age progressed, in line with previous findings (Figure 3A).36 Antennary fucosylated tri-antennary structures (A3Fa; doubly fucosylated tri-antennary glycans) showed a rising pattern similar to their singly fucosylated counterpart (A3F), and are commonly associated with the acute phase proteins α1-antitrypsin and α1-acidic glycoprotein.17 Antennary fucosylation is an important modification that gives rise to (sialyl) LewisX/A determinants. These motifs mediate glycan-lectin interactions in homeostasis, but also occur as glyco-neoantigens in cancerous cells with implications in their metastatic potential.16,72
Another related finding involves the increasing prevalence of bisected structures within di-antennary glycans (A2B) with progressing age (Figure 2A), which may be largely attributable to circulatory IgA and IgG, being known carriers of such glycoforms.17,68,73 It is noteworthy that bisection, at least in the case of IgG, reaches plateau around the age of 60 years, after which notable intersex differences can no longer be observed.74
Of note, genetic, epigenetic, phenotypic, and environmental influences jointly shape the variability of the N-glycome, albeit the highest variability in healthy individuals appear to be brought about by simple demographic descriptors such as age and sex. This highlights the necessity of proper experimental design and large, preferably longitudinal sample sets with well-matched cases and controls in order to find robust disease-associated glycomic signatures.
5. Effect Sizes of Glycosylation Traits across Nine Clinical Studies
In our 2019 investigation, we stated the growing acknowledgment of glycans in the realm of personalized medicine by the identification of approximately 10,000 publications retrieved through the PubMed search term “glycan AND biomarkers”.36 Upon conducting an analogous subsequent search, we observed a surge in publications, exceeding 5,000 within the past 5 years. This trend signifies sustained interest and progressive expansion despite the roadblocks holding the field back from clinical translation.75−77
In this section, we expand on our previous report36 that included colorectal cancer (CRC),45 type 2 diabetes (T2D)37 and inflammatory bowel disease (IBD)38 to 5 more diseases including pancreatic35 and breast cancer46 (PC and BC), multiple myeloma (MM),41 the parasitic infectious disease leishmaniasis (LH),33 and finally an autoimmune liver disease (autoimmune hepatitis; AIH).47 We explored differences in N-glycosylation traits across these diseases obtained with our unified methodological approach (Supplementary Figure and Methods), with the aim to elucidate potential links between the various pathological conditions and glycosylation patterns, that in turn may show promise as clinically translatable diagnostic leads either as standalone biomarkers or by pointing toward protein-specificity (Figure 4, Table S4). To quantify the extent of these changes across the studies we explored effect sizes as a uniform measure. Specifically, we used Cohen’s d as the metric for effect size. Cohen’s d is a statistical measure widely used to quantify the effect size, or the magnitude of difference, between two groups. It calculates the difference between two means (such as in treatment vs control groups) and expresses this difference in standard deviation units. This standardized approach is what makes Cohen’s d exceptionally valuable for comparing effects across different studies or experiments, even when these studies vary in their size, scale or measurement units. Please note that confidence intervals of the estimates offer insight into statistical significance: if the CI does not encompass zero, the effect can be considered statistically significant at the chosen confidence level (95%). Nonetheless, we added p-values for the effect sizes in Table S4.
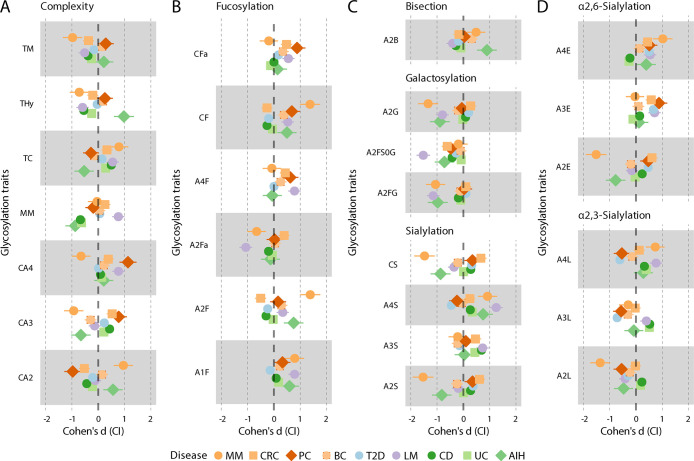
Figure 4.
Forest plots visually illustrating the effect sizes of glycosylation traits observed in the presented studies. The glycosylation traits are grouped based on their structural characteristics. Each horizontal line in the plot represents a different study. Point estimates along these lines show Cohen’s d values–representing the effect size or the magnitude of the differences in glycosylation traits between the case and control groups. Lines extending from these points denote the 95% confidence intervals (CI). Cohen’s d was specifically selected for its capacity to standardize effect sizes across studies, effectively neutralizing the impact of sample size variations. To ensure consistency and comparability across different cohorts, a standardized methodology was applied to recalculate the glycosylation traits. Further details about the specific glycosylation traits and their compositions are available on Figure 1C and in Table S2. Abbreviations in the figure: MM – multiple myeloma,41 CRC – colorectal cancer,45 PC – pancreatic cancer,35 BC – breast cancer,46 T2D – type 2 diabetes,37 LM – leishmaniasis,33 CD – Crohn’s disease,38 UC – ulcerative colitis,38 AIH – autoimmune hepatitis.47 Colors indicate disease categories (orange-red: cancer; blue: metabolic disease; purple: infectious disease; green: autoimmune or inflammatory disease). Shapes were intended to aid visual interpretability. Note that IBD presented on Figure 2 as one category, is divided into the two subcategories CD and UC here.
5.1. Cancers
5.1.1. Multiple Myeloma
A notable distinction in glycan abundance is evident in the cancer types. Multiple myeloma (MM) is a cancer of plasma cells. It displays a TBNG signature that is markedly different from solid tumors (colorectal cancer (CRC), pancreatic cancer (PC) and breast cancer (BC)), which may reflect the fact of MM being a hematological cancer. Of note, high levels of the M protein, which are immunoglobulins secreted by the malignant plasma cells will make a major contribution to the MM TBNG.41 These M proteins, being immunoglobulins, mainly display di-antennary complex-type glycans.78 Accordingly, a number of di-antennary glycosylation traits including A2S, A2G and A2F are largely skewed in MM (Figure 4), which is in agreement with previous studies utilizing capillary electrophoresis and HILIC ultrahigh performance liquid chromatography.79,80 The observed increase in bisection (CB) may be caused by elevated activity of the enzyme responsible for the addition of bisecting GlcNAc reported in MM before.81 Surprisingly, also tetra-antennary glycosylation traits of MM are affected, with elevated levels of both α2,6- (A4E) and α2,3-sialylation (A3L), indicating that TBNG changes in MM exceed the myeloid cell-derived M protein and involve other likely liver-derived glycoproteins.
5.1.2. Pancreatic Cancer
Regarding the three cancers with solid tumors, in contrast to MM, the TBNG for these cancers is predominantly made up of liver-derived proteins and immunoglobulins, with tumor-derived leakage proteins only making a minor direct contribution to the TBNG. PC shows the strongest cancer TBNG signature, with a clear shift of complex-type glycans from di-antennary (CA2) to tri- and tetra-antennary (CA3 and CA4), alongside notable changes in sialylation and fucosylation patterns (Figure 4),35 that follow those previously observed after glycan enrichment from patient sera using a “glycoblotting” approach followed by nonlinkage-specific sialic acid derivatization and MALDI-MS analysis.200 Glycans with any antennarity show a shift in sialic acid linkage usage as evidenced by elevated levels of α2,6-sialylation (A2E, A3E and A4E) and decreased levels of α2,3-sialylation (A2L, A3L and A4L). Various fucosylation traits are elevated, in line with literature data from a single pancreatic cancer patient,82 while the di-antennary, fucosylated, nonsialylated N-glycans show a decreased level of galactosylation (A2FS0G trait). This latter trait can be largely related to IgG glycosylation17 which is known to exhibit decreased galactosylation in various types of solid tumors including PC.83,84 Interestingly, the PC TBNG signatures appear to be largely conserved between nonhereditary and hereditary variants of the disease, and has potential for surveillance of risk populations for early detection of PC.85
5.1.3. Colorectal Cancer
Similar to PC, CRC shows elevated levels of tri- and tetra-antennary glycans on the expense of di-antennaries. Likewise, the CRC shares elevated sialylation with PC, consistent with a previous study,86 but we found this increase to be particularly mediated by A2E, A3E and A4E. Likewise, some of the fucosylation features were in line with PC (Figure 4).45,86 There are, however, some clear TBNG differences between PC and the CRC: CRC shows decreased levels of oligomannosidic N-glycans (TM), in contrast to PC where TMs tend to be elevated. Likewise, CRC shows decreased levels of fucosylation of di-antennary glycans (A2F), comparably to a CE-based TBNG report also laying a link between decreased core fucosylation and fucosyltransferase activity,87 something that is not observed for PC. Interestingly, the CRC TBNG signature shows partial reversion to normal upon surgical removal of the tumor, indicating the potential of TBNG in CRC disease and treatment monitoring.45,87
5.1.4. Breast Cancer
BC shows the weakest TBNG signature of the four cancers which is a remarkable finding on its own (Figure 4).46 Maybe BC is too heterogeneous for a strong, relatively homogeneous TBNG signature, and patients would have to be stratified according to clinical and cellular subtypes of BC for obtaining a more distinctive signature. However, the observed TBNG signatures were largely consistent with prior research, with particular notion on increased (α2,6-)sialylation as well as the elevation of most fucosylation features including those forming the sialyl LewisX epitope at antennae, and increase–albeit minor–of oligomannosidic structures, altogether described as hallmarks of BC progression.88−91 In contrast, another study reported a decrease of oligomannosidic structures in BC.92 The glycobiology of cancers as manifested in TBNG appears to be rather diverse. Mechanistic studies would be needed to understand how the TBNG is shaped in cancer, elucidating the role of tumor secretions and leakage factors in inducing acute phase responses of the liver with accompanying glycosylation changes.
5.2. Metabolic Disease Signature: T2D
For a long time, TBNG studies largely focused on cancer, autoimmune, inflammatory, and infectious diseases. Only recently, the rather strong glycomic signatures of cardiovascular conditions and metabolic disorders received more attention.37,93,94 Obviously, there are pathological commonalities between different disease groups, with inflammatory responses being associated with various types of autoimmune and infectious diseases and cancers, but also metabolic and cardiovascular diseases.
Interestingly, in terms of the TBNG signature, T2D sides with PC and to a lesser extent with CRC regarding sialylation features. Without distinguishing linkage-isomers, these results are in part consistent with previous reports,95,96 but upon the resolution of linkage isomers, linkage-specific sialylation effects are observed, with increased α2,6-sialylation of tri- and tetra-antennary glycans (A3E and A4E) and a concomitant decrease of A3L and A4L (Figure 4).34,37 Interestingly, a genome-wide association study in T2D reported the increased expression of the transcript corresponding to the enzyme responsible for the addition of sialic acids to N-glycans in α2,6-linkage, which is in line with the increase in the relative levels of the corresponding glyco-phenotype.97 Research is needed to define the carriers of these differentially sialylated N-glycans, most likely including major acute-phase proteins derived from the liver. In a next step, regulators of liver glycosylation need to be identified to develop an understanding of the causes of common disease-associated glycosylation changes.
5.3. Autoimmune and Inflammatory Disease Signatures
5.3.1. Inflammatory Bowel Disease
Little is known about blood protein glycosylation in IBD. Crohn’s disease (CD) and ulcerative colitis (UC), which are two IBDs affecting the gastrointestinal tract, show rather similar TBNG signatures as evidenced by their tight clustering in most of the glycomic comparisons displayed in Figure 4. However, this similarity translates to limited effectiveness in using TBNG for distinguishing between CD and UC.38 IgG and IgA glycomes showed a similar, limited performance in discriminating between CD and UC.98,99 The most promising lead for clinical translation of TBNG in IBD is its use for predicting treatment escalations,100 which may already at baseline allow to stratify CD patients according to expected disease course with the promise of a more tailored disease monitoring and treatment.
Of note, in stark contrast to the TBNG similarities shown by UC and CD, they only showed modest resemblance with the CRC signature, indicating distinct systemic glycosylation effects and no clear common TBNG manifestation between these groups of gastrointestinal tract diseases.
5.3.2. Autoimmune Hepatitis
Autoimmune hepatitis (AIH) is a chronic liver autoimmune disorder marked by autoantibodies and elevated total IgG. The disease shows a rather distinct TBNG signature, with high levels of hybrid-type glycans and high levels of bisection of di-antennaries, together with low levels of di-antennary sialylation and a markedly decreased number of mannoses on oligomannosidic structures (glycosylation trait MM). The AIH TBNG signature showed some resemblance with the MM disease signature regarding high levels of di-antennaries (CA2) and low levels of tri-antennaries (CA3) within complex-type glycans, high levels of fucosylation (A2F) and bisection (A2B) together with low levels of α2,3-sialylation (A2L) and α2,6-sialylation (A2E) of di-antennary glycans, and low levels of galactosylation of di-antennary, fucosylated glycans (A2FG). While most of these traits may be attributed to elevated IgG levels with a concomitant skewing of the IgG glycosylation profile, AIH and MM also showed resemblance in displaying high levels of sialylation (A4S), in particular α2,6-sialylation (A4E) of tetra-antennaries.
Currently, diagnosis of AIH relies on a scoring system involving autoantibodies, IgG levels, and liver biopsy results. The invasive nature of liver biopsy, coupled with bleeding risks, underscores the need for noninvasive diagnostic biomarkers in AIH to enhance accuracy and potentially reduce reliance on liver biopsy in the future. Studies on the glycomic dimensions of AIH are lacking, thus further research and replication studies are needed to assess the usefulness of TBNG in the diagnosis of AIH, as well as for the noninvasive diagnosis and staging of AIH-associated liver conditions such as fibrosis and cirrhosis.101
5.4. Infectious Disease Signatures
Visceral leishmaniasis (LM) is a devastating disease caused by a bloodborne protozoic parasite. It often requires bone marrow puncture for diagnosis, and commonly applied antimonial drugs are potentially toxic. The TBNG signature appears to be very strong and further research is needed to assess its potential in the diagnosis of LM. The LM TBNG displays particularly low A2FS0G levels (Figure 4),33 reflecting very low galactosylation of IgG.102 However, it is not unexpected since IgG agalactosylation is typically observed across diseases with an inflammatory component.103 Other prominent features are high levels of α2,3-sialylation of tri- and particularly tetra-antennary glycans (A3L and A4L). Additionally, tetra-antennaries show elevated levels of fucosylation (A4F), and overall antennary fucosylation appears to be elevated (CFa), pointing toward increased levels of sialyl-Lewis structures on corresponding circulatory glycoproteins.
5.5. Summary
In summary, the study of glycan abundance under different pathological conditions reveals distinct and sometimes overlapping TBNG signatures, providing valuable insights into the molecular underpinnings of these diseases. A common physiological trend across various diseases, including cancers, autoimmune disorders, and metabolic disorders (like T2D), is the presence of an inflammatory response, which is reflected in the alterations of glycosylation patterns. For instance, cancer and metabolic diseases exhibit similarities in their TBNG signatures, such as increased α2,6-sialylation of tri- and tetra-antennary glycans. This overarching theme of inflammation-induced glycosylation changes offers a unifying perspective for understanding the systemic effects of these diseases.
Here, we want to bring into consideration that incongruent or divergent findings among studies may stem from factors including disparities in methodology (low-throughput, low resolution, not well-established workflows), a limited range of analyzed glycan features (attributable to, e.g., desialylation or lectin-based enrichment), cohorts with low sample numbers and/or not well-defined clinical characteristics, absence of replication, inconsistent data analysis workflows, or confluence of these elements.
6. Conclusions and Future Perspectives
As a certain role of glycosylation has been observed in every major disease, glycans are considered to be inherently involved in multiple disease pathophysiologies. The breadth of physiological functions driven by glycosylation makes it a promising diagnostic marker and therapeutic target candidate. Hence, development of analytical tools for assessing the glycomic dimension of human diseases becomes more and more widespread, with the aim of obtaining therapeutic and diagnostic leads.104 While there is a lack of standalone technology for fully defining glycans, using a unified analytical approach with sufficient throughput and largely consistent data analysis workflows enables study comparability. Fulfilling these criteria allowed us to explore blood glycomic features in 9 major diseases.
All the studies compiled in the disease TBNG alignment attempt presented in Figure 4 are cohort studies, most of them with a baseline disease (case)-control design, while some of them included patients receiving treatment, including medication. Proper design of case-control studies has to take potential confounders into account including age, sex, their interaction, but also BMI, genetics and life circumstances such as socio-economic status. Clearly, more research is needed to dissect factors influencing glycosylation including TBNG. Regarding the effect of medication, differentiating direct effects of drugs on the TBNG from disease-modifying effect and resulting indirect changes of TBNG proves to be challenging.105 Accordingly, baseline studies with pretreatment sampling are particularly valuable for assessing disease TBNG signatures.
The TBNG signatures of the varied range of diseases, which we intended to compare, turned out to be rather diverse, with the exception of UC and CD that displayed largely converging glycomic signatures. We observed that different cancer types had disparate TBNG signatures. Also, a majority of the diseases had a strong inflammatory component, which, however, did not translate into an apparent, common glycomic signature. On the other hand, some surprising commonalities were observed, with MM and AIH showing a range of very similar, highly skewed glycomic features.
The TBNG studies have to be complemented with glycoproteomic analyses at the glycopeptide level–which may be performed on the same serum and plasma samples. This will shed light on the glycoprotein(s) and glycosylation site(s) that contribute to the observed TBNG signatures. The disease glycoproteomics studies of InterVenn Biosciences have already demonstrated that such glycoproteomic studies can match or outperform released glycan glycomic studies with respect to clinical biomarker potential.106
While current glycoproteomic workflows do provide site- and protein-specific information on glycosylation, they fail to reveal key glycosylation features such as sialic acid linkages and often ambiguous features such as core fucosylation and bisection. Future development of glycoproteomics workflows with inclusion of ion mobility spectroscopy and improvements in MS/MS data interpretation are warranted to enhance the definition of glycan structures from bottom-up glycopeptide-based analyses.107 Additionally, intact protein analysis by MS can provide a valuable contribution to glycoprotein profiling providing an integrated mapping of proteoforms.108
Regarding the relative composition of the glycoproteome, there is a dearth of studies, and information on changes to the glycoproteome in health and disease is still largely lacking. While there is a considerable number of studies using high-end bottom-up glycoproteomics to measure blood protein glycosylation, most of the studies do not achieve an integration of the information to conclude the overall glycosylation of the major blood proteins. Therefore, our knowledge of blood protein glycosylation is still very fragmented, and the changes of the blood glycoproteome with disease are poorly defined. Additionally, limited access to data is hampering attempts to link glycomic and glycoproteomic data for specific diseases.
The TBNG studies show the disease-associated glycosylation changes of plasma cells (specialized B cell progenies) and liver cells, including hepatocytes. To provide further insights into the regulation and functional effects of glycosylation changes of circulatory proteins, clinical glycomics and glycoproteomics studies will have to be complemented by transcriptomics of, and in vitro studies using, e.g., plasma cells retrieved from the circulation, (hepatocyte) cell lines and organoids.
While glycans are implicated in virtually all human diseases, our knowledge of disease-associated glycomic changes and corresponding functional implications is still very restricted. We here explored and compared the TBNG glycomic dimensions of several major human diseases obtained by a standardized MALDI-MS-based methodology, showing a taunting complexity despite the technical limitations of the applied approach with regard to the lack of depth, structural definition, and protein specificity.
It is clear that our exploration of the glycobiology and glyco(proteo)mics layer of human diseases has only started, several successful examples (HelenaBioSciences, GlycanAge) already hallmark the utility of easily and noninvasively accessible blood-derived glycomic biomarkers in precision medicine and personal health and biological age assessment, benefiting from enabling, instrumentally simplistic high-throughput technologies with streamlined sample preparation and data analysis workflows, with likely more examples to follow in the future. Clinical translation of promising glycan-based diagnostic approaches is hypothesized to mitigate the necessity of invasive or other labor- and cost-intensive procedures without sufficient availability and accuracy, thereby facilitating timely interventions and enhancing lifestyle and disease management strategies in the future.
Glossary
Abbreviations
- AIH
Autoimmune Hepatitis
- BC
Breast Cancer
- CD
Crohn’s Disease
- CDG
Congenital Disorders of Glycosylation
- CI
Confidence Interval
- CRC
Colorectal Cancer
- GWAS
Genome-Wide Association Study
- HILIC
Hydrophilic Interaction Liquid Chromatography
- Ig
Immunoglobulin
- IBD
Inflammatory Bowel Disease
- LM
Leishmaniasis
- MALDI
Matrix-Assisted Laser Desorption/Ionization
- MM
Multiple Myeloma
- MODY
Maturity-Onset Diabetes of the Young
- MS
Mass Spectrometry
- PC
Pancreatic Cancer
- T2D
Type 2 Diabetes
- TBNG
Total Blood N-Glycome
- TPNG
Total Plasma N-Glycome
- TSNG
Total Serum N-Glycome
- UC
Ulcerative Colitis
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacsau.4c00043.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. CRediT: Tamas Pongracz conceptualization, investigation, methodology, validation, visualization, writing-original draft, writing-review & editing; Oleg A. Mayboroda conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing-original draft, writing-review & editing; Manfred Wuhrer conceptualization, funding acquisition, investigation, project administration, supervision, validation, writing-original draft, writing-review & editing.
This study was cofunded by the European Union (ERC Synergy, GlycanSwitch, 101071386).
The authors declare no competing financial interest.
Supplementary Material
References
- Varki A. Biological roles of glycans. Glycobiology 2017, 27 (1), 3–49. 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjoldager K. T.; Narimatsu Y.; Joshi H. J.; Clausen H. Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 2020, 21 (12), 729–749. 10.1038/s41580-020-00294-x. [DOI] [PubMed] [Google Scholar]
- Lebrilla C. B.; Liu J.; Wildmalm G.; Prestegard J. H.. Oligosaccharides and Polysaccharides. In Essentials of Glycobiology, 4th ed.; 2022; pp 33–42. [Google Scholar]
- Flynn R. A.; Pedram K.; Malaker S. A.; Batista P. J.; Smith B. A. H.; Johnson A. G.; George B. M.; Majzoub K.; Villalta P. W.; Carette J. E.; et al. Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell 2021, 184 (12), 3109–3124. 10.1016/j.cell.2021.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rini J. M.; Moremen K. W.; Davis B. G.; Esko J. D.. Glycosyltransferases and Glycan-Processing Enzymes. In Essentials of Glycobiology, 4th ed.; 2022; pp 67–78. [Google Scholar]
- Knezevic A.; Gornik O.; Polasek O.; Pucic M.; Redzic I.; Novokmet M.; Rudd P. M.; Wright A. F.; Campbell H.; Rudan I.; et al. Effects of aging, body mass index, plasma lipid profiles, and smoking on human plasma N-glycans. Glycobiology 2010, 20 (8), 959–969. 10.1093/glycob/cwq051. [DOI] [PubMed] [Google Scholar]
- Lauc G.; Essafi A.; Huffman J. E.; Hayward C.; Knezevic A.; Kattla J. J.; Polasek O.; Gornik O.; Vitart V.; Abrahams J. L.; et al. Genomics meets glycomics-the first GWAS study of human N-Glycome identifies HNF1alpha as a master regulator of plasma protein fucosylation. PLoS Genet 2010, 6 (12), e1001256 10.1371/journal.pgen.1001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucic M.; Knezevic A.; Vidic J.; Adamczyk B.; Novokmet M.; Polasek O.; Gornik O.; Supraha-Goreta S.; Wormald M. R.; Redzic I.; et al. High throughput isolation and glycosylation analysis of IgG-variability and heritability of the IgG glycome in three isolated human populations. Mol. Cell Proteomics 2011, 10 (10), M111.010090. 10.1074/mcp.M111.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiding K. R.; Ruhaak L. R.; Uh H. W.; El Bouhaddani S.; van den Akker E. B.; Plomp R.; McDonnell L. A.; Houwing-Duistermaat J. J.; Slagboom P. E.; Beekman M.; et al. Human Plasma N-glycosylation as Analyzed by Matrix-Assisted Laser Desorption/Ionization-Fourier Transform Ion Cyclotron Resonance-MS Associates with Markers of Inflammation and Metabolic Health. Mol. Cell Proteomics 2017, 16 (2), 228–242. 10.1074/mcp.M116.065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharapov S. Z.; Tsepilov Y. A.; Klaric L.; Mangino M.; Thareja G.; Shadrina A. S.; Simurina M.; Dagostino C.; Dmitrieva J.; Vilaj M.; et al. Defining the genetic control of human blood plasma N-glycome using genome-wide association study. Hum. Mol. Genet. 2019, 28 (12), 2062–2077. 10.1093/hmg/ddz054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.; Xie Y.; Li Q.; Artegoitia V. M.; Lebrilla C. B.; Keim N. L.; Adams S. H.; Krishnan S. Diet affects glycosylation of serum proteins in women at risk for cardiometabolic disease. European Journal of Nutrition 2021, 60 (7), 3727–3741. 10.1007/s00394-021-02539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landini A.; Trbojevic-Akmacic I.; Navarro P.; Tsepilov Y. A.; Sharapov S. Z.; Vuckovic F.; Polasek O.; Hayward C.; Petrovic T.; Vilaj M.; et al. Genetic regulation of post-translational modification of two distinct proteins. Nat. Commun. 2022, 13 (1), 1586. 10.1038/s41467-022-29189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moremen K. W.; Tiemeyer M.; Nairn A. V. Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 2012, 13 (7), 448–462. 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull J. E.; Field R. A. Emerging glycomics technologies. Nat. Chem. Biol. 2007, 3 (2), 74–77. 10.1038/nchembio0207-74. [DOI] [PubMed] [Google Scholar]
- Khoury G. A.; Baliban R. C.; Floudas C. A. Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci. Rep 2011, 1, 90. 10.1038/srep00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reily C.; Stewart T. J.; Renfrow M. B.; Novak J. Glycosylation in health and disease. Nat. Rev. Nephrol 2019, 15 (6), 346–366. 10.1038/s41581-019-0129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc F.; Reiding K. R.; Jansen B. C.; Kammeijer G. S.; Bondt A.; Wuhrer M. Human plasma protein N-glycosylation. Glycoconj J. 2016, 33 (3), 309–343. 10.1007/s10719-015-9626-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho S. S.; Reis C. A. Glycosylation in cancer: mechanisms and clinical implications. Nature Reviews Cancer 2015, 15 (9), 540–555. 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- Pinho S. S.; Alves I.; Gaifem J.; Rabinovich G. A. Immune regulatory networks coordinated by glycans and glycan-binding proteins in autoimmunity and infection. Cell Mol. Immunol 2023, 20, 1101–1113. 10.1038/s41423-023-01074-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackstein R.; Hoffmeister K. M.; Stowell S. R.; Kinoshita T.; Varki A.; Freeze H. H.. Glycans in Acquired Human Diseases. In Essentials of Glycobiology, 4th ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, 2022; pp 615–630. [Google Scholar]
- Aizpurua-Olaizola O.; Sastre Toraño J.; Falcon-Perez J. M.; Williams C.; Reichardt N.; Boons G. J. Mass spectrometry for glycan biomarker discovery. TrAC Trends in Analytical Chemistry 2018, 100, 7–14. 10.1016/j.trac.2017.12.015. [DOI] [Google Scholar]
- Pilobello K. T.; Krishnamoorthy L.; Slawek D.; Mahal L. K. Development of a lectin microarray for the rapid analysis of protein glycopatterns. Chembiochem 2005, 6 (6), 985–989. 10.1002/cbic.200400403. [DOI] [PubMed] [Google Scholar]
- Trbojevic-Akmacic I.; Lageveen-Kammeijer G. S. M.; Heijs B.; Petrovic T.; Deris H.; Wuhrer M.; Lauc G. High-Throughput Glycomic Methods. Chem. Rev. 2022, 122 (20), 15865–15913. 10.1021/acs.chemrev.1c01031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuhrer M. Glycomics using mass spectrometry. Glycoconj J. 2013, 30 (1), 11–22. 10.1007/s10719-012-9376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X.; Huang Y.; Cho B. G.; Zhong J.; Gautam S.; Peng W.; Williamson S. D.; Banazadeh A.; Torres-Ulloa K. Y.; Mechref Y. Advances in mass spectrometry-based glycomics. Electrophoresis 2018, 39 (24), 3063–3081. 10.1002/elps.201800273. [DOI] [PubMed] [Google Scholar]
- Bagdonaite I.; Malaker S. A.; Polasky D. A.; Riley N. M.; Schjoldager K.; Vakhrushev S. Y.; Halim A.; Aoki-Kinoshita K. F.; Nesvizhskii A. I.; Bertozzi C. R. Glycoproteomics. Nature Reviews Methods Primers 2022, 2, 48. 10.1038/s43586-022-00128-4. [DOI] [Google Scholar]
- Vreeker G. C. M.; Nicolardi S.; Bladergroen M. R.; van der Plas C. J.; Mesker W. E.; Tollenaar R. A. E. M.; van der Burgt Y. E. M.; Wuhrer M. Automated Plasma Glycomics with Linkage-Specific Sialic Acid Esterification and Ultrahigh Resolution MS. Anal. Chem. 2018, 90 (20), 11955–11961. 10.1021/acs.analchem.8b02391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kailemia M. J.; Xu G.; Wong M.; Li Q.; Goonatilleke E.; Leon F.; Lebrilla C. B. Recent Advances in the Mass Spectrometry Methods for Glycomics and Cancer. Anal. Chem. 2018, 90 (1), 208–224. 10.1021/acs.analchem.7b04202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K.; Han Y.; Yang H.; Lu H.; Tian Z. Mass spectrometry-based qualitative and quantitative N-glycomics: An update of 2017–2018. Anal. Chim. Acta 2019, 1091, 1–22. 10.1016/j.aca.2019.10.007. [DOI] [PubMed] [Google Scholar]
- Vreeker G. C. M.; Bladergroen M. R.; Nicolardi S.; Mesker W. E.; Tollenaar R.; van der Burgt Y. E. M.; Wuhrer M. Dried blood spot N-glycome analysis by MALDI mass spectrometry. Talanta 2019, 205, 120104. 10.1016/j.talanta.2019.06.104. [DOI] [PubMed] [Google Scholar]
- Adamczyk B.; Struwe W. B.; Ercan A.; Nigrovic P. A.; Rudd P. M. Characterization of fibrinogen glycosylation and its importance for serum/plasma N-glycome analysis. J. Proteome Res. 2013, 12 (1), 444–454. 10.1021/pr300813h. [DOI] [PubMed] [Google Scholar]
- Ruhaak L. R.; Uh H. W.; Beekman M.; Hokke C. H.; Westendorp R. G.; Houwing-Duistermaat J.; Wuhrer M.; Deelder A. M.; Slagboom P. E. Plasma protein N-glycan profiles are associated with calendar age, familial longevity and health. J. Proteome Res. 2011, 10 (4), 1667–1674. 10.1021/pr1009959. [DOI] [PubMed] [Google Scholar]
- Porcino G. N.; Bladergroen M. R.; Dotz V.; Nicolardi S.; Memarian E.; Gardinassi L. G.; Nery Costa C. H.; Pacheco de Almeida R.; Ferreira de Miranda Santos I. K.; Wuhrer M. Total serum N-glycans mark visceral leishmaniasis in human infections with Leishmania infantum. iScience 2023, 26 (7), 107021. 10.1016/j.isci.2023.107021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memarian E.; t Hart L. M.; Slieker R. C.; Lemmers R. F. L.; van der Heijden A. A.; Rutters F.; Nijpels G.; Schoep E.; Lieverse A. G.; Sijbrands E. J. G.; et al. Plasma protein N-glycosylation is associated with cardiovascular disease, nephropathy, and retinopathy in type 2 diabetes. BMJ. Open Diabetes Res. Care 2021, 9 (1), e002345 10.1136/bmjdrc-2021-002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreeker G. C. M.; Hanna-Sawires R. G.; Mohammed Y.; Bladergroen M. R.; Nicolardi S.; Dotz V.; Nouta J.; Bonsing B. A.; Mesker W. E.; van der Burgt Y. E. M.; et al. Serum N-Glycome analysis reveals pancreatic cancer disease signatures. Cancer Med. 2020, 9 (22), 8519–8529. 10.1002/cam4.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotz V.; Wuhrer M. N-glycome signatures in human plasma: associations with physiology and major diseases. FEBS Lett. 2019, 593 (21), 2966–2976. 10.1002/1873-3468.13598. [DOI] [PubMed] [Google Scholar]
- Dotz V.; Lemmers R. F. H.; Reiding K. R.; Hipgrave Ederveen A. L.; Lieverse A. G.; Mulder M. T.; Sijbrands E. J. G.; Wuhrer M.; van Hoek M. Plasma protein N-glycan signatures of type 2 diabetes. Biochimica et Biophysica Acta (BBA) - General Subjects 2018, 1862 (12), 2613–2622. 10.1016/j.bbagen.2018.08.005. [DOI] [PubMed] [Google Scholar]
- Clerc F.; Novokmet M.; Dotz V.; Reiding K. R.; de Haan N.; Kammeijer G. S. M.; Dalebout H.; Bladergroen M. R.; Vukovic F.; Rapp E.; et al. Plasma N-Glycan Signatures Are Associated With Features of Inflammatory Bowel Diseases. Gastroenterology 2018, 155 (3), 829–843. 10.1053/j.gastro.2018.05.030. [DOI] [PubMed] [Google Scholar]
- Reiding K. R.; Vreeker G. C. M.; Bondt A.; Bladergroen M. R.; Hazes J. M. W.; van der Burgt Y. E. M.; Wuhrer M.; Dolhain R. Serum Protein N-Glycosylation Changes with Rheumatoid Arthritis Disease Activity during and after Pregnancy. Frontiers in Medicine 2018, 4, 241. 10.3389/fmed.2017.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.; Wuhrer M.; Holst S. Serum sialylation changes in cancer. Glycoconj J. 2018, 35 (2), 139–160. 10.1007/s10719-018-9820-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.; Westhrin M.; Bondt A.; Wuhrer M.; Standal T.; Holst S. Serum protein N-glycosylation changes in multiple myeloma. Biochim Biophys Acta Gen Subj 2019, 1863 (5), 960–970. 10.1016/j.bbagen.2019.03.001. [DOI] [PubMed] [Google Scholar]
- Bladergroen M. R.; Reiding K. R.; Hipgrave Ederveen A. L.; Vreeker G. C. M.; Clerc F.; Holst S.; Bondt A.; Wuhrer M.; van der Burgt Y. E. M. Automation of High-Throughput Mass Spectrometry-Based Plasma N-Glycome Analysis with Linkage-Specific Sialic Acid Esterification. J. Proteome Res. 2015, 14 (9), 4080–4086. 10.1021/acs.jproteome.5b00538. [DOI] [PubMed] [Google Scholar]
- Selman M. H. J.; Hemayatkar M.; Deelder A. M.; Wuhrer M. Cotton HILIC SPE Microtips for Microscale Purification and Enrichment of Glycans and Glycopeptides. Anal. Chem. 2011, 83 (7), 2492–2499. 10.1021/ac1027116. [DOI] [PubMed] [Google Scholar]
- Jansen B. C.; Reiding K. R.; Bondt A.; Hipgrave Ederveen A. L.; Palmblad M.; Falck D.; Wuhrer M. MassyTools: A High-Throughput Targeted Data Processing Tool for Relative Quantitation and Quality Control Developed for Glycomic and Glycoproteomic MALDI-MS. J. Proteome Res. 2015, 14 (12), 5088–5098. 10.1021/acs.jproteome.5b00658. [DOI] [PubMed] [Google Scholar]
- de Vroome S. W.; Holst S.; Girondo M. R.; van der Burgt Y. E. M.; Mesker W. E.; Tollenaar R.; Wuhrer M. Serum N-glycome alterations in colorectal cancer associate with survival. Oncotarget 2018, 9 (55), 30610–30623. 10.18632/oncotarget.25753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreeker G. C. M.; Vangangelt K. M. H.; Bladergroen M. R.; Nicolardi S.; Mesker W. E.; Wuhrer M.; van der Burgt Y. E. M.; Tollenaar R. Serum N-glycan profiles differ for various breast cancer subtypes. Glycoconj J. 2021, 38 (3), 387–395. 10.1007/s10719-021-10001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongracz T.; Biewenga M.; Stoelinga A. E. C.; Bladergroen M. R.; Nicolardi S.; Trouw L. A.; Wuhrer M.; de Haan N.; van Hoek B.. Autoimmune hepatitis displays distinctively high multi-antennary sialylation on plasma N-glycans compared to other liver diseases. Under revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeze H. H. Genetic defects in the human glycome. Nat. Rev. Genet 2006, 7 (7), 537–551. 10.1038/nrg1894. [DOI] [PubMed] [Google Scholar]
- Hennet T.; Cabalzar J. Congenital disorders of glycosylation: a concise chart of glycocalyx dysfunction. Trends Biochem. Sci. 2015, 40 (7), 377–384. 10.1016/j.tibs.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Peanne R.; de Lonlay P.; Foulquier F.; Kornak U.; Lefeber D. J.; Morava E.; Perez B.; Seta N.; Thiel C.; Van Schaftingen E.; et al. Congenital disorders of glycosylation (CDG): Quo vadis?. Eur. J. Med. Genet 2018, 61 (11), 643–663. 10.1016/j.ejmg.2017.10.012. [DOI] [PubMed] [Google Scholar]
- Abu Bakar N.; Lefeber D. J.; van Scherpenzeel M. Clinical glycomics for the diagnosis of congenital disorders of glycosylation. J. Inherit Metab Dis 2018, 41 (3), 499–513. 10.1007/s10545-018-0144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipgrave Ederveen A. L.; de Haan N.; Baerenfaenger M.; Lefeber D. J.; Wuhrer M. Dissecting Total Plasma and Protein-Specific Glycosylation Profiles in Congenital Disorders of Glycosylation. Int. J. Mol. Sci. 2020, 21 (20), 7635. 10.3390/ijms21207635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels H.; Kulkarni P.; van Dael M.; Suppers A.; Willems E.; Zijlstra F.; Kragt E.; Gloerich J.; Schmit P. O.; Pengelley S. Plasma glycoproteomics delivers high-specificity disease biomarkers by detecting site-specific glycosylation abnormalities. J. Adv. Res. 2023, 10.1016/j.jare.2023.09.002. [DOI] [PubMed] [Google Scholar]
- Guillard M.; Morava E.; van Delft F. L.; Hague R.; Korner C.; Adamowicz M.; Wevers R. A.; Lefeber D. J. Plasma N-glycan profiling by mass spectrometry for congenital disorders of glycosylation type II. Clin Chem. 2011, 57 (4), 593–602. 10.1373/clinchem.2010.153635. [DOI] [PubMed] [Google Scholar]
- Yamamoto F. Review: ABO blood group system—ABH oligosaccharide antigens, anti-A and anti-B, A and B glycosyltransferases, and ABO genes. Immunohematology 2004, 20 (1), 3–22. 10.21307/immunohematology-2019-418. [DOI] [PubMed] [Google Scholar]
- Knezevic A.; Polasek O.; Gornik O.; Rudan I.; Campbell H.; Hayward C.; Wright A.; Kolcic I.; O’Donoghue N.; Bones J.; et al. Variability, heritability and environmental determinants of human plasma N-glycome. J. Proteome Res. 2009, 8 (2), 694–701. 10.1021/pr800737u. [DOI] [PubMed] [Google Scholar]
- Kristic J.; Sharapov S. Z.; Aulchenko Y. S. Quantitative Genetics of Human Protein N-Glycosylation. Adv. Exp. Med. Biol. 2021, 1325, 151–171. 10.1007/978-3-030-70115-4_7. [DOI] [PubMed] [Google Scholar]
- Zoldos V.; Horvat T.; Novokmet M.; Cuenin C.; Muzinic A.; Pucic M.; Huffman J. E.; Gornik O.; Polasek O.; Campbell H.; et al. Epigenetic silencing of HNF1A associates with changes in the composition of the human plasma N-glycome. Epigenetics 2012, 7 (2), 164–172. 10.4161/epi.7.2.18918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanabalasingham G.; Huffman J. E.; Kattla J. J.; Novokmet M.; Rudan I.; Gloyn A. L.; Hayward C.; Adamczyk B.; Reynolds R. M.; Muzinic A.; et al. Mutations in HNF1A result in marked alterations of plasma glycan profile. Diabetes 2013, 62 (4), 1329–1337. 10.2337/db12-0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman J. E.; Knezevic A.; Vitart V.; Kattla J.; Adamczyk B.; Novokmet M.; Igl W.; Pucic M.; Zgaga L.; Johannson A.; et al. Polymorphisms in B3GAT1, SLC9A9 and MGAT5 are associated with variation within the human plasma N-glycome of 3533 European adults. Hum. Mol. Genet. 2011, 20 (24), 5000–5011. 10.1093/hmg/ddr414. [DOI] [PubMed] [Google Scholar]
- Kianickova K.; Pazitna L.; Kundalia P. H.; Pakanova Z.; Nemcovic M.; Barath P.; Katrlikova E.; Suba J.; Trebaticka J.; Katrlik J. Alterations in the Glycan Composition of Serum Glycoproteins in Attention-Deficit Hyperactivity Disorder. Int. J. Mol. Sci. 2023, 24 (10), 8745. 10.3390/ijms24108745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornik O.; Wagner J.; Pucic M.; Knezevic A.; Redzic I.; Lauc G. Stability of N-glycan profiles in human plasma. Glycobiology 2009, 19 (12), 1547–1553. 10.1093/glycob/cwp134. [DOI] [PubMed] [Google Scholar]
- Kristic J.; Vuckovic F.; Menni C.; Klaric L.; Keser T.; Beceheli I.; Pucic-Bakovic M.; Novokmet M.; Mangino M.; Thaqi K.; et al. Glycans are a novel biomarker of chronological and biological ages. J. Gerontol A Biol. Sci. Med. Sci. 2014, 69 (7), 779–789. 10.1093/gerona/glt190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura Y.; Endo T. Glycomics and glycoproteomics focused on aging and age-related diseases - Glycans as a potential biomarker for physiological alterations. Biochim. Biophys. Acta 2016, 1860 (8), 1608–1614. 10.1016/j.bbagen.2016.01.013. [DOI] [PubMed] [Google Scholar]
- Parekh R.; Roitt I.; Isenberg D.; Dwek R.; Rademacher T. Age-related galactosylation of the N-linked oligosaccharides of human serum IgG. J. Exp Med. 1988, 167 (5), 1731–1736. 10.1084/jem.167.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobata A. Glycobiology in the field of aging research-introduction to glycogerontology. Biochimie 2003, 85 (1–2), 13–24. 10.1016/S0300-9084(03)00003-8. [DOI] [PubMed] [Google Scholar]
- Dall’Olio F.; Vanhooren V.; Chen C. C.; Slagboom P. E.; Wuhrer M.; Franceschi C. N-glycomic biomarkers of biological aging and longevity: a link with inflammaging. Ageing Res. Rev. 2013, 12 (2), 685–698. 10.1016/j.arr.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Kristic J.; Lauc G.; Pezer M. Immunoglobulin G glycans - biomarkers and molecular effectors of aging. Clin. Chim. Acta 2022, 535, 30–45. 10.1016/j.cca.2022.08.006. [DOI] [PubMed] [Google Scholar]
- Gudelj I.; Lauc G.; Pezer M. Immunoglobulin G glycosylation in aging and diseases. Cell Immunol 2018, 333, 65–79. 10.1016/j.cellimm.2018.07.009. [DOI] [PubMed] [Google Scholar]
- Borelli V.; Vanhooren V.; Lonardi E.; Reiding K. R.; Capri M.; Libert C.; Garagnani P.; Salvioli S.; Franceschi C.; Wuhrer M. Plasma N-Glycome Signature of Down Syndrome. J. Proteome Res. 2015, 14 (10), 4232–4245. 10.1021/acs.jproteome.5b00356. [DOI] [PubMed] [Google Scholar]
- Shikata K.; Yasuda T.; Takeuchi F.; Konishi T.; Nakata M.; Mizuochi T. Structural changes in the oligosaccharide moiety of human IgG with aging. Glycoconj J. 1998, 15 (7), 683–689. 10.1023/A:1006936431276. [DOI] [PubMed] [Google Scholar]
- Varki A. Sialic acids in human health and disease. Trends Mol. Med. 2008, 14 (8), 351–360. 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhaak L. R.; Uh H. W.; Beekman M.; Koeleman C. A.; Hokke C. H.; Westendorp R. G.; Wuhrer M.; Houwing-Duistermaat J. J.; Slagboom P. E.; Deelder A. M. Decreased levels of bisecting GlcNAc glycoforms of IgG are associated with human longevity. PLoS One 2010, 5 (9), e12566 10.1371/journal.pone.0012566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakovic M. P.; Selman M. H.; Hoffmann M.; Rudan I.; Campbell H.; Deelder A. M.; Lauc G.; Wuhrer M. High-throughput IgG Fc N-glycosylation profiling by mass spectrometry of glycopeptides. J. Proteome Res. 2013, 12 (2), 821–831. 10.1021/pr300887z. [DOI] [PubMed] [Google Scholar]
- de Haan N.; Wuhrer M.; Ruhaak L. R. Mass spectrometry in clinical glycomics: The path from biomarker identification to clinical implementation. Clinical Mass Spectrometry 2020, 18, 1–12. 10.1016/j.clinms.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauc G. Precision medicine that transcends genomics: Glycans as integrators of genes and environment. Biochim. Biophys. Acta 2016, 1860 (8), 1571–1573. 10.1016/j.bbagen.2016.05.001. [DOI] [PubMed] [Google Scholar]
- Post M. A.; Lefeber D. J. Clinical glycomics in the diagnostic laboratory. Ann. Transl Med. 2019, 7 (Suppl 6), S220. 10.21037/atm.2019.08.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhrin M.; Kovcic V.; Zhang Z.; Moen S. H.; Nedal T. M. V.; Bondt A.; Holst S.; Misund K.; Buene G.; Sundan A.; et al. Monoclonal immunoglobulins promote bone loss in multiple myeloma. Blood 2020, 136 (23), 2656–2666. 10.1182/blood.2020006045. [DOI] [PubMed] [Google Scholar]
- Kovacs Z.; Simon A.; Szabo Z.; Nagy Z.; Varoczy L.; Pal I.; Csanky E.; Guttman A. Capillary electrophoresis analysis of N-glycosylation changes of serum paraproteins in multiple myeloma. Electrophoresis 2017, 38 (17), 2115–2123. 10.1002/elps.201700006. [DOI] [PubMed] [Google Scholar]
- Mittermayr S.; Le G. N.; Clarke C.; Millan Martin S.; Larkin A. M.; O’Gorman P.; Bones J. Polyclonal Immunoglobulin G N-Glycosylation in the Pathogenesis of Plasma Cell Disorders. J. Proteome Res. 2017, 16 (2), 748–762. 10.1021/acs.jproteome.6b00768. [DOI] [PubMed] [Google Scholar]
- Yoshimura M.; Nishikawa A.; Ihara Y.; Nishiura T.; Nakao H.; Kanayama Y.; Matuzawa Y.; Taniguchi N. High expression of UDP-N-acetylglucosamine: beta-D mannoside beta-1,4-N-acetylglucosaminyltransferase III (GnT-III) in chronic myelogenous leukemia in blast crisis. Int. J. Cancer 1995, 60 (4), 443–449. 10.1002/ijc.2910600404. [DOI] [PubMed] [Google Scholar]
- Nouso K.; Amano M.; Ito Y. M.; Miyahara K.; Morimoto Y.; Kato H.; Tsutsumi K.; Tomoda T.; Yamamoto N.; Nakamura S.; Kobayashi S.; Kuwaki K.; Hagihara H.; Onishi H.; Miyake Y.; Ikeda F.; Shiraha H.; Takaki A.; Nakahara T.; Nishimura S.-I.; Yamamoto K. Clinical utility of high-throughput glycome analysis in patients with pancreatic cancer. J Gastroenterol 2013, 48, 1171–1179. 10.1007/s00535-012-0732-7. [DOI] [PubMed] [Google Scholar]
- Zhao J.; Qiu W.; Simeone D. M.; Lubman D. M. N-linked glycosylation profiling of pancreatic cancer serum using capillary liquid phase separation coupled with mass spectrometric analysis. J. Proteome Res. 2007, 6 (3), 1126–1138. 10.1021/pr0604458. [DOI] [PubMed] [Google Scholar]
- Shih H. C.; Chang M. C.; Chen C. H.; Tsai I. L.; Wang S. Y.; Kuo Y. P.; Chen C. H.; Chang Y. T. High accuracy differentiating autoimmune pancreatitis from pancreatic ductal adenocarcinoma by immunoglobulin G glycosylation. Clin Proteomics 2019, 16, 1. 10.1186/s12014-018-9221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Huang C.; Zhou J.; Zhao K.; Li Y. Causal link between immunoglobulin G glycosylation and cancer: A potential glycobiomarker for early tumor detection. Cell Immunol 2021, 361, 104282. 10.1016/j.cellimm.2021.104282. [DOI] [PubMed] [Google Scholar]
- Levink I. J. M.; Klatte D. C. F.; Hanna-Sawires R. G.; Vreeker G. C. M.; Ibrahim I. S.; van der Burgt Y. E. M.; Overbeek K. A.; Koopmann B. D. M.; Cahen D. L.; Fuhler G. M.; et al. Longitudinal changes of serum protein N-Glycan levels for earlier detection of pancreatic cancer in high-risk individuals. Pancreatology 2022, 22 (4), 497–506. 10.1016/j.pan.2022.03.021. [DOI] [PubMed] [Google Scholar]
- Snyder C. M.; Alley W. R. Jr; Campos M. I.; Svoboda M.; Goetz J. A.; Vasseur J. A.; Jacobson S. C.; Novotny M. V. Complementary Glycomic Analyses of Sera Derived from Colorectal Cancer Patients by MALDI-TOF-MS and Microchip Electrophoresis. Anal. Chem. 2016, 88 (19), 9597–9605. 10.1021/acs.analchem.6b02310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. P.; Ruan C. P.; Wang H.; Hu Z. Q.; Fang M.; Gu X.; Ji J.; Zhao J. Y.; Gao C. F. Identification and assessment of new biomarkers for colorectal cancer with serum N-glycan profiling. Cancer 2012, 118 (3), 639–650. 10.1002/cncr.26342. [DOI] [PubMed] [Google Scholar]
- Kyselova Z.; Mechref Y.; Kang P.; Goetz J. A.; Dobrolecki L. E.; Sledge G. W.; Schnaper L.; Hickey R. J.; Malkas L. H.; Novotny M. V. Breast cancer diagnosis and prognosis through quantitative measurements of serum glycan profiles. Clin Chem. 2008, 54 (7), 1166–1175. 10.1373/clinchem.2007.087148. [DOI] [PubMed] [Google Scholar]
- Alley W. R. Jr; Novotny M. V. Glycomic analysis of sialic acid linkages in glycans derived from blood serum glycoproteins. J. Proteome Res. 2010, 9 (6), 3062–3072. 10.1021/pr901210r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronewitter S. R.; An H. J.; de Leoz M. L.; Lebrilla C. B.; Miyamoto S.; Leiserowitz G. S. The development of retrosynthetic glycan libraries to profile and classify the human serum N-linked glycome. Proteomics 2009, 9 (11), 2986–2994. 10.1002/pmic.200800760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebrehiwot A. G.; Melka D. S.; Kassaye Y. M.; Gemechu T.; Lako W.; Hinou H.; Nishimura S. I. Exploring serum and immunoglobulin G N-glycome as diagnostic biomarkers for early detection of breast cancer in Ethiopian women. BMC Cancer 2019, 19 (1), 588. 10.1186/s12885-019-5817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldova R.; Asadi Shehni A.; Haakensen V. D.; Steinfeld I.; Hilliard M.; Kifer I.; Helland A.; Yakhini Z.; Borresen-Dale A. L.; Rudd P. M. Association of N-glycosylation with breast carcinoma and systemic features using high-resolution quantitative UPLC. J. Proteome Res. 2014, 13 (5), 2314–2327. 10.1021/pr401092y. [DOI] [PubMed] [Google Scholar]
- Memarian E.; Heijmans R.; Slieker R. C.; Sierra A.; Gornik O.; Beulens J. W. J.; Hanic M.; Elders P.; Pascual J.; Sijbrands E.; et al. IgG N-glycans are associated with prevalent and incident complications of type 2 diabetes. Diabetes Metab Res. Rev. 2023, 39 (7), e3685 10.1002/dmrr.3685. [DOI] [PubMed] [Google Scholar]
- Birukov A.; Plavsa B.; Eichelmann F.; Kuxhaus O.; Hoshi R. A.; Rudman N.; Stambuk T.; Trbojevic-Akmacic I.; Schiborn C.; Morze J.; et al. Immunoglobulin G N-Glycosylation Signatures in Incident Type 2 Diabetes and Cardiovascular Disease. Diabetes Care 2022, 45 (11), 2729–2736. 10.2337/dc22-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keser T.; Gornik I.; Vuckovic F.; Selak N.; Pavic T.; Lukic E.; Gudelj I.; Gasparovic H.; Biocina B.; Tilin T.; et al. Increased plasma N-glycome complexity is associated with higher risk of type 2 diabetes. Diabetologia 2017, 60 (12), 2352–2360. 10.1007/s00125-017-4426-9. [DOI] [PubMed] [Google Scholar]
- Adua E.; Memarian E.; Russell A.; Trbojevic-Akmacic I.; Gudelj I.; Juric J.; Roberts P.; Lauc G.; Wang W. High throughput profiling of whole plasma N-glycans in type II diabetes mellitus patients and healthy individuals: A perspective from a Ghanaian population. Arch. Biochem. Biophys. 2019, 661, 10–21. 10.1016/j.abb.2018.10.015. [DOI] [PubMed] [Google Scholar]
- Mahajan A.; Taliun D.; Thurner M.; Robertson N. R.; Torres J. M.; Rayner N. W.; Payne A. J.; Steinthorsdottir V.; Scott R. A.; Grarup N.; et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 2018, 50 (11), 1505–1513. 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc F.; Reiding K. R.; de Haan N.; Koeleman C. A. M.; Hipgrave Ederveen A. L.; Manetti N.; Dotz V.; Annese V.; Wuhrer M. Immunoglobulin A Glycosylation Differs between Crohn’s Disease and Ulcerative Colitis. J. Proteome Res. 2023, 22 (10), 3213–3224. 10.1021/acs.jproteome.3c00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimurina M.; de Haan N.; Vučković F.; Kennedy N. A.; Štambuk J.; Falck D.; Trbojević-Akmačić I.; Clerc F.; Razdorov G.; Khon A.; et al. Glycosylation of Immunoglobulin G Associates With Clinical Features of Inflammatory Bowel Diseases. Gastroenterology 2018, 154 (5), 1320–1333. 10.1053/j.gastro.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubhakar A.; Jansen B. C.; Adams A. T.; Reiding K. R.; Ventham N. T.; Kalla R.; Bergemalm D.; Urbanowicz P. A.; Gardner R. A.; Consortium I.-B.; et al. Serum N-Glycomic Biomarkers Predict Treatment Escalation in Inflammatory Bowel Disease. J. Crohns Colitis 2023, 17 (6), 919–932. 10.1093/ecco-jcc/jjad012. [DOI] [PubMed] [Google Scholar]
- Verhelst X.; Dias A. M.; Colombel J. F.; Vermeire S.; Van Vlierberghe H.; Callewaert N.; Pinho S. S. Protein Glycosylation as a Diagnostic and Prognostic Marker of Chronic Inflammatory Gastrointestinal and Liver Diseases. Gastroenterology 2020, 158 (1), 95–110. 10.1053/j.gastro.2019.08.060. [DOI] [PubMed] [Google Scholar]
- Gardinassi L. G.; Dotz V.; Hipgrave Ederveen A.; de Almeida R. P.; Nery Costa C. H.; Costa D. L.; de Jesus A. R.; Mayboroda O. A.; Garcia G. R.; Wuhrer M.; et al. Clinical severity of visceral leishmaniasis is associated with changes in immunoglobulin g fc N-glycosylation. mBio 2014, 5 (6), e01844 10.1128/mBio.01844-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkunnikova S.; Mijakovac A.; Sironic L.; Hanic M.; Lauc G.; Kavur M. M. IgG glycans in health and disease: Prediction, intervention, prognosis, and therapy. Biotechnol Adv. 2023, 67, 108169. 10.1016/j.biotechadv.2023.108169. [DOI] [PubMed] [Google Scholar]
- National Research Council (US) Committee on Assessing the Importance and Impact of Glycomics and Glycosciences . The National Academies Collection: Reports funded by National Institutes of Health. In Transforming Glycoscience: A Roadmap for the Future; National Academies Press, National Academy of Sciences, 2012. [PubMed] [Google Scholar]
- Singh S. S.; Naber A.; Dotz V.; Schoep E.; Memarian E.; Slieker R. C.; Elders P. J. M.; Vreeker G.; Nicolardi S.; Wuhrer M. Metformin and statin use associate with plasma protein N-glycosylation in people with type 2 diabetes. BMJ. Open Diabetes Res. Care 2020, 8 (1), e001230. 10.1136/bmjdrc-2020-001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai K.; Gupta S.; May F. P.; Xu G.; Shaukat A.; Hommes D. W.; Early Detection of Advanced Adenomas and Colorectal Carcinoma by Serum Glycoproteome Profiling. Gastroenterology 2024, 166 (1), 194–197. 10.1053/j.gastro.2023.09.034. [DOI] [PubMed] [Google Scholar]
- Pett C.; Nasir W.; Sihlbom C.; Olsson B. M.; Caixeta V.; Schorlemer M.; Zahedi R. P.; Larson G.; Nilsson J.; Westerlind U. Effective Assignment of alpha2,3/alpha2,6-Sialic Acid Isomers by LC-MS/MS-Based Glycoproteomics. Angew. Chem., Int. Ed. Engl. 2018, 57 (30), 9320–9324. 10.1002/anie.201803540. [DOI] [PubMed] [Google Scholar]
- Cramer D. A. T.; Franc V.; Caval T.; Heck A. J. R. Charting the Proteoform Landscape of Serum Proteins in Individual Donors by High-Resolution Native Mass Spectrometry. Anal. Chem. 2022, 94 (37), 12732–12741. 10.1021/acs.analchem.2c02215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.