Abstract
Metabolic pathways involved in the formation of cytotoxic end products by Porphyromonas gingivalis were studied. The washed cells of P. gingivalis ATCC 33277 utilized peptides but not single amino acids. Since glutamate and aspartate moieties in the peptides were consumed most intensively, a dipeptide of glutamate or aspartate was then tested as a metabolic substrate of P. gingivalis. P. gingivalis cells metabolized glutamylglutamate to butyrate, propionate, acetate, and ammonia, and they metabolized aspartylaspartate to butyrate, succinate, acetate, and ammonia. Based on the detection of metabolic enzymes in the cell extracts and stoichiometric calculations (carbon recovery and oxidation/reduction ratio) during dipeptide degradation, the following metabolic pathways were proposed. Incorporated glutamylglutamate and aspartylaspartate are hydrolyzed to glutamate and aspartate, respectively, by dipeptidase. Glutamate is deaminated and oxidized to succinyl-coenzyme A (CoA) by glutamate dehydrogenase and 2-oxoglutarate oxidoreductase. Aspartate is deaminated into fumarate by aspartate ammonia-lyase and then reduced to succinyl-CoA by fumarate reductase and acyl-CoA:acetate CoA-transferase or oxidized to acetyl-CoA by a sequential reaction of fumarase, malate dehydrogenase, oxaloacetate decarboxylase, and pyruvate oxidoreductase. The succinyl-CoA is reduced to butyryl-CoA by a series of enzymes, including succinate-semialdehyde dehydrogenase, 4-hydroxybutyrate dehydrogenase, and butyryl-CoA oxidoreductase. A part of succinyl-CoA could be converted to propionyl-CoA through the reactions initiated by methylmalonyl-CoA mutase. The butyryl- and propionyl-CoAs thus formed could then be converted into acetyl-CoA by acyl-CoA:acetate CoA-transferase with the formation of corresponding cytotoxic end products, butyrate and propionate. The formed acetyl-CoA could then be metabolized further to acetate.
Porphyromonas gingivalis (formerly Bacteroides gingivalis), a black-pigmented gram-negative anaerobe, is frequently detected in the lesions of several types of periodontitis (37–39, 57), and its isolation frequency increases in active sites of periodontitis (50). These observations suggest the strong relation of this bacterium with periodontal diseases.
P. gingivalis has several periodontal pathogenic factors, including membrane-associated proteases, immunoactive cellular compounds, and cytotoxic metabolic end products (19, 31). The main cytotoxic end products, butyrate, propionate, and ammonia, have been found to easily penetrate into periodontal tissue, due to their low molecular weights (59), and subsequently to disturb host cell activity and host defense systems at millimolar concentrations (3, 10–12, 24, 40, 42, 49), the concentration levels found in the P. gingivalis culture supernatant and the gingival crevicular fluid of periodontally diseased subjects (7, 41). Among metabolic end products of P. gingivalis, butyrate is considered the most cytotoxic (42). However, only little information is available about the mechanism of cytotoxic end product formation by P. gingivalis.
Several attempts have been made to elucidate the metabolic system of P. gingivalis. Shah and Williams (47) demonstrated that P. gingivalis is capable of degrading aspartate and asparagine to succinate, although P. gingivalis usually produces little succinate (18). Joe et al. (22) reported that this bacterium has glutamate dehydrogenase as an enzyme for glutamate degradation. However, most researchers have concluded that P. gingivalis utilizes mainly peptides instead of single amino acids as sources of energy and cell materials (35, 46, 48, 55, 60, 62). For example, a chemically defined medium for P. gingivalis must be supplemented with a peptide or a protein such as Trypticase (46, 62) or bovine serum albumin (35). Thus, due to the complicated amino acid composition of peptides or proteins, it had been difficult to determine the amino acid metabolic pathway of P. gingivalis. In addition, some enzymes for amino acid metabolism in oral anaerobes are known to be oxygen labile (2, 8, 9, 56), which makes the detection of P. gingivalis metabolic enzyme more difficult.
In this study, we first determined which amino acid moieties in peptides are preferentially utilized by P. gingivalis, and, second, we tested dipeptides of preferentially utilized amino acids, glutamylglutamate and aspartylaspartate, as metabolic substrates for P. gingivalis. Third, on the basis of the detection of metabolic enzymes and the stiochiometric calculations of carbon recovery and the oxidation/reduction ratio during dipeptide metabolism, we determined the metabolic pathways involved in cytotoxic end product formation by P. gingivalis. To all these experimental procedures, we applied careful and strict techniques for anaerobic experiments.
MATERIALS AND METHODS
Microorganism and growth conditions.
P. gingivalis ATCC 33277T was used throughout this study. This bacterium was grown in modified BM medium (56) containing 1% tryptone (Difco, Detroit, Mich.), 1% Proteose Peptone (Difco), 0.5% yeast extract (Difco), 0.5% NaCl, 5 μg of hemin per ml, and 0.5 μg of menadione per ml in 38 mM potassium phosphate buffer (pH 7.0) in an anaerobic chamber (N2, 80%; CO2, 10%; H2, 10%; NHC-type, Hirasawa Works, Tokyo, Japan) at 37°C. Bacterial purity was regularly confirmed by microscopic examination of Gram-stained smears and by culturing on blood agar plates containing hemin and menadione.
Bacterial growth on peptide- and amino acid-based media.
P. gingivalis cells grown to logarithmic growth phase were transferred into five different media: modified BM medium, modified BM medium with the tryptone and Proteose Peptone concentration decreased to 0.25% (1/4-BM medium) and 1/4-BM medium supplemented with 1% tryptone as a mixture of peptides (1/4-BM-T medium), 1% Casamino Acids (Difco) as a mixture of amino acids (1/4-BM-C medium), or 1% Casamino Acids plus 0.05% tryptophan (1/4-BM-CT medium). Bacterial growth was monitored photometrically at 660 nm. Growth in all media reached its maximum within 48 h.
Incubation of washed cells with peptides and amino acids.
The bacterial cells were harvested at the logarithmic growth phase (15 to 18 h after inoculation) by centrifugation. Unless otherwise indicated, the following experiments were carried out in another anaerobic chamber (N2, 90%; H2, 10%; NH-type, Hirasawa Works). During centrifugation and transportation between the anaerobic chambers, the cells were protected from oxygen exposure in double-sealed centrifuge tubes. The bacterial cells were washed twice with 25 mM potassium phosphate buffer (pH 7.0) containing 50 mM NaCl and 5 mM MgCl2 and suspended in the same buffer solution (1.5 to 1.7 mg [dry weight] per ml). The cell suspension (3.6 ml) was incubated at 37°C for 10 min and then mixed with 0.4 ml of 10% tryptone, 10% Casamino Acids, or 10% Casamino Acids plus 0.5% trytophan. After incubation at 37°C for 0, 120, and 240 min, the cell suspension was sampled and taken out of the anaerobic chamber. After centrifugation (10,000 × g, 4°C, 5 min) part of the supernatant (0.5 ml) was mixed with 0.05 ml of 60% perchloric acid and stored at 4°C for analyses of carboxylic acids and ammonia. Another part of the supernatant (0.7 ml) was mixed with 0.7 ml of 12 N HCl and stored at 4°C for analysis of amino acids.
In separate experiments, the cell suspension was mixed with glutamate, aspartate, glutamylglutamate, or aspartylaspartate at a final concentration of 10 mM. After incubation at 37°C for 0, 60, and 120 min, samples of the cell suspension were removed from the anaerobic chamber. After centrifugation (10,000 × g, 4°C, 5 min), part of the supernatant (0.5 ml) was mixed with 0.05 ml of 60% perchloric acid and stored at 4°C for analyses of carboxylic acids and ammonia.
Assay for metabolic end products.
The perchloric acid-acidified supernatant was diluted with 0.2 N HCl. Carboxylic acids, including formic, acetic, propionic, pyruvic, lactic, malic, succinic, butyric, isobutyric, valeric, and isovaleric acids, were analyzed with a carboxylic acid analyzer (model S-3000X; Tokyo Rikakikai, Tokyo, Japan), as described previously (53, 54). Ammonia was assayed enzymatically using glutamate dehydrogenase by the method of Bergmeyer (4).
Assay for amino acids.
The HCl-acidified supernatant (1.0 ml), obtained from the incubation of washed cells with peptide as described above, was placed in a glass ampoule. After the glass ampoule was flushed with nitrogen, it was sealed and then heated at 110°C for 20 h. The amino acids contained in both the HCl-acidified- and the HCl-hydrolyzed-supernatants were labeled with phenyl isothiocyanate and then analyzed by high-performance liquid chromatography, according to the manufacturer's instructions (Wako Pure Chemical Industries, Osaka, Japan). In brief, each supernatant or amino acid standard mixture was dried and mixed with an ethanol-water-triethanolamine (2:2:1) solution. The resultant mixture was dried, mixed with an ethanol-water-triethanolamine-phenyl isothiocyanate (7:1:1:1) solution, and incubated at room temperature for 20 min. The mixture was dried and stored at −80°C until assayed. The dried sample was dissolved in PTC-amino acid mobile phase A (Wako) and then separated in a reverse-phase column (Wakopak WS-PTC; Wako) at 40°C. Eluent solution was PTC-amino acid mobile phase A with a linear gradient of PTC-amino acid mobile phase B (Wako). Separated amino acid derivatives were detected spectrophotometrically at 254 nm.
Glutamine and asparagine contained in the HCl-acidified supernatant were assayed enzymatically by the methods of Lund (26) and Möllering (36), respectively, within 2 h after mixing with HCl.
Assay for metabolic enzymes.
The bacterial cells were harvested and washed as described above and stored as cell pellets at −20°C in a freezer equipped in the NH-type anaerobic chamber. After thawing, the cell pellets were suspended in 40 mM potassium phosphate buffer (pH 7.0) containing 5 mM MgCl2 and oscillated anaerobically by ultrasonication (2 A, 190 W, 4°C, 6 min) as described previously (56). The cell debris and unbroken cells were removed by centrifugation anaerobically (10,000 × g, 4°C, 10 min), and the resultant cell extracts were used for the assay of metabolic enzymes. All the enzyme activities, except dipeptidases, fumarase, oxaloacetate decarboxylase, and acetate kinase, were assayed by reactions coupled to the reduction or oxidation of NAD(P) (ɛ = 6.22 cm−1 at 340 nm), methylviologen (ɛ = 13.0 cm−1 at 600 nm), and p-indonitrotetrazolium (INT) (ɛ = 19 cm−1 at 492 nm) at 35°C with a spectrophotometer placed in the NH-type anaerobic chamber. The detection limit was approximately 1 U per g of protein.
(i) Enzymes involved in glutamate degradation.
The reaction mixture for glutamylglutamate dipeptidase (EC 3.4.13.11) contained 2 mM glutamylglutamate and cell extract in 100 mM potassium phosphate buffer (pH 7.0). The amount of formed glutamate was determined by the method of Lund (26). The reaction mixture for glutamate dehydrogenase (EC 1.4.1.3 and EC 1.4.1.4) contained 15 mM glutamate, cell extracts, and 1 mM NAD, NADP, or oxidized methylviologen in 50 mM triethanolamine-HCl buffer (pH 8.3). The reaction mixture for 2-oxoglutarate oxidoreductase (EC 1.2.4.2 and EC 1.2.7.3) contained 30 mM 2-oxoglutarate, 0.1 mM coenzyme A (CoA), 5 mM MgCl2, cell extracts, and a 1 mM concentration of NAD, NADP, or oxidized methylviologen in 100 mM potassium phosphate buffer (pH 8.0). The activities of succinate-semialdehyde dehydrogenase (EC 1.2.1) and 4-hydroxybutyrate dehydrogenase (EC 1.1.1.61) were assayed by the method of Gharbia and Shah (14). The reaction mixture for reductive reaction of butyryl-CoA oxidoreductase (EC 1.3.99.2) contained 0.2 mM crotonoyl-CoA, cell extracts, and 0.1 mM NADH, NADPH, or reduced methylviologen in 50 mM potassium phosphate buffer (pH 7.0). The mixture for the oxidative reaction contained 0.2 mM butyryl-CoA, cell extracts, and 1 mM NAD, NADP, or oxidized methylviologen in 50 mM potassium phosphate buffer (pH 7.0). The formation of crotonoyl-CoA from 4-hyroxybutyrate was estimated as the reductive activity of butyryl-CoA oxidoreductase in the reaction mixture containing 10 mM 4-hydroxybutyrate, 1 mM acetyl-CoA, cell extracts, and 0.1 mM reduced methylviologen in 50 mM potassium phosphate buffer (pH 7.0). 2-Hydroxyglutarate dehydrogenase (EC 1.1.99.2) in the hydroxyglutarate pathway and 3-methylaspartate ammonia-lyase (EC 4.3.1.2) in the methylaspartate pathway were measured by the method of Gharbia and Shah (14). 4-Aminobutyrate aminotransferase (EC 2.6.1.19) in the aminobutyrate pathway was measured in a reaction mixture containing 1 mM 4-aminobutyrate, 1 mM 2-oxoglutarate, 1 mM NAD, 25 mg of Triton-X per ml, 0.1 mM INT, 0.1 U of diaphorase per ml, 0.1 U of glutamate dehydrogenase per ml, and cell extracts in 32.5 mM triethanolamine–4 mM potassium phosphate buffer (pH 8.6).
(ii) Enzymes involved in aspartate degradation.
The reaction mixture for aspartylaspartate dipeptidase (EC 3.4.13.11) was the same for glutamylglutamate dipeptidase except that glutamylglutamate was replaced by aspartylaspartate. The amount of aspartate formed was determined by the method of Möllering (36). Aspartate ammonia-lyase (EC 4.3.1.1) and fumarate reductase (EC 1.3.1.6 and EC 1.3.99.1), malate dehydrogenase (EC 1.1.1.37 and EC 1.1.1.82), and oxaloacetate decarboxylase (EC 4.1.1.3) were measured as described previously (56). Fumarase (EC 4.2.1.2) was measured as described previously (56) except that MgCl2 was omitted. The reaction mixture for aspartate aminotransferase (EC 2.6.1.1) was the same for 4-aminobutyrate aminotransferase except that 4-aminobutyrate was replaced by aspartate. The reaction mixture for pyruvate oxidoreductase (EC 1.2.4.1 and EC 1.2.7.1) was the same for 2-oxoglutarate oxidoreductase, except that 2-oxoglutarate was replaced by pyruvate.
(iii) Enzymes involved in acyl-CoA metabolism, ATP formation, and oxidation-reduction of NAD, NADP, and methylviologen.
The activity of acyl-CoA:acetate CoA-transferase (EC 2.8.3.1 and EC 2.8.3.8) was estimated by the formation of acetyl-CoA from acyl-CoA and acetate. The reaction mixture contained 200 mM acetate, 1 mM NAD, 10 mM malate, 0.5 U of malate dehydrogenase per ml, 0.2 U of citrate synthase per ml, cell extracts, and 0.1 mM propionyl-, succinyl- or butyryl-CoA. Phosphotransacetylase activity (EC 2.3.1.8) and acetate kinase (EC 2.7.2.1) were measured as described previously (43, 56).
Chemicals and enzyme preparations.
Amino acids, 4-aminobutyrate, l-threo-3-methylaspartate, 2-oxoglutarate, oxaloacetate, malate, fumarate, INT, diaphorase, and methylviologen were purchased from Wako. Glutamylglutamate, aspartylaspartate, CoA, CoA derivatives, succinate-semialdehyde, and 4-hydroxybutyrate were purchased from Sigma Chemical Co., St. Louis, Mo. All the other chemicals and enzyme preparations were obtained from Boehringer Mannheim, Mannheim, Germany.
RESULTS
Bacterial growth response to peptides and amino acids.
P. gingivalis strain ATCC 33277 grew well in modified BM medium, but its growth decreased to approximately one-third in 1/4-BM (Table 1). Bacterial growth recovered in 1/4-BM-T but not in 1/4-BM-C or 1/4-BM-CT, indicating the clear requirement of peptides for the growth of this bacterium.
TABLE 1.
Growth response of P. gingivalis ATCC 33277 to tryptone, Casamino Acids, and Casamino Acids plus tryptophan
| Medium | Maximum growth (optical density at 660 nm)a |
|---|---|
| BM | 1.74 |
| 1/4-BM | 0.60 |
| 1/4-BM-T | 1.27 |
| 1/4-BM-C | 0.59 |
| 1/4-BM-CT | 0.60 |
Mean obtained from three independent experiments.
Metabolic end products from peptides and utilization of amino acid moieties of peptides.
From tryptone, the washed cells of P. gingivalis produced ammonia (26.5 mM in the reaction mixture), butyrate (6.33 mM), acetate (3.26 mM), and propionate (1.51 mM) in addition to small amounts of isobutyrate and isovalerate during a 240-min incubation. On the other hand, only small amounts of end products (<1 mM in reaction mixture) were formed from Casamino Acids or Casamino Acids supplemented with tryptophan.
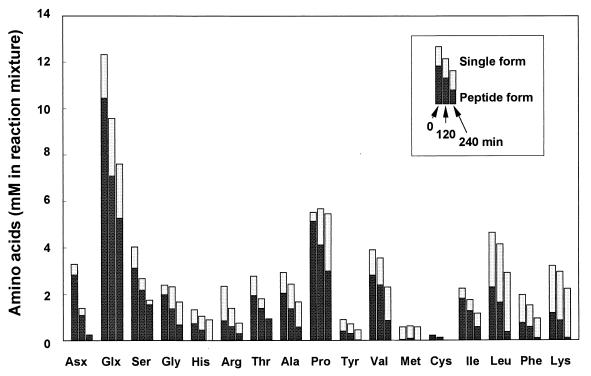
During the incubation, several amino acid moieties of peptides contained in tryptone were utilized by the cells, while single amino acids in tryptone were scarcely consumed (Fig. 1). Among the amino acid moieties in peptides, glutamine/glutamate and asparagine/aspartate were consumed most intensively. Other amino acids except proline were also consumed in peptide form, although their consumption rates were lower than those of glutamine/glutamate and asparagine/aspartate. Proline was converted from a peptide form into a single form, but neither form was utilized significantly.
FIG. 1.
Utilization of single-form and peptide-form amino acids contained in tryptone by the washed cells of P. gingivalis ATCC 33277. Glx, glutamate plus glutamine; Asx, aspartate plus asparagine.
Metabolic end products from glutamylglutamate, aspartylaspartate, glutamate, and aspartate.
Since glutamate and aspartate moieties in the peptide were preferentially utilized, a dipeptide of glutamate or aspartate was then tested as the metabolic substrate of P. gingivalis. The washed cells of P. gingivalis consumed glutamylglutamate and aspartylaspartate and produced significant amounts of end products (Table 2). Butyrate, propionate, and ammonia were mainly produced from glutamylglutamate, while butyrate, succinate, acetate, and ammonia were the main products from aspartylaspartate. Carbon recovery and the oxidation/reduction ratio during dipeptide fermentation were also calculated from the amounts of end products (Table 2). Only a small amount of end product was formed (<0.1 mM in reaction mixture) from glutamate or aspartate.
TABLE 2.
Metabolic end products produced from glutamylglutamate and aspartylaspartate by P. gingivalis ATCC 33277
| Parameter measured | Mean value ± SD for substratea
|
|
|---|---|---|
| Glutamylglutamate | Aspartylaspartate | |
| Metabolic end product concn (mM)b | ||
| Ammonia | 1.57 ± 0.20 | 3.45 ± 0.38 |
| Acetate | 0.02 ± 0.01 | 1.89 ± 0.27 |
| Propionate | 0.47 ± 0.09 | 0.06 ± 0.01 |
| Malate | ND | 0.06 ± 0.03 |
| Succinate | ND | 0.33 ± 0.02 |
| Butyrate | 1.03 ± 0.19 | 0.82 ± 0.13 |
| Carbon recoveryc | 0.96 | 0.84 |
| Oxidation/reduction ratiod | 0.91 | 1.03 |
Mean ± standard deviation obtained from three independent experiments. ND, not detected.
Concentration in reaction mixture of end products formed during a 60-min incubation.
The amounts of utilized dipeptides were assumed to be equal to the amounts of produced ammonia. The amounts of produced carbon dioxide were calculated from [butyrate × 1 + propionate × 2] for glutamylglutamate degradation and [propionate × 1 + acetate × 2] for aspartylaspartate degradation according to the proposed metabolic pathways (Fig. 2).
Calculated from the amounts of end products on the assumption that dipeptides are degraded through the proposed metabolic pathways (Fig. 2).
Enzymes involved in glutamylglutamate degradation.
P. gingivalis cells had a series of enzymes, including glutamylglutamate dipeptidase, glutamate dehydrogenase, 2-oxoglutarate oxidoreductase, succinate-semialdehyde dehydrogenase, 4-hydroxybutyrate dehydrogenase, and butyryl-CoA oxidoreductase, for the degradation of glutamylglutamate to butyryl-CoA (Table 3). Glutamate and 4-hydroxybutyrate dehydrogenases utilized mainly NAD as an electron carrier, while 2-oxoglutarate and butyryl-CoA oxidoreductases utilized only methylviologen. Conversion of 4-hydroxybutyrate into crotonoyl-CoA required acetyl-CoA, but not ATP and CoA, suggesting that acyl-CoA:acetate CoA-transferase functions to transfer CoA from acetyl-CoA to 4-hydroxybutyrate. No activity of 2-hydroxyglutarate dehydrogenase, 3-methylaspartate ammonia-lyase, or 4-aminobutyrate aminotransferase was detected.
TABLE 3.
Metabolic enzymes involved in glutamylglutamate degradation by P. gingivalis ATCC 33277
| Enzyme | Substrate | Electron carrier | Sp act (U/g of protein)a |
|---|---|---|---|
| Glutamylglutamate dipeptidase | Glutamylglutamate | 21.4 ± 5.30 | |
| Glutamate dehydrogenase | Glutamate | NAD | 1,130 ± 128 |
| NADP or MVoxb | ND | ||
| 2-Oxoglutarate oxidoreductase | 2-Oxoglutarate and CoA | NAD or NADP | ND |
| MVox | 11.6 ± 2.96 | ||
| Succinate-semialdehyde dehydrogenase | Succinyl-CoA | NADH | 788 ± 64.3 |
| NADPH or MVredc | ND | ||
| Succinate-semialdehyde | NAD | 1,280 ± 130 | |
| NADP or MVox | ND | ||
| 4-Hydroxybutyrate dehydrogenase | Succinate-semialdehyde | NADH | 2,160 ± 226 |
| NADPH | 330 ± 140 | ||
| MVred | ND | ||
| 4-Hydroxybutyrate | NAD | 325 ± 175 | |
| NADP | 12.3 ± 2.90 | ||
| MVox | ND | ||
| 4-Hydroxybutyrate to crotonoyl-CoA | 4-Hydroxybutyrate and acetyl-CoA | MVred | 148 ± 24.5 |
| −Acetyl-CoA | MVred | 11.7 ± 2.09 | |
| −Acetyl-CoA + ATP + CoA | MVred | 10.8 ± 1.06 | |
| Butyryl-CoA oxidoreductase | Crotonoyl-CoA | NADH | 1.64 ± 0.74 |
| NADPH | ND | ||
| MVred | 2,150 ± 132 | ||
| Butyryl-CoA | NAD, NADP, or MVox | ND |
Mean ± standard deviation obtained from three independent experiments. ND, not detected.
MVox, oxidized methylviologen.
MVred, reduced methylviologen.
Enzymes involved in aspartylaspartate degradation.
P. gingivalis cells had aspartylaspartate dipeptidase and aspartate ammonia-lyase as the main enzymes to convert aspartylaspartate to fumarate (Table 4). This bacterium also had aspartate aminotransferase for the deamination of aspartate to oxaloacetate, although its activity was lower. A series of enzymes, including fumarase, malate dehydrogenase, oxaloacetate decarboxylase, and pyruvate oxidoreductase, was found to be responsible for the oxidative decarboxylation of fumarate to acetyl-CoA. Fumarate reductase was also found to reduce fumarate to succinate (Table 4). Malate dehydrogenase was NAD dependent, while pyruvate oxidoreductase and fumarate reductase utilized methylviologen as an electron carrier.
TABLE 4.
Metabolic enzymes involved in aspartylaspartate degradation by P. gingivalis ATCC 33277
| Enzyme | Substrate | Electron carrier | Sp act (U/g of protein)a |
|---|---|---|---|
| Aspartylaspartate dipeptidase | Aspartylaspartate | 64.1 ± 10.5 | |
| Aspartate ammonia-lyase | Aspartate | 402 ± 242 | |
| Aspartate aminotransferase | Aspartate and 2-oxoglutarate | 3.19 ± 5.52 | |
| Fumarase | Fumarate | 40.5 ± 7.17 | |
| Malate dehydrogenase | Oxaloacetate | NADH | 390 ± 96.9 |
| NADPH or MVredb | ND | ||
| Oxaloacetate decarboxylase | Oxaloacetate | 986 ± 190 | |
| Pyruvate oxidoreductase | Pyruvate and CoA | NAD or NADP | ND |
| MVoxc | 672 ± 435 | ||
| Fumarate reductase | Fumarate | NADH or NADPH | ND |
| MVred | 21.4 ± 5.52 |
Mean ± standard deviation obtained from three independent experiments. ND, not detected.
MVred, reduced methylviologen.
MVox, oxidized methylviologen.
Enzymes involved in acyl-CoA metabolism, ATP formation, and oxidation-reduction of NAD(P) and methylviologen.
P. gingivalis cells had acyl-CoA:acetate CoA-transferase, which transfers CoA from propionyl-, butyryl-, and succinyl-CoAs to acetate, resulting in the formation of acetyl-CoA and the corresponding acids, propionate, butyrate, and succinate (Table 5). This bacterium also had phosphotransacetylase and acetate kinase for the conversion of acetyl-CoA to acetate with the formation of ATP from ADP. Methylviologen:NAD(P) oxidoreductase was detected as an enzyme for electron transfer between NAD(P) and methylviologen (Table 5).
TABLE 5.
Metabolic enzymes involved in acyl-CoA metabolism, ATP formation, and oxidation-reduction of NAD(P) and methylviologen by P. gingivalis ATCC 33277
| Enzyme | Substrate | Electron carrier | Sp act (U/g of protein)a |
|---|---|---|---|
| Acyl-CoA:acetate CoA-transferase | Butyryl-CoA and acetate | 100 ± 7.07 | |
| Propionyl-CoA and acetate | 154 ± 17.8 | ||
| Succinyl-CoA and acetate | 80.7 ± 16.2 | ||
| Phosphotransacetylase | Acetyl-CoA and inorganic phosphate | 4.83 ± 2.35 | |
| Acetate kinase | Acetate and ATP | 9.42 ± 5.07 | |
| Methylviologen:NAD(P) oxidoreductase | NADH and MVoxb | 510 ± 123 | |
| NADPH and MVox | 65.8 ± 19.3 |
Mean ± standard deviation obtained from three independent experiments.
MVox, oxidized methylviologen.
DISCUSSION
P. gingivalis utilized the peptide form rather than the single form of amino acids, especially glutamine/glutamate and asparagine/aspartate moieties of peptides (Fig. 1), indicating that this bacterium preferentially takes up glutamine/glutamate- and asparagine/aspartate-containing peptides. This bacterium can further hydrolyze the incorporated peptides to amino acids and then degrade into cytotoxic end products. Furthermore, the finding of glutamylglutamate and aspartylaspartate utilization by P. gingivalis enabled us to propose the following metabolic pathways through the stoichiometric calculation of consumed dipeptides and end products (Table 2) and the detection of metabolic enzymes (Tables 3 to 5). All the specific activities of enzymes were measured at levels of units per gram of protein, similar to the metabolic enzyme activities found in Bacteroides fragilis (28) and two other black-pigmented gram-negative anaerobes, Prevotella intermedia and Prevotella nigrescens (56).
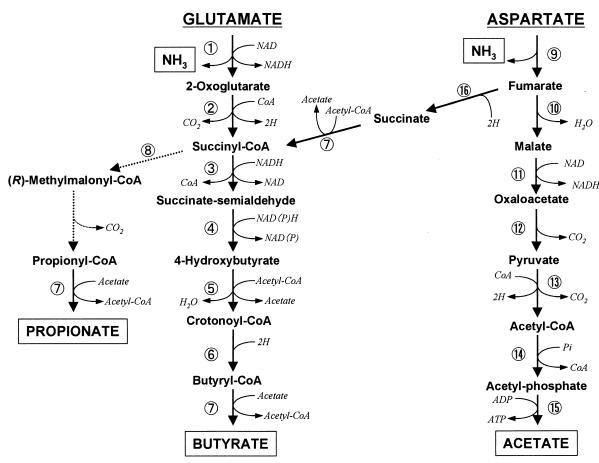
Glutamate derived from peptides is thought to be deaminated and decarboxylated to succinyl-CoA by glutamate dehydrogenase and 2-oxoglutarate oxidoreductase with the production of reducing potential (reactions 1 and 2 in Fig. 2). Subsequently, two-thirds of succinyl-CoA can be converted into butyryl-CoA along with the consumption of the reducing potential by a sequential reaction, including succinate-semialdehyde dehydrogenase, 4-hydroxybutyrate dehydrogenase, and butyryl-CoA oxidoreductase (reactions 3 to 6 in Fig. 2). The butyryl-CoA can further be converted into acetyl-CoA by acyl-CoA:acetate CoA-transferase with the formation of butyrate as the most cytotoxic end product, (reaction 7 in Fig. 2). The formed acetyl-CoA can further be degraded to acetate by phosphotransacetylase and acetate kinase with the production of ATP (reactions 14 and 15 in Fig. 2). One-third of succinyl-CoA could be converted into propionate without the consumption of reducing potential through a sequential reaction, including methylmalonyl-CoA mutase and acyl-CoA:acetate CoA-transferase (reactions 8 to 7 in Fig. 2), as reported for some propionate-producing bacteria (15). The formed acetyl-CoA can also be converted into acetate with ATP production. Methylmalonyl-CoA mutase has been detected in P. gingivalis (21), although this enzyme was not measured in this study. Stoichiometric calculations of carbon recovery and oxidation/reduction ratio during glutamylglutamate degradation (Table 2) support the appropriateness of these pathways. On the basis of these pathways, the metabolic equation for glutamate can be calculated as follows: 3 glutamate → 2 butyrate + propionate + 4 CO2 + 3 NH3.
FIG. 2.
Proposed metabolic pathways for glutamate and aspartate in P. gingivalis. 1, glutamate dehydrogenase; 2, 2-oxoglutarate oxidoreductase; 3, succinate-semialdehyde dehydrogenase; 4, 4-hydroxybutyrate dehydrogenase; 5, enzyme(s) for the conversion of 4-hydroxybutyrate into crotonoyl-CoA; 6, butyryl-CoA oxidoreductase; 7, acyl-CoA:acetate CoA-transferase; 8, methylmalonyl-CoA mutase; 9, aspartate aminotransferase; 10, fumarase; 11, malate dehydrogenase; 12, oxaloacetate decarboxylase; 13, pyruvate oxidoreductase; 14, phosphotransacetylase; 15, acetate kinase; 16, fumarate reductase. Broken lines, not measured but expected pathway.
It is known that Fusobacterium species, including oral strains, metabolize glutamate to butyrate, acetate, and ammonia through the hydroxyglutarate, the methylaspartate, and/or the aminobutyrate pathways (14). Other butyrate-producing bacteria such as Clostridium and Peptostreptococcus species (1, 15) are also reported to have some of these pathways. However, P. gingivalis had no activity of 2-hydroxyglutarate dehydrogenase, 3-methylaspartate ammonia-lyase, and 4-aminobutyrate aminotransferase, the key enzymes of those three pathways. The pathway found in our study, involving glutamate dehydrogenase and 2-oxoglutarate oxidoreductase, seems unique to P. gingivalis.
Aspartate derived from peptides is thought to be deaminated to fumarate by asparate ammonia-lyase (reaction 9 in Fig. 2) and then oxidized to acetate or reduced to propionate and butyrate. In the oxidative degradation, fumarate could be degraded into acetate by a series of enzymes, including fumarase, malate dehydrogenase, oxaloacetate decarboxylase, and pyruvate oxidoreductase (reactions 10 to 15 in Fig. 2). Other oral black-pigmented anaerobes, P. intermedia and P. nigrescens, are reported to have a similar metabolic system for aspartate degradation (56). In the reductive pathway, fumarate could be reduced to succinate by fumarate reductase (reaction 16 in Fig. 2). Although Shah and Williams (47) reported that P. gingivalis had fumarate reductase and produced succinate from aspartate, succinate was not the main end product of this bacterium in our study as well as most previous reports (18, 33, 54). Acyl-CoA:acetate CoA-transferase was found to convert succinate to succinyl-CoA by using acetyl-CoA as a CoA donor, indicating that a part of succinate derived from aspartate can be readily converted to succinyl-CoA (reaction 7 in Fig. 2). A similar conversion of succinate into succinyl-CoA has been proposed for succinate utilization by Clostridium kluyveri (45, 51, 61). Subsequently, the formed succinyl-CoA can further be converted to butyrate or propionate, depending on the intracellular oxidation/reduction balance, as explained above for glutamate degradation. Under the experimental conditions of this study, most succinyl-CoA is thought to be converted into butyrate along with the consumption of surplus reducing potential derived from the oxidative degradation of aspartate to acetate. Stoichiometric calculations of carbon recovery and the oxidation/reduction ratio during aspartylaspartate degradation support the appropriateness of these metabolic pathways (Table 2).
In the pathway of P. gingivalis, oxidative decarboxylation of both pyruvate and 2-oxoglutarate was catalyzed by methylviologen- and CoA-dependent oxoacid oxidoreductases (reactions 2 and 13 in Fig. 2). Although pyruvate oxidoreductase is widely distributed in anaerobically growing bacteria (23, 34, 44) and archaea (5, 6, 27), 2-oxoglutarate oxidoreductase has been detected mainly in hyperthermophilic (16, 29, 30) and methanogenic (58) archaea and photosynthetic bacteria (13). P. gingivalis could be included in rare eubacterial species having 2-oxoglutarate oxidoreductase, such as Helicobacter pylori (17, 20). In most eubacteria, corresponding oxoacid oxidoreductases are known as NAD-dependent dehydrogenases (25, 32, 52).
The reducing potential derived from the oxidative degradation of amino acids must be used immediately for the smooth operation of the overall amino acid metabolism. Methylviologen:NAD(P) oxidoreductase could function as an electron transporter between methylviologen-dependent oxidoreductases and NAD(P)-dependent dehydrogenases during overall amino acid metabolism. Although physiological electron carriers for oxidoreductases were not identified in this study, type b cytochrome and menaquinone are candidates (47).
P. gingivalis can decompose proteins into peptides by highly active extracellular proteases (19, 31) and then incorporate these peptides as shown in this study. This could be advantageous for P. gingivalis to colonize the subgingival area or the sites of periodontitis, where abundant proteins and peptides are supplied from gingival crevicular fluid or inflamed periodontal tissue. This bacterium can further degrade glutamate and aspartate contained in peptides into cytotoxic end products, especially butyrate, propionate, and ammonia, through its unique metabolic pathways (Fig. 2). In addition, the formation of the main end products from glutamate and aspartate implies that the pathways for these amino acids are involved in the main routes of amino acid catabolism in P. gingivalis.
Although an outline of metabolic pathways for glutamate and aspartate has been determined in this study, further study using radioactive tracers and specific enzyme-deficient mutants would be valuable to improve these pathways and elucidate metabolic regulation.
ACKNOWLEDGMENTS
T. Sato was a recipient of the Inoue Postdoctoral Fellowship from the Inoue Foundation for Science. This study was supported in part by the Inamori Foundation (to N.T.) and by Grants-in-Aid for Scientific Research (B) (no. 11470386) from the Ministry of Education, Science, Sports and Culture, Japan (to N.T.).
REFERENCES
- 1.Barker H A. Amino acid degradation by anaerobic bacteria. Annu Rev Biochem. 1981;50:23–40. doi: 10.1146/annurev.bi.50.070181.000323. [DOI] [PubMed] [Google Scholar]
- 2.Barker H A, Kahn J M, Hedrick L. Pathway of lysine degradation in Fusobacterium nucleatum. J Bacteriol. 1982;152:201–207. doi: 10.1128/jb.152.1.201-207.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartold P M, Gully N J, Zilm P S, Rogers A H. Identification of components in Fusobacterium nucleatum chemostat-culture supernatants that are potent inhibitors of human gingival fibroblast proliferation. J Periodontal Res. 1991;26:314–322. doi: 10.1111/j.1600-0765.1991.tb02069.x. [DOI] [PubMed] [Google Scholar]
- 4.Bergmeyer H U. Ammonia. Methods Enzym Anal. 1983;8:454–461. [Google Scholar]
- 5.Blamey J M, Adams M W W. Purification and characterization of pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. Biochim Biophys Acta. 1993;1161:19–27. doi: 10.1016/0167-4838(93)90190-3. [DOI] [PubMed] [Google Scholar]
- 6.Blamey J M, Adams M W W. Characterization of ancestral type of pyruvate ferredoxin oxidoreductase from hyperthermophilic bacterium, Thermotoga maritima. Biochemistry. 1994;33:1000–1007. doi: 10.1021/bi00170a019. [DOI] [PubMed] [Google Scholar]
- 7.Botta G A, Radin L, Costa A, Schito G, Blasi G. Gas-liquid chromatography of the gingival fluid as an aid in periodontal diagnosis. J Periodontal Res. 1985;20:450–457. doi: 10.1111/j.1600-0765.1985.tb00827.x. [DOI] [PubMed] [Google Scholar]
- 8.Buckel W, Barker H A. The pathways of glutamate fermentation by anaerobic bacteria. J Bacteriol. 1974;117:1248–1260. doi: 10.1128/jb.117.3.1248-1260.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claesson R, Edlund M-B, Persson S, Carlsson J. Production of volatile sulfur compounds by various Fusobacterium species. Oral Microbiol Immunol. 1990;5:137–142. doi: 10.1111/j.1399-302x.1990.tb00411.x. [DOI] [PubMed] [Google Scholar]
- 10.Eftimiadi C, Stashenko P, Tonetti M, Mangiante P E, Massara R, Zupo S, Ferrarini M. Divergent effect of the anaerobic bacterial by-product butyric acid on the immune response: suppression of T-lymphocyte proliferation and stimulation of interleukin-1 beta production. Oral Microbiol Immunol. 1991;6:17–23. doi: 10.1111/j.1399-302x.1991.tb00446.x. [DOI] [PubMed] [Google Scholar]
- 11.Eftimiadi C, Tonetti M, Cavallero A, Sacco O, Rossi G A. Short-chain fatty acids produced by anaerobic bacteria inhibit phagocytosis by human lung phagocytes. J Infect Dis. 1990;161:138–142. doi: 10.1093/infdis/161.1.138. [DOI] [PubMed] [Google Scholar]
- 12.Eftimiadi C, Valente S, Mangiante S, Mangiante P E, Niederman R. Short chain fatty acids produced by anaerobic bacteria inhibit adhesion and proliferation of periodontal ligament fibroblasts. Minerva Stomatol. 1993;42:481–485. [PubMed] [Google Scholar]
- 13.Gehring U, Arnon D I. Purification and properties of α-ketoglutarate synthase from a photosynthetic bacterium. J Biol Chem. 1972;247:6962–6969. [PubMed] [Google Scholar]
- 14.Gharbia S E, Shah H N. Pathways of glutamate catabolism among Fusobacterium species. J Gen Microbiol. 1991;137:1201–1206. doi: 10.1099/00221287-137-5-1201. [DOI] [PubMed] [Google Scholar]
- 15.Gottschalk G. Bacterial fermentations. In: Gottschalk G, editor. Bacterial metabolism. 2nd ed. New York, N.Y: Springer-Verlag; 1986. pp. 208–282. [Google Scholar]
- 16.Heider J, Mai X, Adams M W W. Characterization of 2-ketoisovalerate ferredoxin oxidoreductase, a new reversible coenzyme A-dependent enzyme involved in peptide fermentation by hyperthermophilic archaea. J Bacteriol. 1996;178:780–787. doi: 10.1128/jb.178.3.780-787.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman P S, Goodwin A, Johnsen J, Magee K, Veldhuyzen van Zanten S J. Metabolic activities of metronidazole-sensitive and -resistant strains of Helicobacter pylori: repression of pyruvate oxidoreductase and expression of isocitrate lyase activity correlate with resistance. J Bacteriol. 1996;178:4822–4829. doi: 10.1128/jb.178.16.4822-4829.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holdeman L V, Kelly R W, Moore W E C. Genus I. Bacteroides, Castellani and Chalmers 1919, 959AL. In: Krieg N R, editor. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 604–631. [Google Scholar]
- 19.Holt S C, Kesavalu L, Walker S, Genco C A. Virulence factors of Porphyromonas gingivalis. Periodontol 2000. 1999;20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 20.Hughes N J, Clayton C L, Chalk P A, Kelly D J. Helicobacter pylori porCDAB and oorDABC genes encode distinct pyruvate:flavodoxin and 2-oxoglutarate:acceptor oxidoreductases which mediate electron transport to NADP. J Bacteriol. 1998;180:1119–1128. doi: 10.1128/jb.180.5.1119-1128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson C A, Kirszbaum L, Dashper S, Reynolds E C. Cloning, expression and sequence analysis of the genes encoding the heterodimeric methylmalonyl-CoA mutase of Porphyromonas gingivalis W50. Gene. 1995;167:127–132. doi: 10.1016/0378-1119(95)00682-6. [DOI] [PubMed] [Google Scholar]
- 22.Joe A, Murray C S, McBridge B C. Nucleotide sequence of a Porphyromonas gingivalis gene encoding a surface-associated glutamate dehydrogenase and construction of a glutamate dehydrogenase-deficient isogenic mutant. Infect Immun. 1994;62:1358–1368. doi: 10.1128/iai.62.4.1358-1368.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerscher L, Oesterhelt D. Pyruvate: ferredoxin oxidoreductase—new findings on an ancient enzyme. Trends Biol Sci. 1982;7:371–374. [Google Scholar]
- 24.Kurita-Ochiai T, Fukushima K, Ochiai K. Volatile fatty acids, metabolic by-products of periodontopathic bacteria, inhibit lymphocyte proliferation and cytokine production. J Dent Res. 1995;74:1367–1373. doi: 10.1177/00220345950740070801. [DOI] [PubMed] [Google Scholar]
- 25.Lowe P N, Hodgson J A, Perham R N. Dual role of a single multienzyme complex in the oxidative decarboxylation of pyruvate and branched-chain 2-oxoacids in Bacillus subtilis. Biochem J. 1983;215:133–140. doi: 10.1042/bj2150133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lund P. l-Glutamine and l-glutamate: UV-method with glutaminase and glutamate dehydrogenase. Methods Enzym Anal. 1983;8:357–363. [Google Scholar]
- 27.Lunow J, Linder D, Thauer R K. Pyruvate:ferredoxin oxidoreductase from the sulfate-reducing Archaeoglobus fulgidus: molecular composition, catalytic properties, and sequence alignments. Arch Microbiol. 1995;163:21–28. doi: 10.1007/BF00262199. [DOI] [PubMed] [Google Scholar]
- 28.Macy J M, Ljungdahl L G, Gottschalk G. Pathway of succinate and propionate fermentation in Bacteroides fragilis. J Bacteriol. 1978;134:84–91. doi: 10.1128/jb.134.1.84-91.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mai X, Adams M W W. Indolepyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. J Biol Chem. 1994;269:16726–16732. [PubMed] [Google Scholar]
- 30.Mai X, Adams M W W. Characterization of a fourth type of 2-keto acid-oxidizing enzyme from a hyperthermophilic archaeon: 2-ketoglutarate ferredoxin oxidoreductase from Thermococcus litoralis. J Bacteriol. 1996;178:5890–5896. doi: 10.1128/jb.178.20.5890-5896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayrand D, Holt S C. Biology of asaccharolytic black-pigmented Bacteroides species. Microbiol Rev. 1988;52:134–152. doi: 10.1128/mr.52.1.134-152.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCully V, Burns G, Sokatch J R. Resolution of branched-chain oxo acid dehydrogenase complex of Pseudomonas aeruginosa PAO. Biochem J. 1986;233:737–742. doi: 10.1042/bj2330737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKee A S, McDermid A S, Baskerville A, Dowsett A B, Ellwood D C, Marsh P D. Effect of hemin on the physiology and virulence of Bacteroides gingivalis W50. Infect Immun. 1986;52:349–355. doi: 10.1128/iai.52.2.349-355.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meinecke B, Bertram J, Gottschalk G. Purification and characterization of the pyruvate-ferredoxin oxidoreductase from Clostridium acetobutylicum. Arch Microbiol. 1989;152:244–250. doi: 10.1007/BF00409658. [DOI] [PubMed] [Google Scholar]
- 35.Milner P, Batten J E, Curtis M A. Development of a simple chemically defined medium for Porphyromonas gingivalis: requirement for α-ketoglutarate. FEMS Microbiol Lett. 1996;140:125–130. doi: 10.1016/0378-1097(96)00159-0. [DOI] [PubMed] [Google Scholar]
- 36.Möllering H. l-Aspartate and l-asparagine. Methods Enzym Anal. 1983;8:350–357. [Google Scholar]
- 37.Moore L V H, Moore W E C, Cato E P, Smibert R M, Burmeister J A, Best A M, Ranney R R. Bacteriology of human gingivitis. J Dent Res. 1987;66:989–995. doi: 10.1177/00220345870660052401. [DOI] [PubMed] [Google Scholar]
- 38.Moore W E C, Holdeman L V, Cato E P, Smibert R M, Burmeister J A, Palcanis K G, Ranney R R. Comparative bacteriology of juvenile periodontitis. Infect Immun. 1985;48:507–519. doi: 10.1128/iai.48.2.507-519.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore W E C, Holdeman L V, Cato E P, Smibert R M, Burmeister J A, Ranney R R. Bacteriology of moderate (chronic) periodontitis in mature adult humans. Infect Immun. 1983;42:510–515. doi: 10.1128/iai.42.2.510-515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niederman R, Brunkhorst B, Smith S, Weinreb R N, Ryder M I. Ammonia as a potential mediator of adult human periodontal infection: inhibition of neutrophil function. Arch Oral Biol. 1990;35:205–209. doi: 10.1016/0003-9969(90)90159-8. [DOI] [PubMed] [Google Scholar]
- 41.Niederman R, Buyle-Bodin Y, Lu B Y, Robinson P, Naleway C. Short-chain carboxylic acid concentration in human gingival crevicular fluid. J Dent Res. 1997;76:575–579. doi: 10.1177/00220345970760010801. [DOI] [PubMed] [Google Scholar]
- 42.Niederman R, Zhang J, Kashket S. Short-chain carboxylic-acid-stimulated, PMN-mediated gingival inflammation. Crit Rev Oral Biol Med. 1997;8:269–290. doi: 10.1177/10454411970080030301. [DOI] [PubMed] [Google Scholar]
- 43.Nishimura J S, Griffith M J. Acetate kinase from Veillonilla alcalescens. Methods Enzymol. 1981;71:311–316. [Google Scholar]
- 44.Pieulle L, Guigliarelli B, Asso M, Dole F, Bernadac A, Hatchikian E C. Isolation and characterization of the pyruvate-ferredoxin oxidoreductase from the sulfate-reducing bacterium Desulfovibrio africanus. Biochim Biophys Acta. 1995;1250:49–59. doi: 10.1016/0167-4838(95)00029-t. [DOI] [PubMed] [Google Scholar]
- 45.Scherf U, Söhling B, Gottschalk G, Linder D, Buckel W. Succinate-ethanol fermentation in Clostridium kluyveri: purification and characterization of 4-hydroxybutyryl-CoA dehydratase/vinylacetyl-CoA Δ3-Δ2-isomerase. Arch Microbiol. 1994;161:239–245. doi: 10.1007/BF00248699. [DOI] [PubMed] [Google Scholar]
- 46.Seddon S V, Shah H N, Hardie J M, Robinson J P. Chemically defined and minimal media for Bacteroides gingivalis. Curr Microbiol. 1988;17:147–149. [Google Scholar]
- 47.Shah H N, Williams R A D. Catabolism of aspartate and asparagine by Bacteroides intermedius and Bacteroides gingivalis. Curr Microbiol. 1987;15:313–318. [Google Scholar]
- 48.Shah H N, Williams R A D. Utilization of glucose and amino acids by Bacteroides intermedius and Bacteroides gingivalis. Curr Microbiol. 1987;15:241–246. [Google Scholar]
- 49.Singer R E, Buchner B A. Butyrate and propionate: important components of toxic dental plaque extracts. Infect Immun. 1981;32:458–463. doi: 10.1128/iai.32.2.458-463.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slots J, Bragd L, Wikström M, Dahlén G. The occurrence of Actinobacillus actinomycetemcomitans, Bacteroides gingivalis and Bacteroides intermedius in destructive periodontal disease in adults. J Clin Periodontol. 1985;13:570–577. doi: 10.1111/j.1600-051x.1986.tb00849.x. [DOI] [PubMed] [Google Scholar]
- 51.Söhling B, Gottschalk G. Purification and characterization of a coenzyme-A-dependent succinate-semialdehyde dehydrogenase from Clostridium kluyveri. Eur J Biochem. 1993;212:121–127. doi: 10.1111/j.1432-1033.1993.tb17641.x. [DOI] [PubMed] [Google Scholar]
- 52.Sokatch J R, McCully V, Roberts C M. Purification of a branched-chain keto acid dehydrogenase from Pseudomonas putida. J Bacteriol. 1981;148:647–652. doi: 10.1128/jb.148.2.647-652.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi N, Abbe K, Takahashi-Abbe S, Yamada T. Oxygen sensitivity of sugar metabolism and interconversion of pyruvate formate-lyase in intact cells of Streptococcus mutans and Streptococcus sanguis. Infect Immun. 1987;55:652–656. doi: 10.1128/iai.55.3.652-656.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahashi N, Saito K, Schachtele C F, Yamada T. Acid tolerance and acid-neutralizing activity of Porphyromonas gingivalis, Prevotella intermedia and Fusobacterium nucleatum. Oral Microbiol Immunol. 1997;12:323–328. doi: 10.1111/j.1399-302x.1997.tb00733.x. [DOI] [PubMed] [Google Scholar]
- 55.Takahashi N, Schachtele C F. Effect of pH on the growth and proteolytic activity of Porphyromonas gingivalis and Bacteroides intermedius. J Dent Res. 1990;69:1266–1269. doi: 10.1177/00220345900690060801. [DOI] [PubMed] [Google Scholar]
- 56.Takahashi N, Yamada T. Pathways for amino acid metabolism by Prevotella intermedia and Prevotella nigrescens. Oral Microbiol Immunol. 2000;15:96–102. doi: 10.1034/j.1399-302x.2000.150205.x. [DOI] [PubMed] [Google Scholar]
- 57.Tanner A, Maiden M F J, Macuch P J, Murray L L, Kent R L., Jr Microbiota of health, gingivitis, and initial periodontitis. J Clin Periodontol. 1998;25:85–98. doi: 10.1111/j.1600-051x.1998.tb02414.x. [DOI] [PubMed] [Google Scholar]
- 58.Terseegen A, Linder D, Thauer R K, Hedderich R. Structures and functions of four anabolic 2-oxoacid oxidoreductases in Methanobacterium thermoautotrophicum. Eur J Biochem. 1997;244:862–868. doi: 10.1111/j.1432-1033.1997.00862.x. [DOI] [PubMed] [Google Scholar]
- 59.Tonetti M, Eftimiadi C, Damiani G, Buffa P, Buffa D, Botta G A. Short chain fatty acids present in periodontal pockets may play a role in human periodontal diseases. J Periodontal Res. 1987;22:190–191. doi: 10.1111/j.1600-0765.1987.tb01565.x. [DOI] [PubMed] [Google Scholar]
- 60.Wahren A, Gibbons R J. Amino acid fermentation by Bacteroides melaninogenicus. Antonie Leeuwenhoek. 1970;36:149–159. doi: 10.1007/BF02069017. [DOI] [PubMed] [Google Scholar]
- 61.Wolff R A, Urben G W, O'Herrin S M, Kenealy W R. Dehydrogenases involved in the conversion of succinate to 4-hydroxybutanoate by Clostridium kluyveri. Appl Environ Microbiol. 1993;59:1879–1882. doi: 10.1128/aem.59.6.1876-1882.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wyss C. Growth of Porphyromonas gingivalis, Treponema denticola, T. pectinovorum, T. socranskii, and T. vincentii in a chemically defined medium. J Clin Microbiol. 1992;30:2225–2229. doi: 10.1128/jcm.30.9.2225-2229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]