Abstract
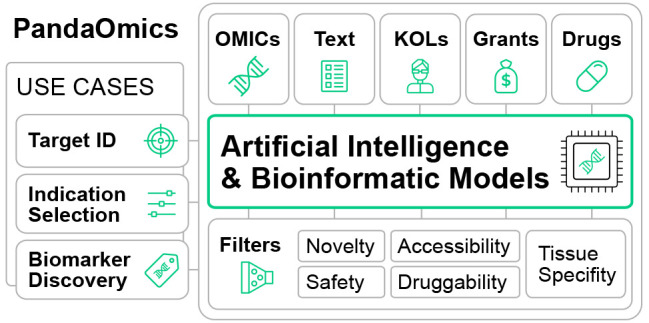
PandaOmics is a cloud-based software platform that applies artificial intelligence and bioinformatics techniques to multimodal omics and biomedical text data for therapeutic target and biomarker discovery. PandaOmics generates novel and repurposed therapeutic target and biomarker hypotheses with the desired properties and is available through licensing or collaboration. Targets and biomarkers generated by the platform were previously validated in both in vitro and in vivo studies. PandaOmics is a core component of Insilico Medicine’s Pharma.ai drug discovery suite, which also includes Chemistry42 for the de novo generation of novel small molecules, and inClinico—a data-driven multimodal platform that forecasts a clinical trial’s probability of successful transition from phase 2 to phase 3. In this paper, we demonstrate how the PandaOmics platform can efficiently identify novel molecular targets and biomarkers for various diseases.
Introduction
Deep learning (DL), a subset of artificial intelligence (AI), has proven to be very effective in speech and image recognition. This is because DL-based architectures are uniquely suited for automatically identifying patterns within complex nonlinear datasets without the need for manual feature engineering. DL methods have recently been adapted to successfully overcome limitations inherent in the standard techniques used for omics data analysis and predicting properties of drugs.1 These adaptations offer exciting possibilities for the development of new methods that efficiently predict novel targets and biomarkers.
Insilico Medicine was one of the first groups to publish a method that uses a deep learning approach to discover new targets.2,3 Since then, the approaches combining classical bioinformatics with AI-driven techniques have been developed and applied in disease mechanism reconstruction and the discovery of new targets.4 Especially encouraging is the recent progress in exploring the targets previously not accessible for small molecule development using AlphaFold and similar approaches. The de novo design of active molecules for such targets has recently been validated in both in vitro and in vivo assays.5,6 Other fast-growing areas in computer-aided drug discovery leveraged in the PandaOmics platform include applying large language models and robotics.7,8 The PandaOmics platform has been routinely and successfully used at Insilico Medicine to drive the therapeutic target discovery process in several therapeutic areas (https://insilico.com/pipeline) and has evolved significantly. In the following section, we describe key features of the PandaOmics platform.
Overview of the Capabilities of the PandaOmics Platform
Dataset Selection and Sample Group Comparison Creation
PandaOmics offers a comprehensive data processing pipeline that facilitates the identification of potential therapeutic targets and biomarkers (Figure 1). By leveraging dynamic omics data, including gene expression, proteomics, and methylation data, the platform conducts a systematic search for relevant datasets. The list of databases used by PandaOmics is provided as Supplementary Table 1. It then assembles a comprehensive data inventory specifically tailored to the disease, condition, or compound of interest. The platform then provides a data exploration interface, allowing researchers to visualize and analyze the compiled data. The interface includes the ability to generate reduced-dimensional plots using popular algorithms such as PCA, tSNE, or UMAP. These plots can be enriched by incorporating metadata, such as disease stage or tissue of origin, assisting in sample group selection. Then, PandaOmics enables the creation of meaningful comparisons between the selected sample groups. Typically, these comparisons involve disease case samples versus paired normal control samples. To ensure a robust and accurate analysis, the platform offers the option to perform batch correction and quality control of the corrected results. In case of all three data types (gene expression, methylation, and proteomics), the analysis is conducted at the gene level, where methylation patterns are mapped into genes, and protein expression is averaged across gene products, providing a comprehensive view of their potential impact on gene function.
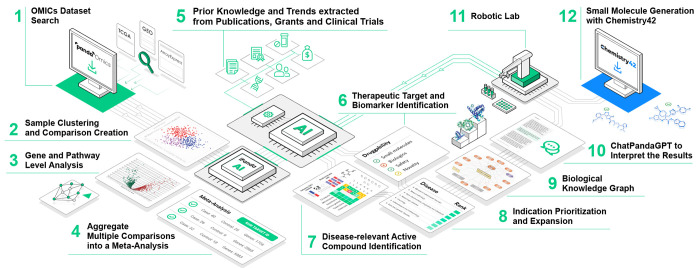
Figure 1.
PandaOmics employs a robust data processing pipeline, starting with dataset selection (1) and sample group comparison (2). It offers gene- and pathway-level analysis (3), including correlation exploration between molecular features and clinical data. The platform combines the results of these steps into meta-analyses (4), enhancing target predictions with diverse data sources including prior knowledge extracted from text data (5). For therapeutic target and biomarker identification (6), it provides a user-friendly interface with 23 disease-specific models. Identification of disease-relevant compounds is also enabled (7). PandaOmics excels in indication prioritization and features a database of precalculated disease meta-analyses (8). It also leverages biological knowledge graph (9) and large language model-based ChatPandaGPT (10) to explain gene-disease associations. Robotic lab (11) for target validation and compound screening forms a feedback loop with the AI core of the platform, streamlining the research process. The target hypotheses identified with PandaOmics serve as an input for Chemistry42 software (12) to perform small molecule generation. The images pertaining to PandaOmics and Chemistry42 have been reproduced with permission from Insilico Medicine IP Limited.
Differentially Expressed Genes, Perturbed Pathways, and Metadata Analysis
PandaOmics also provides gene- and pathway-level data analysis. Biologically relevant pathways are identified using the iPANDA algorithm, which performs pathway activation analyses. Pathways are grouped according to their biological processes such as autophagy or DNA replication, which aid in the identification of key regulatory mechanisms. Furthermore, PandaOmics allows users to explore correlations among the expression of individual genes, pathways, and metadata. This functionality empowers researchers to uncover potential associations between molecular features and clinical or biological characteristics, providing valuable insights into disease mechanisms and potential therapeutic targets and biomarkers.
PandaOmics offers the capability to aggregate multiple disease-relevant comparisons into meta-analyses. Meta-analysis enables the capture of consistent and robust insights across various datasets and incorporates disease-relevant genetic data, including GWAS and information about somatic mutations that drive the pathology. Text data from publications, grant applications, and clinical trials complement the omics data within the meta-analysis. By integrating these diverse data sources, PandaOmics strengthens the insights derived from the omics datasets by placing them in the context of previously published information. This combined knowledge is subsequently employed for target and biomarker prediction.
Therapeutic Target Identification
The meta-analysis page within the PandaOmics platform offers a comprehensive target identification (ID) interface presented as a user-friendly dashboard featuring a ranked list of genes. Each line in the dashboard corresponds to a specific gene, while each column represents a distinct approach or model used to rank the potential targets and important genes. In total, there are 23 disease-specific models employed in the ranking process, broadly categorized as omics-based and text-based approaches. The omics-based models leverage various data types, including gene expression, methylation, proteomics, and genetic information extracted from the meta-analysis. These models are further enhanced by incorporating biological graphs derived from signaling pathway and protein–protein interaction (PPI) network databases and knowledge graphs generated through the analysis of scientific publications. The text-based models depend on extracted relations from publications, clinical trials, and grant applications while considering source credibility and trends.
The omics-based models can be divided into two main groups: bioinformatic approaches and advanced machine learning and graph-based techniques. An example of the first group is the expression score, which relies on the differential expression of disease samples compared to paired normal control samples combined with the expression levels in disease-relevant tissues. This approach provides a straightforward assessment of the gene significance. In contrast, the second group includes more complex methods, such as the heterogeneous graph walk algorithm. This algorithm utilizes a guided random walk-based approach on a heterogeneous graph, where nodes represent genes and diseases and edges represent their associations. The model learns representations of nodes and subsequently identifies gene nodes closely related to the reference disease node, enabling the discovery of potential target genes.
The PandaOmics interface offers rich functionality to refine the ranked list of genes according to various disease-agnostic criteria. These criteria include factors such as druggability by small molecules and therapeutic antibodies, safety considerations, novelty of the target, tissue-specific expression patterns, protein class, biological process involvement, availability of crystal structures, and level of pharmaceutical development. Additionally, users can create their lists of genes for use as filters. Furthermore, all of the models and filters used to rank genes can be toggled on or off based on user preference. To facilitate the process, the interface provides five predefined scenarios for ranking, including associated genes, novel targets for small molecules, novel targets for biologics, targets for repurposing, and trending genes. Users can create and save their own scenarios such as identifying novel targets with genetic evidence. This flexibility allows PandaOmics to serve as a versatile framework for target and biomarker discovery capable of addressing the diverse needs of the pharmaceutical industry. For a more detailed understanding of the models and filters in PandaOmics, descriptions can be found in the PandaOmics user manual, available at https://insilico.com/pandaomics/help. The descriptions of the scores available as of December 2023 are provided in Supplementary Table 2.
Indication Prioritization
The PandaOmics platform offers an indication prioritization and expansion functions that enhances its utility in target discovery and therapeutic development. This feature enables researchers to expand their focus beyond a single disease and explore the potential cross-indication applicability of their target candidates. In terms of the user interface, the indication prioritization function provides a dashboard similar to the target ID feature, featuring the same set of scores that are normalized for cross-disease comparison. Diseases are conveniently grouped based on an internal classification system designed to align with the pipeline divisions of leading pharmaceutical enterprises. This categorization can be structured along major therapeutic domains or specific tissue/organ systems, enhancing user convenience and accessibility.
Crucially, the entire indication prioritization/expansion feature is further streamlined by PandaOmics’ repository of precalculated disease meta-analyses, encompassing over 8000 diseases, with dedicated emphasis on more than 500 manually curated meta-analyses. Manually curated disease meta-analyses involve human-patient-derived data from disease-relevant tissues, controlled for disease, tissue, age, and gender. Only datasets with untreated disease samples and paired normal control samples (minimum three samples) are considered. The original data undergo distribution control, normalization, and outlier detection. The analysis extends to genetic and text data from the GWAS catalog, ClinVar, and Intogen databases, with variant filtration based on confidence scores. This database is pivotal for efficiently executing indication prioritization and expansion, guiding researchers to strategically assess the applications of the selected therapeutic targets.
Compound Identification
Just as with gene ranking, the compound ID module leverages gene expression profiles of compounds and integrates text data to assess their significance. By analyzing the gene expression signatures of compounds and comparing them to known therapeutic targets or disease-associated genes, PandaOmics assigns scores to compounds, enabling their prioritization. This integration of compound analysis seamlessly complements the comprehensive target and biomarker discovery offered by PandaOmics, providing researchers with a powerful tool to identify potential therapeutic compounds alongside gene targets.
Biological Knowledge Graph
PandaOmics goes beyond assisting researchers with the target and biomarker selection step. It also provides comprehensive evidence to support each generated hypothesis. The platform integrates omics data, such as gene–disease associations, with insights derived from the scientific literature through a powerful biological knowledge graph. This knowledge graph is constructed using advanced algorithms for entity recognition and relation extraction, incorporating information about genes, diseases, biological processes, and compounds. In addition, the platform incorporates valuable insights from clinical trial data, providing a deeper understanding of the competitive landscape.
ChatPandaGPT
To further enhance data interpretation, the platform utilizes ChatPandaGPT, a large language model that enables text summarization of omics-driven findings and their contextualization within the published data. This functionality allows researchers to access not only summaries of supporting data for the potential or actual use of specific targets in the context of a given disease but also to obtain answers to their queries within the context provided by the meta-analysis and the knowledge graph. Therefore, ChatPandaGPT may help PandaOmics users to make informed decisions on the target selection by providing textual summaries of the data available within the platform.
Integration with Robotic Platforms
Robotic platforms including cell culture, screening of the compounds and cell knockout/knockdown models, next-generation sequencing, and cell imaging form a powerful synergy with the PandaOmics therapeutic target discovery platform, facilitating rapid and precise discovery of novel targets and biomarkers. By integrating robotic systems into the research process, tasks such as target and compound validation can be executed in a standardized manner, significantly reducing human error and increasing throughput. The robust sequencing and phenotypic data generated by robotic lab can be seamlessly incorporated into PandaOmics, enriching the dataset and enhancing the accuracy of target and biomarker prediction. In turn, the insights and predictions generated by PandaOmics can guide the experimental design and selection of targets for further validation and testing in a robotic lab setting. This iterative process forms a feedback loop, where the findings from the robotic lab inform the PandaOmics analysis, which in turn enables researchers to identify and validate potential therapeutic targets and biomarkers in a highly standardized manner.
Summary of the AI Capabilities of the PandaOmics Platform
In conclusion, the PandaOmics platform presents a comprehensive set of capabilities for therapeutic target discovery and biomarker development. It leverages dynamic omics data and advanced algorithms to facilitate data exploration, pathway analysis, and meta-analysis, strengthening the insights derived from the datasets. The platform offers a user-friendly interface with customizable ranking approaches and filters, allowing researchers to refine their gene lists according to various criteria. Integration with a robotic lab enhances the efficiency and accuracy of target validation and compound screening. By integrating diverse data sources and leveraging advanced language models, PandaOmics empowers researchers to make informed decisions and gain valuable insights into the pursuit of therapeutic targets and biomarkers.
Validation
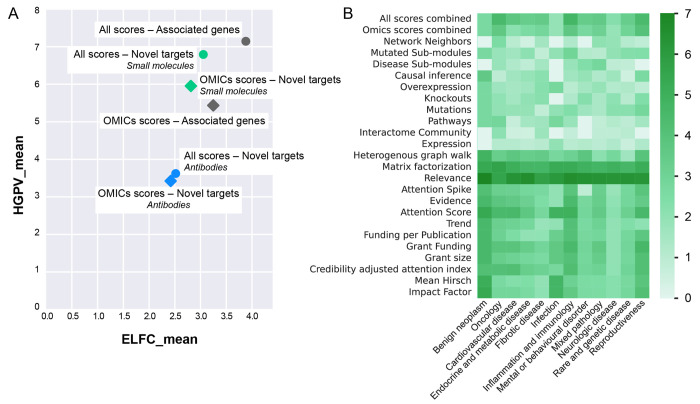
The PandaOmics target discovery engine underwent thorough validation to ensure its effectiveness in identifying novel targets. The validation metrics, log fold change of enrichment (ELFC) and statistical significance of the enrichment based on hypergeometric p-value (HGPV), were used to measure the performance of the PandaOmics predictive models, respectively.9 While ELFC refers to the log-transformed fold change of enrichment showing how much the top of the list was enriched by known targets, HGPV stands for the statistical significance of the effect and shows how likely the same level of enrichment could be achieved from a random list of genes. Higher values of ELFC and HGPV correspond to a higher predictive power of the ranking approach. All PandaOmics models have been validated in ELFC and HGPV coordinates, both in general and across various therapeutic areas.
The average fold enrichment achieved by using all the scores is approximately 15 (slightly below 4 on the logarithmic scale as shown in Figure 2A), meaning that if the random ranking contains only two relevant genes associated with the disease among the top-100, the aggregated PandaOmics ranking contains about 30 associated genes out of the top-100. Similarly, the omics scores alone achieve a fold enrichment of 10 (slightly above 3 on the logarithmic scale). This validation demonstrates that PandaOmics provides a set of hypotheses with solid evidence, saving valuable time for researchers. Nonetheless, the platform is not a magic pill that produces a single ideal hypothesis but instead offers a diverse range of hypotheses with strong underlying evidence, allowing researchers to select the most promising targets for further pursuit in drug development programs. The quality of results obtained through PandaOmics is equally robust across major therapeutic areas such as oncology, inflammation and immunology, cardiometabolic diseases, fibrosis, and other disease areas with well-defined internal molecular mechanisms (Figure 2B). This versatility highlights the platform’s capability to address a wide range of diseases and biological processes, making it a valuable tool for target discovery in diverse medical fields. However, the application of PandaOmics is limited when it comes to diseases with external causes, such as viral and fungal infections, where the underlying molecular mechanisms may not be as well-defined. Overall, the platform’s strong validation and wide applicability make it a valuable resource for researchers seeking to identify potential therapeutic targets in various diseases and biological processes.
Figure 2.
Validation of the Target ID capability of the PandaOmics platform. (A) Evaluation of predefined target ID scenarios: The PandaOmics platform’s predefined target ID scenarios were validated using log fold change of enrichment (ELFC) and statistical significance of the enrichment based on hypergeometric p-value (HGPV) metrics to assess ranking performance. Three scenarios, namely, associated genes, novel targets (small molecules), and novel targets (antibodies), were evaluated in two distinct settings: considering all scores and omics scores only. (B) Performance across therapeutic areas: The figure illustrates the performance in terms of the average fold enrichment on the logarithmic scale (ELFC) of PandaOmics scores across 12 major therapeutic areas. Each of the 23 models available in PandaOmics was separately evaluated for its ability to rank known targets or genes associated with the disease within the top-100 of the list. Results were averaged for each therapeutic area, and the combined performances of all scores and omics scores are also presented.
Case Studies
As of September 2023, the PandaOmics platform and its core signaling pathway analysis algorithm iPANDA were referenced in over 20 scientific papers. The original iPANDA paper describing the pathway analysis algorithm used in PandaOmics platform demonstrates how this algorithm can be applied to develop biomarkers of susceptibility to neoadjuvant therapy in various types of breast cancer.10 The bulk gene expression data deconvolution by the same algorithm was used to estimate the relative abundance of various T cell types in oral squamous cell carcinoma patients from The Cancer Genome Atlas and Chicago Head and Neck Genomics Cohort datasets.11,12 Similar clusters enriched for either CD8+ T cells or Treg cells were later identified using iPANDA to analyze other human solid cancer types that are amenable to immune-based therapy.13
PandaOmics and iPANDA have proven to be valuable tools for biomarker discovery across diverse diseases. These tools were successfully applied to the identification of potential biomarkers associated with androgenic alopecia, the mammalian embryonic–fetal transition, gallbladder cancer, and smoke-induced lung cancer.3,14−16 Application of the platform to therapeutic area selection and the search for biomarkers facilitated the development of bifunctional antibody-ligand traps (Y-traps) for cancer immunotherapy. This study showcases the diversity of therapeutic modalities that can be tackled using the platform.17 Additionally, PandaOmics has demonstrated its capability in novel target discovery, enabling the identification of therapeutic candidates for various diseases and conditions. The identification of potential therapeutic targets for idiopathic pulmonary and kidney fibrosis, aging, glioblastoma multiforme, and head and neck squamous cell carcinoma emphasize the potential of PandaOmics in identifying unique targets for different disease contexts.18−21 Among the recent 2023 case studies, PandaOmics identified CAMMK2, MARCKS, and p62 that were successfully validated in Alzheimer’s Disease cell models and KDM1A as a dual aging/oncology target validated to extend the lifespan of C. elegans.22,23
The case studies also highlight the diversity of diseases and biological mechanisms that can be addressed by PandaOmics and iPANDA. The platform has been applied to study DNA repair disorders, kidney epithelial cell fate specialization, and human muscle aging, among other conditions.24−26 The ability to uncover molecular events and signaling pathways associated with various diseases and biological processes demonstrates the versatility of PandaOmics in providing valuable insights into disease mechanisms and potential therapeutic targets. Below we describe several PandaOmics case studies in more detail.
Targeting the Hallmarks of Aging: Identification of Therapeutic Targets for Age-Related Diseases
In this case study, the AI-powered PandaOmics platform was applied to identify therapeutic targets associated with the aging process. The well-established concept of the hallmarks of aging was utilized to classify the mechanisms driving aging. The hallmarks of aging are a set of interconnected cellular and molecular processes that contribute to the aging phenotype and include factors such as genomic instability, telomere attrition, epigenetic alterations, loss of proteostasis, deregulated nutrient sensing, mitochondrial dysfunction, cellular senescence, and stem cell exhaustion.27 A comprehensive list of targets with varying levels of novelty and evidence was generated through meta-analysis and the application of specific filters and settings. The relevance of inflammation and extracellular matrix stiffness in aging and age-related diseases was highlighted, with many top targets identified playing a role in these processes. Among those one can find the targets corresponding to the approved therapies like KDR, MMPs, JAKs, and TLRs as well as novel targets like HCK or TNIK. Especially of interest that the latter has recently entered phase 2 for idiopathic pulmonary fibrosis.28 Overall, the application of PandaOmics in target discovery across multiple disease areas was demonstrated, revealing high-confidence and novel targets associated with the hallmarks of aging.29
For the analysis, age-associated diseases (AADs) and nonage-associated diseases (NAADs) were selected based on the impact of age on disease onset. Microarray and RNA-seq datasets for these diseases were retrieved and processed by PandaOmics, and meta-analysis was performed for each dataset. Specific filter settings, including druggability, novelty, safety, tissue specificity, target family, and development filters, were applied for target identification. The resulting list of targets was categorized based on their novelty and involvement in the hallmarks of aging.
To identify targets implicated in multiple diseases and associated with aging, the top-100 genes from AADs and NAADs were extracted for each novelty setting. The selected genes from both AADs and NAADs were overlapped, leading to the categorization of these genes into two groups: AAD targets and common targets. A thorough literature review was conducted to assess the association of the obtained genes with the hallmarks of aging, considering their biological functions, pathways, and roles in regulating key aging-related processes. Many of the identified targets were found to be linked to inflammation and extracellular matrix stiffness as well as other hallmarks of aging.
Validation of the AI-derived targets involved comparing them with well-known aging-associated genes and examining their roles in aging-related pathways. The presence of known aging-associated genes within the identified targets served as a further validation. Additionally, the high-confidence targets were compared with aging-related gene databases and clinical trials, demonstrating significant enrichment and potential clinical relevance. Pathway enrichment analysis revealed that the AI-derived targets intersected with key aging-associated pathways including the PI3K-AKT, MAPK, and FOXO signaling pathways. These targets played an important role in regulating various cellular processes involved in aging such as apoptosis, autophagy, cell proliferation, DNA repair, inflammation, and mitochondrial maintenance.
In summary, the application of PandaOmics in identifying aging-associated therapeutic targets was previously showcased. Leveraging the platform’s capabilities, a list of targets associated with the hallmarks of aging and their potential relevance in multiple diseases was successfully generated. PandaOmics proved to be a valuable tool for target discovery across diverse disease areas, offering a comprehensive and systematic approach to uncovering therapeutic targets with implications for aging and age-related diseases.
Target Identification and Validation in ALS
Amyotrophic lateral sclerosis (ALS) is a severe neurodegenerative disease characterized by the progressive degeneration of motor neurons in the brain and spinal cord. Currently, there is a lack of effective therapeutic regimens for ALS, necessitating the need for novel treatment approaches. In this study, the PandaOmics platform was employed to analyze the expression profiles of central nervous system (CNS) samples from public datasets and direct induced pluripotent stem cell (iPSC)-derived motor neurons (diMNs) from Answer ALS. The CNS samples included 237 ALS cases and 91 controls, while the diMN samples consisted of 135 ALS cases and 31 controls.9
Using over 20 AI and bioinformatics models, potential therapeutic targets were ranked by PandaOmics based on their target-disease associations, druggability, developmental state, and tissue specificity. Various scores and filters were utilized by the platform to select high-confidence and novel targets. By customizing the filter settings, 17 high-confidence targets and 11 novel targets, totaling 28 potential therapeutic candidates for ALS, were identified by PandaOmics. These targets were ranked based on their metascores and were selected for further investigation. To validate the relevance of these targets, experiments were conducted using a Drosophila model that mimicked C9orf72-mediated ALS (c9ALS), which is the most common familial ALS subtype. With this model, the efficacy of eight unreported genes (KCNB2, KCNS3, ADRA2B, NR3C1, P2RY14, PPP3CB, PTPRC, and RARA) in rescuing eye neurodegeneration caused by (G4C2)30 repeat expansion was verified.
Furthermore, pathway analysis using PandaOmics’ proprietary iPANDA algorithm revealed dysregulated pathways associated with different stages of ALS development. Multiple dysregulated pathways were identified in CNS and diMN data, providing insights into the underlying pathophysiology of ALS. Pathway clusters associated with the activated innate immune system, programmed cell death, unfolded protein response, and ERBB4 signaling were observed in the CNS fALS healthy cohort. Moreover, several pathways related to RNA metabolism, mitochondrial protein import, and cell cycle regulation were found to be dysregulated in CNS-based disease cohorts.
Overall, the application of PandaOmics in generating and validating a therapeutic target hypothesis for ALS was demonstrated in our study. The AI-driven platform facilitated the identification of potential targets based on comprehensive analyses of expression profiles and pathway dysregulation. The combination of AI-based target discovery and validation using a Drosophila model significantly accelerated the target discovery process, providing new insights into ALS pathophysiology and potential opportunities for therapeutic interventions. The identified targets are available on the ALS.AI platform for further evaluation and validation.
Combining PandaOmics, AlphaFold, and Chemistry42 for Efficient Target Identification and Validation in Hepatocellular Carcinoma
In this case study, the AlphaFold program’s predicted protein structures were utilized to identify and validate a therapeutic target using the PandaOmics and Chemistry42 platforms. The target of interest was selected for the treatment of hepatocellular carcinoma (HCC), a prevalent and challenging cancer type with limited effective treatments. Data analysis and filtering based on text and omics data from multiple datasets were performed by PandaOmics to generate a ranked list of potential targets for HCC. Cyclin-dependent kinase 20 (CDK20) was selected as the initial target due to its strong disease association and limited experimental structure information.5
To identify CDK20 as a therapeutic target for hepatocellular carcinoma (HCC), a systematic search for relevant datasets was conducted by the PandaOmics platform, and a tailored data inventory specific to HCC was assembled. Comparisons were made between disease case samples and paired normal control samples and aggregated in a meta-analysis. The target ID settings of the first-in-class scenario were followed by the PandaOmics platform, which considered the druggability of the protein by small molecules, novelty of the target, exclusion of targets in phase 1 or later stage clinical trials in the past three years or targeted by approved drugs, and exclusion of targets with resolved crystal structures.
Using the predicted structure of CDK20 generated by AlphaFold, structure-based compound generation was employed by Chemistry42 to produce a library of molecules. From this library, seven compounds were synthesized and tested in biological assays. One of the compounds, ISM042-2-001, demonstrated a binding constant (Kd) value of 9.2 ± 0.5 μM (n = 3) in the CDK20 kinase binding assay. This hit compound provided initial evidence of target engagement within just 30 days of target selection and the synthesis of only seven compounds.
Building upon the findings from the initial hit compound, a second round of compound generation was conducted by using AI-powered approaches. Six additional compounds were synthesized and tested, leading to the discovery of a more potent hit molecule, ISM042-2-048. This molecule exhibited an average Kd value of 566.7 ± 256.2 nM (n = 3) and an average IC50 value of 33.4 ± 22.6 nM (n = 3) in binding and inhibitory assays against CDK20. Notably, ISM042-2-048 also demonstrated selective antiproliferation activity in an HCC cell line with CDK20 overexpression, further supporting its potential as a therapeutic candidate.
The successful identification and validation of a hit compound for CDK20 in HCC using AlphaFold-predicted protein structures highlight the power of combining the AI capabilities of PandaOmics and Chemistry42 platforms in the early stages of drug discovery. By leveraging the comprehensive data analysis and molecular generation capabilities of these platforms, a potential therapeutic target was rapidly identified, novel molecules generated, and their binding and inhibitory activities validated. This case study demonstrates the potential of AI applications and the integration of diverse data sources and AI tools for efficient and effective target discovery in drug development.
Combining PandaOmics and FuzDrop for the Identification of Therapeutic Targets Associated with Protein Phase Separation
In pursuit of therapeutic targets for diseases associated with protein phase separation (PPS), the PandaOmics platform was employed in this case study.22 PandaOmics was integrated with FuzDrop, a method predicting the likelihood of proteins undergoing liquid–liquid phase separation to analyze 64 diseases. These diseases were segmented into four quadrants based on their potential for PPS-based therapeutic targeting. Alzheimer’s Disease (AD) fell within the “Promising Priority” quadrant, indicating the potential for PPS-based therapeutic strategies due to the enrichment of PPS-prone disease-associated proteins and pathways. A total of 12 targets were proposed for AD based on PandaOmics analysis, with eight being high-confidence targets and four being low-confidence targets. The target landscape provided a matrix of disease-associated genes encoding PPS-prone proteins, offering detailed profiles for each target, including evidence of PPS, protein localization, the propensity of a protein to undergo spontaneous PPS score, PandaOmics rank score, disease specificity score, and text-based confidence score. Notably, three high-confidence AD targets—SYN1, APC, and YAP1—were found to have corresponding drug candidates in the FDA Clinical Trials database, underscoring their clinical relevance.
To validate the predicted targets, the phase behavior of three high-confidence targets (MARCKS, CAMKK2, and p62/SQSTM1) was investigated in two AD cell models. The study involved SH-SY5Y cells treated with Aβ42 and hiPSC-derived neurons with the APPSwe mutation. The results demonstrated the altered phase behavior of these targets under AD conditions, supporting the validity of the predictions. Specifically, cytoplasmic condensates were observed in MARCKS in response to Aβ42, while CAMKK2 exhibited axonal localization in APPSwe neurons. P62/SQSTM1 condensates increased in both models, indicating a potential therapeutic relevance. This study contributes to the emerging field of PPS-related disease research, demonstrating the application of PandaOmics in prioritizing and validating therapeutic targets. The proposed targets for AD present opportunities for further investigation and development of PPS-based interventions, offering a promising avenue for future therapeutic strategies.
PandaOmics: Integration within the Pharma.AI Ecosystem
PandaOmics plays a crucial role in the Pharma.AI ecosystem by Insilico Medicine, working in tandem with other tools, such as Chemistry42 and inClinico. Chemistry42 is a generative chemistry platform that leverages artificial intelligence to facilitate the automated generation of small molecule compounds for drug discovery. By integration with Chemistry42, PandaOmics utilizes the predicted protein structures generated by AlphaFold to guide the compound generation process, enabling the rapid identification of potential hit molecules for novel targets. inClinico, and on the other hand, is a powerful platform for in silico prediction of clinical trial outcomes, providing a virtual testing environment for potential drug candidates. PandaOmics complements inClinico by providing essential insights into potential therapeutic targets and biomarkers. By leveraging the comprehensive data processing capabilities of PandaOmics, researchers can identify and prioritize targets that show promise for further evaluation in clinical trials using inClinico. This integration enables a seamless transition from target identification to compound generation and virtual testing, streamlining the drug discovery and development process.
Overall, PandaOmics, Chemistry42, and inClinico collectively form the Pharma.AI ecosystem, offering an end-to-end solution for accelerating and optimizing key steps in drug discovery. From target identification and hit molecule generation using PandaOmics and Chemistry42, to virtual testing and simulation using inClinico, these tools work synergistically to enhance the efficiency and effectiveness of drug discovery efforts, ultimately leading to the development of novel therapeutics with improved success rates and reduced costs.
Data Availability Statement
The tool is available by subscription at https://pandaomics.com/. Since the platform allows potential upload of sensitive patient data, the users are vetted to minimize the probability of malicious misuse. Academic users can apply for free access. The free demo version with data upload function disabled is also available through the teachable login at https://insilico-medicine-school.teachable.com/p/target-discovery.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jcim.3c01619.
Author Contributions
∇ Petrina Kamya and Ivan V. Ozerov contributed equally.
The authors declare the following competing financial interest(s): P.K., I.V.O., F.W.P., K.T., T.F., S.C., V.N., X.L., S.L., M.K., D.P., A.A., F.R., and A.Z. work for Insilico Medicine, a commercial artificial intelligence company that developed the PandaOmics platform.
Supplementary Material
References
- Aliper A.; Plis S.; Artemov A.; Ulloa A.; Mamoshina P.; Zhavoronkov A. Deep Learning Applications for Predicting Pharmacological Properties of Drugs and Drug Repurposing Using Transcriptomic Data. Mol. Pharmaceutics 2016, 13 (7), 2524–2530. 10.1021/acs.molpharmaceut.6b00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Feng Y.; Huang J.; Wang T.; Cheng G. A Novel Framework for the Identification of Drug Target Proteins: Combining Stacked Auto-Encoders with a Biased Support Vector Machine. PLoS One 2017, 12 (4), e0176486 10.1371/journal.pone.0176486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M. D.; Labat I.; Sternberg H.; Larocca D.; Nasonkin I.; Chapman K. B.; Singh R.; Makarev E.; Aliper A.; Kazennov A.; Alekseenko A.; Shuvalov N.; Cheskidova E.; Alekseev A.; Artemov A.; Putin E.; Mamoshina P.; Pryanichnikov N.; Larocca J.; Copeland K.; Izumchenko E.; Korzinkin M.; Zhavoronkov A. Use of Deep Neural Network Ensembles to Identify Embryonic-Fetal Transition Markers: Repression of COX7A1 in Embryonic and Cancer Cells. Oncotarget 2018, 9 (8), 7796–7811. 10.18632/oncotarget.23748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pun F. W.; Ozerov I. V.; Zhavoronkov A. AI-Powered Therapeutic Target Discovery. Trends Pharmacol. Sci. 2023, 44 (9), 561–572. 10.1016/j.tips.2023.06.010. [DOI] [PubMed] [Google Scholar]
- Ren F.; Ding X.; Zheng M.; Korzinkin M.; Cai X.; Zhu W.; Mantsyzov A.; Aliper A.; Aladinskiy V.; Cao Z.; Kong S.; Long X.; Man Liu B. H.; Liu Y.; Naumov V.; Shneyderman A.; Ozerov I. V.; Wang J.; Pun F. W.; Polykovskiy D. A.; Sun C.; Levitt M.; Aspuru-Guzik A.; Zhavoronkov A. AlphaFold Accelerates Artificial Intelligence Powered Drug Discovery: Efficient Discovery of a Novel CDK20 Small Molecule Inhibitor. Chem. Sci. 2023, 14 (6), 1443–1452. 10.1039/D2SC05709C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W.; Liu X.; Li Q.; Gao F.; Liu T.; Chen X.; Zhang M.; Aliper A.; Ren F.; Ding X.; Zhavoronkov A. Discovery of Novel and Selective SIK2 Inhibitors by the Application of AlphaFold Structures and Generative Models. Bioorg. Med. Chem. 2023, 91, 117414. 10.1016/j.bmc.2023.117414. [DOI] [PubMed] [Google Scholar]
- Schneider G. Automating Drug Discovery. Nat. Rev. Drug Discovery 2018, 17 (2), 97–113. 10.1038/nrd.2017.232. [DOI] [PubMed] [Google Scholar]
- Urban A.; Sidorenko D.; Zagirova D.; Kozlova E.; Kalashnikov A.; Pushkov S.; Naumov V.; Sarkisova V.; Leung G. H. D.; Leung H. W.; Pun F. W.; Ozerov I. V.; Aliper A.; Ren F.; Zhavoronkov A. Precious1GPT: Multimodal Transformer-Based Transfer Learning for Aging Clock Development and Feature Importance Analysis for Aging and Age-Related Disease Target Discovery. Aging 2023, 15 (11), 4649–4666. 10.18632/aging.204788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pun F. W.; Liu B. H. M.; Long X.; Leung H. W.; Leung G. H. D.; Mewborne Q. T.; Gao J.; Shneyderman A.; Ozerov I. V.; Wang J.; Ren F.; Aliper A.; Bischof E.; Izumchenko E.; Guan X.; Zhang K.; Lu B.; Rothstein J. D.; Cudkowicz M. E.; Zhavoronkov A. Identification of Therapeutic Targets for Amyotrophic Lateral Sclerosis Using PandaOmics - An AI-Enabled Biological Target Discovery Platform. Front. Aging Neurosci. 2022, 14, 914017. 10.3389/fnagi.2022.914017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozerov I. V.; Lezhnina K. V.; Izumchenko E.; Artemov A. V.; Medintsev S.; Vanhaelen Q.; Aliper A.; Vijg J.; Osipov A. N.; Labat I.; West M. D.; Buzdin A.; Cantor C. R.; Nikolsky Y.; Borisov N.; Irincheeva I.; Khokhlovich E.; Sidransky D.; Camargo M. L.; Zhavoronkov A. In Silico Pathway Activation Network Decomposition Analysis (iPANDA) as a Method for Biomarker Development. Nat. Commun. 2016, 7, 13427. 10.1038/ncomms13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarev E.; Schubert A. D.; Kanherkar R. R.; London N.; Teka M.; Ozerov I.; Lezhnina K.; Bedi A.; Ravi R.; Mehra R.; Hoque M. O.; Sloma I.; Gaykalova D. A.; Csoka A. B.; Sidransky D.; Zhavoronkov A.; Izumchenko E. In Silico Analysis of Pathways Activation Landscape in Oral Squamous Cell Carcinoma and Oral Leukoplakia. Cell Death Discov 2017, 3, 17022. 10.1038/cddiscovery.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saloura V.; Izumchenko E.; Zuo Z.; Bao R.; Korzinkin M.; Ozerov I.; Zhavoronkov A.; Sidransky D.; Bedi A.; Hoque M. O.; Koeppen H.; Keck M. K.; Khattri A.; London N.; Kotlov N.; Fatima A.; Vougiouklakis T.; Nakamura Y.; Lingen M.; Agrawal N.; Savage P. A.; Kron S.; Kline J.; Kowanetz M.; Seiwert T. Y. Immune Profiles in Primary Squamous Cell Carcinoma of the Head and Neck. Oral Oncol. 2019, 96, 77–88. 10.1016/j.oraloncology.2019.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao J. L.; Korzinkin M.; Zhavoronkov A.; Ozerov I. V.; Walker M. T.; Higgins K.; Lingen M. W.; Izumchenko E.; Savage P. A. Effector T Cell Responses Unleashed by Regulatory T Cell Ablation Exacerbate Oral Squamous Cell Carcinoma. Cell Rep. Med. 2021, 2 (9), 100399. 10.1016/j.xcrm.2021.100399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatas G. N.; Wu J.; Pappas A.; Mirmirani P.; McCormick T. S.; Cooper K. D.; Consolo M.; Schastnaya J.; Ozerov I. V.; Aliper A.; Zhavoronkov A. An Analysis of Gene Expression Data Involving Examination of Signaling Pathways Activation Reveals New Insights into the Mechanism of Action of Minoxidil Topical Foam in Men with Androgenetic Alopecia. Cell Cycle 2017, 16 (17), 1578–1584. 10.1080/15384101.2017.1327492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbannayya T.; Leal-Rojas P.; Zhavoronkov A.; Ozerov I. V.; Korzinkin M.; Babu N.; Radhakrishnan A.; Chavan S.; Raja R.; Pinto S. M.; Patil A. H.; Barbhuiya M. A.; Kumar P.; Guerrero-Preston R.; Navani S.; Tiwari P. K.; Kumar R. V.; Prasad T. S. K.; Roa J. C.; Pandey A.; Sidransky D.; Gowda H.; Izumchenko E.; Chatterjee A. PIM1 Kinase Promotes Gallbladder Cancer Cell Proliferation via Inhibition of Proline-Rich Akt Substrate of 40 kDa (PRAS40). J. Cell Commun. Signal. 2019, 13 (2), 163–177. 10.1007/s12079-018-00503-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solanki H. S.; Raja R.; Zhavoronkov A.; Ozerov I. V.; Artemov A. V.; Advani J.; Radhakrishnan A.; Babu N.; Puttamallesh V. N.; Syed N.; Nanjappa V.; Subbannayya T.; Sahasrabuddhe N. A.; Patil A. H.; Prasad T. S. K.; Gaykalova D.; Chang X.; Sathyendran R.; Mathur P. P.; Rangarajan A.; Sidransky D.; Pandey A.; Izumchenko E.; Gowda H.; Chatterjee A. Correction: Targeting Focal Adhesion Kinase Overcomes Erlotinib Resistance in Smoke Induced Lung Cancer by Altering Phosphorylation of Epidermal Growth Factor Receptor. Oncoscience 2021, 8, 108–109. 10.18632/oncoscience.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi R.; Noonan K. A.; Pham V.; Bedi R.; Zhavoronkov A.; Ozerov I. V.; Makarev E.; Artemov A. V.; Wysocki P. T.; Mehra R.; Nimmagadda S.; Marchionni L.; Sidransky D.; Borrello I. M.; Izumchenko E.; Bedi A. Bifunctional Immune Checkpoint-Targeted Antibody-Ligand Traps That Simultaneously Disable TGFβ Enhance the Efficacy of Cancer Immunotherapy. Nat. Commun. 2018, 9 (1), 741. 10.1038/s41467-017-02696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C.Breaking Big Pharma’s AI Barrier: Insilico Medicine Uncovers Novel Target, New Drug for Pulmonary Fibrosis in 18 Months; Fierce Biotech: USA, February 24, 2021.
- Hale C.Insilico Medicine’s AI Engines Continue to Churn out New Drug Candidates, Now in Kidney Fibrosis. Fierce Biotech: USA, August 4, 2021.
- Broner E. C.; Trujillo J. A.; Korzinkin M.; Subbannayya T.; Agrawal N.; Ozerov I. V.; Zhavoronkov A.; Rooper L.; Kotlov N.; Shen L.; Pearson A. T.; Rosenberg A. J.; Savage P. A.; Mishra V.; Chatterjee A.; Sidransky D.; Izumchenko E. Doublecortin-Like Kinase 1 (DCLK1) Is a Novel NOTCH Pathway Signaling Regulator in Head and Neck Squamous Cell Carcinoma. Front. Oncol. 2021, 11, 677051. 10.3389/fonc.2021.677051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A.; Harpaz Z.; Ren C.; Shneyderman A.; Veviorskiy A.; Dralkina M.; Konnov S.; Shcheglova O.; Pun F. W.; Leung G. H. D.; Leung H. W.; Ozerov I. V.; Aliper A.; Korzinkin M.; Zhavoronkov A. Identification of Dual-Purpose Therapeutic Targets Implicated in Aging and Glioblastoma Multiforme Using PandaOmics - an AI-Enabled Biological Target Discovery Platform. Aging 2023, 15 (8), 2863–2876. 10.18632/aging.204678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C. M.; Gonzalez Diaz A.; Fuxreiter M.; Pun F. W.; Zhavoronkov A.; Vendruscolo M. Multiomic Prediction of Therapeutic Targets for Human Diseases Associated with Protein Phase Separation. Proc. Natl. Acad. Sci. U. S. A. 2023, 120 (40), e2300215120 10.1073/pnas.2300215120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pun F. W.; Leung G. H. D.; Leung H. W.; Rice J.; Schmauck-Medina T.; Lautrup S.; Long X.; Liu B. H. M.; Wong C. W.; Ozerov I. V.; Aliper A.; Ren F.; Rosenberg A. J.; Agrawal N.; Izumchenko E.; Fang E. F.; Zhavoronkov A. A Comprehensive AI-Driven Analysis of Large-Scale Omic Datasets Reveals Novel Dual-Purpose Targets for the Treatment of Cancer and Aging. Aging Cell 2023, 22, e14017 10.1111/acel.14017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamoshina P.; Volosnikova M.; Ozerov I. V.; Putin E.; Skibina E.; Cortese F.; Zhavoronkov A. Machine Learning on Human Muscle Transcriptomic Data for Biomarker Discovery and Tissue-Specific Drug Target Identification. Front. Genet. 2018, 9, 242. 10.3389/fgene.2018.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mkrtchyan G. V.; Veviorskiy A.; Izumchenko E.; Shneyderman A.; Pun F. W.; Ozerov I. V.; Aliper A.; Zhavoronkov A.; Scheibye-Knudsen M. High-Confidence Cancer Patient Stratification through Multiomics Investigation of DNA Repair Disorders. Cell Death Dis. 2022, 13 (11), 999. 10.1038/s41419-022-05437-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berquez M.; Chen Z.; Festa B. P.; Krohn P.; Keller S. A.; Parolo S.; Korzinkin M.; Gaponova A.; Laczko E.; Domenici E.; Devuyst O.; Luciani A. Lysosomal Cystine Export Regulates mTORC1 Signaling to Guide Kidney Epithelial Cell Fate Specialization. Nat. Commun. 2023, 14 (1), 3994. 10.1038/s41467-023-39261-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C.; Blasco M. A.; Partridge L.; Serrano M.; Kroemer G. The Hallmarks of Aging. Cell 2013, 153 (6), 1194–1217. 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First drug discovered and designed with generative AI enters Phase II trials, with first patients dosed. EurekAlert! https://www.eurekalert.org/news-releases/993844 (accessed 2023–12–20).
- Pun F. W.; Leung G. H. D.; Leung H. W.; Liu B. H. M.; Long X.; Ozerov I. V.; Wang J.; Ren F.; Aliper A.; Izumchenko E.; Moskalev A.; de Magalhães J. P.; Zhavoronkov A. Hallmarks of Aging-Based Dual-Purpose Disease and Age-Associated Targets Predicted Using PandaOmics AI-Powered Discovery Engine. Aging 2022, 14 (6), 2475–2506. 10.18632/aging.203960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The tool is available by subscription at https://pandaomics.com/. Since the platform allows potential upload of sensitive patient data, the users are vetted to minimize the probability of malicious misuse. Academic users can apply for free access. The free demo version with data upload function disabled is also available through the teachable login at https://insilico-medicine-school.teachable.com/p/target-discovery.