Abstract
Whole-genome duplication is a common macromutation with extensive impacts on gene expression, cellular function, and whole-organism phenotype. As a result, it has been proposed that polyploids have “general-purpose” genotypes that perform better than their diploid progenitors under stressful conditions. Here, we test this hypothesis in the context of stresses presented by anthropogenic pollutants. Specifically, we tested how multiple neotetraploid genetic lineages of the mostly asexually reproducing greater duckweed (Spirodela polyrhiza) perform across a favorable control environment and 5 urban pollutants (iron, salt, manganese, copper, and aluminum). By quantifying the population growth rate of asexually reproducing duckweed over multiple generations, we found that across most pollutants, but not all, polyploidy decreased the growth rate of actively growing propagules but increased that of dormant ones. Yet, when considering total propagule production, polyploidy increased tolerance to most pollutants, and polyploids maintained population-level fitness across pollutants better than diploids. Furthermore, broad-sense genetic correlations in growth rate among pollutants were all positive in neopolyploids but not so for diploids. Our results provide a rare test and support for the hypothesis that polyploids are more tolerant of stressful conditions and can maintain fitness better than diploids across heterogeneous stresses. These results may help predict that polyploids may be likely to persist in stressful environments, such as those caused by urbanization and other human activities.
Keywords: urban ecology, urban evolution, Lemnaceae, environmental trade-off, genetic correlation, autopolyploidy
Go big or go home: polyploidy increases tolerances to urban pollutants.
Introduction
Polyploidy (the possession of more than two sets of each chromosome resulting from whole-genome duplication) is a major macromutation that occurs throughout eukaryotes, especially in plants where most have at least one genome duplication in their past (e.g., Wood et al., 2009). Polyploids often show increased genetic diversity, genomic plasticity, and phenotypic novelty relative to their diploid progenitors (Leitch & Leitch, 2008; Soltis et al., 2009; Van de Peer et al., 2020), allowing them to rapidly adapt and/or invade new habitats (e.g., Beest et al., 2012; Pyšek et al., 2023). Interestingly, polyploidy is often associated with historical periods of environmental change (Fawcett et al., 2009; Van de Peer et al., 2017) as well as with harsh or stressful habitats globally (Ehrendorfer, 1980; Rice et al., 2019). Indeed, it has long been proposed that polyploids might have “general-purpose” genotypes that are beneficial across a variety of circumstances (Baker, 1965; Stebbins, 1971). Recently, these ideas have been combined to suggest that polyploids may perform better than diploids in the face of contemporary anthropogenic stressors, such as those found in urban environments (Tossi et al., 2022; Van de Peer et al., 2020; Van Drunen & Johnson, 2022).
Urban environments, with their vast infrastructure, impervious surfaces, and human and industrial pollutants, represent a confluence of anthropogenic forces. Plants in urban habitats often experience increased abiotic stresses, such as elevated temperature, altered water and nitrogen availability, novel chemical pesticides and herbicides, and industrial waste (Parris, 2016; Van Drunen & Johnson, 2022). Contaminants that can be detrimental to plant growth, such as alkaloids (e.g., aluminum), heavy metals (copper, iron, and manganese), and salts (NaCl), among many others, enter soils and waterways through un- or partially treated effluent, atmospheric deposition, and stormwater runoff from impervious surfaces (Hope et al., 2004; Jacobson, 2011; Ruas et al., 2022). While some contaminants are micronutrients (e.g., Mn, Fe) that play essential roles in the plant life cycle, excesses and imbalances can induce cytotoxic and genotoxic effects and reduce plant fitness (Dutta et al., 2018). Because polyploids can be more tolerant than diploids to a wide range of abiotic stresses, e.g., thermal, drought, saline stress, nutrient deficiencies, and excesses (Tossi et al., 2022; Van de Peer et al., 2020), they may have growth advantages in these contaminated environments. Surprisingly, pollution and micronutrient excess are the least studied urban stresses to plants (Ruas et al., 2022) and also the least studied from the perspective of ploidal tolerance differences (Tossi et al., 2022). Thus, an important step toward testing the hypothesis that polyploids will have an advantage in urban habitats via increased stress tolerance (Van Drunen & Johnson, 2022) will be to compare diploid and polyploid fitness under a suite of common urban contaminants.
Stress tolerance can be achieved through various physiological, molecular, and morphological mechanisms (reviewed in Tossi et al., 2022), which may affect whether ploidy alters tolerance to different stressors in a specific or general way. For example, plants have multiple detoxifying and sequestration mechanisms (Dutta et al., 2018). While polyploids were found to have greater tolerance in approximately 90% of the 57 reviewed studies that induced stress through excess salt, drought, heat/cold, nutritional deficiency/excess, or UV-B exposure (Supplemental Tables 1–4 in Tossi et al., 2022), polyploidy can sometimes lead to lower stress tolerance. For instance, Mouhaya et al. (2010) found increased sensitivity to saline stress in natural autotetraploid citrus species. Furthermore, since few studies examine the independent effects of multiple stressors (but see Bafort et al., 2023; Mattingly & Hovick, 2023), the question of whether polyploidy provides general stress tolerance in any given system has not yet been answered. However, in theory, due to increased gene copies, allelic diversity, or network flexibility, polyploids would generally be expected to exhibit higher (De Smet & Van de Peer, 2012; Parisod et al., 2010) tolerance to various stressors, especially where greater enzymatic activity enhances detoxification or increased transmembrane transport upregulates sequestration. Additionally, the larger cell or vacuole size of polyploids (cell size is correlated with genome size and ploidy Doyle & Coate, 2019; Simonin & Roddy, 2018) may allow for greater storage of toxic substances, as observed in hyperaccumulator species (Leitenmaier & Küpper, 2011). Therefore, when the mechanisms of tolerance are the same for two stressors (e.g., both involving the same antioxidant pathway; Alfatih et al., 2020; Dutta et al., 2018), polyploidy may have a similar effect on tolerance for each stressor. In fact, this would result in a positive correlation in tolerance for these stressors in both diploids and polyploids, but polyploids would have higher overall levels due to increased gene products or cell size.
Additionally, if the size or structure of duplicated gene networks changes, as suggested by simulation (Ebadi et al., 2023; Malcom, 2011), this may afford polyploids genetic flexibility (Leitch & Leitch, 2008) to circumvent trade-offs between stress tolerance mechanisms that may constrain diploids (Des Marais & Juenger, 2010). For example, if the mechanisms to cope with toxicity from two stressors involve antagonistic pleiotropy, alternate pathways, or rely on the same resource pool (e.g., Siemens et al., 2012), we may find that diploids show sensitivity to one but not the other (leading to a negative correlation). The greater output of gene products or opportunities for gene network rewiring by the polyploid genome may buffer against these trade-offs, leading to less negative or even overall positive correlation among stress responses for polyploids. Alternatively, if the underlying mechanisms (e.g., uptake limitation vs. antioxidative to detoxify) or genetic determinants (e.g., allelic diversity or gene interactions) vary among stressors, then we might find no universal pattern in the effect of polyploidy on stress tolerance. Thus, it is still an open question as to whether polyploids have “general-purpose” genotypes (Baker, 1965; Stebbins, 1971) that buffer them against a wide range of stressors, including those commonly encountered in urban settings, and whether they show an altered correlation structure across these stressors.
The ultimate expression of stress tolerance is maintaining fitness in the face of a particular stressor (Simms, 2000). A polypoid possessing a general-purpose genotype is predicted to exhibit fitness homeostasis (e.g., lack of fitness plasticity) across multiple environments (Madlung, 2013; Mattingly & Hovick, 2023; Wei et al., 2019). However, most studies comparing stress tolerance between diploids and polyploids compare enzymatic metrics, physiological responses, or fitness proxies of growth or development (e.g., reviewed in Tossi et al., 2022) to a single stressor and have not addressed fitness homeostasis across a range of stressors (but see Mattingly & Hovick, 2023; Wei et al., 2019). Because stress may impact different aspects of plant life history, these may not fully be captured by measuring only physiological responses or biomass change in individuals, whereas fitness integrates all impacts. The most appropriate fitness comparison is to be made at the population level, where diploid and polyploid growth within generations and reproduction among generations contribute to population fitness (e.g., Anneberg et al., 2023a; Selmecki et al., 2015).
Recurrent formation of polyploids from genetically different diploids is a common phenomenon in nature (Kolář et al., 2017; Soltis & Soltis, 1999) and could lead to variation in stress tolerance, even when polyploidy involves one parental genome (autopolyploidy) rather than two genomes (allopolyploidy) (e.g., Bafort et al., 2023; Mattingly & Hovick, 2023; Wei et al., 2019). This variation in stress tolerance may ultimately determine the probability of polyploid establishment under novel or stressful conditions (Soltis & Soltis, 1999; Van Drunen & Johnson, 2022). While the use of multiple sources of natural diploids and polyploids can address this to some degree (e.g., Wei et al., 2019), synthetic neopolyploids are recognized as a powerful tool because they avoid the confounding effects of evolution after duplication that exist in wild polyploid–diploid comparisons (Bomblies, 2020). Furthermore, including multiple genotypes of synthetic autopolyploids allows for evaluating the potential for genetic diversity to contribute to stress tolerance (co)variation in diploids and how these change with polyploidy.
Here we used diploids and neopolyploids of the model plant, the giant duckweed (Spirodela polyrhiza; Lemnaceae). These floating aquatic angiosperms mostly reproduce clonally and have a rapid generation time of 4–5 days (Acosta et al., 2021), making them a proven system for experimental population-level studies (Armitage & Jones, 2019; Hart et al., 2019; Hess et al., 2022; Hitsman & Simons, 2020; Huber et al., 2021; Subramanian & Turcotte, 2020). Here, as in these other studies, we quantify fitness as multigenerational asexual population growth rate. Several diploid duckweed species are being studied for stress responses at the phenomenological and mechanistic levels and used as potential phytoremediators (Chmilar & Laird, 2019; Dalu & Ndamba, 2003; Ekperusi et al., 2019; Huber et al., 2007; O’Brien et al., 2022; Ziegler et al., 2017).
We grew six genetically distinct pairs of diploids and their immediate neotetraploid descendants (Anneberg et al., 2023a) individually in water contaminated with one of five urban pollutants (copper, iron, salt, aluminum, and manganese) and a control to answer these specific questions concerning stress tolerance.
How does environmental pollution alter the relative fitness of diploids and their derived neotetraploids? Does it depend on genetic lineage or pollutant?
Are neotetraploids more stress tolerant than diploids, or does it depend on genetic lineage or pollutant?
Do neotetraploids maintain fitness (lower fitness plasticity) across pollutants better than their progenitor diploids?
Are there genetic trade-offs in fitness across pollutants, and is this altered by polyploidy?
Methods
Study system and the creation of neopolyploids
Spirodela polyrhiza (L.) Schleid is a globally distributed small floating aquatic plant in the family Lemnaceae (Jacobs, 1947). It exists in freshwater ponds, streams, as well as being common in urban parks, drainage ditches, and wastewater collection sites where it encounters numerous pollutants (Dalu & Ndamba, 2003; O’Brien et al., 2022). When reproducing asexually through budding, it can produce actively growing individuals (hereafter referred to as fronds) or, under stress, produce dormant propagules termed turions (Appenroth et al., 1996; Jacobs, 1947). Approximately half of duckweed species produce turions, and these can help survive unfavorable conditions (e.g., winter freezing, low nutrients) by sinking to the bottom of ponds and germinating once conditions improve.
In 2017, S. polyrhiza was sampled from natural and urban ponds in Western Pennsylvania and Eastern Ohio, United States. (Supplementary Table S1). Individuals were used to establish six monoclonal colonies and were confirmed to be unique genotypes using 10 microsatellite loci (Supplementary Table S1; Kerstetter et al., 2023; Xu et al., 2018). They were cultured in conditions that maintain asexual reproduction and formed the initial genetic lineages. Synthetic neotetraploids were created from six genetically distinct diploids using the mitotic inhibitor colchicine (Sigma-Aldrich, CAS: 64-86-8) which occurred in 2019 (for SP.01, SP0.5, SP.07, and SP.11) and 2021 (SP.41 and SP.43). Details of the methodology can be found in Anneberg et al. (2023a), and four of the lineages used here are reported therein. Briefly, after exposing populations of a single diploid genotype to colchicine, we tested ploidy using flow cytometry following Wei et al. (2020). We retained both converted neotetraploids and unconverted diploids from each genetic lineage. These were maintained in quarter-strength growth media described in Appenroth et al. (1996). Before the experiment, each of the 12 sublineages (a diploid and polyploid of each genetic lineage) was grown in common garden conditions for 2 weeks. This consisted of growth in full-strength media under fluorescent lights at room temperature.
Experimental design
We selected five urban pollutants that duckweed populations may commonly encounter (Bhat, 2012; O’Brien et al., 2022; Vo et al., 2018). The concentrations used during the experiment were determined by pilot studies on other duckweed diploid genotypes (Zallek & Turcotte, unpublished). We selected concentrations that reduced population growth but did not kill all the duckweed within a few weeks. Given that our focus is to compare among ploidies rather than among pollutants per se, we did not aim to equalize the negative impacts of each pollutant. Each experimental unit consisted of a 120-ml glass jar (Fisher, United States, # FB02911775) to which 90 ml of quarter-strength autoclaved media was added. Jars were set in plastic trays, and a large plastic lid covered the tray. Pollutant treatments varied in the addition of nothing (control), 0.04 mM of FeCl3 (iron), 40 mM of NaCl (salt), 0.6 mM of MnCl2 (manganese), 0.0025 mM of CuSO4 (copper), or 0.015 mM of Al2(SO4)3 (aluminum). The growth media also contained small quantities of Fe (0.025 mM), Mn (0.013 mM), Na (0.0258 mM), and Cl (0.026 mM) as micronutrients. The levels of pollutants we used were similar or slightly higher than those observed in various studies of urban water bodies (Herngren et al., 2005; Rhodes-Dicker & Passeport, 2019), except for manganese, which required a much higher concentration to impact the duckweed performance more akin to those from mine or industrial discharge (Cravotta & Brady, 2015). During the week of January 23, 2023, four individual duckweed fronds were added to each jar. On days 7, 14, and 21, jars were photographed, and duckweed fronds were manually counted from the photos using Fiji (Schindelin et al., 2012). Turions were counted on day 21. We counted fronds and turions separately because the relative performance of diploids and neopolyploids can depend on which is quantified (Anneberg et al., 2023a).
The experiment was conducted by 836 undergraduate students at the University of Pittsburgh, who were taking a research focus lab course in the spring semester 2023. The students were divided into 42 course sections, with up to 20 students in each section. These sections were taught by 13 instructors across three laboratory classrooms. Genetic lineages were tested across different rooms and instructors, but each section only tested two lineages. Sublineages (diploid and polyploid) of a specific genetic lineage were always tested together. Specifically, students worked in pairs, setting up four jars of a single lineage (e.g., SP.05) in a factorial design of control or a single pollutant crossed with the diploid or polyploid sublineage. This approach led to an unbalanced design with five times as many control jars, but it was important to teach students the importance of controls. Thus, each section set up 24 control jars and four jars of each metal. After removing jars with missing data or major errors (e.g., adding greater than 50% too many duckweed initially), a total of 1,591 experimental units (jars) were analyzed. Each sublineage by pollutant combination (excluding control) had an average of 13.25 (SD = 3.08) replicates.
Statistical analysis
To address the relative performance of ploidies under varied pollutants, we quantified the population growth rate of fronds and turions separately. First, using akaike information criterion (AIC), we found that fronds grew in a manner best described by a linear population growth model, as opposed to exponential or logistic growth (Supplementary Figure S1, ΔAIC of 19,656 and 4,485, respectively). The model had the abundance of fronds as a response variable, and explanatory fixed factors included ploidy, genetic lineage, and pollutant, as well as their interactions. The initial abundance was set as the initial frond number added to each jar (i.e., not estimated by the model). Jar was a random effect that accounted for repeated measures. We tested various random effects (section number, instructor, or room) and variance functions. The best fitting model had jar nested within section number as random effects, and a variance function that increased with the day. Models were fit using the nlme function in the nlme package (Bates & R Core Team, 2023). In addition to the general statistical analysis, which used type III sums of squares, we conducted planned contrasts comparing the growth rates of diploids vs. polyploids within each pollutant using the emmeans package (Russell, 2023).
We tested for differences in turion production using a linear mixed-effects model with turion as the response variable, with fixed factors of ploidy, genetic lineage, and pollutant, as well as their interactions, and section number as a random effect using the lme function in the nlme package. We then conducted planned contrasts as above.
To evaluate ploidal differences in tolerance to pollutants and fitness plasticity across pollutants, we analyzed the total abundance of individuals (fronds and turions combined) to provide an overall assessment of population growth. We decided to combine both types of individuals with equal contributions because their relative impact on fitness depends on phenological, environmental, and demographic conditions (e.g., the severity of winter). To evaluate tolerance and fitness maintenance, we first quantified the growth rate within each jar. We fit a simple linear population growth model to each jar with the initial abundance predetermined by the initial number of individuals added. Then we calculated tolerance as the growth rate in a given pollutant divided by the control, where each pair of jars was measured by a single student and represented one control and one single pollutant of the same lineage and ploidy. We fit a linear mixed-effects model to these tolerance values with ploidy, lineage, pollutant, and their interactions as fixed effects and section number as a random effect. We also conducted planned contrasts comparing diploids to polyploids across all pollutants as well as in each pollutant treatment.
To quantify fitness plasticity in the face of the five pollutants, excluding the control, we also analyzed the variation in growth rate at the jar level. We calculated the Relative Distance Plasticity Index (RDPI, Valladares et al., 2006) for each sublineage (combination of lineage and ploidy). The relative differences in growth rate between two jars are the absolute difference in growth rates divided by the sum of both growth rates. This is calculated for all possible pairings of jars that have the same duckweed sublineage but only between jars that have a different pollutant treatment (e.g., diploid SP.05 iron and salt). Then, the relative differences are summed and divided by the number of pairings to give the RDPI value. RDPI values of 0 imply fitness is maintained across environments (no plasticity), and values of 1 imply maximum plasticity. This process is then repeated for each sublineage. Using these distances, we fit a linear model with lineage and ploidy and their interaction as fixed effects. We then used planned contrasts to compare ploidy within each lineage. We used scripts from Ameztegui (2017) to run our analyses.
We then calculated the broad-sense genetic correlations among pairs of pollutants. First, we fit a mixed-effect model to the jar-level growth rate data that had ploidy, lineage, pollutant, and their interactions as fixed effects, and section number as a random effect. Given that the duckweed reproduces clonally, we calculated genetic correlations among each pair of pollutants using the estimated marginal means from the mixed-effect model. We calculated Pearson’s correlation coefficients and their significance among the six diploid lineages for each possible pair of pollutants and repeated this procedure for the polyploids. We used the cor function in the base package of R as well as the corrplot package (Wei & Simko, 2021). Finally, we used a paired t-test on the correlation coefficients to test whether the correlations differed among diploids and polyploids.
Results
Environmental pollution alters the relative performance of diploids and their derived neotetraploids in ways that depend on the pollutant and genetic lineage
The effect of ploidy on performance quantified as both the growth rate in frond abundance and the total number of turions produced, had pollutant- and genetic lineage-dependent effects (see Supplementary Figure S1 for time series and Supplementary Figure S2 for example photos). A linear mixed-effect model revealed that ploidy, lineage, pollutant, and all interactions significantly impacted the growth rate of fronds (all p < .001; Supplementary Table S2; Figure 1). Although interpretation is complex given the interactions, planned contrasts (Figure 1) revealed that the relative performance of the diploids depended both quantitatively (effect size) and qualitatively (direction of effect) on the pollutants. Of all conditions tested, in the control, the neotetraploids performed worse relative to the diploids as their daily lineage growth rates (3.06) were 37% lower than those of diploids (4.84, see Figure 1 for p-values). In addition, neotetraploids did significantly worse in aluminum (−31%), iron (−30%), and salt (−18%) but the absolute differences in growth rates were smaller (Figure 1). Yet, in manganese, performance did not differ significantly (−8%), and in copper, the polyploids performed significantly better (+18%). While there was variation among genetic lineages, they generally followed these overall trends (Figure 1). In the control, iron, salt, and aluminum diploid populations grew faster than their derived polyploids for all lineages, but the effect sizes varied. But in copper and manganese polluted media, approximately half of the genetic lineages showed polyploids performed better than diploids. Moreover, when we fit models to each pollutant separately, there were significant ploidy by lineage interactions (p < .001) in all stressors except for iron and aluminum (p = .18 and p = .14, respectively).
Figure 1.
Daily frond population growth rates (estimated marginal means ± 1 SE) separated by ploidy and genetic lineage (including an overall average) across the six pollutant treatments (including the control). The table above shows the percentage difference of neopolyploid (4×) in relation to diploid (2×), along with the p-values resulting from the six planned contrasts between diploids and neotetraploids for each pollutant treatment averaged across all lineages. Point positions were dodged for clarity, with circles representing diploids and triangles representing neotetraploids.
In contrast to fronds, turion production was higher in neopolyploids than diploids but was also strongly impacted by genetic lineage and pollutant type (Figure 2; Supplementary Figure S1). Unlike the frond growth rate, the control conditions led to similar total turion production, even though there were generally more fronds in the control treatment. The mixed model revealed strong effects of genetic lineage and its interactions with ploidy and pollutant (all p ≤ .0001; Supplementary Table S3). Yet, averaging across lineages, planned contrasts show that polyploids produce more turions in iron (+196%; Figure 2), aluminum (+131%), copper (+94%), and in control (+29%), whereas they do not differ significantly from diploids in manganese and salt. Pollutant-specific models revealed that all treatments had significant ploidy by lineage interactions (salt p = .049, all others p < .0001). Indeed, salt almost completely prevented turion production for both diploids (estimated marginal means of 0.012) and neopolyploids (0.139), but this was not a significant difference.
Figure 2.
Total turion production (estimated marginal means ± 1 SE) separated by ploidy and genetic lineage (including an overall average) across the six pollutant treatments (including the control). The table above shows the percentage difference of neopolyploid (4×) in relation to diploid (2×), along with the p-values resulting from the six planned contrasts between diploids and neotetraploids for each pollutant treatment. Point positions were dodged for clarity, with circles representing diploids and triangles representing neotetraploids.
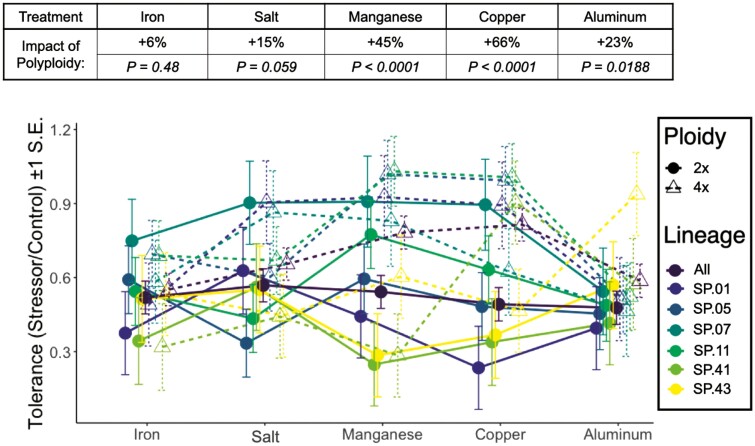
Neotetraploids are often more tolerant of pollutant stress
When tolerance is estimated as population growth under pollutant stress divided by that under control conditions, neopolyploidy generally increases tolerance, although this was also influenced by lineage (Figure 3). Again, interactions between ploidy, lineage, and pollutant type were significant (Supplementary Table S4). A planned contrast averaging across all pollutants and lineages found that polyploids are more tolerant (estimated marginal mean 0.678) than diploids (0.520, p < .0001). This positive impact of neopolyploidy is statistically supported for all pollutants (planned contrasts; +6% to +66%, Figure 3) except for iron. Yet, genetic lineage had a strong impact, with neopolyploids in three lineages (SP.01, SP.05, and SP.11) having higher tolerance than their respective diploids under all stressors, neopolyploids in two lineages (SP.41 and SP.43) had rank order changes across pollutants, whereas neotetraploid SP.07 was uniformly less tolerant.
Figure 3.
Tolerance of each pollutant (stressor/control, estimated marginal means ± 1 SE) combining frond and turion growth across ploidy and genetic lineage (including an overall average). Tolerance values of 1 imply no impact of stressors on fitness. The table above shows the percentage difference of neopolyploid (4×) in relation to diploid (2×), along with the p-values resulting from the five planned contrasts between diploids and neotetraploids for each pollutant. Point positions were dodged for clarity, with circles representing diploids and triangles representing neotetraploids.
Neotetraploids maintain fitness across pollutants better and show fewer genetic trade-offs in fitness under varied pollutants than their progenitor diploids
Polyploids maintained population growth across various stressors better than diploids as evidenced by both lower plasticity (measured as RDPI) and positive broad-sense genetic correlations. Ploidy, lineage, and their interactions significantly impacted fitness plasticity across the five pollutants (Figure 4; Supplementary Table S5, all p < .0001). Overall, polyploids had lower fitness plasticity than diploids (RDPI: 0.268 vs. 0.290), but this varied among lineages. In SP.01, SP.05, SP.11, and SP.41, neotetraploids maintained fitness better than their diploid progenitors. But the opposite was observed with SP.43. Finally, lineage SP.07 had the lowest and least variable fitness plasticity between ploidies (diploid vs. neotetraploid: 0.190 and 0.193, Figure 4).
Figure 4.
Plasticity in fitness across all pollutants (excluding the control) for each lineage and ploidy. Plasticity, quantified as the Relative Distance Plasticity Index (RDPI) of growth rates, combining fronds and turions, among the five pollutants. Values of 0 imply complete fitness maintenance (no plasticity) and 1 imply maximum plasticity. Post hoc Tukey HSD test results are shown above each lineage pair.
Broad-sense genetic correlations in growth rate between pairs of metal pollutants were impacted by neopolyploidy. Correlations ranged from negative to positive in diploids but were all positive in neotetraploids (Figure 5). However, given the small sample size, only four individual correlations had p-values between 0.001 and 0.07 (diploids: copper salt −0.81, p = .05; iron salt −0.77, p = .07; neotetraploids: aluminum-copper 0.96, p = .003; iron-manganese 0.92, p = .011). Nevertheless, across all correlations, a paired t-test revealed that diploids had significantly weaker and more negative correlations than polyploids (difference of −0.53, t = −3.09, df = 9, p = .013).
Figure 5.
Broad-sense genetic correlations of population growth rate between pairs of pollutant environments (excluding the control). The lower triangle of the matrix includes Pearson’s correlations for diploids and the upper triangle for tetraploids. p-values < .075 are shown. Positive (negative) correlations are in blue (red) with a darker shade reflecting the strength of the correlation.
Discussion
Our exploration of the effects of neotetraploidy on duckweed population growth across five common urban pollutants provided support for two hypothesized advantages of polyploidy, both in a general sense and with special regard to urban habitats. We found that, relative to their diploid progenitors, neopolyploid duckweeds (a) are better stress tolerators and (b) show lower fitness plasticity across heterogeneous pollutant environments, thus appearing to have general-purpose genotypes. While the evidence for these generalizations is compelling, important complexities were also revealed. Specifically, ploidal responses varied by genetic lineage as well as by pollutant. Indeed, some stressful conditions reversed the ranking of ploidies. In the following paragraphs, we explore in more depth the implications of both genetic and environment-dependent advantages of neopolyploidy under urban pollution.
Our results demonstrated that stress conditions can upend fitness differences between the ploidies. Large diploid population growth advantages under favorable (control) conditions (Figures 1 and 2) corroborate previous studies with synthetic neopolyploid duckweeds (Anneberg et al., 2023a; Assour et al., 2023). However, here we showed that when exposed to pollutant stress, diploids were more similar to polyploids, and one contaminant (copper) even reversed the direction of the diploid–neopolyploid difference. Similar, pollutant-dependent outcomes have been seen at the organismal level, where salt, low nutrients, and their combination lead to varying responses in two Arabidopsis neotetraploids (Mattingly & Hovick, 2023). Much like our results, Bafort et al. (2023), using a different set of pairs of neotetraploid–diploid duckweed, found the advantage of polyploidy to be environment and genetic lineage specific. Neopolyploids showed an increase in frond surface area at low concentrations of salt. Yet their study did not measure turions. Turion production, typically higher in neopolyploid duckweed, was suppressed by salt (Figure 2). Perhaps this gave rise to greater allocation by neopolyploids to fronds production under these conditions, and lower ploidal difference as well (Figure 1). Taken together, these studies suggest that there are stressful conditions associated with urban activities that may allow neopolyploids to establish and persist locally. Whether neopolyploids are more common in urban environments remains unknown (Van Drunen & Johnson, 2022). Indeed, because neopolyploid duckweeds invest more heavily in allocation to future propagules in duckweeds (turion production) across most pollutant conditions tested, neopolyploids may be even more likely to persist in urban environments with severe winters (Anneberg et al., 2023a).
Consistent with these fitness results, we also found that neopolyploidy leads to higher tolerance across most pollutants and genetic lineages. Relative to control conditions, polyploids are better able to maintain their total population growth rate (fronds and turions; Figure 3). Although this result is influenced by the fact that diploids have higher fitness in control conditions (Figure 1), it may indicate that polyploids are better able to handle stress but at a cost that is only apparent in ideal, uncontaminated growth conditions, which are increasingly rare in the Anthropocene. Whether neopolyploid’s higher tolerance is due to a higher capacity for storage, greater detoxification, or other mechanisms remains to be determined with cellular morphometric and enzymatic studies, such as those reviewed in Tossi et al. (2022).
We found that relative to diploids, neopolyploids had lower plasticity in population growth rate across stress conditions indicating maintenance of fitness across heterogeneous pollutants. While on first glance this may seem counter to other predictions and findings of increased phenotypic plasticity in polyploids (Mattingly & Hovick, 2023; Parisod et al., 2010; Spoelhof et al., 2017; Van de Peer et al., 2017; Wei et al., 2019), but not always (Harms et al., 2021; Kornstad et al., 2022; Walczyk & Hersch-Green, 2023). It is important to note that these other studies focus on plasticity in functional traits, not fitness, and indeed higher trait plasticity is expected to buffer fitness and thus lead to lower fitness plasticity (see Wei et al., 2019). Specifically, high plasticity for functional traits may allow a newly formed polyploid to maintain fitness (low plasticity) in a novel or stressful environment. Here we did not measure specific traits, so we do not know how or what phenotypic changes might contribute to the fitness maintenance seen in polyploids. These could be changes in frond size, thickness, or photosynthetic activity seen in Bafort et al. (2023) in response to salt and cadmium pollution, or changes in the numerous other mechanisms of detoxification of pollutants observed in diploid duckweeds (Ekperusi et al., 2019; Huber et al., 2021). Work to make these mechanism–function connections is a key next step.
Nevertheless, the shifts in broad-sense genetic correlations of fitness across pollutants, from negative to positive (Figure 5), for neopolyploids give rise to the intriguing possibility that neopolyploidy leads to instantaneous buffering from constraints. And while we acknowledge the low power of this test, these seem not to be driven by similar types of pollutants, as the significant pairs were from different classes (alkaloids, heavy metals, and salt) rather than within one. Such an outcome could reflect a global advantage of larger cells (gigas effect; Doyle & Coate, 2019) and thus increased storage of toxic substances by neopolyploids. However, they could also reflect upregulation of shared antioxidant pathways or rewiring of pathways that were otherwise antagonistic (Alfatih et al., 2020; Des Marais & Juenger, 2010; Lu et al., 2020), so we encourage greater exploration of this idea with an increased number of genetic lineages, classes, and concentrations of pollutants while still employing rigorous fitness measures. Lastly, integrated studies of stress tolerance across levels from the cellular to organismal level will be crucial for understanding the polyploidy–stress relationship across systems (Wei et al., 2020).
Across our study, we found a strong impact of the interaction between genetic lineage and ploidy within or among different pollutants. The effect of ploidy on performance, tolerance, and fitness plasticity was all influenced by genetic lineage, either quantitatively or sometimes qualitatively. Even though genomic studies suggest that S. polyrhiza has low genetic diversity (Ho et al., 2019; Xu et al., 2019), both our study with lineages of NE USA and that of Bafort et al. (2023) with lineages from different continents demonstrated pronounced differences among lineages in the effects of polyploidization. This disconnect suggests that functional differences within diploid S. polyrhiza may derive from epigenetic variation that can be altered by autopolyploidization along with genetic variation (Chen, 2007; Huber et al., 2021) and strongly influence stress tolerance and population growth. Regardless of the mechanism, our results join those of several others demonstrating the importance of genetic variation in diploid progenitors on neopolyploid morphology (Wei et al., 2020), population growth (Anneberg et al., 2023a), and response to abiotic (Bafort et al., 2023; Wei et al., 2020) and biotic interactions (Anneberg et al., 2023b; Assour et al., 2023; Forrester et al., 2020). They provide additional support for the idea that the repeated evolution of polyploids may be key to their success (Kolář et al., 2017; Soltis & Soltis, 1999; Wei et al., 2020). Moreover, this type of variation is especially important in the context of eco-evolutionary dynamics that could occur in urban environments (Verrelli et al., 2022) because it not only suggests that the impact of ploidy is modulated by the genetic background of the progenitor diploid, but also indicates that there is ample opportunity for natural selection. Finally, because we have shown here that the degree and direction of ploidal fitness difference varies among the pollutants, it means there is the potential for heterogeneity in ploidal dominance within urban environments depending on specific pollutant environments.
In conclusion, our study corroborates that of others, suggesting that the combination of environmental perturbations and independent polyploidization events may be crucial for the establishment of polyploids (Anneberg et al., 2023a; Fawcett et al., 2009; Wei et al., 2020). This may be even more important in the 21st century as urban stressors may elevate both polyploid production and stress levels (Van Drunen & Johnson, 2022).
Supplementary Material
Acknowledgments
We thank the students of Duckweed Survivor (BioSci 0057, Spring 2023 semester) and instructors (M. Amour, D. Bisi, S. Gess, N. Kanmanii, N. Kaufmann, A. Martin, A. Nigam, K. Parks, B. Reed, J. Robertson, S. Stuckman, K. Wagner, and C. Wiesner) for conducting the experiment, and K. Butela, K. Wozniak, and L. Rzodkiewicz for input on the course development. Additionally, we would like to thank S. Bhattacharya, J. Robertson, and K. Kroeger for preparing duckweed populations for the course, and E. O’Neill, J. Kerstetter, T. Anneberg, and A. Burr for their help in creating and maintaining the diploid–polyploid lineages.
Contributor Information
Martin M Turcotte, Department of Biological Sciences, University of Pittsburgh, Pittsburgh, PA, United States.
Nancy Kaufmann, Department of Biological Sciences, University of Pittsburgh, Pittsburgh, PA, United States.
Katie L Wagner, Department of Biological Sciences, University of Pittsburgh, Pittsburgh, PA, United States.
Taylor A Zallek, Department of Biological Sciences, University of Pittsburgh, Pittsburgh, PA, United States.
Tia-Lynn Ashman, Department of Biological Sciences, University of Pittsburgh, Pittsburgh, PA, United States.
Data and code availability
The data underlying the article and the analysis code are available in the online Supplementary material. The files include:
Supp.Turcotte.et.al.Evol.Letters.pdf (Supplemental figures and tables); Data.Turcotte.et.al.Evol.Letters.csv (CSV file with all raw data); AnalysisCode.Turcotte.et.al.Evol.Letters.R (R script with analyses); and rdpi_matrix.R and RDPI.R (addition dependencies to run the RDPI analysis slightly modified from Ameztegui (2017)).
Author contributions
M.M.T., T.Z., N.K., and K.W. created and deployed the curriculum and helped collect, collate, and clean the data. All authors contributed to the design of the experiment. M.M.T. and T.L.A. conceived the project, interpreted the data, and wrote the manuscript. M.M.T. analyzed the data. All authors contributed to manuscript revisions.
Funding
This work was supported by the University of Pittsburgh through the Dietrich School of Arts and Sciences, the Department of Biogical Sciences, and a Mellon Fellowship to TAZ as well as by NSF grants DEB-1935410 to MMT and DEB-2027604 to TLA.
Conflict of interest: The authors declare no conflict of interest.
References
- Acosta, K., Appenroth, K. J., Borisjuk, L., Edelman, M., Heinig, U., Jansen, M. A. K., Oyama, T., Pasaribu, B., Schubert, I., Sorrels, S., Sree, K. S., Xu, S., Michael, T. P., & Lam, E. (2021). Return of the Lemnaceae: Duckweed as a model plant system in the genomics and postgenomics era. The Plant Cell, 33(10), 3207–3234. 10.1093/plcell/koab189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfatih, A., Wu, J., Jan, S. U., Zhang, Z. -S., Xia, J. -Q., & Xiang, C. -B. (2020). Loss of rice PARAQUAT TOLERANCE 3 confers enhanced resistance to abiotic stresses and increases grain yield in field. Plant, Cell & Environment, 43(11), 2743–2754. 10.1111/pce.13856 [DOI] [PubMed] [Google Scholar]
- Ameztegui, A. (2017). Plasticity: A package for computing plasticity indices. GitHub repository. https://github.com/ameztegui/Plasticity
- Anneberg, T. J., O’Neill, E. M., Ashman, T. -L., & Turcotte, M. M. (2023a). Polyploidy impacts population growth and competition with diploids: Multigenerational experiments reveal key life history tradeoffs. The New Phytologist, 238(3), 1294–1304. 10.1111/nph.18794 [DOI] [PubMed] [Google Scholar]
- Anneberg, T. J., Turcotte, M. M., & Ashman, T. -L. (2023b). Plant neopolyploidy and genetic background differentiates the microbiome of duckweed across a variety of natural freshwater sources. Molecular Ecology, 32(21), 5849–5863. 10.1111/mec.17142 [DOI] [PubMed] [Google Scholar]
- Appenroth, K. J., Teller, S., & Horn, M. (1996). Photophysiology of turion formation and germination in Spirodela polyrhiza. Biologia Plantarum, 38(1), 95–106. [Google Scholar]
- Armitage, D. W., & Jones, S. E. (2019). Negative frequency‐dependent growth underlies the stable coexistence of two cosmopolitan aquatic plants. Ecology, 100(5), e02657. 10.1002/ecy.2657 [DOI] [PubMed] [Google Scholar]
- Assour, H. A., Ashman, T. -L., & Turcotte, M. M. (2023). Neopolyploidy-induced changes in the giant duckweed (Spirodela polyrhiza) alter herbivore preference, performance, and plant population performance. bioRxiv. 2023.11.14.567047. 10.1101/2023.11.14.567047 [DOI] [PubMed] [Google Scholar]
- Bafort, Q., Wu, T., Natran, A., De Clerck, O., & Van de Peer, Y. (2023). The immediate effects of polyploidization of Spirodela polyrhiza change in a strain-specific way along environmental gradients. Evolution Letters, 7(1), 37–47. 10.1093/evlett/qrac003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, H. G. (1965). Characteristics and modes of origin of weeds. In Baker H. G. & Stebbins G. L. (Eds.), Genetics of colonizing species (pp. 147–172). Academic Press. [Google Scholar]
- Bates, J.; R Core Team. (2023). nlme: Linear and nonlinear mixed effects models. R package version 3.1-163. https://CRAN.R-project.org/package=nlme
- Beest, M., Le Roux, J. J., Richardson, D. M., Brysting, A. K., Suda, J., & Kubesová, M., & Pysek, P. (2012). The more the better? The role of polyploidy in facilitating plant invasions. Annals of Botany, 109, 19–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat, D. M. (2012). Assessment of heavy metal pollution in urban pond ecosystems. Universal Journal of Environmental Research and Technology, 2(4), 225–232. [Google Scholar]
- Bomblies, K. (2020). When everything changes at once: Finding a new normal after genome duplication. Proceedings of the Royal Society B: Biological Sciences Biological Sciences, 287(1939), 20202154. 10.1098/rspb.2020.2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z. J. (2007). Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annual Review of Plant Biology, 58(1), 377–406. 10.1146/annurev.arplant.58.032806.103835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmilar, S. L., & Laird, R. A. (2019). Demographic senescence in the aquatic plant Lemna gibba L. (Araceae). Aquatic Botany, 153, 29–32. 10.1016/j.aquabot.2018.11.004 [DOI] [Google Scholar]
- Cravotta, C. A., & Brady, K. B. C. (2015). Priority pollutants and associated constituents in untreated and treated discharges from coal mining or processing facilities in Pennsylvania, USA. Applied Geochemistry, 62, 108–130. [Google Scholar]
- Dalu, J. M., & Ndamba, J. (2003). Duckweed based wastewater stabilization ponds for wastewater treatment (a low cost technology for small urban areas in Zimbabwe). Physics and Chemistry of the Earth, Parts A/B/C, 28(20–27), 1147–1160. 10.1016/j.pce.2003.08.036 [DOI] [Google Scholar]
- De Smet, R., & Van de Peer, Y. (2012). Redundancy and rewiring of genetic networks following genome-wide duplication events. Current Opinion in Plant Biology, 15(2), 168–176. 10.1016/j.pbi.2012.01.003 [DOI] [PubMed] [Google Scholar]
- Des Marais, D. L., & Juenger, T. E. (2010). Pleiotropy, plasticity, and the evolution of plant abiotic stress tolerance. Annals of the New York Academy of Sciences, 1206, 56–79. 10.1111/j.1749-6632.2010.05703.x [DOI] [PubMed] [Google Scholar]
- Doyle, J. J., & Coate, J. E. (2019). Polyploidy, the nucleotype, and novelty: The impact of genome doubling on the biology of the cell. International Journal of Plant Sciences, 180(1), 1–52. 10.1086/700636 [DOI] [Google Scholar]
- Dutta, S., Mitra, M., Agarwal, P., Mahapatra, K., De, S., Sett, U., & Roy, S. (2018). Oxidative and genotoxic damages in plants in response to heavy metal stress and maintenance of genome stability. Plant Signaling & Behavior, 13(8), e1460048. 10.1080/15592324.2018.1460048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebadi, M., Bafort, Q., Mizrachi, E., Audenaert, P., Simoens, P., Van Montagu, M., Bonte, D., & Van de Peer, Y. (2023). The duplication of genomes and genetic networks and its potential for evolutionary adaptation and survival during environmental turmoil. Proceedings of the National Academy of Sciences of the United States of America, 120(41), e2307289120. 10.1073/pnas.2307289120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrendorfer, F. (1980). Polyploidy and distribution. In Lewis W. H. (Ed.), Polyploidy, biological relevance (pp. 45–60). Plenum Press. [Google Scholar]
- Ekperusi, A. O., Sikoki, F. D., & Nwachukwu, E. O. (2019). Application of common duckweed (Lemna minor) in phytoremediation of chemicals in the environment: State and future perspective. Chemosphere, 223, 285–309. 10.1016/j.chemosphere.2019.02.025 [DOI] [PubMed] [Google Scholar]
- Fawcett, J. A., Maere, S., & Van de Peer, Y. (2009). Plants with double genomes might have had a better chance to survive the Cretaceous–Tertiary extinction event. Proceedings of the National Academy of Sciences of the United States of America, 106(14), 5737–5742. 10.1073/pnas.0900906106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester, N. J., Rebolleda-Gómez, M., Sachs, J. L., & Ashman, T. -L. (2020). Polyploid plants obtain greater fitness benefits from a nutrient acquisition mutualism. The New Phytologist, 227(3), 944–954. 10.1111/nph.16574 [DOI] [PubMed] [Google Scholar]
- Harms, N. E., Cronin, J. T., & Gaskin, J. F. (2021). Increased ploidy of Butomus umbellatus in introduced populations is not associated with higher phenotypic plasticity to N and P. AoB PLANTS, 13(4). 10.1093/aobpla/plab045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart, S. P., Turcotte, M. M., & Levine, J. M. (2019). Effects of rapid evolution on species coexistence. Proceedings of the National Academy of Sciences of the United States of America, 116(6), 2112–2117. 10.1073/pnas.1816298116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herngren, L., Goonetilleke, A., & Ayoko, G. A. (2005). Understanding heavy metal and suspended solids relationships in urban stormwater using simulated rainfall. Journal of Environmental Management, 76(2), 149–158. 10.1016/j.jenvman.2005.01.013 [DOI] [PubMed] [Google Scholar]
- Hess, C., Levine, J. M., Turcotte, M. M., & Hart, S. P. (2022). Phenotypic plasticity promotes species coexistence. Nature Ecology & Evolution, 6(9), 1256–1261. 10.1038/s41559-022-01826-8 [DOI] [PubMed] [Google Scholar]
- Hitsman, H. W., & Simons, A. M. (2020). Latitudinal variation in norms of reaction of phenology in the greater duckweed Spirodela polyrhiza. Journal of Evolutionary Biology, 33(10), 1405–1416. 10.1111/jeb.13678 [DOI] [PubMed] [Google Scholar]
- Ho, E. K. H., Bartkowska, M., Wright, S. I., & Agrawal, A. F. (2019). Population genomics of the facultatively asexual duckweed Spirodela polyrhiza. The New Phytologist, 224(3), 1361–1371. 10.1111/nph.16056 [DOI] [PubMed] [Google Scholar]
- Hope, D., Naegeli, M. W., Chan, A. H., & Grimm, N. B. (2004). Nutrients on asphalt parking surfaces in an urban environment. In Wieder R. K., Novák M., & Vile M. A. (Eds.), Biogeochemical investigations of terrestrial, freshwater, and wetland ecosystems across the globe (pp. 371–390). Springer Netherlands. [Google Scholar]
- Huber, J. A., Mark Welch, D. B., Morrison, H. G., Huse, S. M., Neal, P. R., Butterfield, D. A., & Sogin, M. L. (2007). Microbial population structures in the deep marine biosphere. Science, 318(5847), 97–100. 10.1126/science.1146689 [DOI] [PubMed] [Google Scholar]
- Huber, M., Gablenz, S., & Höfer, M. (2021). Transgenerational non-genetic inheritance has fitness costs and benefits under recurring stress in the clonal duckweed Spirodela polyrhiza. Proceedings of the Royal Society B: Biological Sciences, 288(1955), 20211269. 10.1098/rspb.2021.1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, D. L. (1947). An ecological life‐history of Spirodela polyrhiza (greater duckweed) with emphasis on the turion phase. Ecological Monographs, 17(4), 437–469. 10.2307/1948596 [DOI] [Google Scholar]
- Jacobson, C. R. (2011). Identification and quantification of the hydrological impacts of imperviousness in urban catchments: A review. Journal of Environmental Management, 92(6), 1438–1448. 10.1016/j.jenvman.2011.01.018 [DOI] [PubMed] [Google Scholar]
- Kerstetter, J., Reid, A., Armstrong, J., Zallek, T., Hobble, T., & Turcotte, M. (2023). Characterization of microsatellite markers for the duckweed Spirodela polyrhiza and Lemna minor tested on samples from Europe and the United States of America. Genetic Resources, 4(7), 46–55. 10.46265/genresj.alfv3636 [DOI] [Google Scholar]
- Kolář, F., Čertner, M., Suda, J., Schönswetter, P., & Husband, B. C. (2017). Mixed-ploidy species: Progress and opportunities in polyploid research. Trends in Plant Science, 22(12), 1041–1055. 10.1016/j.tplants.2017.09.011 [DOI] [PubMed] [Google Scholar]
- Kornstad, T., Ohlson, M., & Fjellheim, S. (2022). Phenotypic responses to light, water, and nutrient conditions in the allopolyploid Arabidopsis suecica and its parent species A. thaliana and A. arenosa: Does the allopolyploid outrange its parents? Ecology and Evolution, 12(5), e8915. 10.1002/ece3.8915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch, A. R., & Leitch, I. J. (2008). Genomic plasticity and the diversity of polyploid plants. Science, 320(5875), 481–483. 10.1126/science.1153585 [DOI] [PubMed] [Google Scholar]
- Leitenmaier, B., & Küpper, H. (2011). Cadmium uptake and sequestration kinetics in individual leaf cell protoplasts of the Cd/Zn hyperaccumulator Thlaspi caerulescens. Plant, Cell & Environment, 34(2), 208–219. 10.1111/j.1365-3040.2010.02236.x [DOI] [PubMed] [Google Scholar]
- Lu, Q., Liu, J., Chen, L., Yang, D., Shen, J., Li, J., Liston, A., Ashman, T. -L., & Dong, M. (2020). ABA-regulated ploidy-related genes and non-structural carbon accumulation may underlie cold tolerance in tetraploid Fragaria moupinensis. Environmental and Experimental Botany, 179, 104232. [Google Scholar]
- Madlung, A. (2013). Polyploidy and its effect on evolutionary success: Old questions revisited with new tools. Heredity, 110(2), 99–104. 10.1038/hdy.2012.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcom, J. W. (2011). Evolution of competitive ability: An adaptation speed vs. accuracy tradeoff rooted in gene network size. PLoS One, 6(4), e14799. 10.1371/journal.pone.0014799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly, K. Z., & Hovick, S. M. (2023). Autopolyploids of Arabidopsis thaliana are more phenotypically plastic than their diploid progenitors. Annals of Botany, 131(1), 45–58. 10.1093/aob/mcab081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouhaya, W., Allario, T., Brumos, J., Andrés, F., Froelicher, Y., Luro, F., Talon, M., Ollitrault, P., & Morillon, R. (2010). Sensitivity to high salinity in tetraploid citrus seedlings increases with water availability and correlates with the expression of candidate genes. Functional Plant Biology, 37(7), 674–685. 10.1071/fp10035 [DOI] [Google Scholar]
- O’Brien, A. M., Yu, Z. H., Pencer, C., Frederickson, M. E., LeFevre, G. H., & Passeport, E. (2022). Harnessing plant-microbiome interactions for bioremediation across a freshwater urbanization gradient. Water Research, 223, 118926. 10.1016/j.watres.2022.118926 [DOI] [PubMed] [Google Scholar]
- Parisod, C., Holderegger, R., & Brochmann, C. (2010). Evolutionary consequences of autopolyploidy. The New Phytologist, 186(1), 5–17. 10.1111/j.1469-8137.2009.03142.x [DOI] [PubMed] [Google Scholar]
- Parris, K. M. (2016). Ecology of urban environments. John Wiley & Sons Ltd. [Google Scholar]
- Pyšek, P., Lučanová, M., Dawson, W., Essl, F., Kreft, H., Leitch, I. J., Lenzner, B., Meyerson, L. A., Pergl, J., van Kleunen, M., Weigelt, P., Winter, M., & Guo, W. -Y. (2023). Small genome size and variation in ploidy levels support the naturalization of vascular plants but constrain their invasive spread. The New Phytologist, 239(6), 2389–2403. 10.1111/nph.19135 [DOI] [PubMed] [Google Scholar]
- Rhodes-Dicker, L., & Passeport, E. (2019). Effects of cold-climate environmental factors temperature and salinity on benzotriazole adsorption and desorption in bioretention cells. Ecological Engineering, 127, 58–65. 10.1016/j.ecoleng.2018.11.016 [DOI] [Google Scholar]
- Rice, A., Šmarda, P., Novosolov, M., Drori, M., Glick, L., Sabath, N., Meiri, S., Belmaker, J., & Mayrose, I. (2019). The global biogeography of polyploid plants. Nature Ecology & Evolution, 3(2), 265–273. 10.1038/s41559-018-0787-9 [DOI] [PubMed] [Google Scholar]
- Ruas, R. de B, Costa, L. M. S., & Bered, F. (2022). Urbanization driving changes in plant species and communities—A global view. Global Ecology and Conservation, 38, e02243. [Google Scholar]
- Russell, V. L. (2023). emmeans: Estimated marginal means, aka least-squares means. R package version 1.8.4-1. https://CRAN.R-project.org/package=emmeans
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., Tinevez, J. -Y., White, D. J., Hartenstein, V., Eliceiri, K., Tomancak, P., & Cardona, A. (2012). Fiji: An open-source platform for biological-image analysis. Nature Methods, 9(7), 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki, A. M., Maruvka, Y. E., Richmond, P. A., Guillet, M., Shoresh, N., Sorenson, A. L., De, S., Kishony, R., Michor, F., Dowell, R., & Pellman, D. (2015). Polyploidy can drive rapid adaptation in yeast. Nature, 519(7543), 349–352. 10.1038/nature14187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemens, D. H., Duvall-Jisha, J., Jacobs, J., Manthey, J., Haugen, R., & Matzner, S. (2012). Water deficiency induces evolutionary tradeoff between stress tolerance and chemical defense allocation that may help explain range limits in plants. Oikos, 121, 790–800. [Google Scholar]
- Simms, E. L. (2000). Defining tolerance as a norm of reaction. Evolutionary Ecology, 14(4–6), 563–570. 10.1023/a:1010956716539 [DOI] [Google Scholar]
- Simonin, K. A., & Roddy, A. B. (2018). Genome downsizing, physiological novelty, and the global dominance of flowering plants. PLoS Biology, 16(1), e2003706. 10.1371/journal.pbio.2003706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis, D. E., Albert, V. A., Leebens-Mack, J., Bell, C. D., Paterson, A. H., Zheng, C., Sankoff, D., Depamphilis, C. W., Wall, P. K., & Soltis, P. S. (2009). Polyploidy and angiosperm diversification. American Journal of Botany, 96(1), 336–348. 10.3732/ajb.0800079 [DOI] [PubMed] [Google Scholar]
- Soltis, D. E., & Soltis, P. S. (1999). Polyploidy: Recurrent formation and genome evolution. Trends in Ecology & Evolution, 14(9), 348–352. 10.1016/s0169-5347(99)01638-9 [DOI] [PubMed] [Google Scholar]
- Spoelhof, J. P., Soltis, P. S., & Soltis, D. E. (2017). Pure polyploidy: Closing the gaps in autopolyploid research. Journal of Systematics and Evolution, 55(4), 340–352. 10.1111/jse.12253 [DOI] [Google Scholar]
- Stebbins, G. L. (1971). Chromosomal evolution in higher plants. Edward Arnold. [Google Scholar]
- Subramanian, S. K., & Turcotte, M. M. (2020). Preference, performance, and impact of the water‐lily aphid on multiple species of duckweed. Ecological Entomology, 45(6), 1466–1475. 10.1111/een.12932 [DOI] [Google Scholar]
- Tossi, V. E., Martínez Tosar, L. J., Laino, L. E., Iannicelli, J., Regalado, J. J., Escandón, A. S., Baroli, I., Causin, H. F., & Pitta-Álvarez, S. I. (2022). Impact of polyploidy on plant tolerance to abiotic and biotic stresses. Frontiers in Plant Science, 13, 869423. 10.3389/fpls.2022.869423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valladares, F., Sanchez-Gomez, D., & Zavala, M. A. (2006). Quantitative estimation of phenotypic plasticity: Bridging the gap between the evolutionary concept and its ecological applications. Journal of Ecology, 94, 1103–1116. [Google Scholar]
- Van de Peer, Y., Ashman, T. -L., Soltis, P. S., & Soltis, D. E. (2020). Polyploidy: An evolutionary and ecological force in stressful times. The Plant Cell, 33, 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer, Y., Mizrachi, E., & Marchal, K. (2017). The evolutionary significance of polyploidy. Nature Reviews Genetics, 18(7), 411–424. 10.1038/nrg.2017.26 [DOI] [PubMed] [Google Scholar]
- Van Drunen, W. E., & Johnson, M. T. J. (2022). Polyploidy in urban environments. Trends in Ecology & Evolution, 37(6), 507–516. 10.1016/j.tree.2022.02.005 [DOI] [PubMed] [Google Scholar]
- Verrelli, B. C., Alberti, M., Roches, S. D., Harris, N. C., Hendry, A. P., Johnson, M. T. J., Savage, A. M., Charmantier, A., Gotanda, K. M., Govaert, L., Miles, L. S., Rivkin, L. R., Winchell, K. M., Brans, K. I., Correa, C., Diamond, S. E., Fitzhugh, B., Grimm, N. B., Hughes, S., ... Ziter, C. (2022). A global horizon scan for urban evolutionary ecology. Trends in Ecology & Evolution, 37, 1006–1019. [DOI] [PubMed] [Google Scholar]
- Vo, T. M. C., Dao, M. P., & Dao, T. S. (2018). Growth of duckweed upon exposure to aluminum and atrazine in the laboratory conditions. Journal of Vietnamese Environment, 9(2), 106–111. 10.13141/jve.vol9.no2.pp106-111 [DOI] [Google Scholar]
- Walczyk, A. M., & Hersch-Green, E. I. (2023). Investigating the effects of whole genome duplication on phenotypic plasticity: implications for the invasion success of giant goldenrod Solidago gigantea. Oikos, e09990. 10.1111/oik.09990 [DOI]
- Wei, N., Cronn, R., Liston, A., & Ashman, T. -L. (2019). Functional trait divergence and trait plasticity confer polyploid advantage in heterogeneous environments. The New Phytologist, 221(4), 2286–2297. 10.1111/nph.15508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., Du, Z., Liston, A., & Ashman, T. -L. (2020). Genome duplication effects on functional traits and fitness are genetic context and species dependent: Studies of synthetic polyploid Fragaria. American Journal of Botany, 107(2), 262–272. 10.1002/ajb2.1377 [DOI] [PubMed] [Google Scholar]
- Wei, T., & Simko, V. (2021). R package “corrplot”: Visualization of a correlation matrix (Version 0.92). https://github.com/taiyun/corrplot.
- Wood, T. E., Takebayashi, N., Barker, M. S., Mayrose, I., Greenspoon, P. B., & Rieseberg, L. H. (2009). The frequency of polyploid speciation in vascular plants. Proceedings of the National Academy of Sciences of the United States of America, 106(33), 13875–13879. 10.1073/pnas.0811575106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, N., Hu, F., Wu, J., Zhang, W., Wang, M., Zhu, M., & Ke, J. (2018). Characterization of 19 polymorphic SSR markers in Spirodela polyrhiza (Lemnaceae) and cross‐amplification in Lemna perpusilla. Applications in Plant Sciences, 6(5), e01153. 10.1002/aps3.1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S., Stapley, J., Gablenz, S., Boyer, J., Appenroth, K. J., Sree, K. S., Gershenzon, J., Widmer, A., & Huber, M. (2019). Low genetic variation is associated with low mutation rate in the giant duckweed. Nature Communications, 10(1), 1243. 10.1038/s41467-019-09235-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler, P., Sree, K. S., & Appenroth, K. J. (2017). The uses of duckweed in relation to water remediation. Desalination and Water Treatment, 63, 327–342. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying the article and the analysis code are available in the online Supplementary material. The files include:
Supp.Turcotte.et.al.Evol.Letters.pdf (Supplemental figures and tables); Data.Turcotte.et.al.Evol.Letters.csv (CSV file with all raw data); AnalysisCode.Turcotte.et.al.Evol.Letters.R (R script with analyses); and rdpi_matrix.R and RDPI.R (addition dependencies to run the RDPI analysis slightly modified from Ameztegui (2017)).