Abstract
The connection between the physiological control of the ς54-dependent Pu promoter of the TOL plasmid pWW0 of Pseudomonas putida and the stringent response mediated by the alarmone (p)ppGpp has been examined in vivo an in vitro. To this end, the key regulatory elements of the system were faithfully reproduced in an Escherichia coli strain and assayed as lacZ fusions in various genetic backgrounds lacking (p)ppGpp or overexpressing relA. Neither the responsiveness of Pu to 3-methyl benzylalcohol mediated by its cognate activator XylR nor the down-regulation of the promoter by rapid growth were affected in relA/spoT strains to an extent which could account for the known physiological control that governs this promoter. Overexpression of the relA gene [predicted to increase intracellullar (p)ppGpp levels] did, however, cause a significant gain in Pu activity. Since such a gain might be the result of indirect effects, we resorted to an in vitro transcription system to assay directly the effect of ppGpp on the transcriptional machinery. Although we did observe a significant increase in Pu performance through a range of ς54-RNAP concentrations, such an increase never exceeded twofold. The difference between these results and the behavior of the related Po promoter of the phenol degradation plasmid pVI150 could be traced to the different promoter sequences, which may dictate the type of metabolic signals recruited for the physiological control of ς54-systems.
The toluene degradation pathway determined by the TOL plasmid pWW0 of Pseudomonas putida mt-2 is both an enzymatic and regulatory paradigm for the metabolism of recalcitrant compounds in the environment (reviewed in reference 42). The key event in the activation of the whole pathway is the substrate-dependent transcription of the cognate catabolic operons encoded by the plasmid. Expression of the upper-pathway TOL operon for bioconversion of toluene and xylenes into the corresponding carboxylic acids (22) is driven by the ς54-dependent promoter Pu. This promoter is activated at a distance by the enhancer-binding and toluene-responsive protein XylR with the structural assistance of the integration host factor (IHF). In addition, the cells must be in an adequate metabolic status for Pu activity, since an excess of certain carbon sources (10, 11, 24) or rapid growth in rich medium (8, 16, 17, 25, 29) entirely inhibits promoter output in vivo even if the compound to be degraded is present in the culture. This behavior, which is phenomenologically akin to catabolic repression (16, 17, 27, 29), is by no means exclusive to the Pu promoter. Expression of many other biodegradative pathways of Pseudomonas is also inhibited by a number of growth conditions which adjust the activity of specific catabolic promoters to a given metabolic and physiological status (9).
More than one mechanism may cause the down-regulation of Pu in certain growth scenarios. Glucose and other carbohydrates (24) decrease Pu activity through a process involving the ptsN gene (11, 38). However, rapid growth in rich medium also results in a separate negative signal in the system through the control of the activity of ς54 (8). In addition, full activity in vivo of ς54 requires (at least in Escherichia coli) the participation of the FtsH product (6), a protein with both protease and chaperone activities which is involved in the turnover of the heat shock factor ς32 (23) and other regulators. In spite of all these observations, the specific instruments for the physiological control of Pu remain unknown.
A case similar to the Pu promoter of the TOL plasmid is that of the Po promoter of the dimethylphenol (dmp)-degrading pathway of plasmid pVI150 of Pseudomonas sp. strain CF600. Po is a ς54 promoter which is activated at a distance by the phenol-responsive protein DmpR, which has high similarity to XylR (35). Although the sequence of the Pu and Po promoters are different, the upstream activating motifs (UAS) are similar enough to be recognized by both proteins as binding sites (19). On this basis, DmpR and XylR behave more as variants of the same protein than as two distinct proteins. Like Pu, the activation of Po by DmpR is also subjected to a tight metabolic control by certain carbon sources and by the growth rate (47). A recent study by Shingler's laboratory (48) has traced the physiological down-regulation of Po to the need for (p)ppGpp, the signal molecule which triggers the stringent response to amino acid starvation (12). This is a very attractive possibility, since many metabolic conditions are reflected in the intracellular levels of this alarmone molecule (reviewed in reference 12).
The present study was undertaken to examine whether the observed physiological control of the Pu promoter of the TOL plasmid could also be connected to the direct or indirect effects of (p)ppGpp. The results in vivo an in vitro, given below, clearly demonstrate that although Pu activity is indeed stimulated by ppGpp, the effect is insufficient to account for the inhibition of Pu during exponential growth. In addition, our data suggest that the moderate effect of ppGpp on Pu is the result of a direct stimulation of the transcription initiation complex by the alarmone.
MATERIALS AND METHODS
Strains, plasmids, and general procedures.
The strains and plasmids used in this work are listed in Table 1. Recombinant DNA techniques were carried out by published methods (43). All plasmids used in the transcription assays were derived from vector pTE103, which adds a strong T7 terminator downstream of the promoter under study (18). Plasmid pEZ10 carries Pu and has been described previously (7, 36). Plasmid pTE103-Po carries a 481-bp DNA insert spanning positions −471 to +10 of the Po promoter sequence (39). Similarly, plasmid pTE103-PlacUV5 bears an insert with this control promoter between positions −117 and +12. All cloned inserts and DNA fragments were verified through automated DNA sequencing in an Applied Biosystems device. All the supercoiled plasmid DNA templates used for in vitro transcription were prepared with the Qiagen plasmid purification system.
TABLE 1.
Bacterial strains and plasmids used in this study
| E. coli K-12 strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| CC118 | Δ(ara-leu) araD ΔlacX74 galE galK phoA thi-1 | 28 |
| MG1655 | F− λ−, K-12 prototroph | M. Cashel |
| CF1693 | MG1655 ΔrelA251::Km ΔspoT207::Cm | 50 |
| BL21(DE3) | F−ompT gal[dcm] [lon] hsdSB (rB− mB−), λ with the T7 RNA polymerase gene | 46 |
| Plasmids | ||
| pMCP1 | Spr Smr RP4oriT pSP10 oriV, 5.2-kb NotI insert xylR/Pu-lacZ MAD1 (20) | 6 |
| pMCP2 | Spr Smr RP4oriT pSP10oriV, 4.5-kb NotI insert xylRΔA/Pu-lacZ MAD2 (20) | 6 |
| pCNB0118 | Cmr, pSU21 derivative, relA gene, NdeI engineered site at its initiationg codon | 30 |
| pET-19b | Apr, pBR322 derivative; T7/lac promoter expression vector for His-tagged fusions | Novagen, Madison, Wisc. |
| pCNB0209R | Apr, pET-19b derivative with relA inserted in the NdeI site | O. Martínez-Costa and F. Malpartida |
| pTE103 | Apr, transcription template vector with T7 terminator downstream of pUC8 polylinker | 18 |
| pTE103-placUV5 | pTE103 with 129-bp EcoRI-BamHI fragment inserted from pUJ8-lacUV5 (4) | This work |
| pEZ10 | pTE103 with 301-bp EcoRI-BamHI fragment from −208 to +93 of Pu inserted | 36 |
| pTE103-Po | pTE103 with 481-bp EcoRI-BamHI fragment from −471 to +10 of Po inserted | This work |
Growth and induction conditions.
Bacteria were generally grown at 30°C in rich Luria-Bertani (LB) medium (32). M9 minimal medium (43) supplemented with 0.2% succinate was supplemented, where indicated, with 0.2% Casamino Acids. When required, media were also supplemented with 150 μg of ampicillin per ml, 50 μg of streptomycin per ml, 50 μg of spectinomycin per ml, 30 μg of chloramphenicol per ml, and isopropyl-1-thio-β-d-galactopyranoside (IPTG). Promoter activity in vivo was monitored in all cases by assaying the accumulation of β-galactosidase in cells permeabilized with chloroform and sodium dodecyl sulfate (SDS) as described by Miller (32) under the conditions specified in each case. To measure the accumulation of β-galactosidase, overnight cultures of each of the strains under study were diluted twice 50-fold in the same medium to suppress the β-galactosidase accumulated by stationary-phase bacteria. Fresh exponential cells were then further regrown with aeration, and samples were taken at the stages indicated in each experiment. Where needed, the cultures were exposed to saturating vapors of the upper TOL pathway inducer 3-methyl benzylalcohol (1). The β-galactosidase activity values given throughout this paper represent the average of at least three independent experiments in duplicate samples.
Proteins and protein techniques.
Purified factor ς54 and core RNA polymerase (RNAP) from E. coli were the kind gift of B. Magasanik. IHF was obtained from H. Nash. The ς70-RNAP holoenzyme was purchased from Amersham. The XylR variant called XylRΔA is identical to the wild-type protein except for the deletion of its N-terminal module (called the A domain). This variant is fully constitutive and can thus activate transcription from Pu in the absence of any aromatic inducer (20, 36). XylRΔA was purified to apparent homogeneity by metalloaffinity purification of the His-tagged protein as described by Pérez-Martín and de Lorenzo (36). His-tagged RelA in crude extracts was detected as previously described (20). To this end, whole E. coli cells were disrupted in sample buffer containing 2% SDS and 5% β-mercaptoethanol and run in 10% polyacrylamide gels containing SDS (26). Samples were then blotted on a Immobilon-P membrane (Millipore) and probed with a 1:5,000 dilution of an anti-His monoclonal antibody from Clontech. The band corresponding to the protein was identified with anti-mouse immunoglobulin G IgG conjugated to horseradish peroxidase and developed with an H2O2–luminol-luciferin system.
Construction of a His-tagged RelA expression plasmid and purification of the protein.
To obtain a variant of the relA gene of E. coli encoding a product amenable to affinity purification, the NdeI insert of plasmid pCNB0118 (30), which bears the relA sequence, was cloned in His fusion and Plac/lacIq/pT7 expression vector pET19-b (Novagen), to generate plasmid pCNB0209R. Depending on the host strain, the resulting hybrid gene was expressed through the Plac promoter upon addition of IPTG, through the T7 promoter, or through both. This is the case when pCNB0209R was transformed in E. coli BL21(DE3)(pLys) (46), since the strain bears a chromosomal Plac-based system for expression of the T7 polymerase. For purification of the His-tagged RelA protein, an overnight culture of E. coli BL21(DE3) cotransformed with both pLys and pCNB0209R was grown at 37°C in 2YT medium (43), diluted 1:30 in fresh medium, and regrown under the same conditions to an optical density at 600 nm of approximately 0.7. Expression of the His-tagged RelA was then induced by addition of 0.3 mM IPTG, and the culture was given a further incubation for 3 h. The cells were then harvested by centrifugation, washed with ice-cold buffer A (20 mM Tris-acetate [pH 8.5], 5 mM imidazole, 500 mM NaCl, 0.5 mM phenylmethylsulfonyl fluoride), and stored in aliquots of approximately 0.5 g at −70°C. When needed, cells (0.5 g) were suspended in 5 ml of buffer A and disrupted by sonication. After addition of magnesium acetate to 5 mM, the crude lysate was cleared by centrifugation (4,000 × g for 15 min at 4°C). The supernatant was recentrifuged at 25,000 × g for 30 min at 4°C, and the cleared sample was loaded onto a 2-ml Ni2+-nitrilotriacetic acid column (Novagen, Madison, Wis.), which had been precharged with NiSO4 (as recommended by the manufacturer), and equilibrated in A buffer. After the protein sample was loaded, the column was washed with 15 ml of buffer A and then with 20 ml of buffer A containing 60 mM imidazole. Then a 30-ml linear gradient of this metal chelator was run to a final concentration of 1 M imidazole. Fractions (2 ml) were collected at a flow rate of 0.3 ml min−1 and tested for the presence of the desired protein by SDS-polyacrylamide gel electrophoresis analysis and by assays of (p)ppGpp-synthesizing activity (see below). The main peak of His-tagged RelA protein eluted at 250 to 350 mM imidazole. The corresponding fractions were pooled and used for large-scale synthesis of ppGpp. Protein concentrations were determined (5) using bovine serum albumin as the standard. This procedure yielded up to 4 mg of His-tagged RelA per g of cells, with an apparent enzyme purity of ≥98%.
In vitro synthesis and purification of ppGpp.
Preparative-scale synthesis of ppGpp was performed using the His-tagged RelA protein prepared as described above. The standard reaction was carried out at 30°C in a final volume of 5 ml containing 1.5 ml of the His-tagged RelA preparation (stock at 0.1 to 0.3 mg/ml), 2 mM ATP, 2 mM GDP, and 1× buffer RB (50 mM Tris-acetate [pH 8.0], 15 mM magnesium acetate, 60 mM potassium acetate, 30 mM ammonium acetate, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 15% methanol). The progress of the reaction was monitored by ascending thin-layer chromatography on fluorescence-labeled polyethyleneimine cellulose plates, using 1.5 M KH2PO4 (pH 3.4) as chromatographic buffer. When no further increase in the level of ppGpp could be detected (5 to 12 h later), the reaction was stopped by adding formic acid to 1 M. After removal of precipitated protein by centrifugation (2 min at 9,000 × g, the resulting supernatant was diluted at least threefold with 50 mM triethylammonium acetate (pH 7.7) and applied to a 25-ml DEAE-Bio-Gel column (Bio-Rad), previously equilibrated in the same buffer. After being loaded, the column was eluted with 50, 100, and 150 mM triethylammonium acetate (pH 7.7) (25, 25, and 35 ml, respectively) and finally with 150 ml each of 200 and 400 mM triethylammonium acetate (pH 7.7) or with a 300-ml linear gradient of the same buffer to a final triethylammonium acetate concentration of 400 mM. Fractions (5 ml) were collected in each case, and the elution of ppGpp was analyzed by UV spectroscopy and polyethyleneimine thin-layer chromatography. Fractions containing ppGpp were frozen in an ethanol bath, lyophilized, and dissolved in a small volume of water. The ppGpp concentration was determined at 252 nm (ɛ252 = 13,100 M−1 cm−1) (49), and aliquots were frozen and stored at −20°C. The overall yield of ppGpp, calculated on the basis of the GDP initially added to the reaction was ≥80%. The purity of ppGpp (>95%) was assessed by high-performance liquid chromatography analysis in a Waters 600S system fitted with a 996 Photodiode array detector. For analyses, samples were injected into a C18 100 Nucleosil column (4.6 mm by 20 cm) (Sugelabor) and run at a flow rate of 0.7 ml min−1 with a 10-min KH2PO4 (pH 7) linear gradient (5 to 30 mM) in 20 mM tetrabutylammonium bromide–20% methanol (solution A), followed by a 10-min KH2PO4 linear gradient (30 to 100 mM) in solution A and a 5-min isocratic elution with 200 mM KH2PO4 in the same solution.
In vitro transcription assays.
Transcription assays were performed by a published procedure (14). Supercoiled DNA templates were used at 5 nM. Samples (50 μl) of reaction mixtures were placed at 37°C in a buffer consisting of 50 mM Tris-HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 0.1 mM bovine serum albumin, 10 mM dithiothreitol, and 1 mM EDTA. Unless indicated otherwise, each DNA template was premixed with 25 nM core RNAP, 100 nM ς54, 25 nM IHF, and 100 nM XylRΔA. The DNA templates and the proteins were supplemented with purified ppGpp at the concentration indicated in each case and then incubated at 37°C with 4 mM ATP for 20 min to allow open-complex formation. For multiple-round assays, transcription was then initiated by adding a mixture of ATP, CTP, GTP (400 μM each), UTP (50 μM) and 5 μCi of [α-32P]UTP (3,000/mmol). For single-round experiments, heparin (0.1 mg/ml) was added along with the nucleoside triphosphate mixture to prevent reinitiation. After incubation for 10 min at 37°C, the reactions were stopped with an equal volume of a solution containing 50 mM EDTA, 350 nM NaCl, and 0.5 mg of carrier tRNA per ml. Transcription assays with placUV5 templates were carried out with 0.5 nM supercoiled plasmid pTE103-placUV5 mixed with 25 nM ς70-containing E. coli RNAP holoenzyme (Amersham) in the transcription buffer described above and incubated for 15 min at 37°C. The transcription rounds were initiated, as above, by the addition of the same mixture of nucleoside triphosphates, and the mixtures were incubated for 15 min at 37°C. In any case, the mRNA was extracted from the reaction products, precipitated with ethanol, electrophoresed on a denaturing 7 M urea–4% polyacrylamide gel, and visualized by autoradiography. Transcript levels were quantified with a Bio-Rad Molecular Imager FX system.
RESULTS AND DISCUSSION
Reproducing the inhibition of Pu by Casamino Acids in an E. coli reporter system.
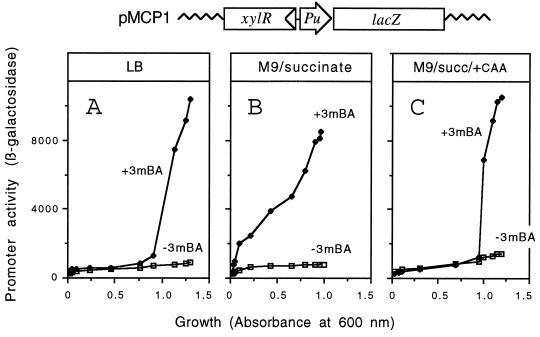
For an adequate genetic assay to examine the physiological conditions that regulate Pu, we employed the low-copy-number plasmid pMCP1 (6), which includes a transcriptional Pu-lacZ fusion and the wild-type xylR gene. As shown in Fig. 1A, this fusion appears to grossly reproduce in E. coli the physiological down-regulation undergone by Pu in rich medium (24), which has been named exponential silencing (8). This is characterized by the lack of significant activity of Pu while cells grow exponentially in LB despite the presence of a XylR effector (such as 3-methyl benzylalcohol) in the medium. In contrast, when the same reporter cells were grown in a minimal mineral medium with a nonrepressive carbon source such as succinate (24), Pu activity was quickly elicited following exposure to the inducer (Fig. 1B). Finally, the presence of 0.2% Casamino Acids in the minimal medium (Fig. 1C) restored the inhibition of Pu during exponential-phase growth of induced cells. The results in Fig. 1 not only validated the use of an E. coli host for the genetic analyses discussed below but also demonstrated the inhibitory effect of an excess of Casamino Acids on the outcome of Pu activity already described in P. putida (29). Since the abundance of amino acids in the medium restrains the stringent response mediated by (p)ppGpp, these results encouraged us to explore the connection between Pu activity and this physiological phenomenon.
FIG. 1.
Physiological control of the Pu promoter of P. putida reproduced in an E. coli host. E. coli strain CC118 harboring plasmid pMCP1 (xylR+/Pu-lacZ), which encodes the wild-type XylR protein, was grown at 30°C in LB rich medium (A), M9 minimal medium with 0.2% succinate (panel B), or M9-succinate medium with 0.2% Casamino Acids (C). At an early growth stage, the cultures were exposed to saturating vapors of the XylR effector 3-methyl benzylalcohol (3mBA), and the accumulation of β-galactosidase activity (expressed in Miller units) was monitored during subsequent growth. The enzymatic activities shown are the average of duplicate samples as explained in the text. Note the repressive effect of Casamino Acids during exponential growth. The organization of the reporter system encoded by pMCP2 is sketched (not to scale) at the top of the figure.
Pu performance in a (p)ppGpp-deficient genetic background.
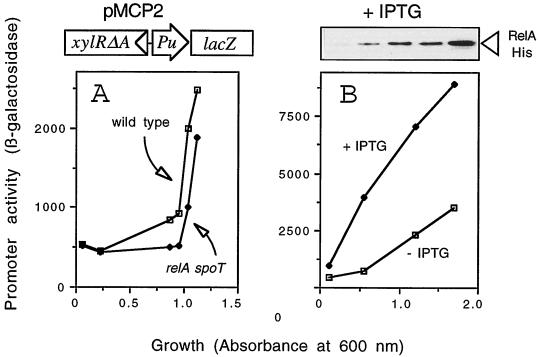
One attractive explanation for the data above described could be that ppGpp is required for Pu performance in vivo. Therefore, the lack of enough intracellular levels of (p)ppGpp brought about by the availability of amino acids in the medium during the earlier stages of growth (12) could inhibit promoter activity. This possibility was easy to test, since production of (p)ppGpp depends of genes relA and spoT in E. coli (50). Should ppGpp be needed for Pu activity, the promoter must become silent in a (p)ppGpp-deficient [(p)ppGpp0] background. To reduce the number of variables, we tested this notion with plasmid pMCP2 as the reporter system. Similarly to pMCP1, pMCP2 also bears a Pu-lacZ fusion, but xylR is present in the form of a truncated gene encoding the variant XylRΔA, with the N-terminal domain deleted. Such a deletion results in the constitutive activity of the protein independently of effector addition (20). XylRΔA makes Pu nonresponsive to aromatic inducers, but the promoter still maintains its metabolic control (8). Therefore, this reporter system reflects the physiological regulation of Pu as a phenomenon different from its activation by XylR inducers. Both the wild-type E. coli MG1655 and its isogenic ΔrelA ΔspoT derivative E. coli CF1693 were transformed with pMCP2, and the accumulation of β-galactosidase was monitored during growth in LB medium. As shown in Fig. 2A, the pattern of LacZ expression in the two strains was very similar, with only relatively minor differences. Not only were β-galactosidase levels comparable, but also Pu displayed the same extent of exponential silencing in the (p)ppGpp0 strain as in its wild-type counterpart. Similar results were obtained when plasmid pMCP1, bearing the wild-type xylR gene, was transformed in the same strains and the cultures were induced by 3-methyl benzylalcohol (data not shown). These results sufficed by themselves to rule out the possibility that the alarmone was the predominant signal which mediates the physiological control of Pu. However, it was also found that the levels of β-galactosidase accumulated by the ΔrelA ΔspoT strain were systematically 20 to 30% lower than those in the ppGpp+ cells; therefore, the signal may be playing a minor role in Pu activity.
FIG. 2.
Pu performance in vivo in the absence of (p)ppGpp or with an excess of the alarmone. (A) The wild-type strain E. coli MG1655 and its ppGpp0 isogenic derivative E. coli CF1693 (relA spoT) harboring the reporter plasmid pMCP2 (xylRΔA+/Pu-lacZ) were grown in LB medium at 30°C and assayed for β-galactosidase activity (expressed in Miller units). The growth rates of the two strains were indistinguishable. There was a moderate decrease in accumulation of the reporter product in the relA spoT host. The reporter system encoded by pMCP2, bearing the constitutive xylR alele xylRΔA, is sketched at the top of the figure. (B) The wild-type strain E. coli MG1655 was cotransformed with pMCP2 (xylRΔA+/Pu-lacZ) and pCNB0209R (expressing a His-tagged RelA product under the control of a lacI/Plac system) (see the text). The cotransformant was grown in LB medium, and the accumulation of β-galactosidase was monitored throughout growth after the addition of 0.1 mM IPTG. The expression levels of the RelA product were revealed by the protein blot, shown at the top of the figure, which was probed with an anti-poly His monoclonal antibody (the last lane corresponds to advanced stationary-phase cells not plotted in the graph). The significant increase in Pu activity upon relA overexpression is evident.
Overexpression of relA improves Pu performance.
To ascertain whether the small effect of (p)ppGpp in Pu detected with the ΔrelA ΔspoT strain could be exacerbated by artificially increasing the intracellular levels of the alarmone, we contransformed compatible plasmids pMCP2 (xylRΔA+/Pu-lacZ) and pCNB0209R in host strain E. coli MG1655. Plasmid pCNB0209R carries a His-tagged variant of the relA gene of E. coli (see Materials and Methods), whose expression is tightly controlled by a lacI/Plac system. Intracellular levels of (p)ppGpp can thus be artificially elevated even in the presence of amino acids by the addition of 0.1 mM IPTG to the medium (44) because of the increased activity of the relA product. Figure 2B shows the course of β-galactosidase accumulation of E. coli MG1655(pMCP2, pCNB0209R) during growth in LB medium with or without the addition of IPTG. It is worth mentioning that under the assay conditions, overproduction of the His-tagged RelA product (as detected with a Western blot assay [Fig. 2B]) did not significantly affect the growth rate. The data in Fig. 2B show that overexpression of RelA, predicted to result in higher intracellular levels of ppGpp, appeared both to increase the overall activity of Pu threefold and to cause an induction pattern devoid of growth stage regulation. These results suggested that the partial dependency of Pu activity on (p)ppGpp indicated by the experiments in Fig. 2 could indeed be genuine, albeit somewhat minor under normal physiological conditions. However, since (p)ppGpp also regulates the intracellular levels of IHF (2) and since the binding sites for this protein are not saturated during exponential growth (34), it is also possible that the (p)ppGpp-dependent increase in Pu activity in vivo reflects an indirect effect on IHF levels. In fact, we have observed that overproduction of IHF also causes an increase of Pu activity and a partial relief of exponential silencing (data not shown). These observations encouraged us to test directly the effect of ppGpp in an in vitro transcription system for Pu as explained below.
ppGpp directly stimulates the ς54-dependent transcriptional machinery of Pu.
To ascertain unequivocally whether the influence of ppGpp on Pu suggested by the data above reflected a direct or an indirect effect, we exploited the in vitro transcription system developed previously in our laboratory for this promoter (7, 36). This includes not only the purified core RNAP, the ς54 factor, and the XylRΔA protein but also the IHF, which is required for the assembly of an optimal geometry of the DNA region (15) and for the recruitment of the enzyme to the promoter sequences (4). In addition, we used the supercoiled DNA template named pEZ10 (36), which contains the promoter region as a 301-bp fragment spanning positions −208 to +93 of Pu, cloned upstream of a strong T7 terminator. We used also the ς70-dependent promoter lacUV5 as a control basically independent of ppGpp (13, 40, 41). This promoter was assembled in the same transcription template as Pu. For the experiments described below, it was of the utmost importance to use preparations of ppGpp which had been purified shortly before use. We had noticed early in this project that this compound is often inactive when shipped from distant origins or after storage for long periods. This problem encouraged us to develop a simplified and very efficient procedure for the production of large amounts of ppGpp. The method, which is described in detail in Materials and Methods, uses an in vitro reaction between ATP and GDP catalyzed by the purified His-tagged RelA protein in solution.
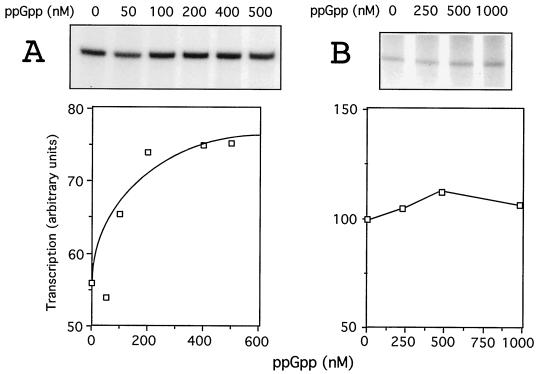
With all the elements under examination in hand, we were finally able to test the effect of ppGpp in a cell-free system. As a first approach, we used the conditions already optimized for the transcription of Pu in vitro (7, 36). To this end, the DNA template was incubated with IHF and XylRΔA, along with subsaturating concentrations of ς54-RNAP and increasing amounts of purified ppGpp. The assays were carried out in the absence of heparin to allow reinitiation; thus, the transcripts were representative of the outcome of multiple transcription rounds. As shown in Fig. 3A, Pu activity was indeed stimulated by increasing the concentrations of ppGpp in the transcription mixtures through the range from 0.05 to 1 μM, with saturation being reached at 0.3 to 0.5 μM. Such an stimulation was, however, moderate, since it never exceeded more than 40 to 60% of the levels of transcripts produced without ppGpp. Under the same conditions, the activity of the control lacUV5 promoter was significantly less affected by ppGpp (Fig. 3B). Although these data provided an explanation for the behavior of Pu in vivo when placed in a strain lacking ppGpp (Fig. 2A), they did not fully account for the very high activity of Pu in vivo upon overproduction of RelA (and thus an increased level of the alarmone [Fig. 2B]). Therefore, some indirect effects of ppGpp on Pu, e.g., an influence on IHF levels (2), are also likely to occur. In addition, the results in Fig. 3 left unanswered two outstanding questions; i.e., how general is the effect of ppGpp in ς54 promoters, and what are the molecular mechanisms involved?
FIG. 3.
Effect of ppGpp on the transcriptional activity of Pu in vitro. (A) Results of multiple-round transcription reactions with 5 nM Pu-containing template pEZ10, 100 nM XylRΔA, 25 nM IHF, 25 nM core RNAP, 100 nM ς54, and increasing concentrations of purified ppGpp as indicated. Under these conditions, pEZ10 produces an mRNA of 394 nucleotides. The levels of transcript found in each case are plotted below the autoradiograph of the gel. The baseline value starts at 50 arbitrary units. (B) Control with the placUV5 promoter. The transcription reaction mixtures contained 5 nM supercoiled pTE103-PlacUV5 template mixed with 25 nM E. coli ς70 RNAP holoenzyme and different amounts of ppGpp. In this system, PlacUV5 produces an mRNA of 313 nucleotides.
Comparison of the responsiveness of Pu and Po to ppGpp.
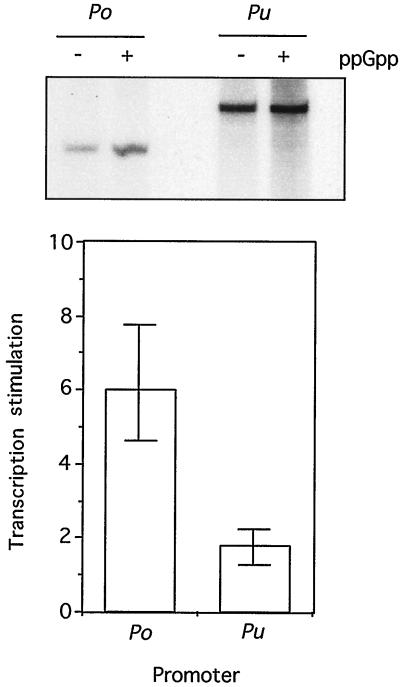
As mentioned above, the ς54 promoter Po drives the expression of a phenol-degrading pathway in Pseudomonas sp. strain CF600 in a fashion very similar to that of Pu for the upper TOL pathway (45, 47). The two promoters have identical UASs for their respective activators, DmpR and XylR (19). Furthermore, we have shown previously (37) that XylRΔA binds the Po enhancer with the same affinity as it binds to the cognate promoter Pu. This is not surprising, since the two proteins have identical recognition surfaces (33) in the helix-turn-helix motif of their DNA-binding D domain (35). This provided an opportunity to examine the effect of ppGpp on a different ς54 template whose only difference was the promoter sequence. To this end, we used the same in vitro transcription setup described above but with the Po sequence from −471 to + 10 as the insert in the pTE103 transcription vector that was used with Pu and with PlacUV5. The result of the experiment is shown in Fig. 4, which gives the data for four independent assays. Under the conditions (300 nM ppGpp) at which we saw the best stimulation of Pu, we detected a dramatic but dissimilar responsiveness of Po to the alarmone. Consistent with the data in Fig. 3A, ppGpp did not increase Pu activity by more than 40 to 60%, an increase based on an already efficient production of transcripts. In contrast, the activity of Po was very low in the absence of ppGpp but was stimulated by four- to eightfold on addition of this compound. These results provided a rationale for the different behavior of Po (48) and Pu in vivo. They also suggested that such differences could be ultimately dictated by specific sequences of each promoter.
FIG. 4.
Comparison of the effects of the ppGpp on the transcriptional activity of Pu and Po in vitro. The result of a typical multiple-round transcription experiment run in parallel with 5 nM Pu-containing template pEZ10 or Po-containing template pTE103-Po is shown at the top of the figure. Assays were carried out under standard conditions (100 nM XylRΔA, 25 nM IHF, 25 nM core RNAP, 100 nM ς54), and 350 nM purified ppGpp was added or omitted as indicated. Note the different sizes of the transcripts arising from Pu (394 nucleotides) or Po (311 nucleotides) due to the different distances from the promoter to the T7 terminator in the vector plasmid (see Materials and Methods). The average stimulation of Pu and Po activity by ppGpp on the basis of four independent experiments is represented below the autoradiograph.
The target of ppGpp in the ς54-dependent transcription machinery.
The mechanism by which ppGpp affects (mostly inhibiting) the performance of many promoters during the stringent response is not yet fully understood and is not devoid of controversy (12, 13). ppGpp seems to bind a distinct site of the β subunit of the ς70 RNAP (13), thereby decreasing the affinity of the sigma factor for the core enzyme (3). Depending on the specific promoter, this results in the enzyme failing to form productive preelongation complexes or to form abortive transcripts (3, 21, 51). However, since ς54 and ς70 are quite different from one another (31), the mechanisms involved in the control of cognate promoters by ppGpp are likely to be dissimilar as well.
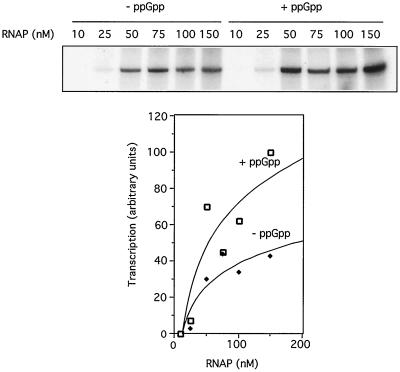
To gain an insight into this issue as it relates to Pu, we performed the experiments in Fig. 5. We tested whether ppGpp had an influence on the formation of the promoter closed complex. To this end, we monitored the effect of ppGpp addition on Pu promoter output under increasing concentrations of ς54-RNAP. If such an effect on promoter binding does occur, the polymerase dose-response curve will be shifted toward the lower concentrations of the enzyme. The results in Fig. 5 clearly indicate that this is not the case here. In addition, we noticed that the affinity of ς54-RNAP for Pu did not change much in response to ppGpp addition, as detected in gel shift assays (M. Carmona, unpublished observations). It thus seems that the effect of the alarmone occurred at a later step in the transcription process. To see whether this step was the formation of the open complex, we used single-round transcription mixtures with ppGpp and increasing amounts of ς54-RNAP but supplemented those mixtures with heparin to avoid reinitiation; therefore, the transcripts reflected the outcome of single transcription rounds. The dose-response curves of the single-round experiments were not dramatically different from those of the multiple rounds in Fig. 5 (data not shown). This further suggested that the effect of ppGpp on Pu occurs at a stage following ς54-RNAP binding, i.e., formation of an open complex and/or elongation. Although the precise mechanism of this phenomenon deserves further investigations, the data above confirm the occurrence of a genuine direct effect of ppGpp in the ς54-dependent transcription machinery.
FIG. 5.
Effect of ppGpp on activation of Pu with increasing concentrations of ς54-RNAP. The upper part of the figure shows the result of multiple-round transcription reactions containing 5 nM Pu-containing plasmid template pEZ10. To this was added 100 nM XylRΔA, 25 nM IHF, and increasing concentrations of ς54-RNAP with or without 300 nM ppGpp as indicated. The intensity of the bands was quantified in a Bio-Rad Molecular Imager FX system and plotted in arbitrary units as a function of RNAP concentration (shown in the lower part of the figure). The effect of ppGpp on the transcriptional output appears to be constant throughout the entire range of enzyme concentrations used, and therefore it does not appear to stimulate binding of the promoter by the ς54-RNAP (see the text for an explanation).
ppGpp contributes to, but does not determine, the physiological control of the Pu promoter.
The work reported above establishes a connection between the behavior of the Pu promoter of the TOL plasmid in vivo and the stringent response mediated by ppGpp. Such an association is, however, relatively minor compared the dramatic dependence of Po, a second ς54-dependent system for which the issue has been examined in detail (48). The differences between the two promoters, Po and Pu, in this respect seem to be authentic even though they share so many genetic elements and physiological behaviors. Although we have not yet made a rigorous comparison between the two systems under all conditions, the published data on Po (48) and the results presented here suggest that most of the physiological down-regulation of Po, but not of Pu, can be attributed to the effect of ppGpp. Since the only major difference between the two promoters is the nucleotide composition of the sequences interspaced within the major shared motifs (UAS, -12/-24), it is tempting to speculate that these sequences may determine the type of metabolic signals which are entered into the promoter. This adds one more nonanticipated degree of regulatory flexibility to ς54-dependent promoters, accounting for their evolutionary success in systems, such as biodegradative pathways, whose expression requires fine-tuning with the overall metabolic status of the cells (9).
ACKNOWLEDGMENTS
We are indebted to V. Shingler for sharing results prior to publication. We gratefully acknowledge B. Magasanik and H. Nash for sending us valuable proteins and C. Montero and M. A. Günther-Sillero for providing technical advice on HPLC analysis.
This research was supported by contracts BIO4-CT97-2040 and QLRT-99-00041 of the EU and by grant BIO98-0808 of the Comisión Interministerial de Ciencia y Tecnología.
REFERENCES
- 1.Abril M A, Michán C, Timmis K N, Ramos J L. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol. 1989;171:6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aviv M, Giladi H, Schreiber G, Oppenheim A B, Glaser G. Expression of the genes coding for the Escherichia coli integration host factor are controlled by growth phase, rpoS, ppGpp and by autoregulation. Mol Microbiol. 1994;14:1021–1031. doi: 10.1111/j.1365-2958.1994.tb01336.x. [DOI] [PubMed] [Google Scholar]
- 3.Bartlett M S, Gaal T, Ross W, Gourse R L. RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrn P1 promoter. J Mol Biol. 1998;279:331–345. doi: 10.1006/jmbi.1998.1779. [DOI] [PubMed] [Google Scholar]
- 4.Bertoni G, Fujita N, Ishihama A, de Lorenzo V. Active recruitment of ς54-RNA polymerase to the Pu promoter of Pseudomonas putida role of IHF and αCTD. EMBO J. 1998;17:5120–5128. doi: 10.1093/emboj/17.17.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–253. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Carmona M, de Lorenzo V. Involvement of the FtsH (HflB) protease in the activity of ς54-promoters. Mol Microbiol. 1999;31:261–270. doi: 10.1046/j.1365-2958.1999.01169.x. [DOI] [PubMed] [Google Scholar]
- 7.Carmona M, de Lorenzo V, Bertoni G. Recruitment of RNA polymerase is a rate-limiting step for the activation of the ς54 promoter Pu of Pseudomonas putida. J Biol Chem. 1999;274:33790–33794. doi: 10.1074/jbc.274.47.33790. [DOI] [PubMed] [Google Scholar]
- 8.Cases I, de Lorenzo V, Pérez-Martín J. Involvement of ς54 in exponential silencing of the Pseudomonas putida TOL plasmid Pu promoter. Mol Microbiol. 1996;19:7–17. doi: 10.1046/j.1365-2958.1996.345873.x. [DOI] [PubMed] [Google Scholar]
- 9.Cases I, de Lorenzo V. Expression systems and physiological control of promoter activity in bacteria. Curr Opin Microbiol. 1998;1:303–310. doi: 10.1016/s1369-5274(98)80034-9. [DOI] [PubMed] [Google Scholar]
- 10.Cases I, de Lorenzo V. Genetic evidence of distinct physiological regulation mechanisms in the ς54Pu promoter of Pseudomonas putida. J Bacteriol. 2000;182:956–960. doi: 10.1128/jb.182.4.956-960.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cases I, Pérez-Martín J, de Lorenzo V. The IIANtr (PtsN) protein of Pseudomonas putida mediates the C source inhibition of the ς54-dependent Pu promoter of the TOL plasmid. J Biol Chem. 1999;274:15562–15568. doi: 10.1074/jbc.274.22.15562. [DOI] [PubMed] [Google Scholar]
- 12.Cashel M, Gentry D R, Hernández V J, Vinella D. The stringent response in Escherichia coli and Salmonella typhimurium. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: American Society for Microbiology; 1996. pp. 1458–1496. [Google Scholar]
- 13.Chaterjii D, Fujita N, Ishihama A. The mediator for stringent control, ppGpp, binds to the beta-subunit of Escherichia coli RNA polymerase. Genes Cells. 1998;3:279–287. doi: 10.1046/j.1365-2443.1998.00190.x. [DOI] [PubMed] [Google Scholar]
- 14.Claverie-Martín F, Magasanik B. Positive and negative effects of DNA bending on activation of transcription from a distant site. J Mol Biol. 1992;227:996–1008. doi: 10.1016/0022-2836(92)90516-m. [DOI] [PubMed] [Google Scholar]
- 15.de Lorenzo V, Herrero M, Metzke M, Timmis K N. An upstream XylR- and IHF-induced nucleoprotein complex regulates the ς54-dependent Pu promoter of TOL plasmid. EMBO J. 1991;10:1159–1167. doi: 10.1002/j.1460-2075.1991.tb08056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duetz W A, Marqués S, de Jong C, Ramos J L, van Andel J G. Inducibility of the TOL catabolic pathway in Pseudomonas putida (pWW0) growing on succinate in continuous culture: evidence of carbon catabolite repression control. J Bacteriol. 1994;176:2354–2361. doi: 10.1128/jb.176.8.2354-2361.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duetz W A, Marqués S, Wind B, Ramos J L, van Andel J G. Catabolite repression of the toluene degradation pathway in Pseudomonas putida harboring pWW0 under various conditions of nutrient limitation in chemostat culture. Appl Environ Microbiol. 1996;62:601–606. doi: 10.1128/aem.62.2.601-606.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elliot T, Geiduschek E P. Defining a bacteriophage T4 late promoter: absence of a −35 region. Cell. 1984;36:211–219. doi: 10.1016/0092-8674(84)90091-6. [DOI] [PubMed] [Google Scholar]
- 19.Fernández S, Shingler V, de Lorenzo V. Cross-regulation by XylR and DmpR activator of Pseudomonas putida suggests that transcriptional control of biodegradative operons evolves independently of catabolic genes. J Bacteriol. 1994;176:5052–5058. doi: 10.1128/jb.176.16.5052-5058.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernández S, de Lorenzo V, Pérez-Martín J. Activation of the transcriptional regulator XylR of Pseudomonas putida by release of repression between functional domains. Mol Microbiol. 1995;16:205–213. doi: 10.1111/j.1365-2958.1995.tb02293.x. [DOI] [PubMed] [Google Scholar]
- 21.Gaal T, Bartlett M S, Ross W, Turnbough C L, Jr, Gourse R L. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 22.Harayama S, Rekik M, Wubbolts M, Rose K, Leppik R, Timmis K N. Characterization of five genes in the upper-pathway operon of TOL plasmid pWW0 from Pseudomonas putida and identification of the gene products. J Bacteriol. 1989;171:5048–5055. doi: 10.1128/jb.171.9.5048-5055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herman C, Thévenet D, D'Ari R, Bouloc P. Degradation of sigma-32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc Natl Acad Sci USA. 1995;92:3516–3520. doi: 10.1073/pnas.92.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holtel A, Marqués S, Möhler I, Jakubzik U, Timmis K N. Carbon source-dependent inhibition of xyl operon expression of Pseudomonas putida TOL plasmid. J Bacteriol. 1994;176:1773–1776. doi: 10.1128/jb.176.6.1773-1776.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hugouvieux N, Köhler T, Rekik M, Harayama S. Growth-phase-dependent expression of the Pseudomonas putida TOL plasmid pWW0 catabolic genes. J Bacteriol. 1990;190:6651–6660. doi: 10.1128/jb.172.12.6651-6660.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.MacGregor C H, Wolff J A, Arora S K, Hyleman P B, Phibbs P V., Jr . Catabolite repression in Pseudomonas aeruginosa. In: Galli E, Silver S, Witholt B, editors. Pseudomonas: molecular biology and biotechnology. Washington, D.C.: American Society for Microbiology; 1991. pp. 198–206. [Google Scholar]
- 28.Manoil C, Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marqués S, Holtel A, Timmis K N, Ramos J L. Transcriptional induction kinetics from the promoters of the catabolic pathways of TOL plasmid pWW0 of Pseudomonas putida for metabolism of aromatics. J Bacteriol. 1994;176:2517–2524. doi: 10.1128/jb.176.9.2517-2524.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martínez-Costa O H, Fernández-Moreno M A, Malpartida F. The relA/spoT-homologous gene in Streptomyces coelicolor encodes both ribosome-dependent (p)ppGpp-synthesizing and -degrading activities. J Bacteriol. 1998;180:4123–4132. doi: 10.1128/jb.180.16.4123-4132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merrick M J. In a class of it own: the RNA polymerase sigma factor ς54 (ςN) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 32.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 33.Morett E, Segovia L. The ς54-bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;178:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murtin C, Engelhorn M, Geiselmann J, Boccard F. A quantitative UV laser footprinting analysis of the interaction of IHF with specific binding sites: re-evaluation of the effective concentration of IHF in the cell. J Mol Biol. 1999;284:949–961. doi: 10.1006/jmbi.1998.2256. [DOI] [PubMed] [Google Scholar]
- 35.North A K, Klose K E, Stedman M, Kustu S. Prokaryotic enhancer-binding proteins reflect eukaryotic-like modularity: the puzzle of nitrogen regulatory protein C. J Bacteriol. 1993;175:4267–4273. doi: 10.1128/jb.175.14.4267-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pérez-Martín J, de Lorenzo V. In vitro activities of an N-terminal truncated form of XylR, a ς54-dependent promoter Pu of Pseudomonas putida. J Mol Biol. 1996;258:575–587. doi: 10.1006/jmbi.1996.0270. [DOI] [PubMed] [Google Scholar]
- 37.Pérez-Martín J, de Lorenzo V. Physical and functional analysis of the prokaryotic enhancer of the ς54-promoters of the TOL plasmid of Pseudomonas putida. J Mol Biol. 1996;258:562–574. doi: 10.1006/jmbi.1996.0269. [DOI] [PubMed] [Google Scholar]
- 38.Postma P W, Lengeler J N, Jacobson R. Phosphoenolpyruvate carbohydrate phosphotransferase system in bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Powlowski J, Shingler V. Genetics and biochemistry of phenol degradation by Pseudomonas sp. CF600. Biodegradation. 1994;5:219–236. doi: 10.1007/BF00696461. [DOI] [PubMed] [Google Scholar]
- 40.Primakoff P. In vivo role of the relA+ gene in regulation of the lac operon. J Bacteriol. 1981;145:410–416. doi: 10.1128/jb.145.1.410-416.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raghavan A, Kameshwari D B, Chatterji D. The differential effects of guanosine tetraphosphate on open complex formation at the Escherichia coli ribosomal protein promoters rplJ and rpsA P1. Biophys Chem. 1998;75:7–19. doi: 10.1016/s0301-4622(98)00185-9. [DOI] [PubMed] [Google Scholar]
- 42.Ramos J L, Marqués S, Timmis K N. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors an plasmid-encoded regulators. Annu Rev Microbiol. 1997;51:341–373. doi: 10.1146/annurev.micro.51.1.341. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Schreiber G, Metzger S, Aizenman E, Roza S, Cashel M, Glaser G. Overexpression of the relA gene in Escherichia coli. J Biol Chem. 1991;266:3760–3767. [PubMed] [Google Scholar]
- 45.Shingler V, Bartilson M, Moore T. Cloning and nucleotide sequence of the gene encoding the positive regulator (DmpR) of the phenol catabolic pathway encoded by pVI150 and identification of DmpR as a member of the NtrC family of transcriptional activators. J Bacteriol. 1993;175:1596–1604. doi: 10.1128/jb.175.6.1596-1604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 47.Sze C, Moore T, Shingler V. Growth-phase dependent transcription of the ς54-dependent Po promoter controlling the Pseudomonas-derived (methyl) phenol dmp operon of pVI150. J Bacteriol. 1996;178:3727–3735. doi: 10.1128/jb.178.13.3727-3735.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sze C C, Shingler V. The alarmone (p)ppGpp mediates physiological-responsive control at the ς54-dependent Po promoter. Mol Microbiol. 1999;31:1217–1228. doi: 10.1046/j.1365-2958.1999.01264.x. [DOI] [PubMed] [Google Scholar]
- 49.Woody A Y M, Woody R W, Malcolm A D B. Effects of ppGpp on transcription by DNA-dependent RNA polymerase from Escherichia coli: circular dichroism, absorption and specific transcription studies. Biochim Biophys Acta. 1987;909:115–125. doi: 10.1016/0167-4781(87)90033-9. [DOI] [PubMed] [Google Scholar]
- 50.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bis-pyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- 51.Zhou Y N, Jin D. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2908–2913. doi: 10.1073/pnas.95.6.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]