Abstract

Research on Parkinson’s disease most often focuses on the ability of the protein α-synuclein (α-syn) to form oligomers and amyloid fibrils, and how such species promote brain death. However, there are indications that α-syn also plays a gene-regulatory role in the cell nucleus. Noncanonical tetrahelical nucleic acids, G-quadruplexes (G4Q), and i-motifs have been shown to play an important role in the control of genomic events. Using the conformation-sensitive single-molecule Förster resonance energy transfer technique we show that monomeric and oligomeric α-syn affect G4Qs and i-motifs in a different way and lead to remodeling of their conformational substates. Aggregated α-syn destabilizes the G4Q leading to unfolding. In contrast, both monomeric and aggregated α-syn enhance folding of the i-motif sequence of telomeric DNA. Importantly, macromolecular crowding is able to partially rescue G4Q from unfolding.
Aggregation and fibrillation of neuronal proteins are one of the hallmarks of neurodegenerative diseases.1,2 The formation of such aggregates induces Parkinson’s disease (PD), Alzheimer’s disease, and multiple system atrophy.3−6 Proteins playing a role in these diseases are prion proteins, tau as well as intrinsically disordered proteins (IDPs) like the small 14 kDa α-synuclein (α-syn), which can induce formation of neurotoxic Lewy bodies or fibrils.7,8 Besides apoptosis, Lewy body aggregates can also reduce formation of dopamine containing vesicles, in that way changing brain activity on a neuronal level.1,9 Nuclear fractions of human dopaminergic neuroblastoma cells contain monomeric and oligomeric forms of α-syn, implicating its significant biochemical role in the nucleus.10−13 Furthermore, monomeric α-syn is known to enhance repairing of double-strand breakage of DNA, potentially rescuing neurons from an apoptotic pathway.7 An aggregation-induced decrease of the α-syn monomer concentration is suspected to raise the death rate of neurons.7 The toxic aggregated form of α-syn detected in PD affected brain was also found to have a higher propensity to interact with DNA.12,14,15 Cherny et al. showed that α-syn interacts with histone-free transcriptionally active DNA segments and reduces transcriptional activity of some genes which respond to environmental stimuli.16 Therefore, studying the conformation of DNAs in the presence of aggregated α-syn has immense importance as the interaction with aggregated α-syn forms can significantly affect DNA replication and transcription along with hastening accumulation of DNA damage.
Noncanonical DNAs play important roles in many biological processes like transcription, translation, and replication.17−19 Among different noncanonical DNAs, guanine-rich (G-rich) sequences, which fold into tetrameric structures, denoted G-quadruplexes (G4Q), have been well characterized in recent years.20−25 On the contrary, cytosine-rich (C-rich) sequences, which form intercalated structures known as i-motifs, are still less explored (Figure 1).21,26 Both structures are observed in the telomeric region of chromosomes.27 Such noncanonical DNA motifs have also been recognized in connection with tumor formation due to their role in promoter regions of different oncogenes.28−31
Figure 1.
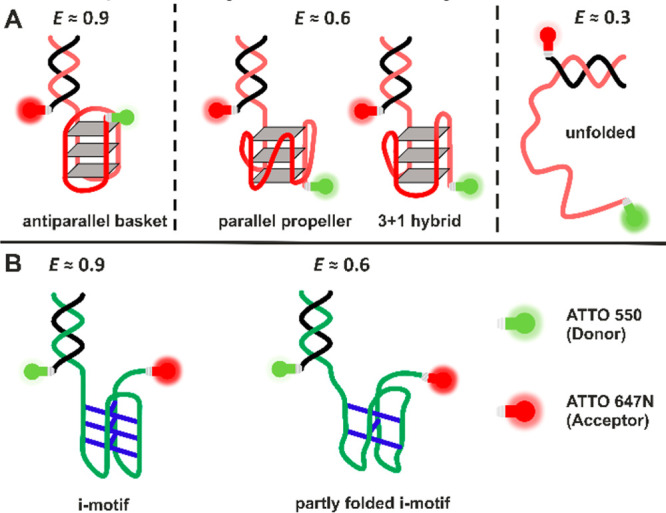
Schematic representation of the different conformations observed for the telomeric G4Q (A) and i-motif (B). G4Qs are stabilized through cyclic Hoogsteen hydrogen-bonding arrangement of 4 guanines with each other and by interactions between cations and the O-6 lone-pair electrons of each guanine. Formation of i-motifs is facilitated by hemiprotonation of cytosines and formation of cytosine–cytosine+ base pairs.17−19
Owing to the lack of a molecular-level understanding of the interaction of α-syn in its native and aggregated state with such noncanonical chromosomal DNA sequences, we carried out corresponding conformation-sensitive single-molecule Förster resonance energy transfer (smFRET) experiments. This method avoids ensemble averaging, which enabled us to elucidate the conformational dynamics of noncanonical DNA structures and how they are affected by the interaction with monomeric and aggregated α-syn. We utilized two different DNA constructs, a telomeric G-quadruplex (hTel G4Q)24,32 of sequence (GGGTTA)3GGG combined with a short double-strand part for the second FRET label, and its complementary i-motif (hTel-i-Mot) of sequence (CCCAAT)3CCC and the same double-strand part for the second FRET label (Figure 1). The α-syn was used at concentrations estimated to be present in brain cells.33
G4Q structures, including hTel, are known to take up several conformations. Depending on monovalent cation type, salt, and osmolyte concentration, as well as on physical parameters like temperature and pressure, antiparallel, parallel, and hybrid conformations have been observed (Figure 1A).24,34,35 Similar studies have been carried out to uncover the conformational space of the hTel-i-Mot sequence, finding a folded i-motif at a low pH of 5, while at neutral pH either random coil or intermediate i-motif structures are suggested (Figure 1B). To mimic cellular crowding effects, measurements have also been carried out in the presence of a macromolecular crowding agent, Ficoll PM 70. For details of sample preparation and technical aspects, please refer to the Supporting Information (SI).
At pH 7.5, the FRET efficiency histograms of the G4Q display two FRET distribution peaks. We assigned the peak at E ≈ 0.9 in the E-histograms to an antiparallel conformation and that at E ≈ 0.6 to a parallel or hybrid conformational state. In the presence of α-syn aggregates, a low FRET efficiency peak emerges at E ≈ 0.3, which corresponds to the unfolded conformation of the G4Q.36,37Figure S1 shows that the conformational states of the G4Q remain unaltered in the presence of monomeric α-syn even up to a concentration as high as 200 μM. On the contrary, in the presence of aggregated α-syn, a drastic change of the E-histogram is observed (Figure 2A), reflecting marked changes in the population of conformational states. Analogous measurements were carried out with a G-quadruplex construct of the c-Myc oncogene promoter region (c-MycG4).21 The effects of monomeric and aggregated α-syn on the parallel c-MycG4 structure were similar, but less prominent, probably owing to its higher stability (Figure S9; for details please refer to the paragraph in the SI). From dynamic light scattering experiments, the size of the oligomers of α-syn added has been determined to vary between about 20 and 400 nm with a maximum at about 130 nm (Figure S3). With increasing concentration of aggregated α-syn, the population of unfolded states (E ≈ 0.3) increases gradually. From 0 to 100 μM aggregated α-syn, the population of unfolded states increases from ∼5% to ∼60% at the expense of the antiparallel conformation whose population is decreasing from ∼80% to ∼20%, keeping the minor population of the parallel conformation (∼25%) essentially unchanged (Figure 2B).
Figure 2.
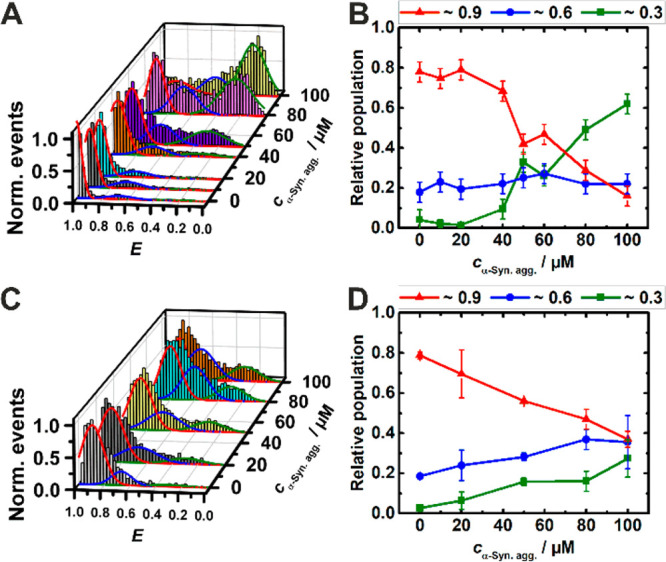
FRET efficiency (E) histogram (A) and relative population (B) of G4Q in buffer with increasing concentration of α-syn aggregates. The G4Q concentration was ∼50 pM. Solutions measured in (C) and (D) contained additional 30 wt % Ficoll. The colors of the relative population of conformers correspond to the Gaussian peaks in the E-histograms (red: antiparallel, blue: parallel, green: unfolded).
As the conformational dynamics of biomolecules may get significantly altered in the crowded in vivo environment of the biological cell when compared to that in dilute buffer medium,37−42 we have also studied the effect of macromolecular crowding on the conformational landscape of these two noncanonical DNA structures in the absence and presence of monomeric and oligomeric α-syn. Figure 2C reveals that the population of conformational states of the G4Q is significantly modulated in the presence of the crowding agent. Addition of 30 wt % Ficoll in the 100 μM α-syn aggregate solution markedly decreases the population of unfolded states of the G4Q, from ∼60% to ∼25%, which is accompanied by a concomitant enrichment of the antiparallel and parallel conformations (Figure 2B and D). This is to say, the crowder is able to largely rescue the folded conformations of the G4Q in the presence of toxic α-syn aggregates.
Generally, telomeric DNA duplexes are found to coexist with G-quaduplexes and i-motifs at acidic pH, and with G-quadruplexes and a (partially or fully) unfolded, random coil-like C-rich strand (C-coil) at neutral pH.42 However, C-rich DNA sequences may also form stable i-motifs at neutral pH in the nuclei and in crowded environments. Quite unexpectedly, compared to the G4Q, the effect of α-syn on the structure of the complementary i-motif sequence differs entirely for both the monomeric and the aggregated state of the protein. Monomeric α-syn induces formation of the i-motif in its folded conformation at pH 7.5, whereas the i-motif is essentially partially folded only in pure buffer at this pH (Figure 3A). The relative population of the folded conformation increases from ∼5% to ∼30% upon addition up to 200 μM monomeric α-syn (Figure 3B). A similar scenario is observed upon addition of the oligomeric protein; here, the conformational transition occurs at much lower concentration (∼100 μM), however (Figure 3C and D). In the presence of the crowding agent, a similar behavior is observed (Figure S2), which implies that α-syn and Ficoll act synergistically in stabilizing the folded structure of the i-motif. Physiological NaCl concentrations lead to no signifiacnt changes of this scenario (see Figures S7 and S8).
Figure 3.
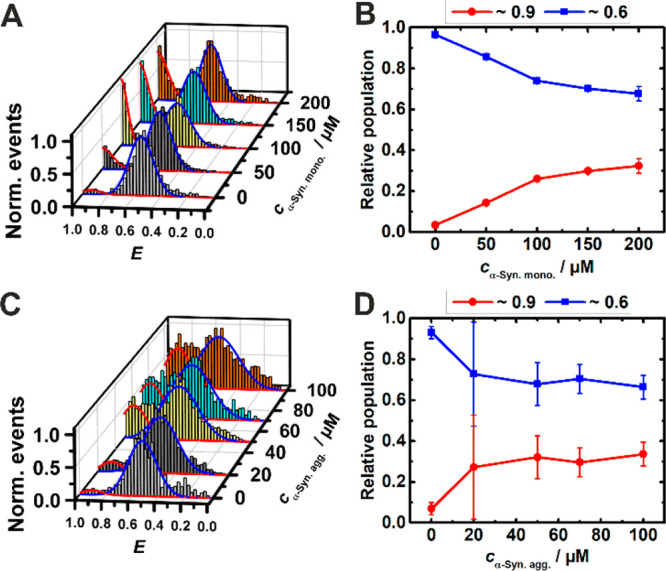
FRET efficiency (E) histogram (A) and relative population (B) of i-motif conformers in buffer with increasing concentration of α-syn monomers. Histogram (C) and relative population (D) of i-motif conformers in buffer with increasing concentration of α-syn aggregates. The i-motif concentration was ∼50 pM. The colors of the relative population of conformers correspond to the Gaussian peaks in the E- histograms (red: fully folded, blue: partly folded).
There is clear evidence that positively charged intrinsically disordered proteins like α-syn can form high-affinity complexes with nucleic acids.44−46 α-Syn is a natively disordered protein under physiological conditions and composed of three distinct regions: an N-terminus that is overall positively charged (+4), a central hydrophobic region so-called NAC (non-Aβ component), which confers the fibrillation potential, and a carboxyl terminus that is highly negatively charged (−12) due to the predominance of negatively charged Asp and Glu residues.47,48 DNA molecules are highly negatively charged on their surface as they are laced with phosphate groups. They can electrostatically interact with the positively charged epsilon amino group of lysine residues located predominantly in the N-terminal and partly in the central region of the α-syn sequence in a nonspecific manner.49 α-Syn aggregates, stabilized by intermolecular β-sheet formation, may have a higher exposed net positive charge, fostering the interaction with DNA.50 Such a view would also be in accordance with the differential binding pattern of HEWL (hen egg white lysozyme) and fibrillar HEWL toward alteration of both DNA and RNA conformations, suggesting also that these nucleic acid–protein interactions are nonspecific in nature.50 Control measurements using monomeric and aggregated HEWL (Figures S7 and S8) are in line with this interpretation. However, the molecular picture of the nature of interactions between such protein aggregates and polyanions is still uncertain.
In the present study, we have seen that monomeric and aggregated α-syn interact quite differently with the different noncanonical DNA structures. At neutral solution conditions, the G4Q is structurally more stable than the i-motif form, which essentially takes on the partially folded state at neutral pH. Thus, monomeric α-syn is unable to alter the conformation of the G4Q but remodels the conformation of the i-motif. However, the codeposition of protein molecules in the form of oligomeric species with a tentatively higher exposed net charge of the aggregated protein ensemble is able to perturb the conformation of G4Q, leading to unfolding of the quadruplex (Figure 4). For such rigid oligomers, the entropy penalty to pay for the interaction with the DNA is also expected to be lower compared to the monomeric α-syn with its random coil-like structure. On the contrary, in the case of the i-motif sequence, α-syn reduces the folding free-energy barrier and shifts the equilibrium toward the folded conformation of the i-motif sequence.
Figure 4.
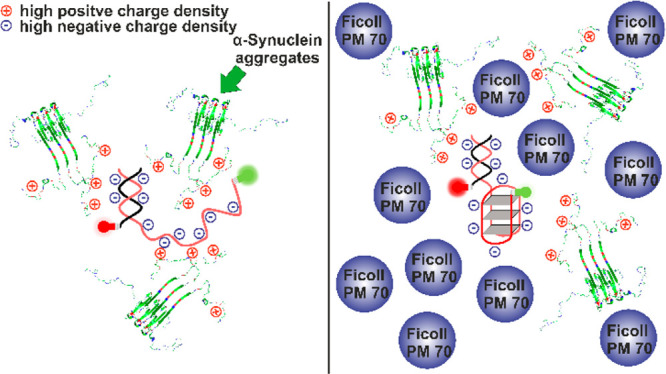
Schematic representation of the interaction of oligomeric α-syn with the telomeric G4Q in the absence (left) and presence (right) of the polymeric crowding agent Ficoll PM 70.
Macromolecular crowding is expected to modulate the energy and conformational landscape of biomolecules, generally favoring more compact conformations by the excluded volume effect.39−41 The extent of exposed solvent accessible surface area is reduced as compared to that in buffer, thereby entropically stabilizing the compact state and shifting the equilibrium toward the folded conformation. High crowder concentrations (30 wt % Ficoll), mimicking the intracellular milieu, result in a strong excluded volume effect, which impacts the conformational dynamics of the noncanonical DNA structures and leads to stabilization of the compact conformation of the G4Q even in the presence of the deteriorating effect of aggregated α-syn. On the other hand, it drives the coiled C-motif to its folded i-motif conformation.
To conclude, for a long time overlooked, α-syn has been found to be present in the cell nucleus, suggesting that it may have additional roles in PD that are related to nuclear gene regulation. Noncanonical DNA structures like G4Qs work closely with several proteins to modulate cellular processes and are known to strongly affect gene expression. Through single-molecule FRET experiments we could show that α-syn at physiologically relevant local concentrations affect single noncanonical DNA structures in a sequence-specific way. Oligomeric α-syn destabilizes the G-quadruplex leading to unfolding. Conversely, the conformational landscape of the i-motif is remodeled, leading to an increase of folded i-motif structures. As G-quadruplexes play a major role as on-and-off switches that modulate polymerase activity, interaction with aggregated α-syn may alter expression profiles of disease-modifying genes with severe pathological consequences.
Acknowledgments
This project received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie Grant Agreement No. 801459 - FP-RESOMUS and was funded by the Deutsche Forschungsgemeinschaft (DFG) under Germany’s Excellence Strategy - EXC 2033-390677874 - RESOLV.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.0c07192.
Experimental procedures of the smFRET, DLS and CD measurements, sample preparation, and further data (PDF)
Author Contributions
‡ J.-M.K. and S.K.M. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Lashuel H. A.; Overk C. R.; Oueslati A.; Masliah E. The many faces of α-synuclein: from structure and toxicity to therapeutic target. Nat. Rev. Neurosci. 2013, 14, 38–48. 10.1038/nrn3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C. A.; Poirier M. A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004, 10, S10–S17. 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Spillantini M. G.; Schmidt M. L.; Lee V. M.; Trojanowski J. Q.; Jakes R.; Goedert M. α-Synuclein in Lewy bodies. Nature 1997, 388, 839–840. 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- Wakabayashi K.; Yoshimoto M.; Tsuji S.; Takahashi H. Alpha-synuclein immunoreactivity in glial cytoplasmic inclusions in multiple system atrophy. Neurosci. Lett. 1998, 249, 180–182. 10.1016/S0304-3940(98)00407-8. [DOI] [PubMed] [Google Scholar]
- Ono K.; Hirohata M.; Yamada M. α-Synuclein Assembly as a Therapeutic Target of Parkinson’s Disease and Related Disorders. Curr. Pharm. Des. 2008, 14, 3247–3266. 10.2174/138161208786404191. [DOI] [PubMed] [Google Scholar]
- Troster A. I. Neuropsychological Characteristics of Dementia with Lewy Bodies and Parkinson’s Disease with Dementia: Differentiation, Early Detection, and Implications for “Mild Cognitive Impairment” and Biomarkers. Neuropsychol. Rev. 2008, 18, 103–119. 10.1007/s11065-008-9055-0. [DOI] [PubMed] [Google Scholar]
- Schaser A. J.; Osterberg V. R.; Dent S. E.; Stackhouse T. L.; Wakeham C. M.; Boutros S. W.; Weston L. J.; Owen N.; Weissman T. A.; Luna E.; Raber J.; Luk K. C.; McCullough A. K.; Woltjer R. L.; Unni V. K. Alpha-synuclein is a DNA binding protein that modulates DNA repair with implications for Lewy body disorders. Sci. Rep. 2019, 9, 1–19. 10.1038/s41598-019-47227-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez V.; Mitra J.; Wang H.; Hegde P. M.; Rao K. S.; Hegde M. L. A multi-faceted genotoxic network of alpha-synuclein in the nucleus and mitochondria of dopaminergic neurons in Parkinson’s disease: Emerging concepts and challenges. Prog. Neurobiol. 2020, 185, 101729. 10.1016/j.pneurobio.2019.101729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh H.; Saleh A.; Yao H.; Cui J.; Shen Y.; Li R. Mini review: linkage between α-Synuclein protein and cognition. Transl. Neurodegener. 2015, 4, 5–10. 10.1186/s40035-015-0026-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Y.; Chase T. N.; Bennett M. C. Muscarinic Receptor Stimulation Induces Translocation of an α-Synuclein Oligomer from Plasma Membrane to a Light Vesicle Fraction in Cytoplasm. J. Biol. Chem. 2001, 276, 28212–28218. 10.1074/jbc.M011121200. [DOI] [PubMed] [Google Scholar]
- Hegde M. L.; Rao K. S. J. Challenges and complexities of α-synuclein toxicity: new postulates in unfolding the mystery associated with Parkinson’s disease. Arch. Biochem. Biophys. 2003, 418, 169–178. 10.1016/j.abb.2003.08.015. [DOI] [PubMed] [Google Scholar]
- Giasson B. I.; Uryu K.; Trojanowski J. Q.; Lee V. M. Mutant and Wild Type Human Alpha-Synucleins Assemble into Elongated Filaments with Distinct Morphologies in Vitro. J. Biol. Chem. 1999, 274, 7619–7622. 10.1074/jbc.274.12.7619. [DOI] [PubMed] [Google Scholar]
- Hegde M. L.; Rao K. S. J. DNA induces folding in α-synuclein: Understanding the mechanism using chaperone property of osmolytes. Arch. Biochem. Biophys. 2007, 464, 57–69. 10.1016/j.abb.2007.03.042. [DOI] [PubMed] [Google Scholar]
- Hegde M. L.; Vasudevaraju P.; Rao K. J. DNA induced folding/fibrillation of alpha-synuclein: new insights in Parkinson’s disease. Front. Biosci., Landmark Ed. 2010, 15, 418–436. 10.2741/3628. [DOI] [PubMed] [Google Scholar]
- Kikuchi A.; Takeda A.; Onodera H.; Kimpara T.; Hisanaga K.; Sato N.; Nunomura A.; Castellani R. J.; Perry G.; Smith M. A.; Itoyama Y. Systemic increase of oxidative nucleic acid damage in Parkinson’s disease and multiple system atrophy. Neurobiol. Dis. 2002, 9, 244–248. 10.1006/nbdi.2002.0466. [DOI] [PubMed] [Google Scholar]
- Cherny D.; Hoyer W.; Subramaniam V.; Jovin T. M. Double-stranded DNA Stimulates the Fibrillation of α-Synuclein in vitro and is Associated with the Mature Fibrils: An Electron Microscopy Study. J. Mol. Biol. 2004, 344, 929–938. 10.1016/j.jmb.2004.09.096. [DOI] [PubMed] [Google Scholar]
- Abou Assi H. A.; Garavís M.; Gonzalez C.; Damha M. J. i-Motif DNA: structural features and significance to cell biology. Nucleic Acids Res. 2018, 46, 8038–8056. 10.1093/nar/gky735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänsel-Hertsch R.; Di Antonio M. D.; Balasubramanian S. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017, 18, 279–284. 10.1038/nrm.2017.3. [DOI] [PubMed] [Google Scholar]
- Rhodes D.; Lipps H. J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y.; Kan Z. Y.; Wang Q.; Yao Y.; Liu J.; Hao Y. H.; Tan Z. Human Telomeric DNA Forms Parallel-Stranded Intramolecular G-Quadruplex in K+ Solution under Molecular Crowding Condition. J. Am. Chem. Soc. 2007, 129, 11185–11191. 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
- Liu L.; Ma C.; Wells J. W.; Chalikian T. V. Conformational preferences of DNA strands from the promotor region of the c-MYC oncogene. J. Phys. Chem. B 2020, 124, 751–762. 10.1021/acs.jpcb.9b10518. [DOI] [PubMed] [Google Scholar]
- Fan Y. H.; Shek Y. L.; Amiri A.; Dubins D. N.; Heerklotz H.; Macgregor R. B. Jr.; Chalikian T. V. Volumetric Characterization of Sodium-Induced G-Quadruplex Formation. J. Am. Chem. Soc. 2011, 133, 4518–4526. 10.1021/ja110495c. [DOI] [PubMed] [Google Scholar]
- Takahashi S.; Sugimoto N. Effect of Pressure on the Stability of G-quadruplex DNA: Thermodynamics Under Crowding Conditions. Angew. Chem., Int. Ed. 2013, 52, 13774–78. 10.1002/anie.201307714. [DOI] [PubMed] [Google Scholar]
- Knop J. M.; Patra S.; Harish B.; Royer C.; Winter R. The Deep Sea Osmolyte Trimethylamine N-Oxide and Macromolecular Crowders Rescue the Antiparallel Conformation of the Human Telomeric G-Quadruplex from Urea and Pressure Stress. Chem. - Eur. J. 2018, 24, 14346–14351. 10.1002/chem.201802444. [DOI] [PubMed] [Google Scholar]
- Di Antonio M.; Ponjavic A.; Radzevičius A.; Ranasinghe R. T.; Catalano M.; Zhang X.; Shen J.; Needham L. M.; Lee S. F.; Klenerman D.; Balasubramanian S. Single-molecule visualization of G-quadruplex in live cells. Nat. Chem. 2020, 12, 832–837. 10.1038/s41557-020-0506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S.; Hossain S. K.S.; Samanta A. Insights into the Folding Pathway of a c-MYC-Promoter-Based i-Motif DNA in Crowded Environments at the Single-Molecule Level. J. Phys. Chem. B 2020, 124, 763–770. 10.1021/acs.jpcb.9b10633. [DOI] [PubMed] [Google Scholar]
- Lipps H. J.; Rhodes D. G-quadruplex structures: In vivo evidence and function. Trends Cell Biol. 2009, 19, 414–422. 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Brooks T. A.; Kendrick S.; Hurley L. Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J. 2010, 277, 3459–3469. 10.1111/j.1742-4658.2010.07759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks T. A.; Hurley L. H. Targeting MYC Expression through G-Quadruplexes. Genes Cancer 2010, 1, 641–649. 10.1177/1947601910377493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X.; Lan W.; Zhang X.; Wu H.; Liu M.; Cao C. Solution structure of all parallel G-quadruplex formed by the oncogene RET promoter sequence. Nucleic Acids Res. 2011, 39, 6753–6763. 10.1093/nar/gkr233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänsel-Hertsch R.; Simeone A.; Shea A.; Hui W. W. I.; Zyner K. G.; Marsico G.; Rueda O. M.; Bruna A.; Martin A.; Zhang X.; Adhikari S.; Tannahill D.; Caldas C.; Balasubramanian S. Landscape of G-quadruplex DNA structural regions in breast cancer. Nat. Genet. 2020, 52, 878–883. 10.1038/s41588-020-0672-8. [DOI] [PubMed] [Google Scholar]
- Tippana R.; Xiao W.; Myong S. G-quadruplex conformation and dynamics are determined by loop length and sequence. Nucleic Acids Res. 2014, 42, 8106–8114. 10.1093/nar/gku464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W.-L.; DeLucia M. W.; Dickson D. W. Synuclein Immunoreactivity in Neuronal Nuclear Inclusions and Neurites in Multiple System Atrophy. Neurosci. Lett. 2004, 354, 99–102. 10.1016/j.neulet.2003.09.075. [DOI] [PubMed] [Google Scholar]
- Lim K. W.; Ng V. C. M.; Martín-Pintado N.; Heddi B.; Phan A. T. Structure of the human telomere in Na+ solution: an antiparallel (2 + 2) G-quadruplex scaffold reveals additional diversity. Nucleic Acids Res. 2013, 41, 10556–10562. 10.1093/nar/gkt771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Villar-Guerra R.; Trent J. O.; Chaires J. B. G-quadruplex secondary structure from circular dichroism spectroscopy. Angew. Chem., Int. Ed. 2018, 57, 7171–7175. 10.1002/anie.201709184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying L.; Green J. J.; Li H.; Klenerman D.; Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 14629–14634. 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tippana R.; Hwang H.; Opresko P. L.; Bohr V. A.; Myong S. Single-molecule imaging reveals a common mechanism shared by G-quadruplex–resolving helicases. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 8448–8453. 10.1073/pnas.1603724113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton L. M.; Barnes C. O.; Li C.; Orans J.; Young G. B.; Pielak G. J. Residue-Level Interrogation of Macromolecular Crowding Effects on Protein Stability. J. Am. Chem. Soc. 2008, 130, 6826–6830. 10.1021/ja8005995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S.; Miyoshi D.; Sugimoto N. Effects of Molecular Crowding on the Structures, Interactions, and Functions of Nucleic Acids. Chem. Rev. 2014, 114, 2733–2758. 10.1021/cr400113m. [DOI] [PubMed] [Google Scholar]
- Ghosh S.; Takahashi S.; Ohyama T.; Endoh T.; Tateishi-Karimata H.; Sugimoto N. Nearest-neighbor parameters for predicting DNA duplex stability in diverse molecular crowding conditions. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 14194–14201. 10.1073/pnas.1920886117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.-X.; Rivas G. N.; Minton A. P. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 2008, 37, 375–397. 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M.; Gnutt D.; Orban A.; Appel B.; Righetti F.; Winter R.; Narberhaus F.; Müller S.; Ebbinghaus S. RNA hairpin folding in the crowded cell. Angew. Chem., Int. Ed. 2016, 55, 3224–3228. 10.1002/anie.201510847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra S.; Schuabb V.; Kiesel I.; Knop J. M.; Oliva R.; Winter R. Exploring the Effects of Cosolutes and Crowding on the Volumetric and Kinetic Profile of the Conformational Dynamics of a Poly dA Loop DNA Hairpin: a Single-molecule FRET Study. Nucleic Acids Res. 2019, 47, 981–996. 10.1093/nar/gky1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A. L.; Watson M.; Wilkins O. G.; Cato L.; Travers A.; Thomas J. O.; Stott K. Highly disordered histone H1–DNA model complexes and their condensates. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, 11964–11969. 10.1073/pnas.1805943115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguado-Llera D.; Goormaghtigh E.; de Geest N.; Quan X.-J.; Prieto A.; Hassan B. A.; Gomez J.; Neira J. L. The Basic Helix-Loop-Helix Region of Human Neurogenin 1 Is a Monomeric Natively Unfolded Protein which Forms a “Fuzzy” Complex upon DNA Binding. Biochemistry 2010, 49, 1577–1589. 10.1021/bi901616z. [DOI] [PubMed] [Google Scholar]
- Holmstrom E. D.; Liu Z.; Nettels D.; Best R. B.; Schuler B. Disordered RNA chaperones can enhance nucleic acid folding via local charge screening. Nat. Commun. 2019, 10, 1–11. 10.1038/s41467-019-10356-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky V. N. A. Protein-Chameleon: Conformational Plasticity of Alpha-Synuclein, a Disordered Protein Involved in Neurodegenerative Disorders. J. Biomol. Struct. Dyn. 2003, 21, 211–234. 10.1080/07391102.2003.10506918. [DOI] [PubMed] [Google Scholar]
- Wu K.-P.; Weinstock D. S.; Narayanan C.; Levy R. M.; Baum J. Structural reorganization of α-synuclein at low pH observed by NMR and REMD simulations. J. Mol. Biol. 2009, 391, 784–796. 10.1016/j.jmb.2009.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S.; Pandey N. K.; Sen S.; Tripathy D. R.; Dasgupta S. Binding of hen egg white lysozyme fibrils with nucleic acids. J. Photochem. Photobiol. B 2013, 127, 52–60. 10.1016/j.jphotobiol.2013.07.015. [DOI] [PubMed] [Google Scholar]
- Calamai M.; Kumita J. R.; Mifsud J.; Parrini C.; Ramazzotti M.; Ramponi G.; Chiti F.; Dobson C. M. Nature and Significance of the Interactions between Amyloid Fibrils and Biological Polyelectrolytes. Biochemistry 2006, 45, 12806–12815. 10.1021/bi0610653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


