Abstract
NDI1 is the unique gene encoding the internal mitochondrial NADH dehydrogenase of Saccharomyces cerevisiae. The enzyme catalyzes the transfer of electrons from intramitochondrial NADH to ubiquinone. Surprisingly, NDI1 is not essential for respiratory growth. Here we demonstrate that this is due to in vivo activity of an ethanol-acetaldehyde redox shuttle, which transfers the redox equivalents from the mitochondria to the cytosol. Cytosolic NADH can be oxidized by the external NADH dehydrogenases. Deletion of ADH3, encoding mitochondrial alcohol dehydrogenase, did not affect respiratory growth in aerobic, glucose-limited chemostat cultures. Also, an ndi1Δ mutant was capable of respiratory growth under these conditions. However, when both ADH3 and NDI1 were deleted, metabolism became respirofermentative, indicating that the ethanol-acetaldehyde shuttle is essential for respiratory growth of the ndi1Δ mutant. In anaerobic batch cultures, the maximum specific growth rate of the adh3Δ mutant (0.22 h−1) was substantially reduced compared to that of the wild-type strain (0.33 h−1). This is consistent with the hypothesis that the ethanol-acetaldehyde shuttle is also involved in maintenance of the mitochondrial redox balance under anaerobic conditions. Finally, it is shown that another mitochondrial alcohol dehydrogenase is active in the adh3Δ ndi1Δ mutant, contributing to residual redox-shuttle activity in this strain.
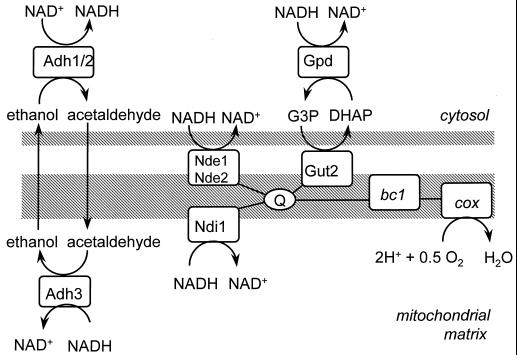
Unlike many other eukaryotes, the yeast Saccharomyces cerevisiae lacks respiratory complex I, which oxidizes mitochondrial NADH and couples this reaction to the generation of a proton motive force. Instead, it has a rotenone-insensitive, non-proton-pumping NADH dehydrogenase. This NADH:ubiquinone oxidoreductase (“internal” NADH dehydrogenase) is encoded by a single gene, NDI1 (6, 17). Ndi1p is localized in the mitochondrial inner membrane, and its active site faces the mitochondrial matrix (17). Mammalian complex I deficiency causes severe cellular disorders, due to impairment of oxidation of NADH by the respiratory chain and to production of superoxide radicals (47). These disorders could be cured successfully by expression of S. cerevisiae NDI1 in complex I-deficient Chinese hamster cells (34). Surprisingly, NDI1 is not essential for respiratory growth of S. cerevisiae on ethanol in shake-flask cultures, even though no residual internal NADH-dehydrogenase activity could be detected in ndi1Δ mutants (17). This observation may be related to the fact that in S. cerevisiae, cytosolic NADH is also a substrate of the respiratory chain. S. cerevisiae harbors two “external” NADH:ubiquinone oxidoreductases, encoded by NDE1 and NDE2 (16, 25, 36). At the protein level, Nde1p and Nde2p reveal 48 and 46% identity to Ndi1p, respectively. Nde1p and Nde2p are also localized in the inner mitochondrial membrane, but their active sites face the cytosol (Fig. 1). Alternatively, oxidation of cytosolic NADH can be coupled to the respiratory chain via the glycerol-3-phosphate shuttle (Fig. 1) (12). In order for the external NADH dehydrogenases and/or the glycerol-3-phosphate shuttle to take over the role of Ndi1p, the redox equivalents of NADH must be shuttled from the mitochondrial matrix to the cytosol, since NADH itself does not readily cross the membrane (45).
FIG. 1.
A scheme of the respiratory chain of Saccharomyces cerevisiae. Adh, alcohol dehydrogenase; bc1, bc1 complex; cox, cytochrome c oxidase; Gpd, soluble glycerol-3-phosphate dehydrogenase; Gut2, membrane-bound glycerol-3-phosphate dehydrogenase; Nde, external NADH dehydrogenase; Ndi1, internal NADH dehydrogenase; Q, ubiquinone; G3P, glycerol-3-phosphate; DHAP, dihydroxy acetone phosphate.
Mammalian cells have redox shuttles, such as the malate-aspartate shuttle (1, 5), that shuttle glycolytic NADH from the cytosol to the mitochondrial matrix, where it can be oxidized by complex I. In S. cerevisiae, in vivo activity of the malate-aspartate shuttle has not been demonstrated unambiguously, but the required enzymes are present (19, 20, 37, 42). Even if a malate-aspartate shuttle is active in S. cerevisiae, it could not explain the growth of the ndi1Δ mutant on ethanol, since it shuttles redox equivalents from the cytosol to the mitochondrial matrix but not in the reverse direction. This is due to the fact that the aspartate-glutamate transporter, one of the components of the shuttle, is driven by the proton motive force (5).
An alternative, reversible redox shuttle, proposed in the literature, is the ethanol-acetaldehyde shuttle (23, 45), consisting of mitochondrial and cytosolic isoenzymes of alcohol dehydrogenase (Fig. 1). Since ethanol and acetaldehyde can diffuse freely across biological membranes, the net result of the ethanol-acetaldehyde shuttle would be the exchange of NADH and H+ for NAD+. S. cerevisiae has at least two cytosolic isoenzymes of alcohol dehydrogenase, encoded by ADH1 and ADH2 (14), and one mitochondrial isoenzyme, encoded by ADH3 (48). In vivo activity of an ethanol-acetaldehyde shuttle, however, has not been demonstrated. Adh1p and Adh2p have other roles as well. Adh1p is primarily involved in alcoholic fermentation (4, 15), while Adh2p has a much higher affinity for ethanol (9) and is mainly involved in consumption of ethanol (4). The physiological function of ADH3 has not been investigated until now. Therefore, the most specific way of investigating the putative ethanol-acetaldehyde shuttle is by deleting ADH3.
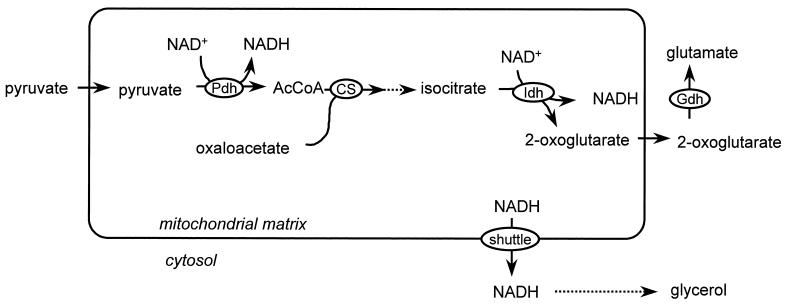
If the ethanol-acetaldehyde shuttle is indeed active in S. cerevisiae mitochondria, it is unlikely to be physiologically important in wild-type cells under aerobic conditions, since S. cerevisiae contains an internal as well as two external NADH dehydrogenases. Under anaerobic conditions, however, a mitochondrial redox shuttle may be essential, as shown by metabolic flux analysis (23). Anaerobically growing S. cerevisiae cells contain only a few large, branched mitochondria (44). Under these conditions the mitochondria do not play a major role in free-energy metabolism. They are essential, however, since important assimilatory reactions are localized in the mitochondria (10, 43). Mitochondrial NADH is generated not only in the tricarboxylic acid (TCA) cycle but also in some of these assimilatory reactions. A major source of mitochondrial NADH is the synthesis of glutamate (Fig. 2) (23). To restore the mitochondrial redox balance under anaerobic conditions, this NADH must be transported to the cytosol, where it can be oxidized by formation of glycerol (38). So far, no experimental evidence is available to confirm a role of the ethanol-acetaldehyde shuttle in anaerobic growth.
FIG. 2.
Proposed physiological role of the ethanol-acetaldehyde redox shuttle under anaerobic conditions (23). In the biosynthesis of amino acids, mitochondrial NADH is generated. Under anaerobic conditions, this NADH may be reoxidized after being shuttled to the cytosol, via formation of glycerol. Pdh, pyruvate-dehydrogenase complex; CS, citrate synthase; Idh, isocitrate dehydrogenase; Gdh, glutamate dehydrogenase.
The aims of the present study were to investigate the physiology of an S. cerevisiae mutant lacking the internal NADH dehydrogenase Ndi1p, the in vivo functioning of the ethanol-acetaldehyde redox shuttle, and its physiological importance during anaerobic growth.
MATERIALS AND METHODS
Yeast strains and maintenance.
The S. cerevisiae strains used in this study are listed in Table 1. Strains were grown to stationary phase in shake-flask cultures on yeast extract-peptone-dextrose medium, containing 10 g of Bacto yeast extract · liter−1, 20 g of peptone (from casein) · liter−1, and 20 g of glucose · liter−1. After addition of sterile glycerol to a final concentration of 20% (vol/vol), 2-ml aliquots were stored in sterile vials at −80°C. These frozen stocks were used to inoculate precultures.
TABLE 1.
S. cerevisiae strains used in this studya
| Strain | Relevant genotype |
|---|---|
| CEN.PK113-7D | MATa URA3 HIS3 LEU2 TRP1 MAL2-8c SUC2 |
| CEN.PK209-1B (ndi1Δ) | MATa URA3 HIS3 LEU2 TRP1 MAL2-8c SUC2 ndi1(81,1420)::loxP-kanMX4-loxP |
| CEN.PK226-1D (adh3Δ) | MATa URA3 HIS3 LEU2 TRP1 MAL2-8c SUC2 adh3(41,1100)::loxP-kanMX4-loxP |
| CEN.PK289-2B (adh3Δ ndi1Δ) | MATa URA3 HIS3 LEU2 TRP1 MAL2-8c SUC2 adh3(41,1100)::loxP-kanMX4-loxP ndi1(81,1420)::loxP-kanMX4-loxP |
The numbers in parentheses indicate the deleted nucleotides (ATG = 1) of the corresponding genes. All strains listed can be obtained from EUROSCARF (European S. cerevisiae Archive for Functional Analysis) at http://www.rz.uni-frankfurt.de/FB/fb16/mikro/euroscarf/index.html.
Construction of null mutants.
Haploid S. cerevisiae null mutants were constructed by replacing the gene(s) of interest with a kanamycin resistance gene (the kanMX module) according to the PCR-based method of Wach et al. (46) as described previously (16). Mating type and replacement of genes by the kanMX module were verified by PCR as described previously. The primers that were used for deletion (S1 and S2) and verification (A1, A4, K1, and K2) are listed in Table 2.
TABLE 2.
Oligonucleotides used for construction of disruption cassettes (S1 and S2) and as primers for analytical PCR of deletion mutants (A1/K1 and A4/K2)a
| Open reading frame | Disruption cassette | Oligonucleotide |
|---|---|---|
| YMR083w/ADH3 | S1 | 5′-ATGTTGAGAACGTCAACATTGTTCACCAGGCGTGTCCAACCAGCTGAAGCTTCGTACGC-3′ |
| YMR083w/ADH3 | S2 | 5′-CGTAACACGCTATTATTTACTAGTATCGACGACGTATCTAGCATAGGCCACTAGTGGATCTG-3′ |
| YMR083w/ADH3 | A1 | 5′-TGCATACTGCCTATTGTCGG-3′ |
| YMR083w/ADH3 | A4 | 5′-CAGCTCATCACCTTGGATCG-3′ |
| YML120c/NDI1 | S1 | 5′-CCTCGACGAATACGCTAGTCAGATTCGCTTCCACCAGATCCAGCTGAAGCTTCGTACGC-3′ |
| YML120c/NDI1 | S2 | 5′-AGAATCATGGACAAGTACAAAATTCTCCATAAGTAGAAGGGCATAGGCCACTAGTGGATCTG-3′ |
| YML120c/NDI1 | A1 | 5′-TGCGTAGGAGGCAGGACC-3′ |
| YML120c/NDI1 | A4 | 5′-CGTCAGCGAGTTATCGTCC-3′ |
| kanMX | K1 | 5′-GGATGTATGGGCTAAATGTACG-3′ |
| kanMX | K2 | 5′-GTTTCATTTGATGCTCGATGAG-3′ |
Cultivation of S. cerevisiae.
Aerobic, glucose-limited chemostat cultivation was performed as described previously (16) at 30°C and pH 5.0. The glucose concentration in the medium reservoir was 7.5 g · liter−1. The cultures were sparged with air at a rate of 0.5 liter · min−1. Cultures were assumed to be at steady state if after at least five volume changes, the biomass density, the CO2 production rate, the O2 consumption rate, and the rate of production of the most important metabolites differed by less than 2% on two consecutive days and if no oscillations of the dissolved oxygen concentration were detectable.
Precultures for batch cultivation were grown to the end of the exponential phase in 500-ml shake flasks containing 100 ml of mineral medium with vitamins (41) and 1% (wt/vol) glucose. The pH was set at 6.0. These cultures were used to inoculate precultures containing 1% glucose at an optical density at 660 nm of 0.1. At exponential phase, these cultures were used to inoculate the final batch cultures.
Aerobic and anaerobic batch cultivations were performed at 30°C in 7-liter laboratory fermentors (Applikon, Schiedam, The Netherlands) at a stirrer speed of 800 rpm. The working volume was 4 liters. The pH was kept at 5.0 by an Applikon ADI 1030 biocontroller via the automatic addition of 2 M KOH or 2 M H2SO4. Aerobic cultures were sparged with air at a flow rate of 4 liters · min−1, controlled by a Brooks 5850S mass-flow controller. Anaerobic cultures were flushed with nitrogen gas (99.999% pure; <5 ppm of O2) at a flow rate of 0.5 liter · min−1. To minimize oxygen diffusion into the cultures, the anaerobic fermentors were fitted with Norprene tubing. The concentration of dissolved oxygen was measured with a Mettler Toledo polarographic electrode and remained above 50% of air saturation in aerobic cultures and below the detection limit in anaerobic cultures. Mineral medium with vitamins and 20 g of glucose · liter−1 was prepared as described by Verduyn et al. (41) but with 0.1 ml of silicone antifoam agent per liter of aerobic medium and 0.15 ml of silicone antifoam agent per liter of anaerobic medium. Anaerobic cultures were supplemented with Tween 80 and ergosterol (40).
Measurement of metabolic fluxes.
CO2 production, O2 consumption, and culture dry weight were measured as described previously (16), except that the sample size for dry-weight determination was adjusted (10 to 50 ml) so as to have always 10 to 30 mg (dry weight) per sample. In chemostat cultures, no significant differences were found between dry-weight samples taken directly from the culture and those taken from the effluent line. The glucose concentration in batch cultures was measured enzymatically with a hexokinase/glucose-6-phosphate dehydrogenase kit (Boehringer Mannheim). Acetate was also determined enzymatically (Boehringer Mannheim). Other metabolites, including glucose in the reservoir medium of chemostat cultures, were determined by high-pressure liquid chromatography on a Waters 2690 separation module with an Aminex HPX-87A column from Bio-Rad at 60°C. The column was eluted with 0.5 g of sulfuric acid · liter−1 at a flow rate of 0.6 ml · min−1. Organic acids were detected by a Waters 2487 dual λ absorbance detector at 214 nm. Ethanol, glycerol, and glucose were detected by a Waters 2410 refractive index detector. Enzymatic analysis of ethanol (40) and glycerol (Boehringer Mannheim) was performed to confirm high-pressure liquid chromatography analysis.
Isolation of mitochondria and measurement of oxygen consumption.
The isolation of mitochondria has been described in detail (16) and proceeded essentially as follows. Cell walls were degraded by treatment with Zymolyase. Spheroplasts were disrupted by subjecting them to 10 strokes in a cooled Potter-Elvehjem homogenizer in hypotonic medium. Cytosolic and mitochondrial fractions were separated by differential centrifugation. When mitochondria were used for oxygen consumption measurements, they were spun down gently for 10 min at 7,800 × g; if they were used for enzyme localization studies, they were spun down at 31,000 × g.
Oxygen consumption by isolated mitochondria was measured as described previously (16) at 30°C with a Clark-type oxygen electrode. Reactions were started with ethanol (5 mM), succinate (5 mM), l-glycerol 3-phosphate (5 mM), or l-malate plus pyruvate (5 mM concentrations of each). NADH was generated in situ by addition of 5 mM glucose, 0.2 mM NAD+, and 1.5 U of Bacillus megaterium glucose dehydrogenase (Sigma) · ml−1, because commercial NADH preparations are contaminated with ethanol (16, 28). Oxygen uptake rates were calculated based on a dissolved oxygen concentration of 236 μM in air-saturated buffer at 30°C. Respiratory control values were determined by addition of 0.25 mM ADP (3). Protein concentrations of mitochondrial preparations were measured according to the Lowry method and corrected for bovine serum albumin that was added to the buffer.
Measurement of enzyme activities.
Glucose-6-phosphate dehydrogenase activity was measured according to the method of Postma et al. (29).
NAD+-linked isocitrate dehydrogenase activity was measured according to the method of Bruinenberg et al. (2).
Alcohol dehydrogenase activity in mitochondrial preparations was measured spectrophotometrically with an assay modified from that of Postma et al. (29). The assay mixture contained 0.1 M potassium phosphate, pH 7.0, and 1 mM NAD+. The reaction was started by addition of 100 mM ethanol or 25 mM pentanol. A distinction was made between the activity inside the mitochondria and that outside or adhering to the mitochondria. To measure the activity outside the mitochondria, isolated mitochondria were osmotically stabilized by addition of 0.65 M sorbitol. NADH oxidation by intact mitochondrial membranes, which would interfere with the alcohol dehydrogenase assay, was inhibited by addition of 1 mM KCN. The activity of alcohol dehydrogenase towards ethanol oxidation is much higher at pH 9, which was used in the original assay (29), but at this pH the mitochondria are damaged and internal and external activities cannot be distinguished. Therefore, a suboptimal pH of 7.0 was used. To measure the internal alcohol dehydrogenase activity, mitochondria were disrupted subsequently by addition of 0.1% Triton X-100. It was verified that both ethanol and NAD+ were required to measure any alcohol dehydrogenase activity. No background activity of NADH oxidase was detectable with either intact or disrupted mitochondria.
RESULTS
Ndi1p is the only internal NADH dehydrogenase.
In S. cerevisiae grown in shake-flask cultures on ethanol as the sole carbon source, Ndi1p is the only functional internal NADH dehydrogenase (17). Since ethanol is a nonfermentable substrate, S. cerevisiae exhibits necessarily respiratory growth under these conditions. This is not the case in shake-flask cultures on glucose, due to oxygen limitation and repression of respiratory enzymes by excess glucose, which lead to alcoholic fermentation (8, 11). In aerobic, glucose-limited cultures at low dilution rates, however, S. cerevisiae exhibits respiratory growth. Since the residual glucose concentration is low under these conditions, repression of respiratory enzymes is relieved. Whether Ndi1p is also the only internal NADH dehydrogenase in the strain used in this study, CEN.PK113-7D, when grown aerobically under glucose-limited conditions, was investigated.
Mitochondria were isolated from aerobic, glucose-limited chemostat cultures grown at a dilution rate of 0.10 h−1. Under these conditions wild-type cells did not exhibit alcoholic fermentation and the ndi1Δ mutant produced only little ethanol (0.09 mmol · g [dry weight]−1 · h−1). The physiological characteristics of these cultures are discussed in more detail in the following paragraph. Mitochondria isolated from ndi1Δ cultures oxidized external NADH and l-glycerol 3-phosphate at a rate similar to that of mitochondria isolated from wild-type cultures (Table 3). Succinate oxidation by ndi1Δ mitochondria even appeared to be elevated relative to that by wild-type mitochondria (Table 3). Mitochondria isolated from ndi1Δ cultures, however, oxidized neither malate plus pyruvate nor ethanol at a detectable rate (Table 3). When the latter substrates are metabolized by mitochondria, NADH is generated in the mitochondrial matrix and oxidized by the internal NADH dehydrogenase. Therefore, this result confirmed that the ndi1Δ strain also lacked significant internal-NADH-dehydrogenase activity when grown in aerobic, glucose-limited cultures.
TABLE 3.
Oxygen consumption rates of mitochondria isolated from S. cerevisiae CEN.PK113-7D (wild type) and the ndi1Δ mutanta
| Substrate | CEN.PK113-7D (NDI1)
|
CEN.PK209-1B (ndi1Δ)
|
||
|---|---|---|---|---|
| O2 consumption rate (μmol · mg of protein−1 · min−1) | RC | O2 consumption rate (μmol · mg of protein−1 · min−1) | RC | |
| NADH (pure) | 0.22 ± 0.06 | 3.0 ± 0.3 | 0.27 ± 0.06 | 2.5 ± 0.2 |
| Malate + pyruvate | 0.11 ± 0.01 | 1.7 ± 0.1 | <0.01 | |
| Ethanol | 0.10 ± 0.02 | 1.4 ± 0.1 | <0.01 | |
| l-Glycerol 3-phosphate | 0.18 ± 0.04 | 2.3 ± 0.1 | 0.22 ± 0.04 | 1.9 ± 0.2 |
| Succinate | 0.10 ± 0.02 | 1.6 ± 0.1 | 0.26 | 1.7 |
Mitochondria were isolated from steady-state, aerobic, glucose-limited chemostat cultures at a dilution rate of 0.10 h−1 at 30°C and at pH 5.0. The glucose concentration in the reservoir medium was 7.5 g · liter−1. Values are averages obtained from two independent mitochondrial isolations plus or minus standard deviations (ςn-1), except for the rate of succinate oxidation by ndi1Δ mitochondria, which was measured once. Data for the wild-type strain are from reference 16 and were obtained according to the same procedures. Oxygen consumption rates were measured in the presence of ADP. The respiratory control (RC) is the oxygen consumption rate in the presence of ADP divided by that in the absence of ADP.
In vivo redox shuttle activity in ndi1Δ cultures.
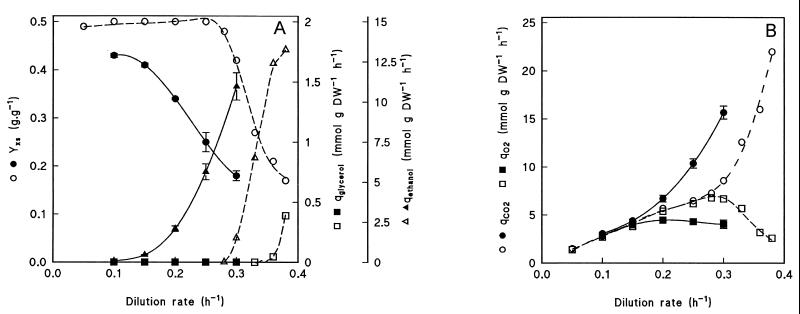
Wild-type and ndi1Δ strains were grown in aerobic, glucose-limited chemostat cultures at a dilution rate of 0.10 h−1. In steady-state chemostat cultures the dilution rate equals the specific growth rate, μ. Wild-type S. cerevisiae did not exhibit alcoholic fermentation at low dilution rates, as can be concluded from the absence of ethanol in culture supernatants, the high biomass yield on glucose (0.49 g · g−1), and a respiratory quotient (i.e., the ratio of the specific rates of CO2 production and oxygen consumption) close to 1 (Fig. 3 and Table 4). At a dilution rate of 0.10 h−1, the biomass yield of the ndi1Δ strain (0.43 g · g−1) was slightly lower than that of the wild type, but only 0.089 mmol of ethanol was produced per g (dry weight) per h, and the respiratory quotient was close to 1 (Fig. 3 and Table 4), indicating that growth of the ndi1Δ mutant was almost completely respiratory. For comparison, anaerobic cultures, which grow fully fermentatively, have a biomass yield on glucose of 0.10 g · g−1 and produce between 8 and 8.5 mmol of ethanol per g (dry weight) per h at the same dilution rate (39, 40).
FIG. 3.
Physiology of S. cerevisiae CEN.PK209-1B (ndi1Δ) and CEN.PK113-7D (wild type) in aerobic, glucose-limited chemostat cultures. The steady-state biomass yield on glucose (YXS) and the specific rates of ethanol (qethanol), glycerol (qglycerol), CO2 (qCO2), and O2 (qO2) production were measured for the ndi1Δ mutant (solid lines, closed symbols) and for the wild-type CEN.PK113-7D (dashed lines, open symbols) at various dilution rates. At dilution rates of 0.35 h−1 or higher, the ndi1Δ strain washed out of the cultures and no steady state was reached. DW, dry weight.
TABLE 4.
Growth and product formation of S. cerevisiae CEN.PK113-7D (wild type) and the isogenic adh3Δ, ndi1Δ, and adh3Δ ndi1Δ mutants in aerobic, glucose-limited chemostat cultures at a dilution rate of 0.10 h−1a
| Parameter | CEN.PK113-7D (ADH3 NDI1) | CEN.PK209-1B (ADH3 ndi1Δ) | CEN.PK226-1D (adh3Δ NDI1) | CEN.PK289-2B (adh3Δ ndi1Δ) |
|---|---|---|---|---|
| Biomass yield (g of biomass · g of glucose−1) | 0.49 ± 0.01 | 0.43 ± 0.00 | 0.48 ± 0.01 | 0.29 ± 0.01 |
| qO2 (mmol · g [DW]−1 · h−1) | 2.7 ± 0.0 | 2.9 ± 0.1 | 2.9 ± 0.1 | 2.7 ± 0.1 |
| qCO2 (mmol · g [DW]−1 · h−1) | 2.8 ± 0.1 | 3.1 ± 0.1 | 2.9 ± 0.0 | 3.9 ± 0.1 |
| Respiratory quotient (qCO2/qO2) | 1.01 ± 0.03 | 1.07 ± 0.01 | 1.02 ± 0.03 | 1.46 ± 0.13 |
| qethanol (mmol · g [DW]−1 · h−1) | ND | 0.089 ± 0.007 | ND | 1.1 ± 0.2 |
| qacetate (mmol · g [DW]−1 · h−1) | ND | 0.007 ± 0.001 | ND | 0.039 ± 0.00 |
Values are averages obtained from two independent, steady-state cultures, plus or minus standard deviations (ςn-1). ND, not detectable. q indicates a specific rate of production (for CO2, ethanol, and acetate) or consumption (O2). DW, dry weight.
Completely respiratory growth of S. cerevisiae on glucose requires the activity of the TCA cycle. Usually the TCA cycle is thought to take place inside the mitochondria and, in that case, all NADH would be produced in the mitochondrial matrix. However, cytosolic isoenzymes of isocitrate dehydrogenase (13) and malate dehydrogenase (19) have been identified in S. cerevisiae. Furthermore, although the pyruvate-dehydrogenase complex is confined to the mitochondria, there is a cytosolic bypass via pyruvate decarboxylase, acetaldehyde dehydrogenase, and acetyl-coenzyme A synthetase (30). 2-Oxoglutarate dehydrogenase, however, is an exclusively mitochondrial enzyme in S. cerevisiae (32), implying that at least in this reaction, intramitochondrial NADH is produced. For respiratory growth of the ndi1Δ mutant, it is required that this NADH be shuttled to the cytosol, where it can be oxidized by the external NADH dehydrogenases or by the glycerol-3-phosphate shuttle.
Adh3p is involved in the reoxidation of mitochondrial NADH in the ndi1Δ mutant.
Our hypothesis was that the respiratory growth of the ndi1Δ mutant is due to the activity of the putative ethanol-acetaldehyde shuttle (45), which transfers redox equivalents from the mitochondria to the cytosol. To test this hypothesis, adh3Δ and adh3Δ ndi1Δ deletion mutants were constructed and grown in aerobic, glucose-limited chemostat cultures. The growth characteristics of the adh3Δ mutant were almost identical to those of the wild-type strain (Table 4). In contrast, the adh3Δ ndi1Δ strain exhibited a reduced biomass yield on glucose, an increased ethanol production, and an increased respiratory quotient compared to both the adh3Δ and the ndi1Δ mutants (Table 4), indicative of respirofermentative growth. Apparently the mitochondrial isoenzyme of alcohol dehydrogenase Adh3p is involved in a pathway that can take over the role of Ndi1p. This demonstrates that Adh3p is involved in the shuttling of mitochondrial NADH to the cytosol, where it can be reoxidized by the external NADH dehydrogenases, Nde1p and Nde2p (Fig. 1). Although the adh3Δ ndi1Δ double mutant converted a large part of the glucose to ethanol, its biomass yield on glucose (0.29 g · g−1) was still much higher than that of a purely fermentative culture (0.10 g · g−1). Therefore, enzymes other than Adh3p may also be capable of shuttling NADH to the cytosol.
Redox shuttles cannot fully replace Ndi1p at high growth rates.
In chemostat cultures, the dilution rate, which at steady state equals the specific growth rate, can be varied. Above, it was shown that NDI1 was not essential for respiratory growth at low dilution rates. The question arises of whether the ethanol-acetaldehyde shuttle can also sustain high rates of respiratory glucose dissimilation. To investigate this, the ndi1Δ mutant was cultivated at increasing growth rates and, consequently, at increasing rates of glucose dissimilation.
When the wild-type strain was cultivated in aerobic, glucose-limited cultures, growth was completely respiratory at dilution rates below 0.30 h−1. At a dilution rate of 0.30 h−1, metabolism became respirofermentative, as indicated by the onset of ethanol production and the decreasing biomass yield (Fig. 3). When the ndi1Δ mutant was grown under the same conditions, a gradual shift from respiratory to fermentative growth was observed above a dilution rate of 0.10 h−1 (Fig. 3), showing that NDI1 is required to maintain a completely respiratory growth up to a dilution rate of 0.30 h−1. Apparently, at high specific growth rates, redox shuttles do not have a sufficient capacity to shuttle all mitochondrial NADH that would be formed during completely respiratory growth into the respiratory chain.
Mitochondrial alcohol dehydrogenase Adh3p is important for anaerobic growth on glucose.
Intramitochondrial NADH is formed not only during respiratory pyruvate dissimilation but also during the biosynthesis of amino acids (Fig. 2). Under anaerobic conditions, this biosynthetic NADH cannot be reoxidized by the respiratory chain. Nissen et al. (23) proposed that under anaerobic conditions an ethanol-acetaldehyde shuttle transfers excess mitochondrial NADH to the cytosol, where the redox balance can be restored by production of glycerol (38). To test this hypothesis, wild-type and adh3Δ cells were cultivated in aerobic and anaerobic batch fermentors on glucose.
Under aerobic conditions, deletion of ADH3 affected neither the maximum specific growth rate nor the biomass yield (Table 5). The specific rate of ethanol production also was not strongly affected by the deletion. Other by-products were produced at low rates, which were only slightly influenced by deletion of ADH3. Apparently, the respiratory chain was sufficiently active to oxidize any excess mitochondrial NADH.
TABLE 5.
Growth and product formation of S. cerevisiae CEN.PK113-7D (wild type) and the isogenic adh3Δ mutant in exponentially growing aerobic and anaerobic batch cultures on glucosea
| Parameter | Results in aerobic culture for:
|
Results in anaerobic culture for:
|
||
|---|---|---|---|---|
| CEN.PK113-7D (ADH3) | CEN.PK226-1D (adh3Δ) | CEN.PK113-7D (ADH3) | CEN.PK226-1D (adh3Δ) | |
| Maximum specific growth rate (h−1) | 0.38 ± 0.01 | 0.37 ± 0.01 | 0.33 ± 0.02 | 0.22 ± 0.02 |
| Biomass yield (g of biomass · g of glucose−1) | 0.14 ± 0.00 | 0.15 ± 0.00 | 0.10 ± 0.00 | 0.10 ± 0.01 |
| Rate of consumption (mmol · g [DW]−1 · h) of glucose | ||||
| Rate of production (mmol · g [DW]−1 · h) of: | 15.7 ± 0.7 | 13.8 ± 0.5 | 19.1 ± 1.1 | 12.6 ± 0.5 |
| Ethanol | 22.3 ± 0.9 | 19.4 ± 0.9 | 28.8 ± 0.6 | 19.2 ± 0.5 |
| Glycerol | 0.97 ± 0.21 | 0.56 ± 0.03 | 3.50 ± 0.60 | 2.34 ± 0.25 |
| Acetate | 0.52 ± 0.08 | 0.74 ± 0.11 | 0.32 ± 0.04 | 0.27 ± 0.01 |
| Lactate | 0.12 ± 0.01 | 0.10 ± 0.00 | 0.32 ± 0.01 | 0.14 ± 0.04 |
| Pyruvate | 0.12 ± 0.01 | 0.13 ± 0.03 | 0.11 ± 0.01 | 0.12 ± 0.02 |
| Succinate | <0.1 | <0.1 | 0.12 ± 0.03 | 0.14 ± 0.07 |
Strains were grown at 30°C, pH 5.0, and the initial glucose concentration in the medium was 20 g · liter−1. The concentration of dissolved oxygen remained above 50% of air saturation in aerobic cultures and below the detection limit in anaerobic cultures. The cultures experienced a lag phase of at most 2 h and then grew exponentially for 8 to 11 h (depending on the growth rate) almost until depletion of glucose. Values are averages obtained from two independent cultures, plus or minus standard deviations (ςn-1). DW, dry weight.
Under anaerobic conditions, the maximum specific growth rate of the adh3Δ mutant was substantially lower than that of the wild-type strain (Table 5). The biomass yield on glucose was not affected by the mutation. The formation rates of the major fermentation products, ethanol and glycerol, decreased proportionally to the maximum specific growth rate. Consequently, the amounts of these metabolites produced per amount of biomass formed were identical in the two strains, and the distribution of the main fluxes was not changed by the mutation.
These results support the hypothesis that the ethanol-acetaldehyde shuttle is important for the reoxidation of mitochondrial NADH under anaerobic conditions but not under aerobic conditions.
Residual mitochondrial alcohol dehydrogenase activity in the adh3Δ ndi1Δ strain.
Above, the importance of the ethanol-acetaldehyde redox shuttle under anaerobic conditions and in a mutant lacking the NDI1 gene was demonstrated. It remained elusive, however, why the biomass yield of the adh3Δ ndi1Δ strain growing aerobically at a dilution rate of 0.10 h−1 was higher than that of a completely fermentative culture (i.e., why growth of this strain was still partially respiratory) and why the adh3Δ strain was growing at all under anaerobic conditions. These results strongly suggest that an additional redox shuttle across the mitochondrial membrane is active. This may be a completely different shuttle or another mitochondrial alcohol dehydrogenase. The latter possibility was tested. Mitochondria were isolated from a steady-state, aerobic, glucose-limited adh3Δ ndi1Δ culture. The cytosolic marker enzyme, glucose-6-phosphate dehydrogenase, was found exclusively in the cytosolic (supernatant) fraction. Of the mitochondrial marker, NAD+-linked isocitrate dehydrogenase, 82% was recovered in the mitochondrial (pellet) fraction and 22% was recovered in the cytosolic fraction, giving an overall recovery of 104%. The mitochondrial fraction contained only 1% of the total cellular activity of ethanol dehydrogenase, while 104% was found in the cytosolic fraction, giving an overall recovery of 105%. From this result it cannot be concluded that there is intramitochondrial alcohol dehydrogenase activity in the adh3Δ ndi1Δ strain, since the mitochondrial fraction may be contaminated with cytosolic alcohol dehydrogenases. Therefore, a distinction was made between the intramitochondrial and extramitochondrial activity in the mitochondrial fraction. First, the mitochondria were subjected to an additional washing step. Subsequently, alcohol dehydrogenase activity was measured in the presence of sorbitol to stabilize the mitochondria. This represents the activity outside of the mitochondria and possibly some activity released by broken mitochondria. Finally, Triton X-100 was added to release the truly intramitochondrial activity. Mitochondria isolated from an aerobic, glucose-limited culture of the ndi1Δ strain released 1.09 μmol of ethanol dehydrogenase · min−1 · mg of protein−1 after addition of Triton X-100 (Table 6). Mitochondria from the adh3Δ ndi1Δ strain released 0.31 μmol · min−1 · mg of protein−1 (Table 6), still 28% of the activity released from the ndi1Δ mitochondria. It was verified that this result was not due to an activation of the enzyme by Triton X-100. In fact, when alcohol dehydrogenase activity was released via disruption of the mitochondria by sonication in the absence of sorbitol, the enzyme activity was somewhat inhibited by Triton X-100 (data not shown). With pentanol as the substrate, the ndi1Δ mitochondria released a small but clearly detectable alcohol dehydrogenase activity (Table 6), consistent with the fact that Adh3p has a high affinity for pentanol (9). The adh3Δ ndi1Δ mitochondria had lost all pentanol-dependent activity (Table 6), indicating that the unknown mitochondrial alcohol dehydrogenase is specific for ethanol.
TABLE 6.
Intramitochondrial alcohol dehydrogenase activity in the ndi1Δ and adh3Δ ndi1Δ strainsa
| Time of measurement | Alcohol dehydrogenase activity (μmol · min−1 · mg of mitochondrial protein−1)
|
|||
|---|---|---|---|---|
|
ndi1Δ with:
|
adh3Δ ndi1Δ with:
|
|||
| Ethanol | Pentanol | Ethanol | Pentanol | |
| Before addition of Triton X-100 | 0.08 ± 0.03 | <0.01 | 0.09 ± 0.02 | <0.01 |
| After addition of Triton X-100 | 1.17 ± 0.04 | 0.06 ± 0.01 | 0.40 ± 0.01 | <0.01 |
| Latency | 1.09 ± 0.03 | 0.06 ± 0.01 | 0.31 ± 0.01 | <0.01 |
Mitochondria were isolated from a steady-state, aerobic chemostat culture at a dilution rate of 0.10 h−1 and washed once by resuspending and centrifuging. The alcohol dehydrogenase activity outside of or adhering to the mitochondria was measured in the presence of sorbitol and KCN, with either ethanol or pentanol as the substrate. Subsequently, in the same assay, intramitochondrial activity was released by addition of Triton X-100. Results in this table were obtained with one preparation of mitochondria from each strain. Essentially the same results were obtained with an independent preparation of mitochondria from another culture.
DISCUSSION
Respiratory growth of the ndi1Δ mutant. This paper constitutes the first experimental evidence for in vivo functioning of an ethanol-acetaldehyde redox shuttle across the mitochondrial membrane, as first proposed by Von Jagow and Klingenberg (45). This shuttle is responsible for the hitherto unexplained ability of an ndi1Δ mutant of S. cerevisiae to exhibit respiratory growth (17). The ndi1Δ mutant lacked the internal NADH dehydrogenase, which couples the oxidation of mitochondrial NADH to the respiratory chain.
Even though at a low dilution rate growth of the ndi1Δ mutant was essentially respiratory, its biomass yield on glucose (0.43 g · g−1) was somewhat lower than that of the wild-type strain (0.49 g · g−1). This may be due to redirection of pyruvate metabolism via the pyruvate-dehydrogenase bypass. In contrast to pyruvate dehydrogenase, which produces mitochondrial NADH, the bypass converts pyruvate to acetyl-coenzyme A via cytosolic pyruvate decarboxylase, acetaldehyde dehydrogenase, and acetyl-coenzyme A synthetase (30). Cytosolic NADPH is then produced instead of mitochondrial NADH. The reaction catalyzed by acetyl-coenzyme A synthetase requires ATP, which should lead to a slight decrease of the biomass yield. In agreement with this, the biomass yield on glucose of an S. cerevisiae mutant lacking pyruvate dehydrogenase activity altogether was 0.44 g · g−1 (31).
Physiological relevance of the ethanol-acetaldehyde shuttle under anaerobic conditions.
Under anaerobic conditions, growth of the adh3Δ mutant was slower than that of the wild-type strain. This is consistent with a role for the ethanol-acetaldehyde shuttle in the transport of mitochondrial NADH formed in biosynthetic reactions to the cytosol. The question arises whether there are other explanations for the slower anaerobic growth of the adh3Δ mutant. It is possible that Adh3p is required to remove toxic acetaldehyde from the mitochondria. This is not very likely, however, since acetaldehyde diffuses across biological membranes and the cytosolic alcohol dehydrogenase Adh1p can remove it. The more likely explanation is that Adh3p is required to reoxidize mitochondrial NADH. Under anaerobic conditions, S. cerevisiae grows completely fermentatively. The cytosolic alcohol dehydrogenase Adh1p then works in the direction of ethanol production. This precludes the functioning of a full shuttle (Fig. 1). Therefore, the mitochondrial ethanol produced by Adh3p must be secreted. The acetaldehyde that is used by the mitochondrial alcohol dehydrogenase has to be generated in the cytosol by pyruvate decarboxylase. To maintain a closed cytosolic redox balance, one glycerol molecule must be formed for each molecule of acetaldehyde entering the mitochondria. In this way, the use of mitochondrial alcohol dehydrogenase couples the reoxidation of intramitochondrial NADH to glycerol formation, which is the primary redox sink in anaerobic S. cerevisiae cultures (38).
Other mitochondrial redox shuttles in S. cerevisiae.
If Ndi1p and Adh3p were the only enzymes that oxidize mitochondrial NADH, it might be expected that the adh3Δ ndi1Δ mutant should grow completely fermentatively under aerobic conditions, or not at all, and that the adh3Δ mutant should not grow under anaerobic conditions. Yet the adh3Δ ndi1Δ mutant exhibited respirofermentative growth in glucose-limited chemostat cultures, and the adh3Δ mutant could still grow under anaerobic conditions, albeit more slowly than the wild type. There are several possible explanations for these observations.
First, we have shown a residual mitochondrial alcohol dehydrogenase activity in the adh3Δ ndi1Δ mutant. The yeast genome contains several alcohol dehydrogenase homologues of which neither the function nor the localization is known (18, 24). According to the program PSORT II (21, 22), four of these, Yal060w, Yal061w, Adh4, and Ydl114w, have a substantial probability (30 to 45%) of being mitochondrial.
Apart from other mitochondrial alcohol dehydrogenases, there may be other types of redox shuttles. Most of the well-studied redox shuttles work in the reverse direction, shuttling NADH from the cytosol to the mitochondrial matrix (5). Recently it was suggested that the malate-oxaloacetate shuttle could export NADH from the mitochondrial matrix to the cytosol (26). All components of this shuttle are present in S. cerevisiae: a mitochondrial and a cytosolic malate dehydrogenase (19, 37) and an oxaloacetate transporter across the mitochondrial inner membrane, Oac1p, which can catalyze the electroneutral exchange of oxaloacetate for malate (27). During respirofermentative growth of the adh3Δ ndi1Δ mutant, however, the malate-oxaloacetate shuttle can work only in the direction of import of NADH into the mitochondria. This is due to the fact that mitochondrial malate dehydrogenase is part of the TCA cycle and operates in the direction of production of oxaloacetate and NADH under these conditions. In anaerobically growing S. cerevisiae, mitochondrial malate dehydrogenase has been proposed to work in the reverse direction (23) and, under these conditions, it might contribute to the export of NADH from the mitochondria.
Finally, a possible explanation for the anaerobic growth of the adh3Δ mutant is the existence of an alternative route of glutamate production which is not coupled to formation of intramitochondrial NADH. The cytosolic pyruvate dehydrogenase bypass has been mentioned. Another source of mitochondrial NADH is NAD+-linked isocitrate dehydrogenase. This may be circumvented by the use of the NADP+-linked isoenzyme, Idp1p, which is sufficient for growth of S. cerevisiae without glutamate (49). Yet even if glutamate synthesis could proceed completely in the cytosol, it is only one of several processes contributing to the mitochondrial NADH synthesis (23), and it remains to be seen whether mitochondrial NADH production can be avoided altogether.
Relevance of the ethanol-acetaldehyde shuttle for higher eukaryotes.
The ethanol-acetaldehyde shuttle is special, because it is reversible and only driven by a gradient of the [NADH]/[NAD+] ratio across the mitochondrial membrane. Most mitochondrial redox shuttles described in the literature transport NADH into the mitochondria and are not readily reversible, since active transport steps are involved (5). Since export of NADH from the mitochondria is important for anaerobic growth, it may be speculated that the ethanol-acetaldehyde shuttle is of more general importance for eukaryotes that can survive long-term hypoxia. A prerequisite is the presence of mitochondrial and cytosolic alcohol dehydrogenase and the ability to synthesize acetaldehyde via pyruvate decarboxylase. Examples of organisms that survive under hypoxic conditions by alcoholic fermentation are goldfish (Carassius auratus) (35) and rice (Oryza sativa) (7, 33). It remains to be investigated whether they depend on a functional ethanol-acetaldehyde shuttle for anaerobic survival.
ACKNOWLEDGMENTS
This work was financially supported by the Dutch Ministry of Economic Affairs (EET program).
We thank Simon de Vries, Karin Overkamp, and our colleagues at DSM Bakery Ingredients for stimulating discussions. We are grateful to Bird Engineering BV, Schiedam, The Netherlands, for a gift of alcohol oxidase used in colorimetric alcohol assays.
REFERENCES
- 1.Borst P. Hydrogen transport and transport metabolites. In: Karlson P, editor. Funktionelle und morphologische Organisation der Zelle. Berlin, Germany: Springer-Verlag; 1963. pp. 137–162. [Google Scholar]
- 2.Bruinenberg P M, Van Dijken J P, Scheffers W A. An enzymic analysis of NADPH production and consumption in Candida utilis. J Gen Microbiol. 1983;129:965–971. doi: 10.1099/00221287-129-4-965. [DOI] [PubMed] [Google Scholar]
- 3.Chance B, Williams G R. Adv. Enzymol. 1956;17:65–134. doi: 10.1002/9780470122624.ch2. [DOI] [PubMed] [Google Scholar]
- 4.Ciriacy M. Alcohol dehydrogenases. In: Zimmermann F K, Entian K-D, editors. Yeast sugar metabolism. Lancaster, Pa: Technomic Publishing Company; 1997. pp. 213–223. [Google Scholar]
- 5.Dawson A G. Oxidation of cytosolic NADH formed during aerobic metabolism in mammalian cells. Trends Biochem Sci. 1979;4:171–176. [Google Scholar]
- 6.De Vries S, Van Witzenburg R, Grivell L A, Marres C A M. Primary structure and import pathway of the rotenone-insensitive NADH-ubiquinone oxidoreductase of mitochondria from Saccharomyces cerevisiae. Eur J Biochem. 1992;203:587–592. doi: 10.1111/j.1432-1033.1992.tb16587.x. [DOI] [PubMed] [Google Scholar]
- 7.Fan T W, Higashi R M, Lane A N. Monitoring of hypoxic metabolism in superfused plant tissues by in vivo1H NMR. Arch Biochem Biophys. 1986;251:674–678. doi: 10.1016/0003-9861(86)90377-2. [DOI] [PubMed] [Google Scholar]
- 8.Gancedo J M. Yeast carbon catabolite repression. Microbiol Mol Biol Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganzhorn A J, Green D W, Hershey A D, Gould R M, Plapp B V. Kinetic characterization of yeast alcohol dehydrogenases. Amino acid residue 294 and substrate specificity. J Biol Chem. 1987;262:3754–3761. [PubMed] [Google Scholar]
- 10.Gbelská Y, Subík J, Svoboda A, Goffeau A, Kovác L. Intramitochondrial ATP and cell functions: yeast cells depleted of intramitochondrial ATP lose the ability to grow and multiply. Eur J Biochem. 1983;130:281–286. doi: 10.1111/j.1432-1033.1983.tb07148.x. [DOI] [PubMed] [Google Scholar]
- 11.Lagunas R. Misconceptions about the energy metabolism of Saccharomyces cerevisiae. Yeast. 1986;2:221–228. doi: 10.1002/yea.320020403. [DOI] [PubMed] [Google Scholar]
- 12.Larsson C, Påhlman I L, Ansell R, Rigoulet M, Adler L, Gustafsson L. The importance of the glycerol 3-phosphate shuttle during aerobic growth of Saccharomyces cerevisiae. Yeast. 1998;14:347–357. doi: 10.1002/(SICI)1097-0061(19980315)14:4<347::AID-YEA226>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 13.Loftus T M, Hall L V, Anderson S L, McAlister-Henn L. Isolation, characterization, and disruption of the yeast gene encoding cytosolic NADP-specific isocitrate dehydrogenase. Biochemistry. 1994;16:9661–9667. doi: 10.1021/bi00198a035. [DOI] [PubMed] [Google Scholar]
- 14.Lupiañez J A, Machado A, Nunez De Castro I, Mayor F. Succinic acid production by yeasts grown under different hypoxic conditions. Mol Cell Biochem. 1974;3:113–116. doi: 10.1007/BF01659183. [DOI] [PubMed] [Google Scholar]
- 15.Lutsdorf U, Megnet R. Multiple forms of alcohol dehydrogenase in Saccharomyces cerevisiae. Arch Biochem Biophys. 1968;126:933–944. doi: 10.1016/0003-9861(68)90487-6. [DOI] [PubMed] [Google Scholar]
- 16.Luttik M A H, Overkamp K M, Kötter P, De Vries S, Van Dijken J P, Pronk J T. The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH. J Biol Chem. 1998;273:24529–24534. doi: 10.1074/jbc.273.38.24529. [DOI] [PubMed] [Google Scholar]
- 17.Marres C A M, De Vries S, Grivell L A. Isolation and inactivation of the nuclear gene encoding the rotenone-insensitive internal NADH:ubiquinone oxidoreductase of mitochondria from Saccharomyces cerevisiae. Eur J Biochem. 1991;195:857–862. doi: 10.1111/j.1432-1033.1991.tb15775.x. [DOI] [PubMed] [Google Scholar]
- 18.Mewes H W, Albermann K, Bahr M, Frishman D, Gleissner A, Hani J, Kleine K, Maierl A, Oliver S G, Pfeiffer F, Zollner A. Overview of the yeast genome. Nature. 1997;387:7–65. doi: 10.1038/42755. [DOI] [PubMed] [Google Scholar]
- 19.Minard K I, McAlister-Henn L. Isolation, nucleotide sequence analysis, and disruption of the MDH2 gene from Saccharomyces cerevisiae: evidence for three isozymes of yeast malate dehydrogenase. Mol Cell Biol. 1991;11:370–380. doi: 10.1128/mcb.11.1.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morin P J, Subramanian G S, Gilmore T D. AAT1, a gene encoding a mitochondrial aspartate aminotransferase in Saccharomyces cerevisiae. Biochim Biophys Acta. 1992;1171:211–214. doi: 10.1016/0167-4781(92)90124-i. [DOI] [PubMed] [Google Scholar]
- 21.Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 22.Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:879–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nissen T L, Schulze U, Nielsen J, Villadsen J. Flux distribution in anaerobic, glucose-limited continuous cultures of Saccharomyces cerevisiae. Microbiology. 1997;143:203–218. doi: 10.1099/00221287-143-1-203. [DOI] [PubMed] [Google Scholar]
- 24.Oliver S G, van der Aart Q J, Agostoni-Carbone M L, Aigle M, Alberghina L, Alexandraki D, Antoine G, Anwar R, Ballesta J P, Benit P, et al. The complete DNA sequence of yeast chromosome III. Nature. 1992;357:38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- 25.Overkamp K M, Bakker B M, Kötter P, Van Tuijl A, De Vries S, Van Dijken J P, Pronk J T. In vivo analysis of the mechanisms for oxidation of cytosolic NADH by Saccharomyces cerevisiae mitochondria. J Bacteriol. 2000;182:2823–2830. doi: 10.1128/jb.182.10.2823-2830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmieri L, Palmieri F, Runswick M J, Walker J E. Identification by bacterial expression and functional reconstitution of the yeast genomic sequence encoding the mitochondrial dicarboxylate carrier protein. FEBS Lett. 1996;399:299–302. doi: 10.1016/s0014-5793(96)01350-6. [DOI] [PubMed] [Google Scholar]
- 27.Palmieri L, Vozza A, Agrimi G, De Marco V, Runswick M J, Walker J E. Identification of the yeast mitochondrial transporter for oxaloacetate and sulfate. J Biol Chem. 1999;32:22184–22190. doi: 10.1074/jbc.274.32.22184. [DOI] [PubMed] [Google Scholar]
- 28.Patchett R A, Jones C W. The apparent oxidation of NADH by whole cells of the methylotrophic bacterium Methylophilus methylotrophus. A cautionary tale. Antonie Leeuwenhoek. 1986;52:387–392. doi: 10.1007/BF00393466. [DOI] [PubMed] [Google Scholar]
- 29.Postma E, Verduyn C, Scheffers W A, Van Dijken J P. Enzymic analysis of the Crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl Environ Microbiol. 1989;55:468–477. doi: 10.1128/aem.55.2.468-477.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pronk J T, Steensma H Y, Van Dijken J P. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast. 1996;12:1607–1633. doi: 10.1002/(sici)1097-0061(199612)12:16<1607::aid-yea70>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 31.Pronk J T, Wenzel T J, Luttik M A H, Klaassen C M, Scheffers W A, Steensma H Y, Van Dijken J P. Energetic aspects of glucose metabolism in a pyruvate-dehydrogenase negative mutant of Saccharomyces cerevisiae. Microbiology. 1994;140:601–610. doi: 10.1099/00221287-140-3-601. [DOI] [PubMed] [Google Scholar]
- 32.Repetto B, Tzagoloff A. In vivo assembly of yeast mitochondrial α-ketoglutarate dehydrogenase complex. Mol Cell Biol. 1991;11:3931–3939. doi: 10.1128/mcb.11.8.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rivoal J, Ricard B, Pradet A. Purification and partial characterization of pyruvate decarboxylase from Oryza sativa L. Eur J Biochem. 1990;194:791–797. doi: 10.1111/j.1432-1033.1990.tb19471.x. [DOI] [PubMed] [Google Scholar]
- 34.Seo B B, Kitajima-Ihara T, Chan E K L, Scheffler I E, Matsuno-Yagi A, Yagi T. Molecular remedy of complex I defects: rotenone-insensitive internal NADH-quinone oxidoreductase of Saccharomyces cerevisiae mitochondria restores the NADH oxidase activity of complex I-deficient mammalian cells. Proc Natl Acad Sci USA. 1998;95:9167–9171. doi: 10.1073/pnas.95.16.9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shoubridge E A, Hochachka P W. Ethanol: novel end product of vertebrate anaerobic metabolism. Science. 1980;209:308–309. doi: 10.1126/science.7384807. [DOI] [PubMed] [Google Scholar]
- 36.Small W C, McAlister-Henn L. Identification of a cytosolically directed NADH dehydrogenase in mitochondria of Saccharomyces cerevisiae. J Bacteriol. 1998;180:4051–4055. doi: 10.1128/jb.180.16.4051-4055.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson L M, Sutherland P, Steffan J S, McAlister-Henn L. Gene sequence and primary structure of mitochondrial malate dehydrogenase from Saccharomyces cerevisiae. Biochemistry. 1988;27:8393–8400. doi: 10.1021/bi00422a015. [DOI] [PubMed] [Google Scholar]
- 38.Van Dijken J P, Scheffers W A. Redox balances in the metabolism of sugars by yeast. FEMS Microbiol Rev. 1986;32:199–224. [Google Scholar]
- 39.Van Hoek, W. P. M., J. P. Van Dijken, and J. T. Pronk. Regulation of fermentative capacity and levels of glycolytic enzymes in chemostat cultures of Saccharomyces cerevisiae. Enzyme Microb. Technol., in press. [DOI] [PubMed]
- 40.Verduyn C, Postma E, Scheffers W A, Van Dijken J P. Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J Gen Microbiol. 1990;136:395–403. doi: 10.1099/00221287-136-3-395. [DOI] [PubMed] [Google Scholar]
- 41.Verduyn C, Postma E, Scheffers W A, Van Dijken J P. Effects of benzoic acid on metabolic fluxes in yeasts: a continuous culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 42.Verleur N, Elgersma Y, Van Roermund C W T, Tabak H F, Wanders R J A. Cytosolic aspartate aminotransferase encoded by the AAT2 gene is targeted to the peroxisomes in oleate grown Saccharomyces cerevisiae. Eur J Biochem. 1997;247:972–980. doi: 10.1111/j.1432-1033.1997.00972.x. [DOI] [PubMed] [Google Scholar]
- 43.Visser W, Van der Baan A A, Batenburg-van der Vegte W, Scheffers W A, Krämer R, Van Dijken J P. Involvement of mitochondria in the assimilatory metabolism of anaerobic Saccharomyces cerevisiae cultures. Microbiology. 1990;140:3039–3046. doi: 10.1099/13500872-140-11-3039. [DOI] [PubMed] [Google Scholar]
- 44.Visser W, Van Spronsen E A, Nanninga N, Pronk J T, Kuenen J G, Van Dijken J P. Effects of growth conditions on mitochondrial morphology in Saccharomyces cerevisiae. Antonie Leeuwenhoek. 1995;67:243–253. doi: 10.1007/BF00873688. [DOI] [PubMed] [Google Scholar]
- 45.Von Jagow G, Klingenberg M. Pathways of hydrogen in mitochondria of Saccharomyces carlsbergensis. Eur J Biochem. 1970;12:583–592. doi: 10.1111/j.1432-1033.1970.tb00890.x. [DOI] [PubMed] [Google Scholar]
- 46.Wach A, Brachat A, Poehlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 47.Wallace D C. Diseases of the mitochondrial DNA. Annu Rev Biochem. 1992;61:1175–1212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- 48.Young E T, Pilgrim D. Isolation and DNA sequence of ADH3, a nuclear gene encoding the mitochondrial isozyme of alcohol dehydrogenase in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:3024–3034. doi: 10.1128/mcb.5.11.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao W N, McAlister-Henn L. Expression and gene disruption analysis of the isocitrate dehydrogenase family in yeast. Biochemistry. 1996;35:7873–7878. doi: 10.1021/bi9605189. [DOI] [PubMed] [Google Scholar]