Abstract
The eight genes which encode the (F1Fo) H+-ATPase in Lactococcus lactis subsp. cremoris MG1363 were cloned and sequenced. The genes were organized in an operon with the gene order atpEBFHAGDC; i.e., the order of atpE and atpB is reversed with respect to the more typical bacterial organization. The deduced amino acid sequences of the corresponding H+-ATPase subunits showed significant homology with the subunits from other organisms. Results of Northern blot analysis showed a transcript at approximately 7 kb, which corresponds to the size of the atp operon. The transcription initiation site was mapped by primer extension and coincided with a standard promoter sequence. In order to analyze the importance of the H+-ATPase for L. lactis physiology, a mutant strain was constructed in which the original atp promoter on the chromosome was replaced with an inducible nisin promoter. When grown on GM17 plates the resulting strain was completely dependent on the presence of nisin for growth. These data demonstrate that the H+-ATPase is essential for growth of L. lactis under these conditions.
The (F1Fo) H+-ATPase complex plays an important role in the free energy metabolism of virtually all living cells. The structures of F1Fo-ATPase complexes from different sources are very similar and consist of two parts: a membrane integral part, Fo, which forms a proton channel, and a soluble part, F1, which contains the catalytic site for ATP hydrolysis. In bacteria, the enzyme is located in the cytoplasmic membrane, where it catalyzes the interconversion of ATP and the transmembrane proton gradient. Depending on the particular organism and on the conditions for growth, the enzymes function in the direction of either ATP synthesis or ATP hydrolysis (14). In organisms which contain a respiratory chain, such as Escherichia coli and Bacillus subtilis, the primary role of the enzyme is to synthesize ATP driven by the proton gradient that results from respiration, when these organisms are supplied with an electron acceptor. In organisms that lack a respiratory chain, or in the absence of electron acceptors, the enzyme generates a transmembrane proton gradient, and this process is then driven by ATP hydrolysis. The anaerobic bacterium Lactococcus lactis also possesses an F1Fo-ATPase complex. This bacterium lacks the respiratory chain, and the enzyme here is involved in the extrusion of protons driven by ATP hydrolysis to generate the necessary driving force for solute transport and to maintain an acceptable intracellular pH value (21, 38). The latter function is supported by the fact that the activity of the F1Fo-ATPase in these anaerobic bacteria is enhanced at low external pH (2, 23).
The anaerobic bacteria have an alternative route to generate a proton gradient across the cytoplasmic membrane, namely, through end product excretion. In the so-called energy recycling model, which was first demonstrated by Michels et al. (27), it was suggested that carrier-mediated excretion of end products can occur in symport with protons, and this contributes to the generation of the transmembrane proton gradient. This mechanism has been thoroughly investigated in Lactococcus lactis by Otto et al. (30), and ten Brink et al. (40), who demonstrated that the energy recycling by lactate efflux makes a significant contribution to the generation of the proton gradient in this organism, particularly at high external pH and low external lactate concentrations. An interesting question is then whether this contribution would be sufficient to allow growth of L. lactis in the absence of the H+-ATPase.
In this paper we report the cloning, sequencing, and characterization of the genes that encode the H+-ATPase in L. lactis subsp. cremoris MG1363. A mutant strain was constructed in which the expression of H+-ATPase on the chromosome is under control of the nisA promoter. The strain was completely dependent on nisin for growth on GM17 plates, which demonstrates that the H+-ATPase is an essential enzyme for growth of L. lactis.
MATERIALS AND METHODS
Bacterial strains.
The plasmid-free L. lactis subsp. cremoris strain MG1363 (16) was used to study the atp operon in L. lactis. E. coli K-12 strain BOE270 is highly competent with respect to transformation and was derived from strain MT102, which is an hsdR derivative of strain MC1000 [araD139 (ara-leu)7679 galU galK (lac)174 rpsL thi-1] (7). BOE270 was used as a host for plasmids in the cloning procedures and for propagation of plasmid DNA in E. coli.
Oligonucleotides and enzymes.
Oligonucleotides were obtained from Hobolth DNA Synthesis (Hillerød, Denmark). Restriction enzymes (Gibco BRL, Pharmacia), Taq and Pfu DNA polymerases (Pharmacia and AH Diagnostics, respectively), calf intestine alkaline phosphatase (Pharmacia), and T4 DNA ligase (Gibco BRL) were used as recommended by the manufacturers.
Sequencing and sequence analysis of the H+-ATPase operon.
The DNA sequencing was carried out either by the dideoxy nucleotide chain termination method (33) with [α-33P]ddNTP (500 Ci/mmol) (Pharmacia) or by autosequencing by capillary electrophoresis with the Dye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer).
The alignments of DNA and amino acid sequences were performed on the BLAST server at the National Center for Biotechnology Information (NCBI). The numbers given below refer to the numbering used in the GenBank sequence.
Transformation.
Cells of E. coli were made competent by the Ca2+ method (32). Plasmid DNA was used to transform the cells by a standard transformation procedure (28), and the transformation mixtures were plated at 30°C on Luria-Bertani agar plates supplemented with either ampicillin (100 μg/ml) or erythromycin (200 μg/ml). Cells of L. lactis (16) were made competent by growth in GM17 medium containing 1% glycine and resuspended in 10% glycerol and 0.5 M sucrose as described by Holo and Nes (18). Plasmid DNA was used to transform the cells by electroporation (18), and the cells were allowed to regenerate in SGM17 medium for 2 h and then plated onto Schmidt-Ruppin plates containing the appropriate selective antibiotic.
Cloning of the atp operon from L. lactis subsp. cremoris MG1363.
Fragments of the atp operon were cloned as PCR products or by the plasmid rescue technique (see below). Chromosomal DNA from L. lactis MG1363 was used as a template for amplification of DNA. Several primer sets were used to amplify different regions of the atp operon (Fig. 1). Here we took advantage of the fact that in an unrelated project, the first part of the atp operon from the closely related bacterium L. lactis subsp. lactis B1014 was accidentally discovered in a clone from a gene library, which allowed us to design primers for the amplification of the genes that encode the Fo part of the enzyme complex. PCR amplification was carried out in a total volume of 100 μl and in the presence of 0.4 mM concentrations of each deoxynucleoside triphosphate (Boehringer), 3 to 5 μM concentrations of each primer DNA, 0.1 μg of chromosomal DNA, 2.5 U of Taq polymerase, and the buffer recommended by the manufacturer (Pharmacia). The reactions were carried out for 25 cycles (1 min at 94°C for denaturing, 1 min at 55°C for annealing, and 2 min at 72°C extension step) by use of a DNA thermal cycler. The resulting PCR products were cloned in pMOSBlue (Amersham) and sequenced. To confirm the correctness of the cloned product, the sequence was also determined directly on the PCR products.
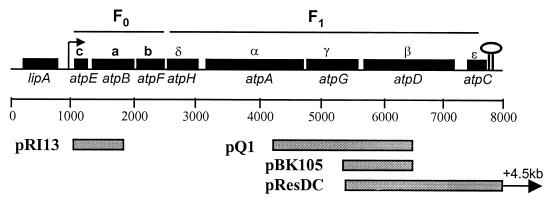
FIG. 1.
Genetic organization of the L. lactis atp operon and DNA fragments used in this work. The open reading frames are shown as boxes, and the designations of the atp genes are shown below the boxes in italic letters. The designations of the H+-ATPase subunits are shown above the boxes. The arrow indicates the direction of transcription of the atp operon, and the stem loop indicates the putative terminator. The cloned fragments, which are referred to in the text, are indicated in the boxes below the scale.
Cloning of atpC by plasmid rescue.
Plasmid pQ1, which harbors the DNA sequence from position 4177 to position 6394 (the C-terminal part atpG of the product and the N-terminal part atpD of the product) and was obtained by cloning a PCR fragment obtained with primers 3987 (5′-TTGGTGGTGGATCAATGACGGC) and 3991 (5′-TTNCCNTCACGAGTACGNTCNCC), was inserted into pMOSBlue. This plasmid was used to construct a plasmid for cloning the remaining part of the atp operon by the plasmid rescue technique as follows. A 3.2-kb EcoRI fragment from pCP12 carrying the erm gene and the strong artificial constitutive promoter CP12 (19) was cloned into pQ1 digested with EcoRI, resulting in pBK105, in which the atpGD′ genes (sequence from position 5268 to 6394) had been placed under control of the CP12 promoter. The plasmid pBK105 was then used to transform L. lactis MG1363 to erythromycin resistance (2 μg/ml). This plasmid is unable to replicate in L. lactis, and only cells with the plasmid integrated into the chromosome will become resistant to erythromycin. If the plasmid integrates into the atp operon by a Campbell-type event, the genes atpDC will come under control of the CP12 promoter. Chromosomal DNA of some transformants was prepared, and the appropriate integration of pBK105 was verified by PCR techniques. The chromosomal DNA was digested with SalI, ligated at a low DNA concentration, and transformed in E. coli, which resulted in pRESDC, in which approximately 4.5 kb downstream of the atp operon was cloned. Plasmid pRESDC was more extensively characterized and sequenced.
Primer extension.
Total RNA was extracted from exponentially growing L. lactis (30°C, optical density at 600 nm [OD600] = 0.5) in GM17 (1% glucose) by the FastRNA kit, BLUE (Bio 101), as recommended by the manufacturer.
Total RNA (10 μg) and 33P-labeled primer (10 pmol) were heated for 2 min at 80°C in 5 μl of hybridization buffer (100 mM KCl, 50 mM HEPES, pH 7.0), followed by a gradual cooling to 30°C over a 60-min period. Three microliters of a solution containing 250 mM Tris-HCl (pH 8.4), 20 mM MgCl2, 20 mM dithiothreitol (DTT), 0.1 mM concentrations of each deoxynucleoside triphosphate, and 0.75 U of avian myeloblastosis virus reverse transcriptase (Life Sciences)/μl was added, and the mixture was incubated at 40°C for 30 min. The extension product was precipitated with ethanol and resuspended in 6 μl of formamide loading buffer, preheated at 85°C for 3 min, and loaded onto a polyacrylamide gel with a set of dideoxy sequencing reactions (33) prepared on a PCR product as a marker. The sequence of the primer used in the 5′-3′ direction was 5′-GACCGATAGCAATTGCTCC-3′ (primer 5264).
Northern blotting.
A single-stranded RNA probe labeled with [α-32P]CTP was derived from a PCR product (primer 5883, 5′-CAACGTGTCCTTCAACGC, and primer T7atpC, 5′-TAATACGACTCACTATAGATAAACCACACCAGCAGGGG), which contains atp′DC′ (position 6918 to 7462) and the T7 promoter, by in vitro transcription using T7 RNA polymerase (Promega). A total RNA preparation (12 μg) was dried in a vacuum drier and resuspended in 4.5 μl of H2O, 2 μl of 5× formaldehyde gel running (FGR) buffer (0.1 M MOPS [morpholinepropanesulfonic acid] [pH 7], 40 mM sodium acetate, 5 mM EDTA), 3.5 μl of formaldehyde (final concentration, 7% [vol/vol]), and 10 μl of formamide (final concentration, 50% [vol/vol]). The RNA molecules were denatured by incubation for 15 min at 60°C and separated by electrophoresis in a 1.2% (wt/vol) agarose gel containing 2.2% formaldehyde, which was run at 5 V/cm with FGR buffer as the electrophoresis buffer. The gel was then washed in H2O for 20 min at room temperature. The RNA was transferred to a Zeta-Probe GT membrane (Bio-Rad) by overnight capillary blotting with 50 mM NaOH as the transfer buffer. The membrane was air dried and prehybridized for 2 h at 42°C in hybridization buffer (1 mM NaCl, 4 mM Na4P2O7, 5× Denhardt's solution, 1% sodium dodecyl sulfate [SDS], 10% [wt/vol] polyethylene glycol 6000, 50 mM Tris-HCl [pH 7.5], 50% [vol/vol] formamide) before the α-32P-labeled riboprobe was added. After overnight hybridization at 42°C, the membrane was washed twice for 5 min at room temperature in 2× SSC, twice at 30 min at 65°C in 0.2× SSC–1% SDS, and twice for 30 min at 65°C in 0.1× SSC before being used for autoradiography (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The 0.24- to 9.5-kb RNA ladder from Gibco BRL was used as a molecular size standard.
Replacement of the chromosomal atp promoter in L. lactis by the nisin-inducible nisA promoter.
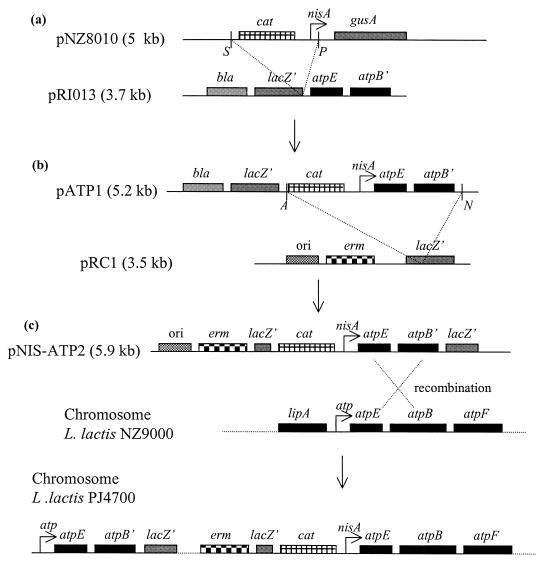
A PCR fragment that harbors the DNA sequence from position +998 to position 1850 (the atpEB′ genes) was amplified using Taq polymerase. After polishing the DNA ends with Pfu polymerase, the fragment was cloned into the SfrII site on the vector pCR-Script Amp SK(+) (Stratagene) (Fig. 2). A plasmid was isolated in which the fragment was inserted in the orientation opposite to that of lacZ (pRI13). A 1.5-kb SalI-PstI fragment from pNZ8010 (12) that carries the cat-194 gene and the nisA promoter was then cloned into pRI13 digested with SalI-PstI, which yielded the plasmid pATP1, in which the atpEB′ genes had been placed under the control of the (nisin-inducible) nisA promoter. A 2.4-kb ApaI-NotI fragment from pATP1, which contains the cat-194 gene, the nisA promoter, and the atpEB′ genes, was cloned into pRC1 digested with ApaI-NotI, which gave rise to plasmid pNIS-ATP2. pRC1 is a 3.5-kb derivative of pBluescript II KS in which the bla gene has been replaced by the ermAM genes to allow for selection of erythromycin resistance in L. lactis (25). The strain NZ9000 (12) is a derivative of strain MG1363 (16) in which the nisR and nisK genes (required for induction of the nisA promoter) are integrated into the pepN locus on the chromosome. Plasmid pNIS-ATP2 was introduced into strain NZ9000 with selection for erythromycin resistance (2 μg/ml) on plates that contained nisin (5 ng/ml). Since this plasmid is unable to replicate in L. lactis, only cells in which the plasmid has integrated into the chromosome should become resistant to erythromycin. If the plasmid integrates into the atpEB locus, the transcription of the entire atp operon will be placed under the control of the nisA promoter. The clones were verified by PCR with primers positioned upstream of the nisA promoter and immediately downstream of position 1850.
FIG. 2.
Cloning strategy used in the replacement of the native atp promoter with the nisin-inducible nisA promoter. (a) A 1.5-kb SalI-PstI fragment from pNZ8010 (12) carrying the cat-194 gene and the nisA promoter was cloned into pRI13 digested with SalI-PstI (pATP1). (b) A 2.4-kb ApaI-NotI fragment from pATP1, containing the cat-194 gene, the nisA promoter, and the atpEB′ genes, was then cloned into pRC1 digested with ApaI-NotI (pNIS-ATP2). (c) Plasmid pNIS-ATP2 was integrated into the atp operon in L. lactis strain NZ9000 with selection for erythromycin resistance (2 μg/ml) on plates containing nisin (5 ng/ml), resulting in replacement of the native atp promoter with the inducible nisA promoter. The designation of the genes is shown above the boxes in italic letters. See Materials and Methods for further details. S, SalI; P, PstI; A, ApaI; N, NotI.
Nucleotide sequence accession number.
The sequence of the lipA gene, the sequence of the atp operon of L. lactis subsp. cremoris strain MG1363, and the sequence downstream of the atp operon (8,912 bp) have been deposited in the NCBI data bank with the accession no. AF059739, and the numbers used in the present paper refer to the numbering used in this sequence.
RESULTS AND DISCUSSION
The genes encoding the H+-ATPase in L. lactis.
The genes encoding the subunits of the H+-ATPase were cloned on a series of overlapping fragments, and the complete sequence of the atp operon was determined and analyzed for the presence of open reading frames (Fig. 1). Within a 7-kb region we identified eight open reading frames with putative ribosome binding sites. The deduced amino acid sequences of the eight gene products of the L. lactis atp operon were aligned with the corresponding amino acid sequences from other organisms, and the sequences of the L. lactis ATPase subunits showed good homology with those of other bacteria (Table 1). The homologies were particularly high between L. lactis, Streptococcus mutans, and Streptococcus bovis, which confirms the close evolutionary relationships of these bacteria. Among the ATPase subunits, the α, β, and γ subunits from the cytoplasmic domain, F1, were especially highly conserved. The consensus nucleotide-binding domains, Walker motifs A (GXXXXGKT) and B (l-hydrophobic-hydrophobic-hydrophobic-d) (1, 42), were also conserved in the deduced sequences of the α and β subunits. Significantly lower homologies were seen for the subunits of the membrane-bound domain, Fo. The δ subunit, a part of the F1 domain, exhibited the lowest subunit homology in the comparison.
TABLE 1.
Homology between the deduced amino acid sequences of the eight L. lactis atp gene products and ATPase subunits from other bacteria
| Source of ATPasea | % Identity (% similarity) of subunits (gene, size [aa])b
|
|||||||
|---|---|---|---|---|---|---|---|---|
| c (atpB, 71) | a (atpE, 237) | b (atpF, 168) | δ (atpH, 175) | α (atpA, 500) | γ (atpG, 289) | β (atpD, 469) | ɛ (atpC, 141) | |
| B. megaterium | 45 (62) | 38 (58) | 32 (62) | 20 (67) | 76 (87) | 48 (67) | 70 (79) | 31 (53) |
| E. coli | 34 (72) | 27 (61) | 33 (55) | 22 (48) | 52 (85) | 35 (59) | 64 (90) | 26 (55) |
| S. mutans | 56 (76) | 47 (65) | 46 (71) | 31 (57) | 84 (92) | 59 (75) | 79 (85) | 53 (69) |
| Streptococcus faecalis | 38 (71) | 51 (69) | 41 (64) | 27 (53) | 82 (91) | 59 (72) | 78 (86) | 52 (68) |
| S. bovis | 54 (76) | 45 (63) | 48 (72) | 34 (58) | 84 (92) | 63 (76) | 78 (83) | 53 (69) |
| PS3 | 40 (65) | 35 (57) | 32 (59) | 24 (44) | 74 (86) | 46 (66) | 72 (81) | 34 (52) |
| Synechococcus sp. | 39 (68) | 31 (52) | 28 (51) | 24 (47) | 62 (78) | 32 (52) | 59 (71) | 35 (58) |
References of bacteria are as follows: B. megaterium (6), E. coli (43), S. mutans (37), S. faecalis (36), S. bovis DDBJ/EMBL/GenBank database accession no. AB009314), thermophilic bacterium PS3 (29), Synechococcus sp. (9).
The genes encode the respective subunits, and the sizes of the subunits are given in amino acids (aa).
The order of the genes was found to be atpE, atpB, atpF, atpH, atpA, atpG, atpD, and atpC, which encode the subunits c, a, b, δ, α, γ, β, and ɛ, respectively (Fig. 1). This organization is virtually identical to what is found in most bacteria (34, 35, 36, 43), though the c and a subunits were reversed in this instance, as has also been observed for other Streptococcus species (13, 37). The functional implications, if any, of this gene reversal in L. lactis are not known.
Three of the genes (atpF, atpA, and atpD) appeared to use UUG as the initiation codon instead of the more frequently used AUG start codon, and it was indeed shown previously that L. lactis can initiate translation at the initiation codons UUG and GUG (41). The gene products encoded by atpF, atpA, and atpD should be produced two, three, and three times more frequently, respectively, than the other gene products, and it is therefore somewhat surprising that the more highly expressed genes, atpF, atpA, and atpD, would use the UUG start codon.
The gene encoding the b subunit, atpF, overlaps with the Shine-Dalgarno sequence of the gene for the δ subunit, atpH, which suggests translational coupling between these genes. Interestingly, such overlap was also reported for atpF and atpH of the atp operon in Anabeana sp. strain PCC 7120 (26), Bacillus megaterium (6), Enterococcus hirae (36), and Clostridium thermoaceticum (10).
In most bacteria, such as E. coli (43) and B. megaterium (6), the atp operon starts with the gene atpI as the first structural gene, but such a gene appears to be absent in L. lactis (although we cannot rule out the possibility that atpI is positioned elsewhere on the chromosome). The function of the polypeptide encoded by the atpI gene in these organisms is unknown; the polypeptide is not an essential part of the H+-ATPase complex, and the atpI gene has been demonstrated to be dispensable for growth (17, 20).
We also determined the sequences up- and downstream of the atp operon in L. lactis. Preceding the first gene in the atp operon, atpE, the lipA gene, which encodes an esterase, was identified (G. Fernandes et al., submitted for publication). Downstream of atpC there was a long noncoding region before the next open reading frame. A homology search at NCBI showed no homology of the putative polypeptide to known proteins.
Transcription of the atp genes.
A standard promoter with −35 (TTGACA) and −10 (TAGAAT) consensus boxes separated by 17 nucleotides was identified in the region upstream of atpE (Fig. 3). The presence of the −35 and −10 consensus sequences suggests that the promoter is recognized by the L. lactis ς39 transcription factor (3), and primer extension analysis confirmed the existence of a transcript that corresponds to this promoter (Fig. 3). In comparison with other lactococcal promoters, the similarities were particularly high between the atp promoter and the rrnA promoter (rRNA operon) (8). The region upstream of the −35 region of the atp promoter sequence (position 853 to 963) has a higher A+T content (75%) than the average value reported for L. lactis DNA (62.8%), which may contribute to the activity of the atp promoter, due to curvature of the A+T-rich sequences (5, 15).
FIG. 3.
The atp promoter and the transcriptional initiation site for the atp operon. (a) Determination of the transcription initiation site of the atp operon. Primer extension analysis was carried out using primer 5264 labeled by 33P as described in Materials and Methods. A sequence ladder was made by sequencing with primer 5264 on a PCR product as described in Materials and Methods. (b) Comparison of the promoter region of related organisms. Letters in bold indicate conserved bases. The putative −35 and −10 consensus boxes of the atp promoter for L. lactis upstream of atpE are underlined. The transcription initiation site (TS) at +1 bp is indicated with an asterisk. Note the extensive homology, particularly around position +1.
Comparison of the promoter region with atp promoters from related bacteria (Fig. 3) showed homology, not only in the −35 and −10 boxes but also directly preceding the −10 box and around the transcription start site. The region upstream of the atp promoter contains several inverted and direct repeats. Such repeats were also observed in Enterococcus faecalis (36), and it was suggested that they may be involved in the regulation of the expression of the atp operon at low external pH (2, 22, 23) in order to keep the intracellular pH at an acceptable level.
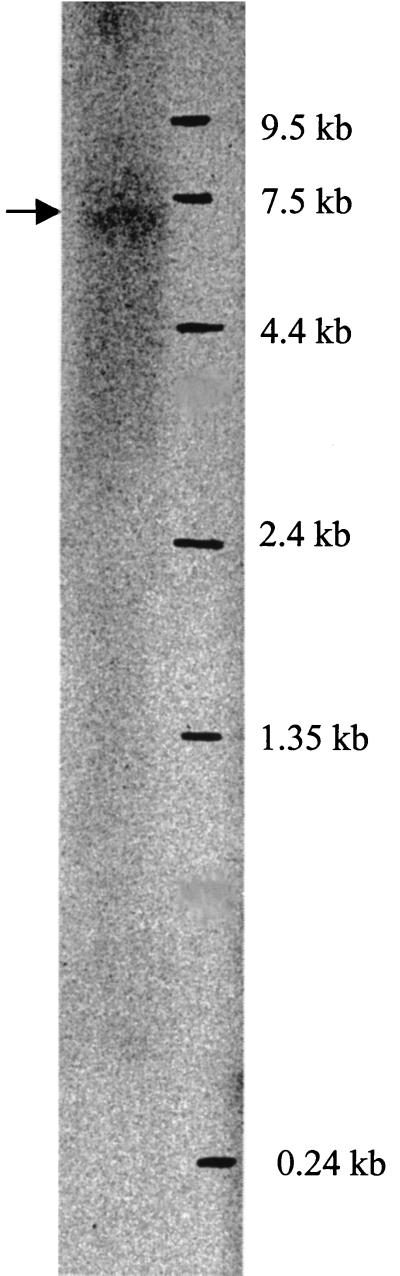
The size of the atp mRNA was determined by Northern blot analysis (Fig. 4), which identified mRNA at approximately 7 kb, which demonstrates that the eight genes are transcribed as a single polycistronic message. Other transcripts could not be identified in the present analysis, in which the 3′ end of the atp operon was used as a probe. But smaller transcripts might still occur if other probes are employed.
FIG. 4.
Northern blot analysis. Total RNA was extracted from L. lactis, and Northern blot analysis was performed as described in Material and Methods. A ribonucleotide probe labeled with [α-32P]CTP containing the C-terminus-encoding part of atpD and the N-terminus-encoding part of atpD (position 7915 to 8459) was used as a probe. The 0.24- to 9.5-kb RNA ladder from Gibco BRL was used as a molecular size standard.
An inverted repeat in the region immediately after atpC was recognized, followed by a T-string (7 bp), a structure that resembles a rho-independent terminator (31). The location of a terminator at this position is also supported by the transcript size found in the Northern analysis.
The F1Fo-ATPase is essential for growth of L. lactis.
In the anaerobic bacterium L. lactis, the role of the H+-ATPase is to maintain the electrochemical proton gradient across the cytoplasmic membrane, and it has been proposed that the H+-ATPase functions to regulate the internal pH (4, 11, 21, 24). Is the H+-ATPase then essential for growth? In principle, the anaerobic bacteria have the option to generate a proton gradient through carrier-mediated excretion of end products in symport with protons (30).
The electrochemical proton gradient (Δp) is composed of an electrical component, the transmembrane potential difference (Δψ), and a chemical component, the transmembrane pH difference (ΔpH). The magnitude of the energy produced by lactate excretion depends strongly on the H+-lactate stoichiometry (n) during the excretion process. If n is 1, the excretion process is electrochemically neutral and only a chemical gradient of protons (ΔpH) can be generated. If n is 2, the translocation is electrogenic and both a ΔpH and a membrane potential (Δψ) can be formed. At high pH (6.8) and a low external lactate concentration (<5 mM), ten Brink and Konings determined the H+-lactate stoichiometry (n) in L. lactis to be 1.9 (39). Thus, in principle the contribution of H+-lactate efflux may suffice so that the H+-ATPase would be dispensable for growth under these conditions.
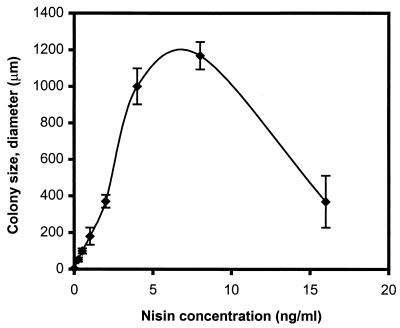
One way to test how important the H+-ATPase is for growth of L. lactis would be to replace the chromosomal atp promoter with an inducible promoter. In order to replace the original atp promoter with an inducible nisin promoter (12), a plasmid, pNIS-ATP2, was constructed, which carries the atpE gene and part of the atpB gene under the control of the nisA promoter. This plasmid, which cannot replicate in L. lactis, was integrated into the chromosome of L. lactis as described in Materials and Methods (Fig. 2). The resulting strain contained an inducible nisin promoter upstream of the entire chromosomal atp operon. When the strain was grown at 30°C on GM17 plates (buffered at pH 7) with different concentrations of nisin, we observed that at very low nisin concentrations the growth of the strain decreased dramatically and in the absence of nisin, growth was completely abolished (Fig. 5). This demonstrates that the H+-ATPase is essential for growth of L. lactis under these conditions, presumably because it is essential for maintaining the proton gradient necessary for solute transport and for maintaining the cytoplasmic pH at an acceptable level. This is also in agreement with the observation that the activity of the F1Fo-ATPase in related anaerobic bacteria is enhanced at low external pH (2, 23).
FIG. 5.
Colonies of strain L. lactis PJ4700, in which the native atp promoter had been replaced by a nisA promoter. The strain was streaked on GM17 plus 2 μg of erythromycin/ml at various nisin concentrations (0, 0.25, 0.5, 1, 2, 4, 8, and 16 ng of nisin/ml), and the graph illustrates the average diameter of colonies obtained with the different nisin concentrations.
ACKNOWLEDGMENTS
We thank Regina Shürmann for excellent technical assistance and Inge Knudsen and Raino K. Hansen for having cloned and sequenced a part of the atp operon. We are also grateful to Allan K. Nielsen for his support with the primer extension and Northern blot analysis and to Lene Kragelund for her kind assistance with the autosequencing at Chr. Hansen A/S.
This work was supported by The Danish Academy of Technical Sciences (ATV) and Chr. Hansen A/S.
REFERENCES
- 1.Abrahams J P, Laslie A G W, Lutter R, Walker J E. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 2.Abrams A, Jensen C. Altered expression of the H+ ATPase in Streptococcus faecalis membranes. Biochem Biophys Res Commun. 1984;122:151–157. doi: 10.1016/0006-291x(84)90452-2. [DOI] [PubMed] [Google Scholar]
- 3.Araya T, Ishinashi N, Shimamura S, Tanaka K, Takahashi H. Genetic and molecular analysis of the rpoD gene from Lactococcus lactis. Biosci Biotechnol Biochem. 1993;57:88–92. doi: 10.1271/bbb.57.88. [DOI] [PubMed] [Google Scholar]
- 4.Bender W A, Marquis R E. Acid tolerance, proton permeabilities, and membrane ATPases of oral streptococci. Infect Immun. 1986;53:331–338. doi: 10.1128/iai.53.2.331-338.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bracco L, Kortlarz D, Kolb A, Diekmann S, Buc H. Synthetic curved DNA sequences can act as transcriptional activators in Escherichia coli. EMBO J. 1989;8:4289–4296. doi: 10.1002/j.1460-2075.1989.tb08615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brusilow W S, Scarpetta M A, Hawthorne C A, Clark W P. Organization and sequence of the genes coding for the proton-translocating ATPase of Bacillus megaterium. J Biol Chem. 1989;264:1528–1533. [PubMed] [Google Scholar]
- 7.Casabadan M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 8.Chiaruttini C, Millet M. Gene organization, primary structure and RNA processing analysis of a ribosomal RNA operon in Lactococcus lactis. J Mol Biol. 1993;230:57–76. doi: 10.1006/jmbi.1993.1126. [DOI] [PubMed] [Google Scholar]
- 9.Cozens A L, Walker J E. The organization and sequence of the genes for ATP synthase subunits in the cyanobacterium Synechococcus 6301. Support for an endosymbiotic origin of chloroplasts. J Mol Biol. 1987;194:359–383. doi: 10.1016/0022-2836(87)90667-x. [DOI] [PubMed] [Google Scholar]
- 10.Das A, Ljungdahl L G. Composition and primary structure of the F1Fo ATP synthase from the obligately anaerobic bacterium Clostridium thermoaceticum. J Bacteriol. 1997;179:3746–3755. doi: 10.1128/jb.179.11.3746-3755.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dashper S G, Reynolds E C. pH regulation by Streptococcus mutans. J Dent Res. 1992;71:1159–1165. doi: 10.1177/00220345920710050601. [DOI] [PubMed] [Google Scholar]
- 12.de Ruyter P G, Kuipers O P, de Vos W M. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenoll A, Munoz R, Garcia E, de la Campa A D. Molecular basis of the optochin-sensitive phenotype of pneumococcus: characterization of the genes encoding the Fo complex of the Streptococcus pneumoniae and Streptococcus oralis H+-ATPases. Mol Microbiol. 1994;12:587–598. doi: 10.1111/j.1365-2958.1994.tb01045.x. [DOI] [PubMed] [Google Scholar]
- 14.Futai M, Kanazawa H. Structure and function of proton-translocating adenosine triphosphatase (F1Fo): biochemical and molecular biological approaches. Microbiol Rev. 1983;47:285–312. doi: 10.1128/mr.47.3.285-312.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gartenberg M R, Crothers D M. Synthetic DNA bending sequences increase the rate of in vitro transcription initiation at the Escherichia coli promoter. J Mol Biol. 1991;219:217–230. doi: 10.1016/0022-2836(91)90563-l. [DOI] [PubMed] [Google Scholar]
- 16.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gay N J. Construction and characterization of an Escherichia coli strain with a uncI mutation. J Bacteriol. 1984;158:820–825. doi: 10.1128/jb.158.3.820-825.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holo H, Nes I F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen P R, Hammer K. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl Environ Microbiol. 1998;64:82–87. doi: 10.1128/aem.64.1.82-87.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen P R, Michelsen O. Carbon and energy metabolism of atp mutants of Escherichia coli. J Bacteriol. 1992;174:7635–7641. doi: 10.1128/jb.174.23.7635-7641.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi H. A proton-translocating ATPase regulates pH of the bacterial cytoplasm. J Biol Chem. 1985;260:72–76. [PubMed] [Google Scholar]
- 22.Kobayashi H, Murakami N, Unemoto T. Regulation of the cytoplasmic pH in Streptococcus faecalis. J Biol Chem. 1982;257:13246–13252. [PubMed] [Google Scholar]
- 23.Kobayashi H, Suzuki T, Kinoshita N, Unemoto T. Amplification of the Streptococcus faecalis proton-translocating ATPase by a decrease in cytoplasmic pH. J Bacteriol. 1984;158:1157–1160. doi: 10.1128/jb.158.3.1157-1160.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi H, Suzuki T, Unemoto T. Streptococcal cytoplasmic pH is regulated by changes in amount and activity of a proton-translocating ATPase. J Biol Chem. 1986;261:627–630. [PubMed] [Google Scholar]
- 25.Le Bourgeois P, Lautier M, Mata M, Ritzenthaler P. New tools for the physical and genetic mapping of Lactococcus strains. Gene. 1992;111:109–114. doi: 10.1016/0378-1119(92)90610-2. [DOI] [PubMed] [Google Scholar]
- 26.McCarn D F, Whitaker R A, Alam J, Vrba J M, Curtis S E. Genes encoding the alpha, gamma, delta, and four Fo subunits of ATP synthase constitute an operon in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1988;170:3448–3458. doi: 10.1128/jb.170.8.3448-3458.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michels P A, Michels J P, Boonstra J, Konings W N. Generation of an electrochemical proton gradient in bacteria by the excretion of metabolic end products. FEMS Microbiol Lett. 1979;5:357–364. [Google Scholar]
- 28.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 29.Ohta S, Yohda M, Ishizuka M, Hirata H, Hamamoto T, Otawara-Hamamoto Y, Matsuda K, Kagawa Y. Sequence and overexpression of subunits of adenosine triphosphate synthase in thermophilic bacterium PS3. Biochim Biophys Acta. 1988;933:141–155. doi: 10.1016/0005-2728(88)90064-3. [DOI] [PubMed] [Google Scholar]
- 30.Otto R, Sonnenberg A S, Veldkamp H, Konings W N. Generation of an electrochemical proton gradient in Streptococcus cremoris by lactate efflux. Proc Natl Acad Sci USA. 1980;77:5502–5506. doi: 10.1073/pnas.77.9.5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santana M, Ionescu M S, Vertes A, Longin R, Kunst F, Danchin A, Glaser P. Bacillus subtilis F1Fo ATPase: DNA sequence of the atp operon and characterization of atp mutants. J Bacteriol. 1994;176:6802–6811. doi: 10.1128/jb.176.22.6802-6811.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saraste M, Gay N J, Eberle A, Runswick M J, Walker J E. The atp operon: nucleotide sequence of the genes for the γ, β, and ɛ subunits of Escherichia coli ATP synthase. Nucleic Acids Res. 1981;9:5287–5296. doi: 10.1093/nar/9.20.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shibata C, Ehara T, Tomura K, Igarashi K, Kobayashi H. Gene structure of Enterococcus hirae (Streptococcus faecalis) F1Fo-ATPase, which functions as a regulator of cytoplasmic pH. J Bacteriol. 1992;174:6117–6124. doi: 10.1128/jb.174.19.6117-6124.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith A J, Quivey R G, Jr, Faustoferri R C. Cloning and nucleotide sequence analysis of the Streptococcus mutans membrane-bound, proton-translocating ATPase. Gene. 1996;183:87–96. doi: 10.1016/s0378-1119(96)00502-1. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki T, Kobayashi H. Regulation of the cytoplasmic pH by a proton-translocating ATPase in Streptococcus faecalis (faecium). A computer simulation. Eur J Biochem. 1989;180:467–471. doi: 10.1111/j.1432-1033.1989.tb14669.x. [DOI] [PubMed] [Google Scholar]
- 39.ten Brink B, Konings W N. The electrochemical proton gradient and lactate concentration gradient in Streptococcus cremoris grown in batch culture. J Bacteriol. 1982;152:682–686. doi: 10.1128/jb.152.2.682-686.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.ten Brink B, Otto R, Hansen U P, Konings W N. Energy recycling by lactate efflux in growing and nongrowing cells of Streptococcus cremoris. J Bacteriol. 1985;162:383–390. doi: 10.1128/jb.162.1.383-390.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van de Guchte M, Kok J, Venema G. Gene expression in Lactococcus lactis. FEMS Microbiol Rev. 1992;88:73–92. doi: 10.1111/j.1574-6968.1992.tb04958.x. [DOI] [PubMed] [Google Scholar]
- 42.Walker J E, Saraste M, Runswick J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker J E, Saraste M, Gay J N. The unc operon: nucleotide sequence, regulation and structure of ATP-synthase. Biochim Biophys Acta. 1984;768:164–200. doi: 10.1016/0304-4173(84)90003-x. [DOI] [PubMed] [Google Scholar]