Abstract
Background
Prostate cancer develops through malignant transformation of the prostate epithelium in a stepwise, mutation-driven process. Although activator protein-1 transcription factors such as JUN have been implicated as potential oncogenic drivers, the molecular programs contributing to prostate cancer progression are not fully understood.
Methods
We analyzed JUN expression in clinical prostate cancer samples across different stages and investigated its functional role in a Pten-deficient mouse model. We performed histopathological examinations, transcriptomic analyses and explored the senescence-associated secretory phenotype in the tumor microenvironment.
Results
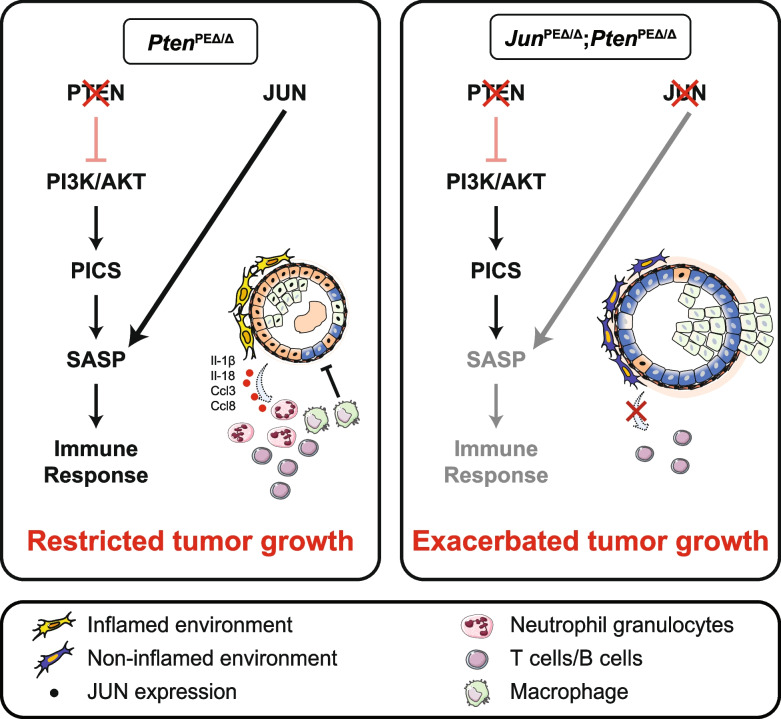
Elevated JUN levels characterized early-stage prostate cancer and predicted improved survival in human and murine samples. Immune-phenotyping of Pten-deficient prostates revealed high accumulation of tumor-infiltrating leukocytes, particularly innate immune cells, neutrophils and macrophages as well as high levels of STAT3 activation and IL-1β production. Jun depletion in a Pten-deficient background prevented immune cell attraction which was accompanied by significant reduction of active STAT3 and IL-1β and accelerated prostate tumor growth. Comparative transcriptome profiling of prostate epithelial cells revealed a senescence-associated gene signature, upregulation of pro-inflammatory processes involved in immune cell attraction and of chemokines such as IL-1β, TNF-α, CCL3 and CCL8 in Pten-deficient prostates. Strikingly, JUN depletion reversed both the senescence-associated secretory phenotype and senescence-associated immune cell infiltration but had no impact on cell cycle arrest. As a result, JUN depletion in Pten-deficient prostates interfered with the senescence-associated immune clearance and accelerated tumor growth.
Conclusions
Our results suggest that JUN acts as tumor-suppressor and decelerates the progression of prostate cancer by transcriptional regulation of senescence- and inflammation-associated genes. This study opens avenues for novel treatment strategies that could impede disease progression and improve patient outcomes.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12943-024-02022-x.
Keywords: Prostate cancer, AP-1 transcription factors, JUN, Senescence, SASP, Immune infiltration
Background
Prostate cancer (PCa) is one of the most frequently diagnosed malignancies in men worldwide [1]. Its significance lies not only in its prevalence but also in the potential to progress to aggressive forms that resist conventional treatments and lead to high mortality rates [2]. The complex molecular programs that determine the routes of PCa progression are still incompletely understood. On the molecular level, the dysregulation of the phosphoinositide 3-kinase (Pl3K) and androgen receptor (AR) pathways has been implicated in the pathology of PCa [3]. The constitutive activation of the Pl3K cascade, which is caused by mutations in the tumor-suppressor gene and Pl3K antagonist Phosphate and tensin homologue (PTEN), was identified in 20% of primary PCa tumors and represents a major oncogenic driver [4]. The current standard treatment for primary advanced-stage PCa is the administration of anti-androgens to deprive the tumor of dihydrotestosterone. PCa inevitably escapes androgen deprivation by relapsing into castration resistant PCa (CRPC), which is associated with loss of PTEN tumor-suppressor activity in 50% of cases. The characteristic dissemination of CRPC into local and distant regions such as bone, is correlated with poor survival [3–5].
In a previously described mouse model, the abrogation of Pten in prostate epithelium (PE) caused activation of a p53-mediated senescence program [6–8]. The emergence of senescence in cancer is considered a double-edged sword: it either confers anti-tumorigenic effects when originating from tumor cells or results in pro-tumorigenic outcomes when the tumor microenvironment (TME) is affected [9]. This phenomenon is mainly attributed to the induction of a senescence-associated secretory phenotype (SASP), characterized by the secretion of soluble signaling factors, proteases and extracellular matrix proteins [10]. In particular, pro-inflammatory cytokines such as IL-6, IL-1, TNF-α, CCL3 and CCL8 attract innate immune cells to the vicinity of the tumor site. As a collective, all components of SASP aid in creation of a pro-tumorigenic microenvironment and ultimately advance tumor progression depending on the tissue context. IL-6 and its downstream effector signal transducer and activator of transcription 3 (STAT3) are known to regulate apoptosis, angiogenesis, proliferation and differentiation, making them promising therapeutic targets in PCa [11]. However, our group has recently challenged active IL-6/STAT3 signaling as a tumor driver in PCa, as loss of Stat3 unexpectedly resulted in increased tumor burden and was accompanied by a bypass of PTEN-loss induced cellular senescence (PICS) in a Pten-deficient PCa mouse model [12, 13].
Besides the hyperactivation of PI3K/AKT and amplification of AR signaling, other mechanisms driving the progression of PCa include the activation of activator protein-1 (AP-1) mediated gene expression [14]. AP-1 transcription factors (TF) such as JUN, were initially considered as proto-oncogenes [15] and deregulation of AP-1 family members was observed in several cancers [16]. Previous studies have suggested that JUN modulates hepatocellular tumorigenesis as a regulator of cell cycle genes and has co-activator and repressor functions in the regulation of AR in the prostate [17–19]. Recent evidence suggests tumor-suppressive functions for several members of the AP-1 TF family and their regulators [17, 20]. For example, the JUN-activating JUN N-terminal kinase (JNK) has previously been identified as a potent tumor-suppressor in a murine PCa model [21]. JUNB, which is also activated by JNK has been associated with growth limiting properties in PCa and its activation may explain the mechanism of JNK’s tumor-suppression [22]. A recent study provides novel insights how the tumor-suppressive functions of AP-1 might be exerted, as JUN was particularly implicated as pioneering factor in bookmarking the enhancers of genes associated with the induction of the senescence program [23].
Here we investigated the role of Jun in a murine model of Pten-loss driven neoplasia of the PE and surveyed the consequence of JUN-deficiency in tumor development and senescence.
Methods
Mouse strains and animal work
To establish the PCa mouse model used in this study, we bred a Pten knockout prostate cancer mouse strain (PtenPEΔ/Δ) [24] with a Jun-floxed (Junfl/fl) [25] mouse strain. The PtenPEΔ/Δ mouse strain was originally established by crossing PtenEx4/Ex5-floxed mice [26] and heterozygous transgenic Probasin (Pb) Cre mice [27]. Pb Cre transgenic mice express the Cre recombinase under the Probasin promoter restricted to PE cells of sexually mature mice [27]. To minimalize tumor burden for breeding animals, heterozygous PtenPEΔ/+ males were used for breeding. The resulting genotypes of experimental animals are: PbCre+/+ (wildtype (wt)), PbCretg/+;Junfl/fl (JunPEΔ/Δ); PbCretg/+;Ptenfl/fl (PtenPEΔ/Δ); PbCretg/+;Junfl/fl;Ptenfl/fl (JunPEΔ/Δ;PtenPEΔ/Δ). For all experiments, mice were sacrificed at 19-weeks of age, with the exception of animals used for the Kaplan–Meier survival analysis and for metastasis analysis (39-weeks of age).
Histological staining
Hematoxylin and eosin (H&E), immunohistochemistry (IHC) and immunofluorescence (IF) stainings were performed on 2 µm sections of formalin-fixed paraffin embedded (FFPE) tissue. H&E staining was done according to routine diagnostic protocols. Details of IHC staining for the different markers are indicated in Supplementary Table 6 and all slides were counterstained with hematoxylin.
For the EpCAM IF staining, slides were dewaxed and heated in pH 6 citrate buffer. After blocking with 2% bovine serum albumin (Roth 8076.4), the slides were incubated in primary antibody (EpCAM, Elab Science, E-AB-70132, dilution 1:300) overnight. Next, slides were incubated for 1 h at room temperature in secondary antibody (Goat anti-Rabbit IgG Alexa Fluor 488, Dilution 1:500) and stained with DAPI.
Human tissue microarray analysis
The generation of human tissue microarrays (TMAs) of healthy and tumor prostate tissues was previously described [28]. The TMAs were stained with an antibody for JUN (Supplementary Table 6) and analysed by trained pathologists. The JUN levels were determined by combining the staining intensity with the percentage of positive cells and graded into absent (0), low-grade (1), medium-grade (2) and high-grade (3). We next stratified TMA samples according to Gleason scores, resulting in three groups (healthy: no Gleason score; low Gleason: Gleason score 5–6; and high Gleason: Gleason score 7–9) and analysed JUN expression for all groups. For the Kaplan–Meier analysis, patients were grouped into absent (0) (JUNabsent) and present (1–3) (JUNpresent) JUN expression and correlated with biochemical recurrence (BCR) data.
Whole slide scan analysis
Analysis of IHC staining was performed with QuPath (version 0.3.2) [29]. First, regions of interest were annotated, excluding non-prostate tissue such as urethra, seminal vesicles and ductus deferens. Cell detection was performed with the StarDist extension [30] for the NIMP-R14 staining and the built-in watershed cell detection plugin for F4/80, CD79b, JUN, Granzyme B and phosphorylated (p)STAT3. Parameters were chosen individually for each staining. Thereafter, smoothed features were calculated with a FWHM radius of 25 µm. The tissue was then classified into tumor/epithelium and stroma using an object classifier, trained individually for each staining. A threshold was set for the mean DAB optical density value, categorizing cells into positive or negative. For pSTAT3, multiple thresholds were set and cells were classified into 1*, 2* and 3* positive to calculate the H-score. The H-score was calculated by multiplying the percentage of cells by their respective intensity value and ranged from 1 to 300. Analysis was performed by a single investigator and evaluated by two independent pathologists. For quantification of Ki67 levels of tumor and non-tumor samples, we defined four circular regions of interest with a radius of 150 µm. Within each region, we manually counted the positive epithelial cells and used QuPath to detect the negative cells. Percentage of p21CIP1/WAF1 positive epithelial cells was estimated by a blinded pathologist. For Galactosidase beta 1 (GLB1), regions of interest were annotated and categorized into positive and negative areas using a stringent pixel threshold for the DAB optical density value. The threshold was adjusted to detect the granular expression pattern. Results shown are from the anterior prostate.
Statistical analysis for immunohistochemistry
Measurements were exported as TSV files and imported into GraphPad PRISM (version 9.5.0). Significance was determined using an ordinary one-way ANOVA with Tukey’s multiple comparisons tests for 3 or more groups. Graphs were created and formatted in GraphPad PRISM.
Protein extraction and immune blotting
Protein extraction from frozen prostate samples and immune blotting was performed as previously described [31]. Briefly, 15–20 µg of protein lysate was separated via SDS-PAGE, transferred onto nitrocellulose membranes (Amersham) and blocked with 5% milk in 1 × TBS /0.1% Tween-20 or with 5% BSA in 1 × TBS /0.1% Tween-20 for 1 h according to manufacturer’s antibody datasheets. Membranes were incubated with primary antibodies against pJUNS73 (CST 9164), JUN (CST 9165), pAKTS473 (CST 4060), AKT (CST 4691), EpCAM (Elab Science, E-AB-70132), β-ACTIN (CST 4967), NLRP3 (CST 15101), Pro-IL-1β (R&D Systems, AF-401-NA) and β-TUBULIN (CST 2146 and CST 2128) at 4 °C overnight. TGX stain free technology (Bio-Rad), β-ACTIN or β-TUBULIN were used as loading controls.
Magnetic cell sorting, library preparation and RNA sequencing
The preparation of sequencing libraries and subsequent RNA sequencing (RNA-seq) was performed as previously described [32]. Briefly, prostates of 19-week-old mice were dissected, processed to yield a single cell suspension and EpCAM (CD326) positive cells were isolated by magnetic cell sorting (Magnisort®, Thermo Fisher Scientific) using anti-CD326-biotin (13–5791–82, eBioscience). EpCAM positive cells were collected by centrifugation at 300 xg for 5 min at 4 °C and stored at -80 °C until further use. High-quality RNA, as assessed by 4200 TapeStation System (Agilent) was used for library preparation according to the manufacturer’s instructions.
RNA sequencing data analysis
Single-end 75 bp reads sequencing of libraries was performed at CEITEC, Centre for Molecular Medicine (Brno, Czech Republic) as previously described [32]. Genes with a false discovery rate (FDR) FDR-adjusted p-value < 0.05 and log2 fold change ≥ 1 or ≤ -1 were considered significantly up- or downregulated.
Kaplan–Meier survival analyses of public datasets
To assess whether expression levels of JUN, PTEN, IL1B, CCL3 and CCL8 affected survival capabilities of human PCa patients, we applied the KM plotter tool (https://kmplot.com/analysis/) which computed probabilities of RFS based on the TCGA-PRAD study [33]. Output data were used for re-plotting of survival curves and performing of cox-regression analyses with R packages “Survival” and “Survminer” and R-script “ggsurvplot”. Combined KM plotter output was used for calculation of subgroups as stratified by expression levels of both genes, such as JUN and PTEN. Relapse-free survival (RFS) analysis was based on survival data of n = 333 PCa patients. Groups were automatically separated and the calculated, best performing and most significant threshold was used as a cut-off. Hazard ratios and p-values were retrieved from Cox-regression analyses.
Results
JUN levels discriminate progression states in prostate cancer dependent on PTEN
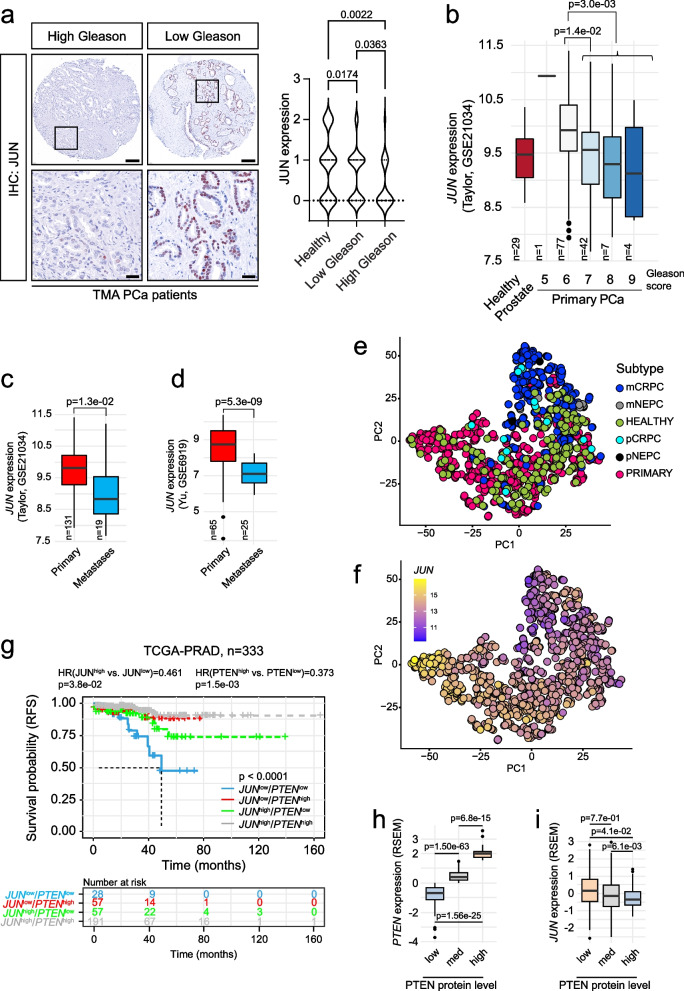
To clarify the role of AP-1 TFs in PCa progression, we investigated the level of the master factor JUN in tissue microarrays (TMA) of low and high progressive human prostate tumors by immunohistochemistry (IHC). We performed semi-quantitative analysis and categorized each tumor based on JUN levels from 0 (absent), 1 (low-grade), 2 (medium-grade) to 3 (high grade) (Fig. 1a). Patients were divided into present (n = 29 + 6 censored subjects) (JUNpresent) and absent (n = 32 + 8 censored subjects) (JUNabsent) cohorts and correlated with biochemical recurrence (BCR) data (Supplementary Fig. 1a). PCa progression is marked by histological changes of the tumor architecture and is categorized by Gleason scoring [34]. We observed a gradual decrease of JUN protein abundance from healthy tissue to primary tumors (low Gleason; Gleason score 5–6), reaching the lowest JUN expression state in advanced tumor stages (high Gleason; Gleason score 7–9) (Fig. 1a). The correlation between JUN protein and patient BCR status revealed a significantly (p = 1.8e-02) diminished BCR-free survival in patients with low JUN, whereas high JUN levels were associated with increased survival probability (Supplementary Fig. 1a). We next mined a publicly available transcriptome dataset ([35]; n = 140) and stratified PCa patients into high-risk and low-risk groups as defined by the prognostic index and characterized by a significant difference in relapse-free survival (RFS) using the SurvExpress webtool [36] (p = 4e-04) (Supplementary Fig. 1b). We investigated JUN mRNA expression in the high- and low-risk groups and found significantly (p = 1.3e-30) higher JUN among low-risk patients compared to the high-risk group (Supplementary Fig. 1c). To explore JUN levels in advanced stages of PCa, we used the Taylor dataset [35], comprising primary tumors of different progression stages and Gleason scores (n = 131) as well as healthy prostate tissue (n = 29). Compared to healthy tissue, we observed higher levels of JUN in early disease stages with Gleason scores 5–6 and significantly decreased expression of JUN in high grade tumors (p = 3e-03; Gleason scores 7–9) (Fig. 1b). Concordantly, JUN was highly expressed in primary tumors (n = 131; n = 65) but significantly lower expressed in PCa metastases (n = 19; n = 25) as observed in two independent datasets (Fig. 1c-d; p = 1.3e-02; [35]; p = 5.3e-09; [37]). We next investigated levels of JUN, JUNB and FOS and observed a comparable regulation (Supplementary Fig. 1d-e). Metastatic CRPC and neuroendocrine PCa (NEPC) present aggressive tumor subtypes that emerge under androgen deprivation therapy and are associated with poor prognosis. We compared levels of JUN and its related TFs FOS and JUNB in primary (n = 715) and metastatic (n = 320) PCa [38], including CRPC and NEPC (Fig. 1e-f, Supplementary Fig. 1f). The tumor-subtype and stage-dependent expression of JUN was highly significant when comparing healthy and primary (p = 2.8e-05), primary and metastatic CRPC (p = 2.6e-43) and primary and metastatic NEPC (p = 5.3e-04) (Supplementary Fig. 1g), suggesting JUN as a potential marker of progressive subtypes of PCa. In addition, our survey revealed higher levels of JUN in primary PCa associated with low Gleason scores than healthy prostates (Supplementary Fig. 1g-h), suggesting a gradual change of JUN levels in PCa development and progression. Our data implicate that JUN and other AP-1 factors except MAF and MAFB may act as suppressors rather than drivers of PCa which was reflected by hazard ratios (HR) calculated from RFS (Supplementary Fig. 1i).
Fig. 1.
JUN levels are correlated with prostate cancer progression stages. a Left panel: Representative immunohistochemistry (IHC) images of tissue microarrays (TMAs) investigating human prostate tumors (n = 60) with high or low Gleason scores stained for JUN protein. Scale bars indicate 150 µm (top row) and 30 µm (bottom row), images are presented in 16.8 × (top row) and 80.0 × magnification (bottom row). The area used for the higher magnification is indicated by the rectangle. Right panel: Violin plot showing JUN expression divided in absent (0), low-grade (1), medium-grade (2) and high-grade (3) in healthy (no Gleason score), low Gleason (Gleason score 5–6) and high Gleason (Gleason score 7–9) TMA samples. b JUN mRNA levels in high (Gleason score ≥ 7) and low (Gleason score < 7) grade human prostate tumors. Data were retrieved from [35]. Significance was determined by an unpaired, two-sided t-test or one-sided Anova. c High and low JUN levels significantly (p = 1.3e-02) discriminate primary prostate tumors (n = 131) (red) and metastases (n = 19) (blue). Data were retrieved from [35]. d High and low JUN levels significantly (p = 5.3e-09) discriminate primary prostate tumors (n = 65) (red) and metastases (n = 25) (blue). Data were retrieved from [37]. Significances in c-d were determined by an unpaired, two-sided t-test. e Principal component analysis (PCA) of prostate tumors of different developmental stages comprising normal prostate tissue, primary tumors and primary (p) and metastatic (m) CRPC and NEPC tumors. Datasets from [38]. f Overlay of JUN expression with PCA clustering from e). JUN levels are color coded from high expression (yellow) to low expression (blue). g Kaplan–Meier survival analysis of TCGA-PRAD [33] tumors (n = 333) assessing levels of JUN and PTEN. Hazard ratios (HR) were determined by Cox-regression analysis: HR(JUNhigh vs. JUNlow) = 0.461, p = 3.8e-02 and HR(PTENhigh vs. PTENlow) = 0.307, p = 1.5e-03. Statistical testing was done with a logrank test. h Co-analysis between PTEN expression (RNA-Seq by Expectation–Maximization (RSEM) and PTEN protein level reverse-phase protein array (RPPA)). i Co-analysis between PTEN protein level (RRPA) and JUN expression (RSEM)
Mutations in the tumor suppressor PTEN are considered as main drivers of oncogenic transformation and malignancy in PCa [4]. As PTEN loss is highly correlated with increasing Gleason score and associated with activation of several downstream processes, primarily via hyperactivation of PI3K/AKT and inactivation of AR signaling [4], we next investigated synergistic effects of additional JUN alterations. We applied the KMplot tool to assess RFS of PCa patients ([33]; n = 333) that were stratified into four risk groups JUNhigh/PTENhigh, JUNhigh/PTENlow, JUNlow/PTENhigh and JUNlow/PTENlow. We observed that patients featuring low levels of JUN and PTEN showed the lowest survival probability whereas patients with high JUN and PTEN expression presented with the most favorable prognosis. In contrast to singular JUN depletion, downregulation of PTEN alone resulted in intermediate survival probabilities confirming its role as main oncogenic driver in PCa (Fig. 1g). Finally, we surveyed reverse-phase protein array (RPPA) data of the TCGA-PRAD cohort [33] and observed that PTEN protein correlated well with PTEN mRNA levels whereas we identified an inverse relationship between PTEN and JUN levels (Fig. 1h-i). Although loss of JUN alone is not sufficient to cause significant changes in survival probability, our data suggest that the absence of PTEN promotes JUN to a survival-determining factor in PCa patients.
Genetic depletion identifies a tumor-suppressive role of JUN in prostate cancer development
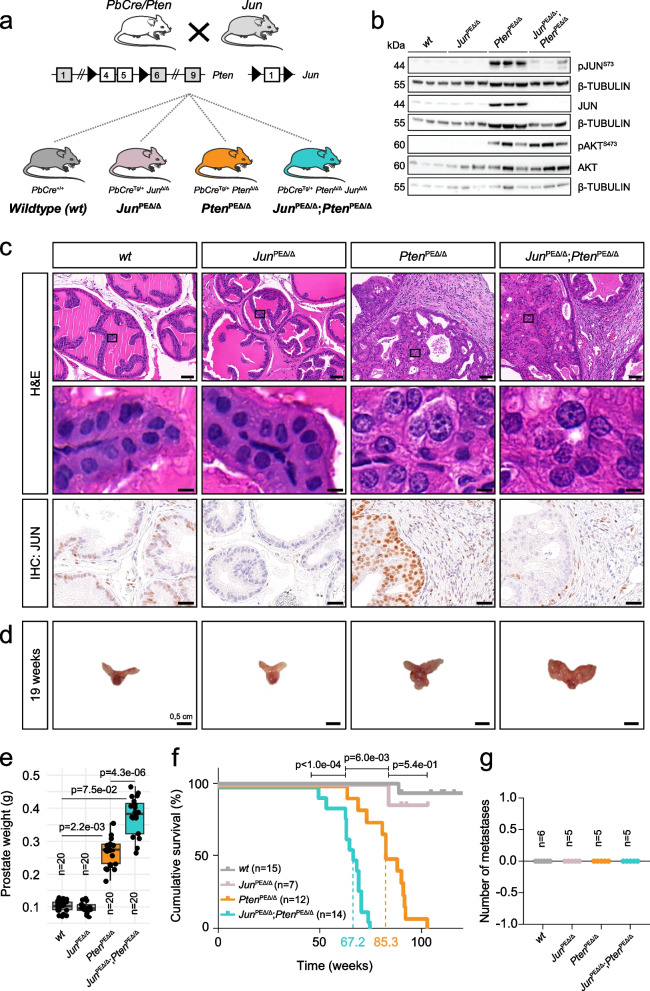
As patients presenting with low expression of JUN and PTEN showed severely reduced survival rates, we next sought to elucidate the mechanistic role of JUN in the development of PTEN-deficient PCa and employed a Pten floxed murine model of PCa (Fig. 2a) [26, 39]. The homozygous deletion of murine Pten via the Probasin (Pb) Cre recombinase [27] mirrored 20% of all primary human PCa cases with homozygous loss of PTEN (Fig. 2a, PbCre/Pten). The PE of homozygous mutants developed hyperplasia that progressed into prostate adenocarcinoma between 12 and 29-weeks of age [39]. We inter-crossed a floxed Jun mouse strain where the sole exon is flanked by loxP sites [25] (Fig. 2a, Jun) to generate 4 individual genotypes. This enabled comparison of prostate tissue of wildtype (wt) mice to either Jun (JunPEΔ/Δ), Pten (PtenPEΔ/Δ) or Jun/Pten (JunPEΔ/Δ; PtenPEΔ/Δ) double knockout mice (Fig. 2a, colored F1 mice). We examined protein extracts of whole prostates and observed a significant increase in levels of phosphorylated (S73) and total JUN in PtenPEΔ/Δ, whereas notable JUN expression was absent in wt prostates (Fig. 2b). We also confirmed efficient Cre-mediated deletion of Jun alone (JunPEΔ/Δ) and in combination with Pten (JunPEΔ/Δ; PtenPEΔ/Δ) (Fig. 2b). As a verification of functional Pten deletion, we detected robust activation of the PI3K/AKT pathway in PtenPEΔ/Δ and JunPEΔ/Δ; PtenPEΔ/Δ mice as assessed by analysis of phosphorylated AKT (pAKTS473) levels (Fig. 2b).
Fig. 2.
Jun-deficiency fosters the progression of Pten-loss induced tumors. a Top: Schematic representation of mouse models used in the study. Homozygous loss of Pten or Jun was achieved by a Probasin promoter-controlled Cre recombinase (PbCre)-mediated ablation of floxed exons 4 and 5 (Pten) or exon 1 (Jun). Bottom: established and investigated genetic models. Wildtype (PbCre+/+; wt) and mice with single knockout of Pten (PbCretg/+; PtenPEΔ/Δ) and Jun (PbCretg/+; JunPEΔ/Δ) were compared with double knockout (PbCretg/+; JunPEΔ/Δ;PtenPEΔ/Δ). PE = prostate epithelium; tg = transgene; Δ = knockout. b Western blot analysis of phosphorylated (pJUNS73 and pAKTS473) and total JUN and AKT. β-TUBULIN served as loading control. Protein lysates of entire organs (n = 3 biological replicates) from 19-week-old wt, PtenPEΔ/Δ, JunPEΔ/Δ and JunPEΔ/Δ;PtenPEΔ/Δ were investigated. c Top row: H&E stainings of 19-week-old wt, PtenPEΔ/Δ, JunPEΔ/Δ and JunPEΔ/Δ;PtenPEΔ/Δ prostates. Scale bars indicate 60 µm (top row) and 2 µm (second row), images are presented in 40.0 × (top row) and 600.0 × magnification (second row). Black rectangles represent the area used for the zoom image below. Bottom row: IHC with an antibody against JUN in 19-week-old prostates of all four experimental groups. Scale bars indicate 30 µm; images are presented in 100.0 × magnification. d Macroscopic images of 19-week-old dissected prostates of wt, PtenPEΔ/Δ, JunPEΔ/Δ and JunPEΔ/Δ;PtenPEΔ/Δ mice. e Box plot showing the weights of prostates in grams between wt, PtenPEΔ/Δ, JunPEΔ/Δ and JunPEΔ/Δ;PtenPEΔ/Δ 19-week-old animals (n = 20). Significance was determined with an unpaired, two-sided t-test. f Kaplan–Meier survival analysis of wt, PtenPEΔ/Δ, JunPEΔ/Δ and JunPEΔ/Δ;PtenPEΔ/Δ animals. Biological replicates are indicated and the cumulative survival (%) is shown. Statistical significance was calculated with a logrank test. g Organs (heart, lung, liver, spleen, kidney, lymph nodes and brain) of 39-week-old wt, PtenPEΔ/Δ, JunPEΔ/Δ and JunPEΔ/Δ;PtenPEΔ/Δ mice were stained with H&E and analysed for metastatic lesion formation. The number of metastases detected in each tissue are shown
To investigate the morphological architecture of prostates upon Jun deletion in the PCa mouse model, we analyzed histological sections by hematoxilin and eosin (H&E) staining (Fig. 2c, top panel). Both wt and JunPEΔ/Δ animals showed physiological growth patterns and morphology, characteristic for the respective prostate lobes. In PtenPEΔ/Δ and JunPEΔ/Δ; PtenPEΔ/Δ prostates, we observed hyperplastic epithelium growing in cribriform patterns into the lumen. Both groups showed anisocytosis, anisokaryosis and alterations in nucleus-to-cytoplasmic ratios, but largely without invasion of the stroma.
Next, we analyzed JUN levels in prostates of all genotypes. Supporting our immunoblot results, IHC revealed increased levels of total JUN predominantly in the PE of PtenPEΔ/Δ mice and absence in epithelial cells of JunPEΔ/Δ and JunPEΔ/Δ; PtenPEΔ/Δ (Fig. 2c, bottom panel). We assessed the effects of Jun deficiency on tumor burden and survival by morphological and survival analyses. Macroscopically, prostates from PtenPEΔ/Δ and JunPEΔ/Δ; PtenPEΔ/Δ mice were notably enlarged as compared to wt or JunPEΔ/Δ prostates (Fig. 2d). This finding was corroborated by prostate weight analysis (Fig. 2e). The additional deletion of Jun on the Pten-deficient background resulted in even higher prostate weights, hinting at JUN’s potential function as a tumor-suppressor in murine PCa development. We performed a Kaplan–Meier survival analysis where overall survival or the occurrence of the discontinuation criteria according to the guidelines of the 3Rs principles were defined as the endpoint of the experiments (Fig. 2f) [40]. We observed comparable survival probabilities of wt and JunPEΔ/Δ mice (p = 5.4e-01) but a significantly decreased survival of PtenPEΔ/Δ (mean survival 85.3 weeks, p = 6e-03) as compared to wt mice. Remarkably, the survival of PtenPEΔ/Δ mice was significantly (p < 1e-04) reduced by the additional deletion of Jun. JunPEΔ/Δ; PtenPEΔ/Δ mice exhibited a mean survival of 67.2 weeks. Despite the significantly reduced survival rates in JunPEΔ/Δ; PtenPEΔ/Δ mice, we did not detect metastatic lesions in the analysed genotypes (Fig. 2g). We therefore conclude that Jun-deficiency alone is not sufficient to induce prostate tumorigenesis, but causes a significant increase in tumor burden and a significant reduction in overall survival in combination with Pten knockout. The results of our murine PCa model reinforce our observations from human PCa samples, suggesting that JUN acts as a tumor-suppressor in PCa.
To determine whether aberrant cellular proliferation contributes to enhanced tumor growth in JunPEΔ/Δ; PtenPEΔ/Δ-deficient prostates, we assessed the number of Ki67+ epithelial cells by IHC. Although we noticed higher Ki67 levels in JunPEΔ/Δ; PtenPEΔ/Δ tumors by trend, the difference was not significant (p = 1.3e-01) when compared to PtenPEΔ/Δ prostates (Supplementary Fig. 2a). To investigate the effects of JUN ablation in vitro, we utilized the CRISPR/Cas9 technology in the human PCa cell lines DU145 (PTEN wildtype) and PC3 (PTEN mutated). We designed three individual guide RNAs for the JUN locus (Supplementary Fig. 2b) and used lentiviral transduction of empty vector (EV) and guide RNA (G1, G12, G14) plasmids. We identified varying efficiencies of JUN knockout in bulk cultures of DU145 and PC3 cell lines (Supplementary Fig. 2c, e) and no significant differences in cellular proliferation (Supplementary Fig. 2d, f). We confirmed the results of unchanged proliferation in single clones of both cell lines which were selected according to complete loss of JUN protein (Supplementary Fig. 2g-j). The in vivo and in vitro results indicate that proliferation may not be the primary biological process influenced by JUN during PCa progression.
Transcriptome profiling reveals JUN-mediated alterations in senescence-associated secretion and immune response
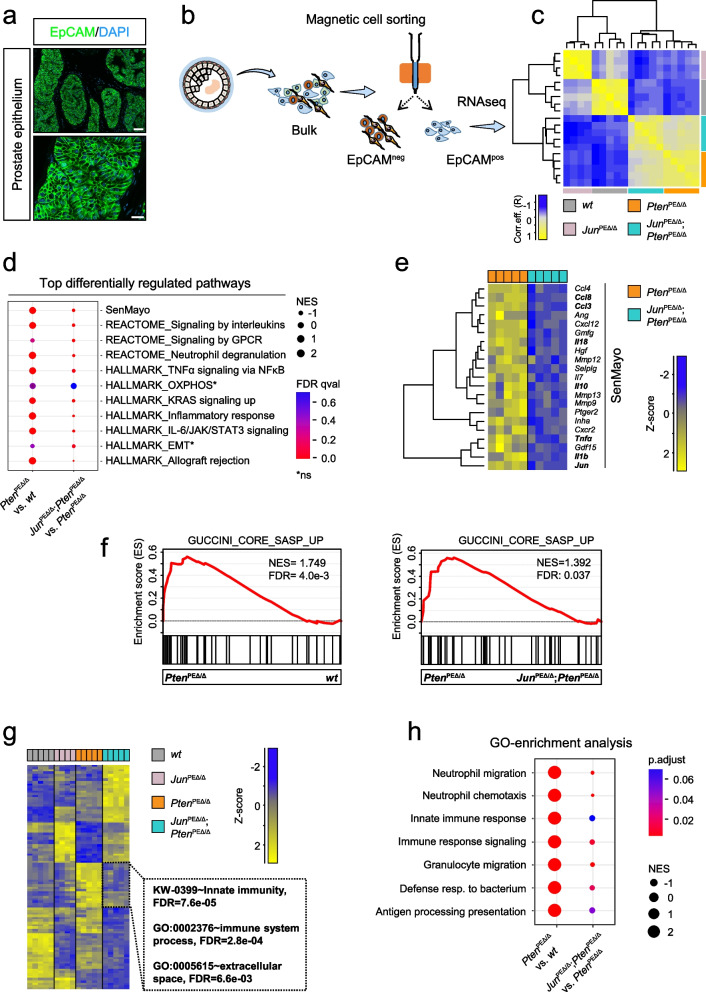
To elucidate the tumor cell-specific molecular programs regulated by JUN in vivo, we performed transcriptome profiling of PE cells across all four experimental murine groups (Fig. 2a). To obtain a homogenous epithelial fraction, we enriched prostate lysates for the Epithelial cell adhesion molecule (EpCAM) showing a uniform expression in PE cells (Fig. 3a, Supplementary Fig. 3a) via magnetic cell separation [32] (Fig. 3b, Supplementary Fig. 3b). The correlation analysis revealed high congruence between JunPEΔ/Δ; PtenPEΔ/Δ and PtenPEΔ/Δ tumor and wt and JunPEΔ/Δ samples (Fig. 3c).
Fig. 3.
Transcriptome profiling of genetic models reveals a JUN-dependent regulation of innate immunity. a Representative immunofluorescence (IF) image of a wt murine prostate for the epithelial marker EpCAM (green). DAPI (blue) is shown as a nuclear stain. Top image: 40.0 × magnification, scale bar represents 60 µm; Bottom image: 147.5 × magnification, scale bar represents 20 µm. b Overview of sample preparation for transcriptome profiling of wt, PtenPEΔ/Δ, JunPEΔ/Δ and JunPEΔ/Δ;PtenPEΔ/Δ prostate samples of 19-week-old animals. An antibody against the epithelial marker EpCAM was used to separate single cell suspensions of minced and digested prostates into EpCAM positive (pos) and negative (neg) fractions by magnetic cell sorting. EpCAMpos cells were used for RNA-seq expression profiling. c Heat map showing correlation analysis of tumor samples described in b) regarding global similarity of samples. The Pearson correlation coefficient (R) is shown (color coded). d Gene onthology (GO)-enrichment analysis of differentially expressed genes (DEGs) showing the top differentially regulated pathways between PtenPEΔ/Δ and JunPEΔ/Δ;PtenPEΔ/Δ. Significance as shown by FDR is color coded, enriched (positive normalized enrichment score (NES)) or depleted (negative NES) processes are indicated. Asterisk represents non-significant pathways (ns). e Heat map showing SenMayo genes most significantly (p ≤ 1e-02) regulated among PtenPEΔ/Δ and JunPEΔ/Δ;PtenPEΔ/Δ prostates. f GSEA enrichment analysis using the Guccini_core_SASP gene set in PtenPEΔ/Δ versus JunPEΔ/Δ;PtenPEΔ/Δ and PtenPEΔ/Δ versus wt animals. g Heat map representation of wt, PtenPEΔ/Δ, JunPEΔ/Δ and JunPEΔ/Δ;PtenPEΔ/Δ samples showing DEGs. “Innate immunity”, FDR = 7.64e-05; “Immune system”, FDR = 2.77e-04 and “Extracellular space”, FDR = 6.60e-03 related processes most discriminated the groups. Genotypes and expression levels are color coded. h GO-enrichment analysis of DEGs showing the regulation of innate immune cells such as neutrophil granulocytes. Significance as shown by p-value is color coded, enriched (positive NES) or depleted (negative NES) processes are indicated. Shown are the signaling pathways enriched in PtenPEΔ/Δ tumors compared to wt (left side) and JunPEΔ/Δ;PtenPEΔ/Δ tumors compared to PtenPEΔ/Δ (right side)
We next performed a comparative analysis of JunPEΔ/Δ; PtenPEΔ/Δ and PtenPEΔ/Δ prostate samples to discern JUN-dependent programs potentially contributing to PCa formation. Our survey revealed 1706 (p.adjust < 5e-02) differentially expressed genes (DEGs) with top 102 genes being up- (log2fold change ≥ 1) and top 91 genes downregulated (log2fold change ≤ -1; Supplementary Table 1). DAVID analysis of top genes showed increased “innate immunity” and “immune system processes” but decreased secretory-, extracellular matrix- and immune-related processes. Notably, Jun ranked among the top 10 downregulated genes confirming the successful knockout in epithelial cells (Supplementary Table 1). Gene set enrichment analysis (GSEA) revealed immune system-related processes, IL-6/STAT3 signaling and senescence-associated gene signatures among the most enriched processes in PtenPEΔ/Δ prostates which were significantly depleted in JunPEΔ/Δ; PtenPEΔ/Δ (Fig. 3d). Our previous work suggested that activation of IL-6/STAT3 signaling and of the downstream acting p19ARF–MDM2–p53 axis contributed to senescence in PtenPEΔ/Δ prostates [12]. We therefore investigated the enrichment level of different senescence signatures including “oncogene-induced senescence” (OIS), “SASP” signatures and the novel “SenMayo” gene signature, consisting of 125 previously identified senescence/SASP-associated factors. SenMayo genes are transcriptionally regulated by senescence and allow identification of senescent cells across tissues [41]. SenMayo genes were significantly (qval = 2.40e-02) enriched in PtenPEΔ/Δ prostates and depleted (qval = 2.64e-02) in JunPEΔ/Δ; PtenPEΔ/Δ tumors (Fig. 3d). Among the depleted SenMayo genes in Jun-deficient PtenPEΔ/Δ prostates, we identified chemokines such as Ccl3, Ccl4 and Ccl8, along with pro-inflammatory cytokines such as Il1b and Tnfa (Fig. 3e). As these secreted cytokines and chemokines represent well described SASP factors, we next investigated a SASP core gene signature previously described in a Pten-deficient prostate model [42]. Using GSEA, we indeed detected enrichment of the SASP core signature in Pten-deficient prostates which was reverted in JunPEΔ/Δ; PtenPEΔ/Δ animals (Fig. 3f). To investigate further aspects of JUN-dependent regulation of senescence in Pten-deficient murine prostates, we stained formalin-fixed paraffin embedded (FFPE) material with the senescence markers p16INK4A, p21CIP1/WAF1 and Galactosidase beta 1 (GLB1) (Supplementary Fig. 3c). We did not observe differences in the amount of p16INK4A positive cells between PtenPEΔ/Δ and JunPEΔ/Δ; PtenPEΔ/Δ tumors, but found significant changes in staining patterns. While we detected prominent nuclear staining in PtenPEΔ/Δ samples, JunPEΔ/Δ; PtenPEΔ/Δ revealed predominantly cytoplasmic localization, hinting at a potential inactivation of p16INK4A via nuclear export [43]. In wt and JunPEΔ/Δ prostates, we observed a weak lobe-dependent expression pattern of p21CIP1/WAF1. Conversely, in PtenPEΔ/Δ and JunPEΔ/Δ; PtenPEΔ/Δ samples, p21CIP1/WAF1 was expressed in each individual epithelial cell, with no discernible difference between the two groups. GLB1 staining displayed its characteristic granular expression pattern prompting us to quantify percentage of positive area however we found no significant difference between PtenPEΔ/Δ and JunPEΔ/Δ; PtenPEΔ/Δ groups. Apart from changes in the p16 staining pattern, we found no significant deregulation of the classic senescence-associated cell cycle markers, implicating that JUN affects the SASP but not senescence-associated cell cycle arrest.
As our results suggest JUN-dependent activation of the IL-6/STAT3 axis and our previous study connected loss of activated STAT3 in Pten-deficient PCa to increased tumor burden via disruption of senescence [12], we sought to analyze STAT3 tyrosine 705 (Y705) phosphorylation (pSTAT3Y705) in the Jun-deficient background. We indeed detected reduced levels of pSTAT3Y705 in both stroma (p = 5.0e-04) and epithelial cells (p < 1.0e-04) of JunPEΔ/Δ; PtenPEΔ/Δ compared to PtenPEΔ/Δ tumors (Supplementary Fig. 3d upper panel, Supplementary Fig. 3e) while total STAT3 levels remained constant (Supplementary Fig. 3d, lower panel). Our findings provide evidence that loss of JUN accompanied by reduced activation of STAT3 bypasses SASP and subsequently amplifies the tumor load in JunPEΔ/Δ; PtenPEΔ/Δ animals. We suggest an interplay of JUN and STAT3 mediating senescence-associated secretion of inflammatory factors in PCa in vivo, reinforcing JUN’s proposed function as a pioneering factor of senescence [23].
JUN deficiency in the PCa mouse model leads to downregulated chemotaxis of innate immune cells
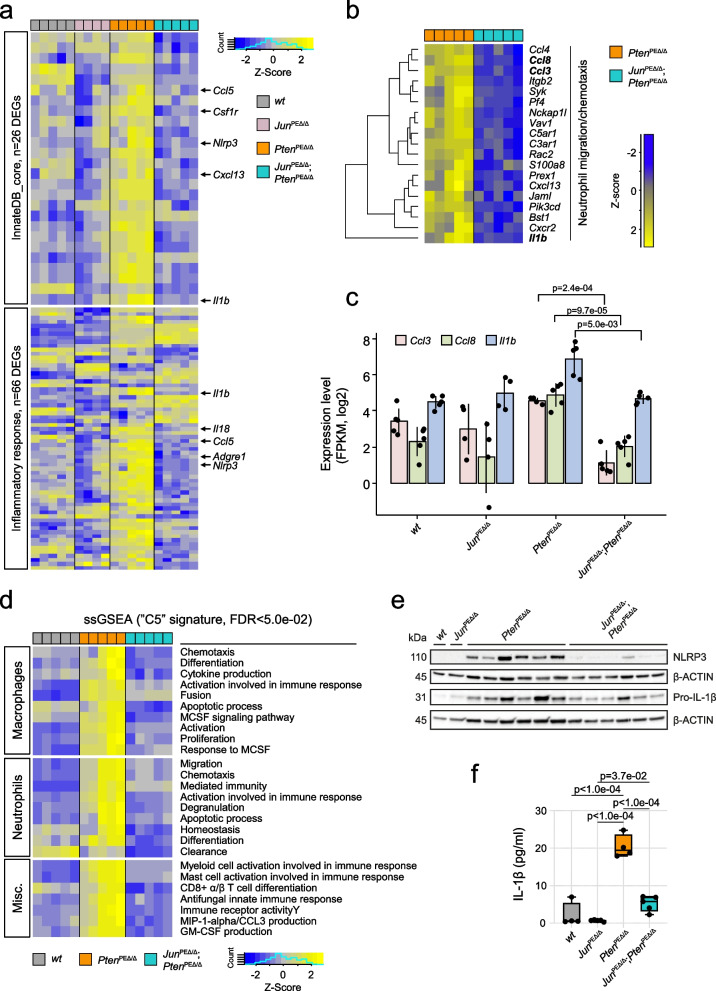
We next compared JunPEΔ/Δ; PtenPEΔ/Δ and PtenPEΔ/Δ prostate samples to uncover additional JUN-dependent biological processes involved in PCa formation. A stringent selection identified ~100 significantly deregulated genes (padj ≤ 1.0e-03, FClog2 ≤ -1.2; n = 59/ FClog2 ≥ 1.2; n = 46; Supplementary Table 3) and uncovered innate immunity and other immune system-related processes as most distinguishing between JunPEΔ/Δ; PtenPEΔ/Δ and PtenPEΔ/Δ prostate tumors (Fig. 3g). Amongst the innate immunity and immune system cluster, gene onthology (GO)-enrichment analysis indeed confirmed immune system-related signatures that were activated in PtenPEΔ/Δ and significantly reduced by Jun-deficiency (Fig. 3h). Innate immunity-related processes are complex and encompass more than 2000 publicly available human and mouse annotated genes [44]. We defined a core immunity-related signature by GSEA applying 645 innate immunity-related genes and investigated the enrichment specifically in PtenPEΔ/Δ prostates. The analysis revealed 111 genes, of which 26 were significantly (p < 1.0e-03) differentially expressed between PtenPEΔ/Δ and JunPEΔ/Δ; PtenPEΔ/Δ prostates (Fig. 4a, top panels, Supplementary Fig. 4a, Supplementary Table 4). Using the “Hallmark Inflammatory response” signature, we uncovered a similar pattern as the majority of genes from both signatures were significantly (p < 1.0e-03) elevated in PtenPEΔ/Δ and depleted in JunPEΔ/Δ; PtenPEΔ/Δ prostates (Fig. 4a, bottom panels, Supplementary Fig. 4a). Hence, the homozygous loss of Pten was accompanied by inflammation and inflammatory response likely driven by increased levels of Il1b, Nlrp3 and chemokines such as Ccl5.
Fig. 4.
JUN expression determines the level of immune cell infiltration of Pten-loss driven tumors. a Heat map showing JUN-dependent regulation of genes related to innate immunity (upper panel) and inflammatory response (lower panel) in wt, JunPEΔ/Δ, PtenPEΔ/Δ and JunPEΔ/Δ;PtenPEΔ/Δ prostates. JUN-dependent core factors such as Il1b, Nlrp3 and Ccl5 are highlighted. b Heat map presenting the JUN-dependent regulation of genes involved in migration and chemotaxis of neutrophil granulocytes in PtenPEΔ/Δ and JunPEΔ/Δ;PtenPEΔ/Δ prostates. Genotypes and expression levels in a-b are color coded. c Expression levels (log2, FPKM) of Ccl3, Ccl8 and Il1b are significantly (Ccl3, p = 2.4e-04; Ccl8, p = 9.7e-05 and Il1b, p = 5.0e-03) reduced in EpCAM+ cells of JunPEΔ/Δ;PtenPEΔ/Δ prostates. Significance was determined by an unpaired two-sided t-test. d Single-sample GSEA analysis using the M5 signature of Broad Institute’s molecular signature database (MsigDB) revealing enrichment of macrophage- and neutrophil-associated properties in PtenPEΔ/Δ compared to JunPEΔ/Δ;PtenPEΔ/Δ prostates. e Western blot analysis of NLRP3 and non-cleaved Pro-IL-1β in all four experimental groups in biological replicates. β-ACTIN served as loading control. f Multiplex immunoassay of homogenized prostate samples of 19-week-old wt, JunPEΔ/Δ, PtenPEΔ/Δ and JunPEΔ/Δ;PtenPEΔ/Δ animals for analysis of IL-1β levels in pico grams (pg)/ml of indicated biological replicates. Statistical testing was done with one-way Anova, significant p-values are indicated
Cells of the innate immune system, including neutrophil granulocytes, mast cells and macrophages serve as the primary defense against infections and consequently recruit T and B cells to infection sites [45]. Among the DEGs of PtenPEΔ/Δ versus JunPEΔ/Δ; PtenPEΔ/Δ prostates, we identified neutrophil movement-specific gene signatures that play a crucial role in the recruitment of immune cells (Fig. 4b) [46]. We observed that cytokines involved in chemotaxis of immune cells such as Ccl3, Ccl8 and Il1b were significantly deregulated between groups (Fig. 4c). To further dissect the potentially involved immune cell subsets, we conducted single sample GSEA using the M5 ontology gene sets signature from the molecular signature database (MsigDB). We identified enrichment of macrophage- and neutrophil-specific gene signatures characterized by cellular activities such as migration, activation/differentiation and enhanced expression indicating production of MIP1α/CCL3 and GM-CSF. Moreover, single sample GSEA revealed processes related to other immune cell subsets such as mast cells, myeloid cells and CD8+ T cells that were significantly enriched in PtenPEΔ/Δ compared to wt prostates and depleted in JunPEΔ/Δ; PtenPEΔ/Δ (Fig. 4d). This implicates JUN in the control of inflammatory states during PCa progression. We validated the JUN-dependent regulation of IL-1β, TNF-α and NLRP3, all involved in the regulation of inflammatory response processes by immunoblot and cytokine analyses (Fig. 4e-f and Supplementary Fig. 4b).
To further examine the apparent shifts in immune system-related transcriptomic signatures, we assessed granulocytic or lymphocytic cell infiltrations based on microscopic characteristics in H&E staining of all four genotypes (Supplementary Fig. 4c). We detected no or low-grade infiltration by inflammatory cells in wt and JunPEΔ/Δ specimens. In contrast, PtenPEΔ/Δ mouse prostates exhibited increased levels of high- and middle-grade infiltrations, which were significantly mitigated in JunPEΔ/Δ; PtenPEΔ/Δ prostates. Increased immune cell infiltration of PtenPEΔ/Δ prostates as identified by histo-pathological analysis therefore supported the results of transcriptome profiling. This highlights the importance of JUN in the regulation of inflammation by affecting the secretion of pro-inflammatory cytokines in Pten-deficient PCa.
Epithelial JUN deficiency modulates the migration of innate immune cells from the periphery
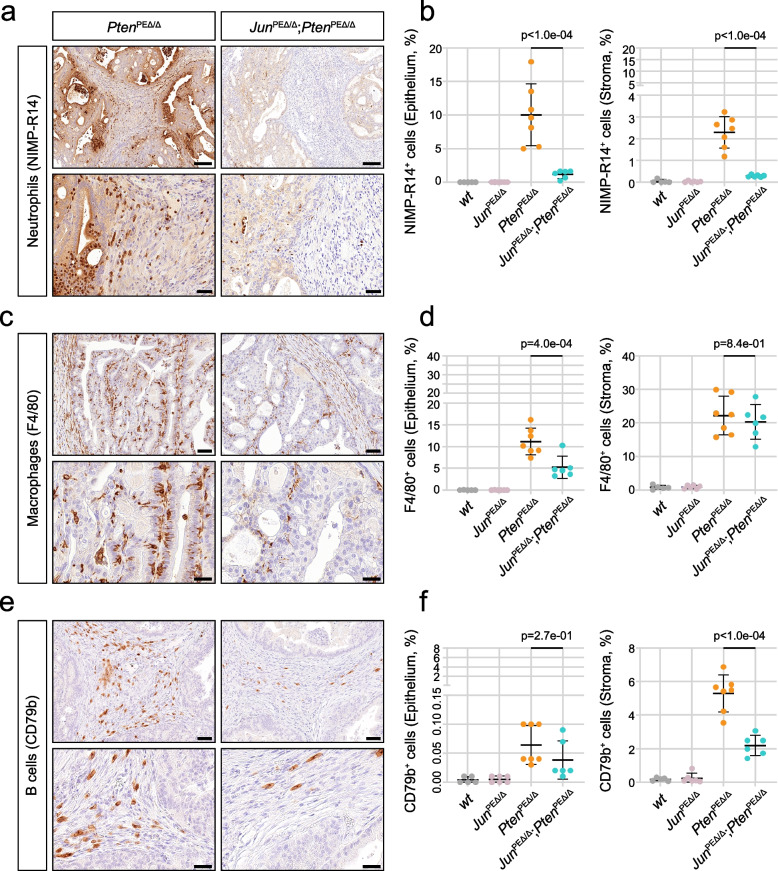
To investigate the distribution and abundance of infiltrating immune cells, we performed IHC stainings. Neutrophils and inflammatory monocytes were stained using the antibody clone NIMP-R14, which targets the specific cell surface markers and differentiation antigens Ly-6G and Ly-6C (Fig. 5a). In PtenPEΔ/Δ prostates, we observed high numbers of neutrophils migrating from the blood vessels across the stroma into the epithelium, where they predominantly accumulated, and subsequently advanced into the lumen. In JunPEΔ/Δ; PtenPEΔ/Δ prostates, we detected significantly (p < 1.0e-04) less neutrophils in the stroma and epithelium, but the migration patterns remained consistent with PtenPEΔ/Δ tumors (Fig. 5b). In contrast, macrophages, stained by the marker F4/80 were primarily located in the stroma, with no significant differences between the groups (Fig. 5c-d). We observed significantly (p = 4.0e-04) less macrophages infiltrating the epithelium in prostates with additional deficiency of Jun. In conclusion, PtenPEΔ/Δ displayed a highly immune infiltrated phenotype, which was substantially reverted in prostates with additional deficiency of Jun. This observation suggests that JUN may be essential for tumor cell recognition by innate and consequently adaptive immune cells.
Fig. 5.
Histological analysis of infiltrating immune cells reveals downregulated innate immune response in JunPEΔ/Δ;PtenPEΔ/Δ prostates. a Representative images of IHC stainings of NIMP-R14, a pan-marker of neutrophil granulocytes, indicating high neutrophil infiltration of PtenPEΔ/Δ prostates, reverted by the additional loss of Jun in JunPEΔ/Δ;PtenPEΔ/Δ prostates. Top row: 20.0 × magnification, scale bar represents 150 µm; Bottom row: 63.0 × magnification, scale bar represents 40 µm. b Quantification of NIMP-R14+ neutrophils in epithelium (left) and stroma (right). A significantly decreased (p < 1e-04) infiltration of neutrophils in tumors and adjacent stroma of JunPEΔ/Δ;PtenPEΔ/Δ prostates is evident. c Representative images of IHC stainings for the pan-marker of macrophages F4/80. A high infiltration of PtenPEΔ/Δ prostates and adjacent stroma by macrophages is evident and reverted by the additional loss of Jun in JunPEΔ/Δ;PtenPEΔ/Δ prostates. Top row: 40.0 × magnification, scale bar represents 60 µm; Bottom row: 100.0 × magnification, scale bar represents 30 µm. d Quantification of F4/80+ macrophages in epithelium (left) and stroma (right). A significantly decreased (p = 4e-04) infiltration of macrophages in tumors but not adjacent stroma (p = 8.3e-01) of JunPEΔ/Δ;PtenPEΔ/Δ prostates is evident. e Representative images of IHC stainings of B cell infiltration using the pan-marker CD79b. A high infiltration of stroma adjacent to PtenPEΔ/Δ prostates by CD79b+ B cells is evident and reverted by the additional loss of Jun in JunPEΔ/Δ;PtenPEΔ/Δ prostates. Top row: 40.0 × magnification, scale bar represents 60 µm; Bottom row: 100.0 × magnification, scale bar represents 30 µm. f Quantification of B cells in epithelium (left) and stroma (right). B cell infiltration as observed in the stroma of PtenPEΔ/Δ prostates was significantly decreased (p < 1e-04) in JunPEΔ/Δ;PtenPEΔ/Δprostates. Statistical significance between PtenPEΔ/Δ and JunPEΔ/Δ;PtenPEΔ/Δ groups are indicated in b, d and f
Neutrophils attract T cells to the site of inflammation via secretion of chemokines such as CCL2 and CCL5 [47, 48]. We utilized multiplex IHC to discern the T cell subsets, employing a marker panel consisting of CK/CD3/CD4/CD8/CD45/PD-1/DAPI. We observed various T cell subpopulations (T helper (CD4+) and cytotoxic T cells (CD8+), PD-1 positive and negative) mainly in the stroma and to a lesser degree in the epithelium (Supplementary Fig. 5a-b). To further investigate active cytotoxic T cells and natural killer cells, we performed Granzyme B IHC (Supplementary Fig. 5c-d). We did not observe a significant effect of Jun deficiency on any of the investigated populations. Additionally, we investigated the infiltration of B cells, stained by CD79b. B cells were found almost exclusively in the stroma, with significantly less infiltration in JunPEΔ/Δ; PtenPEΔ/Δ compared to PtenPEΔ/Δ prostates (Fig. 5e-f). In summary, IHC validated the JUN-dependent modulation of the immune cell compartment, particularly affecting innate immune cells. This phenotype was likely provoked by a JUN-dependent regulation of neutrophil attracting chemokines such as IL-1β.
Increased expression of SASP factors is correlated with prolonged survival in prostate cancer
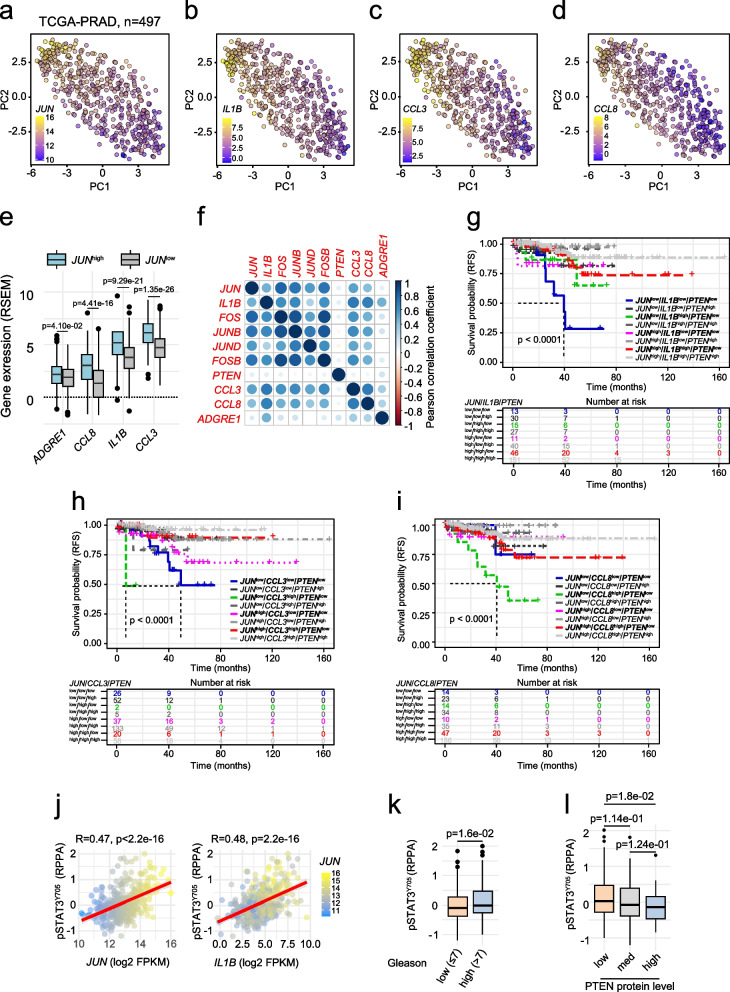
To translate our findings to the human disease, we investigated a potential association of JUN and SASP factors. We compared the levels of JUN, IL1B, CCL3 and CCL8 in patient data (TCGA-PRAD [33]) by PCA and found high IL1B, CCL3 and CCL8 mRNA in tumors expressing high levels of JUN. In contrast, tumors expressing low JUN levels revealed equally low amounts of IL1B, CCL3 and CCL8 (Fig. 6a-d). Upon separation of TCGA-PRAD tumors by using mean JUN expression as cut-off in JUN high and low expressing groups, we indeed confirmed enrichment of SASP factors and neutrophil marker ADGRE1 in JUN high subgroups (Fig. 6e). We next performed a Pearson correlation analysis and detected a significant but weak positive (R ≤ 0.64; p ≤ 0.05) association of IL1B, CCL3 and CCL8 with JUN and additional AP-1 factors such as JUND, JUNB, FOS and FOSB (Fig. 6f).
Fig. 6.
Expression of immune cell-attracting chemokines CCL3 and CCL8 correlates with levels of JUN in patient datasets. a-d Principal component analysis (PCA) representation of human PCa illustrating expression levels of JUN, IL1B, CCL3 and CCL8. Expression levels are color coded from high (yellow) to low (blue). e Box plots indicating significant enrichment of ADGRE1 (F4/80, p = 4.10e-02), CCL8 (p = 4.41e-16), IL1B (p = 9.29e-21) and CCL3 (p = 1.35e-26) in JUNhigh and JUNlow separated groups. f Pearson correlation of indicated AP-1 factors, PTEN, CCL3, CCL8, IL1B and ADGRE1. Strength of correlation is color coded. g-i Kaplan–Meier survival analyses of TCGA-PRAD tumors (n = 333) assessing the effect of IL1B (g), CCL3 (h) and CCL8 (i) on RFS in the context of PTEN and JUN. j Correlation of JUN (left) and IL1B (right) expression to amount of phosphorylated STAT3 (pSTAT3Y705) in the TCGA-PRAD cohort (n = 352). k Box plot of reverse-phase protein array (RPPA) data representing reduced levels of pSTAT3Y705 (p = 1.6e-02) in high risk PCa of Gleason scores > 7 (range 8–10) compared to low risk (Gleason scores ≤ 7). l Box plot of RPPA data representing gradually decreasing levels of pSTAT3Y705 (p = 1.8e-02) in PTEN low, medium and high tumors. Dataset used for Fig. 6 is TCGA-PRAD [33]
Assuming that JUN mediates tumor-suppressor activity via positive regulation of SASP factors required for the recruitment of immune cells, we expected that high levels of SASP factors may be associated with favorable prognosis. Hence, we asked whether cytokine expression may act in concert with JUN to influence patient survival. We stratified patients according to their PTEN, JUN and IL1B (Fig. 6g) CCL3 (Fig. 6h) or CCL8 (Fig. 6i) levels and compared the RFS between groups. As expected, PTENhigh groups (presented in gray shades) showed overall favorable outcomes and did not significantly differ from one another. In groups where all marker genes were lowly expressed we generally detected the worst prognosis while groups with a singular lowly expressed marker showed intermediate prognosis. CCL8 did not follow the observed IL1B and CCL3 profiles suggesting that this chemokine does not act in concert with JUN in the absence of PTEN. Finally, we investigated a potential relationship between JUN and STAT3 activation and explored STAT3’s role in immune modulation in TCGA-PRAD data [33]. We correlated RPPA of pSTAT3Y705 with levels of JUN and IL1B. We observed a weak but significant correlation of JUN (R = 0.47, p < 2.2e-16) and IL1B (R = 0.48, p = 2.2e-16) with pSTAT3Y705 (Fig. 6j). As observed for JUN, PCa exhibiting a high (> 7) Gleason score showed reduced levels of pSTAT3Y705 (p = 1.6e-02) when compared to low risk tumors (Gleason score ≤ 7) (Fig. 6k). To correlate the main oncogenic driver PTEN to pSTAT3Y705, we grouped patients according to varying PTEN levels and observed a dose dependent decrease of pSTAT3Y705 (Fig. 6l). These results were analogous to our previous findings where we detected gradually reduced JUN expression in PTEN medium and high expressing tumors (Fig. 1i). These data might hint at interconnected mechanisms of both transcriptional regulators. In summary, we propose that levels of JUN and STAT3 potentially orchestrated via PTEN, determine progression stages of human prostate tumors by modulating the immune response through regulation of cytokines and interleukins as identified in the Jun-deficient murine PCa model.
Discussion
PCa is among the most frequently diagnosed malignancies in men worldwide and a significant number of patients progress to advanced and lethal stages. The mortality linked to metastatic PCa highlights the pressing need to elucidate its intricate mechanisms and pinpoint viable therapeutic interventions. Despite this urgency, the cellular mechanisms and environmental contexts that control PCa development and progression remain incompletely understood. Loss of PTEN is evident in 20% of primary human prostate carcinomas and escalates in 50% of metastatic CRPC [4]. Comparable to the human situation, Pten loss leads to the formation of precancerous lesions in PE cells in mouse models [49, 50]. Aggressive carcinomas develop only in the presence of additional mutations [51], such as abnormal expression of ERG [52], loss of IL-6/STAT3 functionality [12, 13], dysfunction of the methyltransferase Kmt2c [32] or activation of the RAS/MAPK cascade [51, 53]. While several studies indicate that augmented JUN expression drives PCa progression [14, 54], the functional role of JUN and AP-1 TFs in PCa remains controversial. Intriguingly, genetic disruption of JunB in vivo accelerated the progressive phenotype of Pten-deficient PCa [22]. A recent study using in vivo CRISPR to achieve combinatorial deletion of Pten and Fos in the PE led to increased tumorigenesis potentially via upregulation of Jun [55]. In contrast, we did not observe compensatory functions of other AP-1 family members in our genetically engineered mouse model or the analysis of human patient data. We therefore speculate that Fos deletion might be more susceptible to influencing levels of other AP-1 members than Jun or that the upregulation of JUN might arise due to differential CRISPR targeting efficiencies of Pten and Fos in the chosen model. An alternative explanation for the increased tumorigenesis observed upon Fos knockout in PCa is provided by evidence that FOS stimulates the trans-activation properties of JUN but represses its AR co-activator function [56]. Loss of Fos might therefore predominantly favor Jun’s pro-proliferative co-activator function and weaken its anti-proliferative trans-activator function [57]. Our results similarly point towards a context dependent tumor-suppressive role, rather than a driving function of JUN in PCa progression.
In the present study, our focus was to delineate the role of JUN in PCa. We first examined JUN levels in clinical PCa samples and analyzed JUN patterns across varying progression stages from three publicly available datasets [35, 37, 38]. We found that JUN expression increased in tumors relative to normal prostates, however we did not observe the same effect when we compared healthy to low Gleason stages in the TMA data. It remains to be elucidated whether this discrepancy can be explained by differences in protein versus RNA levels or the PTEN mutation status in early stages of malignant transformation. Importantly, the levels of JUN, FOS and JUNB and the levels of JUN in the TMA dataset were all significantly decreased with progression of PCa. This suggests that high JUN levels may protect from development of progressive disease, a hypothesis further supported by the increased survival rates of patients harboring high JUN expressing tumors. By co-integrating the tumor suppressor PTEN in our survival analysis, we found that levels of PTEN and JUN determine survival probabilities of PCa and revealed the worst prognosis in JUNlow/PTENlow PCa but highest RFS in JUNhigh/PTENhigh tumors. Encouraged by these findings, we studied the functional role of JUN in a murine PCa model, characterized by homozygous loss of Pten (PtenPEΔ/Δ) [26, 39]. Mirroring the early prostatic intraepithelial neoplasia (PIN) stages of human PCa, JUN was significantly upregulated in PtenPEΔ/Δ prostates. Consistent with human patient data, depletion of Jun alone had no effect on the morphological architecture and growth of the prostate. Epithelial cells of PtenPEΔ/Δ prostates developed hyperplasia, subsequently forming prostate adenocarcinoma and rapidly progressing upon additional deletion of Jun. The aggressive phenotype observed in JunPEΔ/Δ; PtenPEΔ/Δ prostates resulted in decreased survival of mice and increased prostate weight and size. We did not detect signs of severe organ dysfunction, systemic inflammation or metastatic disease (Fig. 2g).
The TME is a dynamic system characterized by chronic inflammation and participation of diverse host components, but plays a pivotal role in cancer progression [58]. Within the TME, immune cells such as tumor-associated macrophages (TAM) and tumor-associated neutrophils (TAN) both foster cancer progression or combat tumor cells, underscoring their dual roles in tumorigenesis [59–61]. Central to this environment is the SASP, where senescent cells release a plethora of inflammatory mediators. SASP-driven effects often culminate in the immune-mediated clearance of potential tumorigenic cells, a process termed “senescence surveillance” [62, 63]. Our histopathologic examination of PtenPEΔ/Δ PCa samples revealed significant enrichment of neutrophils and macrophages that infiltrated the tumors and adjacent stroma. Concurrent deletion of Jun strikingly reduced tumor infiltration with neutrophils and macrophages and accelerated tumor growth. Transcriptomic analyses of PtenPEΔ/Δ and JunPEΔ/Δ; PtenPEΔ/Δ prostates revealed a JUN-dependent modulation of SASP-associated genes, but we did not identify compensatory upregulation of other AP-1 members as it has been described upon inactivation of FOS [64]. To investigate further aspects of senescence and address senescence-associated cell cycle arrest, we conducted IHC stainings for p16INK4a, p21CIP/WAF1 and GLB1. We did not observe quantitative differences in expression of these classical senescence markers indicating that JUN is involved in the regulation of SASP, but not senescence-associated cell cycle arrest. The regulation of SASP without affecting cell cycle arrest has been previously demonstrated. This was evidenced by the association of the chromatin reader BRD4 with recruitment to enhancer regions activating SASP genes in senescent cells [65]. Recent findings have implicated AP-1 and in particular JUN as pioneering factors on a specific enhancer landscape essential for the execution of senescence-controlling programs [23]. In line with these results, we propose that loss of Pten coupled with an increase in JUN likely instigates a JUN-driven SASP phenotype.
SASP involves the expression and secretion of inflammatory cytokines such as CCL3, CCL8, IL-1β and TNF-α [32, 66] which subsequently recruit immune cells such as neutrophils, macrophages and T cells [10, 67, 68]. As we observe downregulated secretion of IL-1β and TNF-α in JunPEΔ/Δ; PtenPEΔ/Δ prostates, we suggest that Jun depletion in Pten-deficient prostates disrupts SASP. This impedes the recruitment of neutrophils and macrophages, as well as tumor cell clearance by macrophages and dendritic cells [9, 62]. We thus propose JUN as a key regulator of SASP. Our results support a previous study where JUN depletion was linked to diminished inflammatory responses and reverting the senescent/SASP phenotype of RAS-OIS fibroblasts to a proliferating phenotype [23]. Furthermore, GM-CSF, a direct JUN target has been shown to amplify macrophage and neutrophil immune responses [69] and modulate pro-inflammatory cytokine secretion such as TNFα and IL-6 [70].
Another intriguing mechanism of JUN-dependent modulation of the immune phenotype in PCa may depend on STAT3 levels. Our previous work identified activation of STAT3 and a p19ARF–MDM2–p53 axis to induce senescence upon Pten depletion [12]. Consistently, Jun loss was associated with decreased IL-6-JAK-STAT3 signaling, evidenced by significantly reduced pSTATY705 levels in JunPEΔ/Δ; PtenPEΔ/Δ prostates. ENCODE database exploration [71] revealed mutual promoter binding sites for JUN and STAT3 suggesting a potential JUN-STAT3 interplay in impacting senescence pathways in PCa (https://maayanlab.cloud/Harmonizome/dataset/CHEA+Transcription+Factor+Targets). This interplay is supported by results of a STAT3 binding analysis in CD4+ T cells, which suggests that STAT3 directly regulates the expression of Jun and Fos and may potentially function in a positive feedback loop [72]. Therefore, therapeutic activation of STAT3 potentially causes SASP factor modulation and may elevate JUN levels in tumors, thereby restricting tumor progression and enhancing PCa patient survival.
Conclusions
In summary, our data suggest that JUN functions as a pivotal regulator of SASP and survival in PTEN-deficient PCa, orchestrating the recruitment dynamics of TAMs and TANs within the TME. Given the indispensable role of robust SASP in immune surveillance of preneoplastic anomalies, its therapeutic modulation presents intricate challenges. Our recent investigations have shown the potential of the antidiabetic agent metformin, which curtails multiple pro-inflammatory SASP components by inhibiting NF-κB nuclear translocation [73]. Metformin increases STAT3 in advanced PCa cases, leading to significant tumor growth attenuation, underscored by reduced mTORC1/CREB and AR levels in a PCa murine model [13]. The interplay between JUN, STAT3 and PTEN might represent a key mechanism that could be exploited for therapeutic advances.
Supplementary Information
Acknowledgements
This research was supported using resources of the VetCore Facilities (VetImaging, Genomics and Transcriptomics) of the University of Veterinary Medicine Vienna. We acknowledge the Core Facility Genomics and Core Facility Bioinformatics supported by the NCMG research infrastructure (LM2023067 funded by MEYS CR) for their support with the bioinformatic analysis of scientific data presented in this paper. We would like to thank Anton Jäger, Medical University of Vienna for macroscopic image generation and support. We thank Romana Sickha for technical assistance.
Authors’ information
Torben Redmer, Martin Raigel and Christina Sternberg contributed equally to this work as first authors. Sabine Lagger and Lukas Kenner contributed equally to this work as last authors.
Abbreviations
- AP-1
Activator protein-1
- AR
Androgen receptor
- BCR
Biochemical recurrence
- BRD4
Bromodomain containing protein 4
- CRPC
Castration resistant prostate cancer
- DEG
Differentially expressed gene
- EpCAM
Epithelial cell adhesion molecule
- EV
Empty vector
- FDR
False discovery rate
- FFPE
Formalin-fixed paraffin embedded
- GLB1
Galactosidase beta 1
- GO
Gene ontology
- GSEA
Gene set enrichment analysis
- H&E
Hematoxylin and eosin
- HR
Hazard ratio
- IF
Immunofluorescence
- IHC
Immunohistochemistry
- JNK
JUN N-terminal kinase
- MsigDB
Molecular signature database
- NEPC
Neuroendocrine prostate cancer
- NES
Normalized enrichment score
- OIS
Oncogene induced senescence
- PbCre
Probasin Cre
- PCa
Prostate cancer
- PCA
Principal component analysis
- PE
Prostate epithelium
- PI3K
Phosphoinositide 3-kinase
- PICS
PTEN-loss induced cellular senescence
- PIN
Prostatic intraepithelial neoplasia
- PTEN
Phosphate and Tensin Homologue
- RFS
Relapse-free survival
- RNA-seq
RNA sequencing
- RPPA
Reverse-phase protein array
- SASP
Senescence-associated secretory phenotype
- STAT3
Signal transducer and activator of transcription 3
- TAM
Tumor-associated macrophage
- TAN
Tumor-associated neutrophil
- TCGA-PRAD
Cancer Genome Atlas Prostate Adenocarcinoma
- TF
Transcription factor
- TMA
Tissue microarray
- TME
Tumor microenvironment
Authors’ contributions
Conceptualization: TR, MR, CS, SL, LK. Formal Analysis: TR, KT, MO, MB, HAN, VB. Funding Acquisition: LK, SP. Investigation: TR, MR, CS, RZ, CP, DL, AA, TL, PK, SH, MS, SS, MO, MB, HAN, IG, OM, SL. Methodology: MR, CS, RZ, CP, AA, TL, SMi, MT, NSH, BT, VB, BS, GE, SL. Project administration: TR, MR, CS, SL, LK. Resources: TR, MR, CS, RM, JT, SMa, FA, SP, JLP, GE, SL, LK. Supervision: SL, LK. Validation: TR, MR, CS, KT, PK, SH, MS, MO, FS, BT, VB, OM, GE. Visualization: TR, MR, CS, RZ, CP, AA, MB, HAN, TL. Writing – original draft: SL, TR, MR. Writing – review & editing: TR, MR, CS, TL, SS, RM, SMa, FA, BS, OM, GE, SL, LK. All authors read and approved the final manuscript.
Funding
LK acknowledges the support from MicroONE, a COMET Modul under the lead of CBmed GmbH, which is funded by the federal ministries BMK and BMDW, the provinces of Styria and Vienna, and managed by the Austrian Research Promotion Agency (FFG) within the COMET—Competence Centers for Excellent Technologies—program. Financial support was also received from the Austrian Federal Ministry of Science, Research and Economy, the National Foundation for Research, Technology and Development, and the Christian Doppler Research Association, as well as Siemens Healthineers. LK was also supported by European Union Horizon 2020 Marie Sklodowska-Curie Doctoral Network grants (ALKATRAS, n. 675712; FANTOM, n. P101072735 and eRaDicate, n. 101119427) as well as BM Fonds (n. 15142), the Margaretha Hehberger Stiftung (n. 15142), the Christian-Doppler Lab for Applied Metabolomics (CDL-AM), and the Austrian Science Fund (grants FWF: P26011, P29251, P34781 as well as the International PhD Program in Translational Oncology IPPTO 59.doc.funds). OM is supported by the Austrian Science Fund (FWF) project (P32579) and LK, OM, GE and SP European Union Horizon 2020 Marie Sklodowska-Curie Doctoral Network grants ALKATRAS, n. 675712; FANTOM, n. P101072735. Additionally, this research was funded by the Vienna Science and Technology Fund (WWTF), grant number LS19-018. LK, OM, GE and SP are members of the European Research Initiative for ALK-Related Malignancies (www.erialcl.net). SM and BS received funding from FWF DocFund DOC32-B28 and SFB F6101. CS was supported by grant Nr. 70112589 from the Deutsche Krebshilfe, Bonn, Germany. SP received funding from Next Generation EU for the project National Institute for Cancer Research (Program EXCELES, Nr. LX22NPO5102).
Availability of data and materials
The RNA-seq dataset supporting the conclusions of this article is available in the GEO repository with the accession number GSE242433.
Declarations
Ethics approval and consent to participate
Institutional Review Board Statement: The use of clinical material was approved by the Research Ethics Committee of the Medical University Vienna, Austria (1877/2016) and conducted in adherence to the Declaration of Helsinki protocols. Patient consent was waived due to the completely anonymized, retrospective nature of the study.
All animal studies were reviewed and approved by the Federal Ministry Republic of Austria for Education, Science and Research and conducted according to regulatory standards (BMWFW-66.009/0144-WF/II/3b/2014 and the amendments BMWFW-66.009/0063-WF/V/3b/2017, BMWFW-66.009/0137-WF/V/3b/2019, BMWFW-66.009/0359-V/3b/2019, 2020–0.016.881, 2020–0.659.052 and 2022–0.325.014).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Torben Redmer, Martin Raigel, Christina Sternberg, Sabine Lagger and Lukas Kenner shared contribution.
Contributor Information
Torben Redmer, Email: torben.redmer@vetmeduni.ac.at.
Sabine Lagger, Email: sabine.lagger@vetmeduni.ac.at.
Lukas Kenner, Email: lukas.kenner@meduniwien.ac.at.
References
- 1.Gandaglia G, Leni R, Bray F, Fleshner N, Freedland SJ, Kibel A, Stattin P, Van Poppel H, La Vecchia C. Epidemiology and Prevention of Prostate Cancer. Eur Urol Oncol. 2021;4:877–892. doi: 10.1016/j.euo.2021.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Berenguer CV, Pereira F, Câmara JS, Pereira JAM. Underlying Features of Prostate Cancer-Statistics, Risk Factors, and Emerging Methods for Its Diagnosis. Curr Oncol. 2023;30:2300–2321. doi: 10.3390/curroncol30020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan ME, Li J, Xu HE, Melcher K, Yong E. Androgen receptor: structure, role in prostate cancer and drug discovery. Acta Pharmacol Sin. 2015;36:3–23. doi: 10.1038/aps.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jamaspishvili T, Berman DM, Ross AE, Scher HI, De Marzo AM, Squire JA, Lotan TL. Clinical implications of PTEN loss in prostate cancer. Nat Rev Urol. 2018;4:222–234. doi: 10.1038/nrurol.2018.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z, Trotman LC, Shaffer D, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung SH, Hwang HJ, Kang D, Park HA, Lee HC, Jeong D, Lee K, Park HJ, Ko YG, Lee JS. mTOR kinase leads to PTEN-loss-induced cellular senescence by phosphorylating p53. Oncogene. 2019;38:1639–1650. doi: 10.1038/s41388-018-0521-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorgoulis V, Adams PD, Alimonti A, et al. Cellular Senescence: Defining a Path Forward. Cell. 2019;179:813–827. doi: 10.1016/j.cell.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Schosserer M, Grillari J, Breitenbach M. The Dual Role of Cellular Senescence in Developing Tumors and Their Response to Cancer Therapy. Front Oncol. 2017;7:315584. doi: 10.3389/fonc.2017.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coppé J-P, Desprez P-Y, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Culig Z, Puhr M. Interleukin-6 and prostate cancer: Current developments and unsolved questions. Mol Cell Endocrinol. 2018;462:25–30. doi: 10.1016/j.mce.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Pencik J, Schlederer M, Gruber W, et al. STAT3 regulated ARF expression suppresses prostate cancer metastasis. Nat Commun. 2015;6:7736. doi: 10.1038/ncomms8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pencik J, Philippe C, Schlederer M, et al. STAT3/LKB1 controls metastatic prostate cancer by regulating mTORC1/CREB pathway. Mol Cancer. 2023;22:133. doi: 10.1186/s12943-023-01825-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ouyang X, Jessen WJ, Al-Ahmadie H, et al. Activator protein-1 transcription factors are associated with progression and recurrence of prostate cancer. Cancer Res. 2008 doi: 10.1158/0008-5472.CAN-07-6055. [DOI] [PubMed] [Google Scholar]
- 15.Vogt PK. Fortuitous convergences: the beginnings of JUN. Nat Rev Cancer. 2002;2:465–469. doi: 10.1038/nrc818. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Bergami P, Lau E, Ronai Z. Emerging roles of ATF2 and the dynamic AP1 network in cancer. Nat Rev Cancer. 2010;10:65–76. doi: 10.1038/nrc2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 18.Cai C, Hsieh CL, Shemshedini L. c-Jun has multiple enhancing activities in the novel cross talk between the androgen receptor and Ets variant gene 1 in prostate cancer. Mol Cancer Res. 2007;5:725–735. doi: 10.1158/1541-7786.MCR-06-0430. [DOI] [PubMed] [Google Scholar]
- 19.Bubulya A, Chen SY, Fisher C, Zheng Z, Shen X, Shemshedini L. c-Jun Potentiates the Functional Interaction between the Amino and Carboxyl Termini of the Androgen Receptor. J Biol Chem. 2001;276:44704–44711. doi: 10.1074/jbc.M107346200. [DOI] [PubMed] [Google Scholar]
- 20.Shaulian E. AP-1 - The Jun proteins: Oncogenes or tumor suppressors in disguise? Cell Signal. 2010;22:894–899. doi: 10.1016/j.cellsig.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 21.Hübner A, Mulholland DJ, Standen CL, et al. JNK and PTEN cooperatively control the development of invasive adenocarcinoma of the prostate. Proc Natl Acad Sci. 2012;109(30):12046 LP–12051. doi: 10.1073/pnas.1209660109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomsen MK, Bakiri L, Hasenfuss SC, Wu H, Morente M, Wagner EF. Loss of JUNB/AP-1 promotes invasive prostate cancer. Cell Death Differ. 2015;22:574–582. doi: 10.1038/cdd.2014.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martínez-Zamudio RI, Roux P-F, de Freitas JANLF, et al. AP-1 imprints a reversible transcriptional programme of senescent cells. Nat Cell Biol. 2020;22:842–855. doi: 10.1038/s41556-020-0529-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birbach A, Eisenbarth D, Kozakowski N, Ladenhauf E, Schmidt-Supprian M, Schmid JA. Persistent inflammation leads to proliferative neoplasia and loss of smooth muscle cells in a prostate tumor model. Neoplasia. 2011;13:692–703. doi: 10.1593/neo.11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behrens A, Sibilia M, David J-P, Möhle-Steinlein U, Tronche F, Schütz G, Wagner EF. Impaired postnatal hepatocyte proliferation and liver regeneration in mice lacking c-jun in the liver. EMBO J. 2002;21:1782–1790. doi: 10.1093/emboj/21.7.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki A, Yamaguchi MT, Ohteki T, et al. T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity. 2001 doi: 10.1016/S1074-7613(01)00134-0. [DOI] [PubMed] [Google Scholar]
- 27.Wu X, Wu J, Huang J, Powell WC, Zhang J, Matusik RJ, Sangiorgi FO, Maxson RE, Sucov HM, Roy-Burman P. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mech Dev. 2001;101:61–69. doi: 10.1016/S0925-4773(00)00551-7. [DOI] [PubMed] [Google Scholar]
- 28.Oberhuber M, Pecoraro M, Rusz M, et al. STAT 3‐dependent analysis reveals PDK 4 as independent predictor of recurrence in prostate cancer. Mol Syst Biol. 2020;16(4):e9247. doi: 10.15252/msb.20199247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bankhead P, Loughrey MB, Fernández JA, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017 doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt U, Weigert M, Broaddus C, Myers G. Cell Detection with Star-convex Polygons. 2018 doi: 10.1007/978-3-030-00934-2_30. [DOI] [Google Scholar]
- 31.Ding Z, Wu CJ, Chu GC, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011 doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Limberger T, Schlederer M, Trachtová K, et al. KMT2C methyltransferase domain regulated INK4A expression suppresses prostate cancer metastasis. Mol Cancer. 2022;21:89. doi: 10.1186/s12943-022-01542-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research Network TCGAR The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humphrey PA. Gleason grading and prognostic factors in carcinoma of the prostate. Mod Pathol. 2004 doi: 10.1038/modpathol.3800054. [DOI] [PubMed] [Google Scholar]
- 35.Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aguirre-Gamboa R, Gomez-Rueda H, Martínez-Ledesma E, Martínez-Torteya A, Chacolla-Huaringa R, Rodriguez-Barrientos A, Tamez-Peña JG, Treviño V. SurvExpress: An Online Biomarker Validation Tool and Database for Cancer Gene Expression Data Using Survival Analysis. PLoS ONE. 2013;8:1–9. doi: 10.1371/journal.pone.0074250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu YP, Landsittel D, Jing L, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004 doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 38.Bolis M, Bossi D, Vallerga A, et al. Dynamic prostate cancer transcriptome analysis delineates the trajectory to disease progression. Nat Commun. 2021;12:7033. doi: 10.1038/s41467-021-26840-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S, Gao J, Lei Q, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4:209–221. doi: 10.1016/S1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 40.Tannenbaum J, Bennett BT. Russell and Burch’s 3Rs then and now: The need for clarity in definition and purpose. J Am Assoc Lab Anim Sci. 2015;54:120–132. [PMC free article] [PubMed] [Google Scholar]
- 41.Saul D, Kosinsky RL, Atkinson EJ, et al. A new gene set identifies senescent cells and predicts senescence-associated pathways across tissues. Nat Commun. 2022 doi: 10.1038/s41467-022-32552-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guccini I, Revandkar A, D’Ambrosio M, et al. Senescence Reprogramming by TIMP1 Deficiency Promotes Prostate Cancer Metastasis. Cancer Cell. 2021;39:68–82.e9. doi: 10.1016/j.ccell.2020.10.012. [DOI] [PubMed] [Google Scholar]
- 43.Nilsson K, Landberg G. Subcellular localization, modification and protein complex formation of the cdk-inhibitor p16 in Rb-functional and Rb-inactivated tumor cells. Int J Cancer. 2006;118:1120–1125. doi: 10.1002/ijc.21466. [DOI] [PubMed] [Google Scholar]
- 44.Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, Winsor GL, Hancock REW, Brinkman FSL, Lynn DJ InnateDB: systems biology of innate immunity and beyond-recent updates and continuing curation. 10.1093/nar/gks1147 [DOI] [PMC free article] [PubMed]
- 45.Marshall JS, Warrington R, Watson W, Kim HL. An introduction to immunology and immunopathology. Allergy, Asthma Clin Immunol. 2018;14:49. doi: 10.1186/s13223-018-0278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sionov RV, Fridlender ZG, Granot Z. The Multifaceted Roles Neutrophils Play in the Tumor Microenvironment. Cancer Microenviron. 2015 doi: 10.1007/s12307-014-0147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reichel CA, Puhr-Westerheide D, Zuchtriegel G, Uhl B, Berberich N, Zahler S, Wymann MP, Luckow B, Krombach F. C-C motif chemokine CCL3 and canonical neutrophil attractants promote neutrophil extravasation through common and distinct mechanisms. Blood. 2012 doi: 10.1182/blood-2012-01-402164. [DOI] [PubMed] [Google Scholar]
- 48.Metzemaekers M, Gouwy M, Proost P. Neutrophil chemoattractant receptors in health and disease: double-edged swords. Cell Mol Immunol. 2020;17:433–450. doi: 10.1038/s41423-020-0412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang SI, Parsons R, Ittmann M. Homozygous deletion of the PTEN tumor suppressor gene in a subset of prostate adenocarcinomas. Clin Cancer Res. 1998;4(3):811–5. [PubMed] [Google Scholar]
- 50.Wise HM, Hermida MA, Leslie NR. Prostate cancer, PI3K, PTEN and prognosis. Clin Sci. 2017;131(3):197–210. doi: 10.1042/CS20160026. [DOI] [PubMed] [Google Scholar]
- 51.Baker SJ, Reddy EP. Understanding the temporal sequence of genetic events that lead to prostate cancer progression and metastasis. Proc Natl Acad Sci U S A. 2013;110:14819–14820. doi: 10.1073/pnas.1313997110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carver BS, Tran J, Gopalan A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009 doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mulholland DJ, Kobayashi N, Ruscetti M, Zhi A, Tran LM, Huang J, Gleave M, Wu H. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-11-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thakur N, Gudey SK, Marcusson A, Fu JY, Bergh A, Heldin CH, Landstrom̈ M, TGFβ-induced invasion of prostate cancer cells is promoted by c-Jun-dependent transcriptional activation of Snail1. Cell Cycle. 2014 doi: 10.4161/cc.29339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Udayappan UK, Casey PJ. c-Jun Contributes to Transcriptional Control of GNA12 Expression in Prostate Cancer Cells. Molecules. 2017 doi: 10.3390/molecules22040612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tillman K, Oberfield JL, Shen X-Q, Bubulya A, Shemshedini L. c-Fos Dimerization with c-Jun Represses c-Jun Enhancement of Androgen Receptor Transactivation. Endocrine. 1998;9:193–200. doi: 10.1385/ENDO:9:2:193. [DOI] [PubMed] [Google Scholar]
- 57.Chen S-Y, Cai C, Fisher CJ, Zheng Z, Omwancha J, Hsieh C-L, Shemshedini L. c-Jun enhancement of androgen receptor transactivation is associated with prostate cancer cell proliferation. Oncogene. 2006;25:7212–7223. doi: 10.1038/sj.onc.1209705. [DOI] [PubMed] [Google Scholar]
- 58.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 59.Galdiero MR, Bonavita E, Barajon I, Garlanda C, Mantovani A, Jaillon S. Tumor associated macrophages and neutrophils in cancer. Immunobiology. 2013;218:1402–1410. doi: 10.1016/j.imbio.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 60.Qian B-Z, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun B, Qin W, Song M, Liu L, Yu Y, Qi X, Sun H. Neutrophil Suppresses Tumor Cell Proliferation via Fas /Fas Ligand Pathway Mediated Cell Cycle Arrested. Int J Biol Sci. 2018;14:2103–2113. doi: 10.7150/ijbs.29297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takasugi M, Yoshida Y, Ohtani N. Cellular senescence and the tumour microenvironment. Mol Oncol. 2022;16:3333–3351. doi: 10.1002/1878-0261.13268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Riedel M, Berthelsen MF, Cai H, et al. In vivo CRISPR inactivation of Fos promotes prostate cancer progression by altering the associated AP-1 subunit Jun. Oncogene. 2021;40:2437–2447. doi: 10.1038/s41388-021-01724-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tasdemir N, Banito A, Roe J-S, et al. BRD4 Connects Enhancer Remodeling to Senescence Immune Surveillance. Cancer Discov. 2016;6:612–629. doi: 10.1158/2159-8290.CD-16-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muñoz-Espín D, Serrano M. Cellular senescence: From physiology to pathology. Nat Rev Mol Cell Biol. 2014;15:482–496. doi: 10.1038/nrm3823. [DOI] [PubMed] [Google Scholar]
- 67.Alexander E, Hildebrand DG, Kriebs A, Obermayer K, Manz M, Rothfuss O, Essmann F, Schulze-Osthoff K. IκBζ is a regulator for the senescence-associated secretory phenotype in DNA damage- and oncogene-induced senescence. J Cell Sci. 2013;126:3738–3745. doi: 10.1242/jcs.128835. [DOI] [PubMed] [Google Scholar]
- 68.Freund A, Orjalo AV, Desprez PY, Campisi J. Inflammatory networks during cellular senescence: causes and consequences. Trends Mol Med. 2010 doi: 10.1016/j.molmed.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lotfi N, Thome R, Rezaei N, Zhang G-X, Rezaei A, Rostami A, Esmaeil N. Roles of GM-CSF in the Pathogenesis of Autoimmune Diseases: An Update. Front Immunol. 2019;10:452989. doi: 10.3389/fimmu.2019.01265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mausberg AK, Jander S, Reichmann G. Intracerebral granulocyte-macrophage colony-stimulating factor induces functionally competent dendritic cells in the mouse brain. Glia. 2009;57:1341–1350. doi: 10.1002/glia.20853. [DOI] [PubMed] [Google Scholar]
- 71.Luo Y, Hitz BC, Gabdank I, et al. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 2020;48:D882–D889. doi: 10.1093/nar/gkz1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Durant L, Watford WT, Ramos HL, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32:605–615. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moiseeva O, Deschênes-Simard X, St-Germain E, Igelmann S, Huot G, Cadar AE, Bourdeau V, Pollak MN, Ferbeyre G. Metformin inhibits the senescence-associated secretory phenotype by interfering with IKK / NF -κ B activation. Aging Cell. 2013;12:489–498. doi: 10.1111/acel.12075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq dataset supporting the conclusions of this article is available in the GEO repository with the accession number GSE242433.