Abstract
Detailed characterization of human pancreatic islets is key to elucidating the pathophysiology of all forms of diabetes, especially type 2 diabetes (T2D). However, access to human pancreatic islets is limited. Pancreatic tissue for islet retrieval can be obtained from brain-dead organ donors or from patients undergoing pancreatectomy, often referred to as ‘living donors’. Different protocols for human islet procurement can substantially impact islet function. This variability, coupled with heterogeneity between individuals and islets, results in analytical challenges to separate genuine disease pathology or differences between human donors from experimental noise. There are currently no international guidelines for human donor phenotyping, islet procurement and functional characterisation. This lack of standardization means that substantial investments from multiple international efforts towards improved understanding of diabetes pathology cannot be fully leveraged. In this Perspective, we overview the status of the field of human islet research, highlight the challenges and propose actions which could accelerate research progress and increase understanding of T2D to slow its pandemic spreading.
Introduction
The development of protocols for isolation of human islets and their improvement for treatment of type 1 diabetes (T1D) by islet transplantation have enhanced access to isolated human islets for research1. There are now multiple academic and commercial entities around the world which provide human islets for research, some of which also generate and make available data characterising this important mini-organ for diabetes2–7. Some of these initiatives have now taken first steps to characterize and compare islets from individuals with and without type 2 diabetes (T2D)2,7–14. However, questions persist in the field regarding the importance and accuracy of metabolic phenotyping of these donors and the modalities for islet retrieval and downstream analysis. Reaching a consensus on these unresolved and often controversial topics is important since information gathered thus far has lacked consistency.
In this perspective article we confront our different views on current practices to obtain and study human islets with the intent to identify both strengths and limitations of each approach, as well as strategies to temper such limitations and accelerate efforts to deliver robust insights into T2D pathogenesis. We limit our discussion to the procurement and study of adult islets given that access to fetal and neonatal material presents additional challenges and is performed in very few centres.
Pancreas Procurement for Human Islet Research: The Donors
Human islets can be obtained from various sources. Pancreatic tissue samples from autopsy are useful for the histological study of islet cell morphology, morphometry and (indirectly) turnover, but lack of functional data, limited molecular information and possible autolysis of the pancreatic tissues remain problematic15. Laparoscopic-assisted biopsy samples have occasionally been obtained from individuals with T1D16,17, but ethical constraints due to the risk of complications restrict the applicability of this approach. In recent years, most information on human islets and the insulin-producing beta cells within the islet has come from two sources: islets isolated from organ donors or pancreatic fragments from pancreatectomized patients, henceforth referred to as living donors (FIGURE 1).
Fig. 1. Comparative analysis of living and organ donors as the source of pancreatic tissue for islet studies.
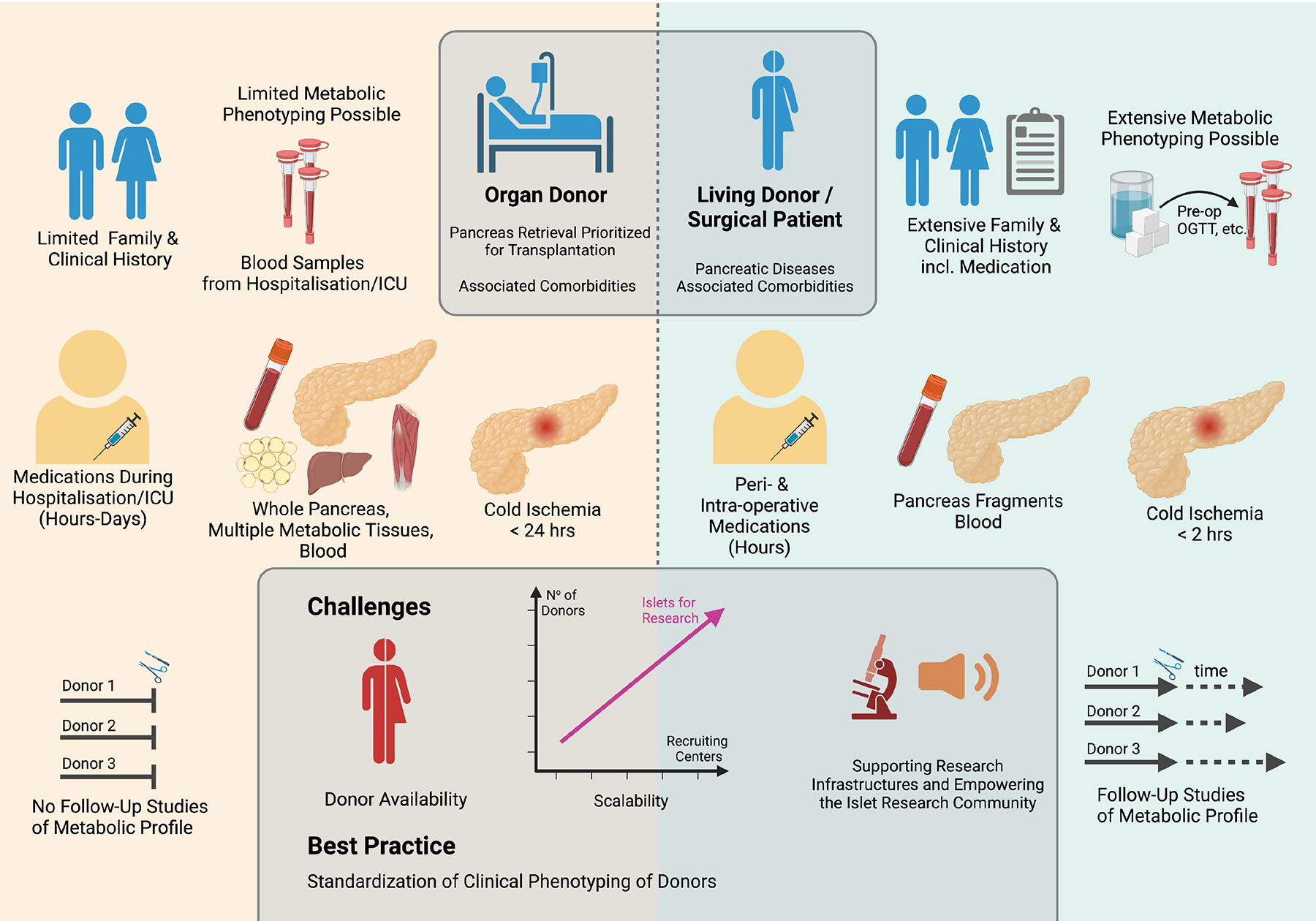
The schematic representation illustrates some key opportunities and limitations specifically associated with either organ donors (left) or living donors (right). These range from clinical and laboratory profiling of the donor, amount and properties of the retrieved pancreatic tissue, possibilities for analysis of other organs and follow-up investigations of the same donor. The lower central box highlights challenges in the field, such as the need to increase the number of donors and recruiting centers by increasing public awareness and funding to support the infrastructures required for these activities. Best practice requires the standardization of the clinical phenotyping of the donors. This figure was created with BioRender.com.
Organ donors
Most information on human islet cells has been generated from islets isolated from the pancreas of brain-dead multi-organ donors2,3,7–15,18. Information on clinical and family history on these subjects is necessarily more limited than in the case of living donors. In addition to body mass index, age and sex of the donor, the glycemic status may be inferred based on the medical history and, if available, by HbA1c level, which provides a measure of the glycemic control during the last 3 months prior to death. Like other biochemical parameters, such as insulin or C-peptide concentrations, glycemia and HbA1c may be affected, at least in part, by the clinical status of the patient, including pharmacological treatment, large blood loss due to trauma, and blood transfusion. The integrity of the exocrine pancreas may be inferred by markers, such as amylase and lipase.
This approach involves the resection of the whole pancreatic gland, thereby enabling the enzymatic digestion and isolation of many islets, which could be then widely distributed to many laboratories for comprehensive imaging, functional, biochemical, (epi)genomic and transcriptional analyses (FIGURE 2 and FIGURE 3). This isolation strategy is different from studies of pancreatic fragments or slices from living donors where the limited available tissue is studied using standardized experimental approaches in just a few laboratories and where an integrated team of clinical investigators, surgeons, pathologists, and scientists work collaboratively. However, enzymatic isolation and culturing of islets as well as dissociation and sorting of single islet cells may affect their molecular composition and function19,20.
Fig. 2. Models for recruitment of donors and pancreatic tissue for islet isolation and characterization.
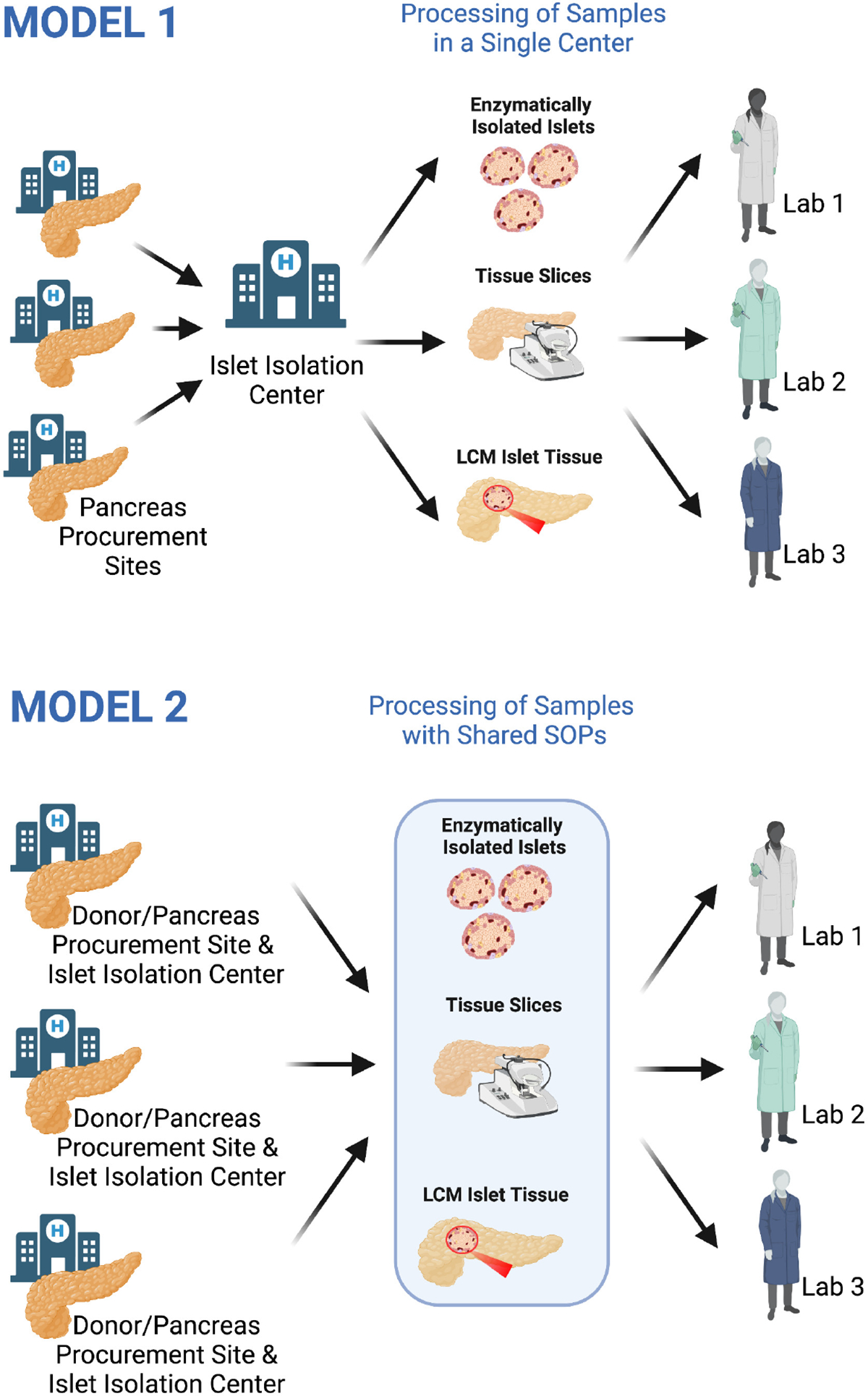
Model 1 (top) relies on a single isolating centre obtaining whole pancreas organs or pancreatic fragments from donors for tissue processing. Enzymatic isolated islets, pancreatic tissue slices and sections or islet fragments obtained by laser capture microdissection are then distributed to individual laboratories, which independently perform assays in parallel, some of which may be duplicated. Data analysis is restricted to the assays performed by a single laboratory, leading to the generation and publication of complementary, or potentially overlapping datasets, which are then deposited in public databases post-publication. This approach, which prevailed until recently, offers the opportunity to integrate different datasets from different groups and verify the reproducibility of data, but also entails a greater risk for redundancy and it is not well suited in cases where a finite amount of pancreatic tissue is available, as in the case of surgical samples from living donors. Model 2 (bottom) follows a more centralized workflow, with multiple sites each being responsible for the recruitment of living and/or organ donors, their phenotyping, and the retrieval of the islets/islet fragments by enzymatic isolation, laser capture microdissection or the generation of pancreatic tissue slices/sections are carried out according to shared standardised operating procedures. Biological samples are further distributed to different laboratories, each well-qualified for the performance of a specific predefined assay. Collected clinical, laboratory and meta-data arising from the analysis are then jointly integrated and stored in a database accessible to all members of the network. Following their joint publication and deposition in public databases, data become available to the whole community. This figure was created with BioRender.com.
Fig 3. Models for islet isolation and characterization.
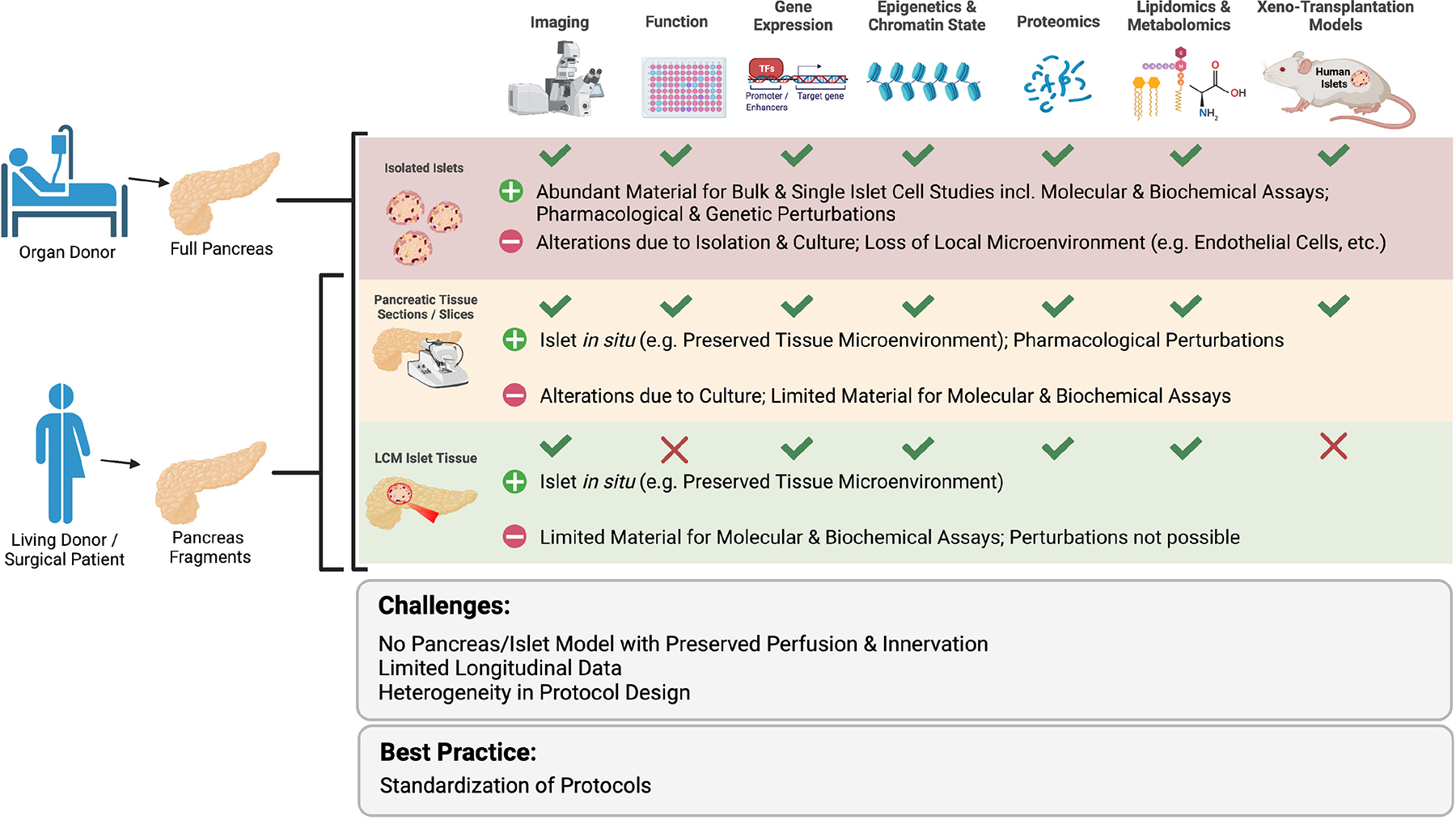
Whole pancreas from organ donors can be used for enzymatic isolation of islets, to obtain tissue slices and sections as well as islet fragments upon laser capture microdissection. In the case of surgical samples from living donors, the limited tissue availability precludes the enzymatic isolation of islets in numbers adequate in quality and quantity. As indicated, each of these different sources of islet material is best suited for a given set of analysis and comes with specific advantages (+) and limitations (−) relative to the others. Challenges and best practices common to all approaches are indicated. Xenotransplantation of human pancreatic tissue slices in immunodeficient mice is a potential future opportunity. This figure was created with BioRender.com.
Pancreatic tissue slices and sections as well as laser microdissected islet portions for in situ studies can also be obtained from organ donors (FIGURE 3). Samples of other tissues involved in the inter-organ metabolic crosstalk (like e.g., fat, skeletal muscle, and liver) may also be accessible. Since brain death is associated with a systemic inflammatory response21, histology of the retrieved gland should be performed to assess the degree of inflammatory cell infiltration, which according to some studies, but not others, may be increased with prolonged life support15. The cold ischemia of pancreases from organ donors before islets can be analysed is usually longer than that for the surgical samples from living donors. The threshold beyond which the pancreatic tissue is no longer suitable for islet studies is uncertain. For pancreas transplantation, the recommendation is for pancreatic grafts to be preserved for <12 hours, although islets are routinely isolated for research after preservation times of up to 24 hours22. More recently, the transplantation of pancreas and islets from non-heart-beating donors has been proposed23. In a few cases, the ex-vivo function of islets isolated from heart-beating and non-beating donors has been compared, and no major differences emerged24. Thus, the collection of islets from the latter should be encouraged, to improve access to human pancreatic islets.
Living Donors
Living donors, most of whom have undergone a pancreatectomy due to pancreatic ductal adenocarcinoma, can be metabolically phenotyped prior to surgery, whilst their detailed clinical and family history, including information on pharmacological treatments, can be collected7,14,15,25. Whether pre-surgical neoadjuvant chemotherapy affects islet cells and glucose homeostasis remains to be investigated. Measurements of pre-surgical fasting glucose and HbA1c levels combined with an oral glucose tolerance test (OGTT) may provide an opportunity to distinguish living donors with euglycemia from those with impaired fasting glycemia, impaired glucose tolerance or T2D at the time of surgery7,14,25. Type 3c diabetes (i.e. diabetes secondary to the underlying exocrine pancreatic disorder) is suspected in cases of recent onset hyperglycemia, typically <1 year before the onset of symptoms leading to pancreatectomy. Although more challenging to perform in patients with cancer, hyperinsulinemic-euglycemic and hyperglycemic clamps provide additional routes to assess insulin secretion and sensitivity in vivo25,26. In addition, follow-up studies after surgery may be conducted, contributing to the understanding of the mechanisms leading to the onset of T2D and means to (potentially) predict the disease27,28.
A concern associated with the study of islet cells of living donors with cancer or pancreatitis is the potential molecular and clinical changes induced by these pathologies on islet cells or systemic metabolism (e.g., weight loss, altered oral intake). Therefore, islets in the “healthy” pancreatic tissue from the surgical margins should be histologically examined to exclude their infiltration by cancer cells. In non-diabetic donors an influence of cancer may also be revealed by the expression of genes generally not expressed in islet cells29. There is some evidence, however, that gene expression of retrieved islets is more influenced by the procedure for islet isolation, (i.e., enzymatic vs. laser capture microdissection) than by the presence of a concomitant pancreatic disorder, but more information is needed7. The influence on systemic metabolism from the malignancy on islets is not known. Although the amount of tissue obtained upon surgery is limited (FIGURE 2), portions of living donor Islets in situ can be thoroughly investigated either upon their laser capture microdissection or within fresh tissue slices and sections of fixed specimens7,14,21,25,30 (FIGURE 3).
Models for islet procurement
A major limitation of research on T2D with human islets is the cross-sectional rather than prospective characterisation of both the living and organ donor cohorts from which islets or tissue are obtained. It is inherent in the design of these experiments that only one measurement time-point is obtainable, which offers challenges in drawing conclusions regarding disease course. For example, it is not possible to tell whether patients with T2D have reduced beta cell mass because they were born with fewer/smaller islets or because there was a gradual decrease in their number over time.
For all approaches, islet heterogeneity constitutes a challenge, some of which might be attributable to genetic and epigenetic factors, which differ among human subjects much more than in syngeneic animal models exposed to the same environment. However, significant “noise” is also likely to result from the different protocols and reagents used by different labs for islet retrieval and investigation, including for live isolated islets and tissue slices and the time interval between their acquisition and processing. This variability speaks again to the value of implementing standard operating procedures to investigate the molecular profiles of islets or islet cells, whether upon enzymatic digestion or laser capture microdissection, which would be highly desirable and a major goal for our research community. In this respect the adoption of a centralised model where basic molecular and functional profiling coupled with genotyping of the donors are performed could be very beneficial, cost effective and reduce redundancy (FIGURE 2).
The anatomical origin of the islets, which in the case of living donors with cancer is dominantly in the head of the pancreas, seems instead to be less critical, as recent studies in both living and organ donors did not identify major differences between islets from the head or the body and tail of the pancreas7,14. However, >90% of PP cells are in the islets derived from the ventral pancreatic bud, and are therefore almost exclusively found in the posterior head and uncinate portions of the gland32.
Generally, two important caveats exist: 1) for the living donor approach it is assumed that islets in the surgical fragment represent islets in the entire pancreas; and 2) for the organ donor approach isolated islets are assumed to reflect the combination of islets from all pancreas regions, without loss of “fragile” islets/cells. The limited amount of tissue available from the “healthy” resection margins of surgical specimens precludes the reliable isolation of islets by enzymatic digestion. Hence, validation experiments requiring genetic manipulations are currently only pursuable in enzymatically isolated islets, i.e. not amenable for islets of living donors. However, physiological measurements, such as static and dynamic insulin secretion evoked by glucose and other segretagogues as well as measurements of the cytoplasmic calcium concentration ([Ca2+]i) can also be carried out in fresh pancreatic tissue slices. Therefore, these analyses could be a way in the future to compare and contrast living vs. organ donor islets.
Characterising the Human Islet in Health and Disease
Access to human islets for research continues to differ between countries and in almost all cases involves a cost which varies depending on funding models and whether islets are sourced from academic centers or commercial suppliers. Some commercial suppliers (e.g. inSphero - https://insphero.com/products/islet/) will provide so called “pseudo islets”, made by re-aggregating a defined number of single islet cells after their dissociation, with validated responsiveness to glucose and other insulin secretagogues.
An important consideration when designing an experiment requiring human islets is whether one is seeking to determine a physiological or a pathophysiological process. Whereas a limited number of islet preparations may be sufficient to establish the role of certain factors and pathways in the regulation of insulin secretion, the elucidation of whether these pathways become disrupted in T2D is much more challenging and requires many islet preparations from donors matched for age, sex, BMI, and possibly other clinical and laboratory parameters.
T2D is heterogeneous and progressive as is exemplified by marked differences in the clinical characteristics of the patients33. As both tissue retrieval and processing impact gene expression and islet function, experiments seeking to establish differences between tissue from donors with and without T2D are susceptible to artifacts unrelated to pathophysiology. The challenges here are exemplified by the very limited overlap in gene expression changes across the studies which have been performed7,10,13,14. The islet itself consists of multiple cell types, each with a precise role to play in governing how glucose homeostasis is achieved34. The 3-dimensional microenvironment and communication between the different islet cell types is also important for correct function19.
Current models for Islet Isolation, distribution and characterisation
Defining the pathophysiological changes in the islet which underlie T2D requires the careful study of large numbers of well-characterised donors with sufficient available tissue for multiple standardized assays to permit phenotyping of both isolated and in situ cells using molecular imaging and “omics” ideally at the level of single cells. Reproducibility of single-cell omics approaches, however, mainly due to technological limitations such as high variability for low expressed genes, amplification bias and the low number of cells sequenced remains an issue20.
Islets from donors with diabetes are rarely available through commercial routes or through clinical islet isolation centers as they ordinarily decline these organs due to a lack of suitability for transplant. Dedicated programs to obtain islets for research from living or organ donors exist, which require resources to support infrastructures and coordination between organ procurement and islet isolation centers (FIGURE 2).
The characterization of islets obtained through these initiatives varies but falls into two general models. In the first model after processing material is distributed to many independent labs and each research team works independently to generate data on the material, whilst in the second centralized model there are several different strategies to coordinate activity. These include pipelines where all assays are performed locally, co-operatives of several designated centers or a hybrid model where a core set of assays (e.g., measurement of glucose-stimulated insulin secretion and histology) are run locally and then complemented by data deposited by researchers who have received the islets through a distribution program. A recent review article by Walker and colleagues provides an excellent summary of these efforts34.
Both approaches have strengths and weaknesses, and much depends on the funding available to support the infrastructure and the centralised existence of expertise and resources34. Efforts to improve coordination of ‘core-assays’ across programs are encouraged. The NIDDK Integrated Islet Distribution Program (IIDP) performs both functional (Human Islet Phenotyping Program, HIPP) and genetic assays (Human Islet Genetic Initiative, HIGI) centrally4. Researchers who obtain islets from this program can access these data for these islets. These pipelines are coordinated with similar operations in the Human Pancreas Atlas Program (HPAP) and the Alberta Islet Core (AIC) which facilitates the alignment of assays and coordinated adoption of new platforms5,35. Unlike the IIDP and AIC, the HPAP program does not distribute islets but makes the data from islet analysis available to all investigators.
In Europe, several countries have islet isolation and characterization programs, including Italy, Switzerland, Belgium, France and the UK2,6,7. Some of them have used procedures standardized within European Community funded consortia (e.g., IMIDIA, RHAPSODY, INNODIA, T2DSystems). The Nordic Network for Clinical Islet transplantation provides islets for research across Scandinavia3. In Germany with the support of the German Center for Diabetes Research, but also the EU-IMI RHAPSODY and INNODIA consortia, a research program obtaining islets from living donors has been initiated. The limited amount of tissue available through this route will require a centralised analytical approach. Currently, an international program for the study of islets from living donors does not yet exist. In our view its establishment would provide critical infrastructure for research on human islets.
Standardising Islet Characterisation Efforts
The complexity of establishing a standard set of assays can be exemplified by considering the fundamental measurement of insulin secretion. It seems logical to propose that the concentrations of glucose and other secretagogues should stay close to physiological levels but when measuring multiple hormones or disentangling their influence this may not be optimal. Research groups have historically established their own pipelines and can understandably be reluctant to transition from a protocol which has served them well for years to one about which they have no “back-catalog” of data to draw upon.
One way to move towards consensus here would be for major stakeholders (e.g., the American Diabetes Association (ADA), the Human Islet Research Network (HIRN) and the European Association for the Study of Diabetes [EASD]) to work jointly to propose standardisation of a core set of assays that are performed on all human islets studied for research. These guidelines would standardise concentrations for glucose and a battery of other secretagogues that target specific pathways known or suspected to be of relevance to T2D (e.g., sulfonylureas, GLP-1, membrane depolarization with arginine or high extracellular K+). These should also include recommendations for the analysis and presentation of the reported data36. The IIDP and Alberta Islet Core programs as well as partners of the German Center for Diabetes Research already exchange islet samples to allow their characterisation through multiple pipelines. This exchange could be extended to other programs to provide a reference set of donors which have been studied across efforts. The islet auto-antibody field has for some time run workshops to evaluate assays performed in different centres and reach consensus on guidelines for their use37.
Generating a complete picture of the T2D human islets from DNA through RNA and protein to lipid, metabolites and function is an increasingly possible endeavor (FIGURE 3). Integrating individual level multi-omic data, often now at single-cell resolution, provides opportunities to distinguish cause from effect and to understand the role of different islet cell types on disease phenotype. The number of assays which can be performed on a single donor however is dependent on the tissue requirements for each assay and although many omic-assays are now possible at single cell resolution (e.g., scRNA-seq, snATAC-seq, single-cell proteomics) there are limitations (e.g., evaluation of low abundance transcripts and proteins).
For many omic-assays the standardization of protocols and analytical pipelines are in their infancy. As exemplified by the shift from bulk to single-cell sequencing, the rapid adoption of new technologies either by pivoting to new improved platforms, such as those enabling measurements of absolute rather than relative protein, lipid and metabolite quantities, or the inclusion of additional assays to the phenotyping pipeline can add enormous value. Given the importance of the islet micro-environment the emergence of spatial ‘omics’ holds the promise to increase our understanding on the role of local molecular regulation on islet pathology.
Data Analysis, Integration and Federation - Are We FAIR?
The principles of FAIR are that data should be Findable, Accessible, Interoperable and Reusable38. Although several international teams have established independent biorepositories of multi-omic data on human islets (e.g., HPAP, RHAPSODY), and even taken steps to add to and amalgamate existing data (e.g., TIGER, INSPIRE), it has proved challenging to bring these data together in any formal joint analysis2,12,14. Efforts to both meta-analyse2 and re-process12 through a single analytical pipeline have been explored. Despite previous calls for standardization of functional and genomic assays and improved reporting of meta-data, many existing cohorts still fall short on these basic principles. This lack of standardization is an enormous loss to the field as individually these collections are underpowered for most analyses but collectively, they have potential to make significant headway in understanding disease heterogeneity and diabetes pathogenesis. Both the INSPIRE and TIGER efforts are testament to this and demonstrate both the impact of larger sample sizes and the advantages of standardisation2,12.
Setting up for Success
Establishing the necessary infrastructure to facilitate data accessibility and interoperability starts with organ procurement (FIGURE 4). Assigning a unique identifier to any living or organ donor through a centralized route not only provides traceability but ensures that duplicate samples deposited in multiple biobanks can be identified and dealt with appropriately in any analysis. Where data are deposited and how they can be accessed also influence their reusability.
Fig 4. Schematic representation of data lifecycle management needs and challenges.
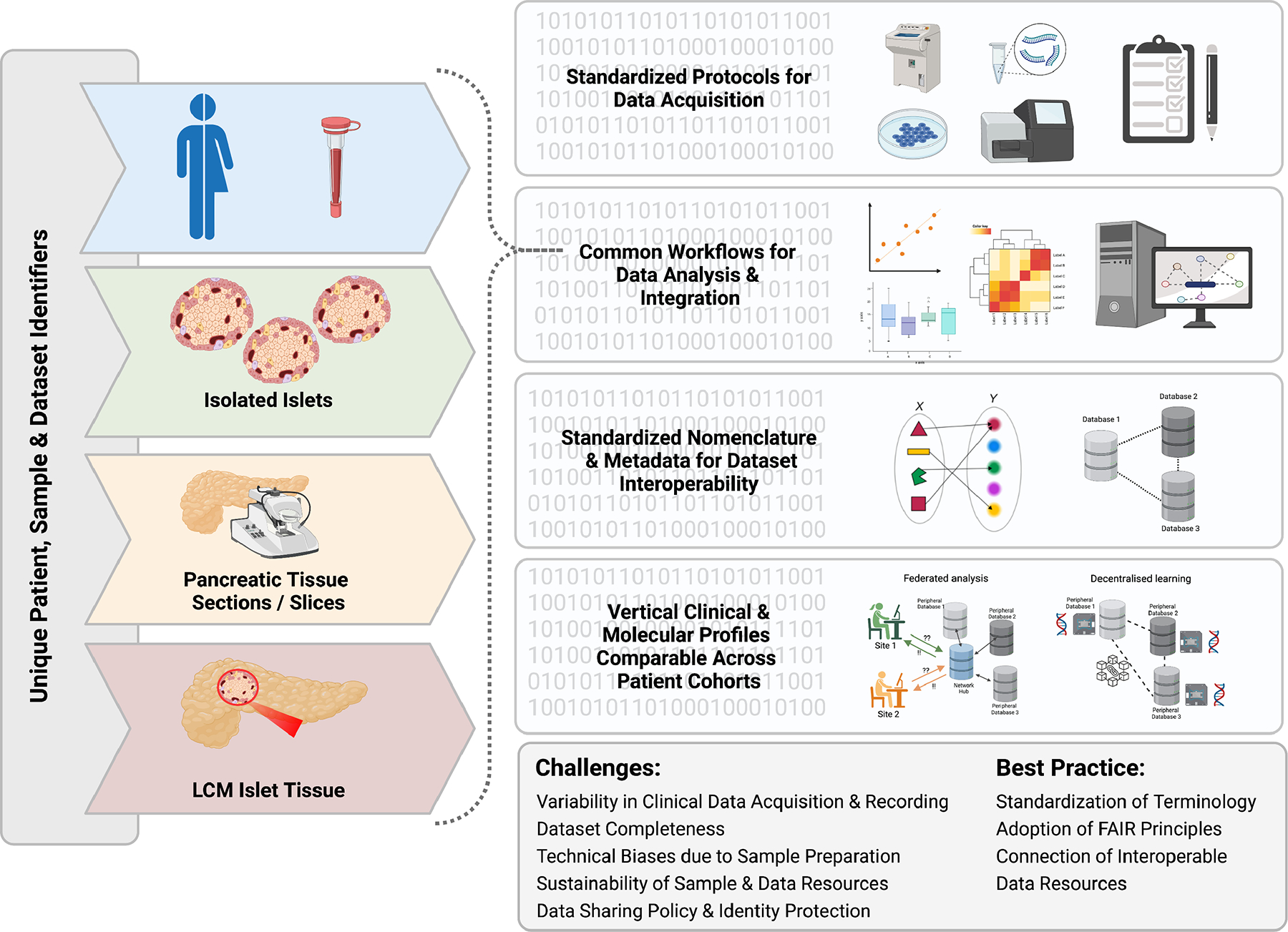
(left) Islet preparations are assigned a unique sample and donor identifier prior to data acquisition to ensure that each sample is unique and can be traced back to its origin (right). Data should be acquired using standardized protocols and be processed using common workflows for each technology to minimize batch and other confounding factors, ensuring data of the same type can be compared. Clinical information, sample metadata and molecular, functional, and imaging results need to be described according to international semantic standards to ensure interoperability between datasets. This enables them to be either integrated into a single database or be components of a federated network of interoperable datasets (as shown). Finally, standardized acquisition, processing and description of terminologies and pipelines enables clinical and laboratory data to be aligned and analyzed across donor cohorts, increasing statistical power to link findings to disease outcomes or molecular pathways. This can be done either by accessing a single database containing the combined data, or (as shown) via federated networks enabling interactive analysis of distributed databases (Federated analysis) or decentralised learning strategies such as Swarm learning where AI models can be learnt through exchange of model parameters between distributed databases. Current challenges and best practice are indicated. This figure was created with BioRender.com.
Distinguishing between platforms which offer users the chance to “browse” data and their interpretation and those which offer access to unprocessed data for analysis is important. HPAP provides registered users with opportunities to both visualise results on individual donors and download raw data (https://hpap.pmacs.upenn.edu). The TIGER resource (http://tiger.bsc.es/ ) offers users the opportunity to visualise processed published islet gene expression data across multiple cohorts in the context of other tissues and provides details of islet expression quantitative loci for each gene. Other consortia, such as RHAPSODY, make their raw data available upon publication.
The legal frameworks for data sharing vary between countries, and even within a country are often open to the interpretation of an individual institutional review board. Therefore, it is not always possible to openly share detailed clinical and genomic data, which is a significant barrier for their combined analysis. In such cases these barriers can be overcome with the use of federated networks, where the data remain on local servers under the control of data custodians, but end users are able to access and analyse the data at a distance. Such a system has been implemented for diabetes research in the EU-IMI RHAPSODY project, where non-disclosive cross-cohort analysis can be performed at a distance without sharing of individual-level data39. The Common Metabolic Disease Accelerated Medicines Partnership (CMD-AMP) (https://hugeamp.org) is a role model for the use of a single portal democratising access to valuable data generated with public funds to the broadest community of researchers accelerating research in both the public and private sectors.
With the growing number of data sets, particularly those at single-cell resolution, it is increasingly possible to use Artificial Intelligence (AI)-based approaches to identify disease state- or cell state-defining classifiers. Swarm Learning is an example of such an approach, where global AI models can be learnt from distributed datasets in a secure non-disclosive environment40. As more and more data are collected an overall picture of diseased and healthy islets can be built. This system should enable not only remote AI but also the visualisation and comparison of the datasets available.
As always, the independent validation of these classifiers (e.g., disease vs health in beta cells) across independent datasets remains important. For this to work, relevant clinical data need to be formatted in the same way (e.g., OMOP CDM), aligned to standardized vocabularies/ontologies (e.g., SNOMED, LOINC, RxNorm), and associated -omics data aligned to common standards to enable cross-cohort analysis. In the EU, EHDEN (https://www.ehden.eu) is creating a network of standardized clinical datasets for federated analysis. It would thus be beneficial to align to and learn from such initiatives to make clinical and -omics data from islets accessible for analysis through distributed networks when open data sharing is not possible.
Conclusions
Current therapies for T2D are not disease modifying, rather they target the manifestations of this disease, primarily hyperglycemia, and not the underlying mechanisms causing beta cell dysfunction, which remain unknown. Hence, research on the physiology and pathophysiology of human islets, albeit challenging, continues to be of paramount importance. To increase the possibilities of uncovering causal mechanisms for diabetes, and the potential to address them prior to disease onset, changes to current operational models are required. As described above these include the availability of additional islet sources, more powerful and reliable technological platforms for the standardized generation of quantitative islet data, implementation of more transparent and coordinated interaction models for the sharing of samples and the integration of datasets. Concerted efforts are especially timely in view of the recent identification of different prediabetes and T2D clusters35,41, which argue for a more precise molecular taxonomy, including that of the islets. In Box 1 we distill a series of recommendations for the field which we are confident can be achieved through the concerted effort of all stakeholders, including the scientific community, funding agencies, and the public.
BOX 1: Recommended actions.
Establishment of an international task force to bring existing efforts together to reach consensus of standardisation of methods.
Establish an international network for the collection and study of islets in surgical samples from living donors according to standardized protocols.
Development of national and international integrated teams for pancreas donor identification and characterization, organ or tissue procurement or processing, and islet or tissue distribution.
Standardized operating procedures for organ retrieval, processing and procurement of isolated islets or pancreatic fragments.
Standardized assessment of islet function and its adoption across all centres. This could include the exchange of samples and their characterisation by multiple centres.
Deposition of raw and meta-data generated on islet preparations received by individual investigators from isolation centres into shared data repositories before or after publication.
Standardized molecular phenotyping of islets/pancreatic fragments as well as fresh pancreatic tissue slices from organ and living donors.
Establishment and enhancement of international collaborations and communication for procurement, analysis, data sharing (e.g., hands-on training courses, workshops, consensus statements).
Development of international pipelines for data accessibility, sharing and integration.
Regular evaluation and refinement of all protocols to ensure they are aligned with best practices and technological advancements.
Efforts to link electronic health records to organ donors to improve clinical history and demographic information.
Acknowledgments
We are most grateful to all islet donors and their families for their support in this endeavor. We thank Dr. Frank J Moller for helping us in the preparation of the figures. A.L.G. is a Wellcome Senior Fellow in Basic Biomedical Science. A.L.G. is funded by the Wellcome (200837) and National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (U01-DK105535; U01-DK085545, UM1DK126185, U01DK123743, U24DK098085) and the Stanford Diabetes Research Center (NIDDK award P30DK116074). P.M. is supported by the Italian Ministry of University and Research, PRIN 2017, Molecular and Pathophysiological Heterogeneity of Autoimmune Diabetes: Implication for Precision Medicine, 2017KAM2R5_005. A.C.P. is funded by the Human Islet Research Network (RRID:SCR_014393), the Human Pancreas Analysis Program (RRID:SCR_016202), DK106755, DK123716, DK112232, DK112217, DK20593 (Vanderbilt Diabetes Research and Training Center), The Leona M. and Harry B. Helmsley Charitable Trust, and the Department of Veterans Affairs (BX000666). PR is funded by the Medical Research Council (MRC), the Swedish Research Council and The Leona M. and Harry B. Helmsley Charitable Trust. Ma.Sa. is supported by grants from the National Institutes of Health R01DK078803, R01DK068471, R01DK114427, R01DK122607, U01DK120429, U01HG012059, and UH3DK122639. Mi.So. is funded by the BMBF-German Center for Diabetes Research (DZD e.V.), the Deutsche Forschungsgemeinschaft (DFG) grants SO 818/10-1 and IRTG2251, and the DFG-ANR program SO 818/6-1. This work is also supported with funds by the Innovative Medicines Initiative 2 Joint Undertaking under grant agreements No 115881 (RHAPSODY) to MI.So., P.M., and M.I, and No. 115797 (INNODIA) and No. 945268 (INNODIA HARVEST) to Mi.So and P.M. This Joint Undertaking receives support from the Union’s Horizon 2020 research and innovation programme, “EFPIA”, “JDRF” and “The Leona M. and Harry B. Helmsley Charitable Trust”. This work is further supported by the Swiss State Secretariat for Education‚ Research and Innovation (SERI) under contract number 16.0097-2. The opinions expressed and arguments employed herein do not necessarily reflect the official views of these funding bodies.
Footnotes
Conflicts of Interest
ALG and ACP are members of the Human Pancreas Atlas Program (HPAP), ALG is a member of the Integrated Islet Distribution Program. ALG and P.M are members of the Horizon 2020 funded T2DSystems. ALG, ACP, Ma.Sa are members of the Human Islet Research Network. P.M. M.I. and Mi.So. are members of Rhapsody and IMIDIA. Through their membership of these consortia the authors have received financial support as outlined in the acknowledgements.
References
- 1.Rickels MR & Robertson RP Pancreatic Islet Transplantation in Humans: Recent Progress and Future Directions. Endocr Rev 40, 631–668 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso L et al. TIGER: The gene expression regulatory variation landscape of human pancreatic islets. Cell Rep 37, 109807 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asplund O et al. Islet Gene View - a tool to facilitate islet research. bioRxiv, 435743 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brissova M et al. The Integrated Islet Distribution Program answers the call for improved human islet phenotyping and reporting of human islet characteristics in research articles. Diabetologia 62, 1312–1314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaestner KH, Powers AC, Naji A & Atkinson MA NIH Initiative to Improve Understanding of the Pancreas, Islet, and Autoimmunity in Type 1 Diabetes: The Human Pancreas Analysis Program (HPAP). Diabetes 68, 1394–1402 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marchetti P et al. Fostering improved human islet research: a European perspective. Diabetologia 62, 1514–1516 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solimena M et al. Systems biology of the IMIDIA biobank from organ donors and pancreatectomised patients defines a novel transcriptomic signature of islets from individuals with type 2 diabetes. Diabetologia 61, 641–657 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fadista J et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc Natl Acad Sci U S A 111, 13924–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosengren AH et al. Reduced insulin exocytosis in human pancreatic β-cells with gene variants linked to type 2 diabetes. Diabetes 61, 1726–33 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taneera J et al. Identification of novel genes for glucose metabolism based upon expression pattern in human islets and effect on insulin secretion and glycemia. Hum Mol Genet 24, 1945–55 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Taneera J et al. A systems genetics approach identifies genes and pathways for type 2 diabetes in human islets. Cell Metab 16, 122–34 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Viñuela A et al. Genetic variant effects on gene expression in human pancreatic islets and their implications for T2D. Nat Commun 11, 4912 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marselli L et al. Persistent or Transient Human beta Cell Dysfunction Induced by Metabolic Stress: Specific Signatures and Shared Gene Expression with Type 2 Diabetes. Cell Rep 33, 108466 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Wigger L et al. Multi-omics profiling of living human pancreatic islet donors reveals heterogeneous beta cell trajectories towards type 2 diabetes. Nat Metab 3, 1017–1031 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Marchetti P, Suleiman M & Marselli L Organ donor pancreases for the study of human islet cell histology and pathophysiology: a precious and valuable resource. Diabetologia 61, 770–774 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krogvold L, Edwin B, Buanes T et al. Pancreatic biopsy by minimal tail resection in live adult patients at the onset of type 1 diabetes: experiences from the DiViD study. Diabetologia 57, 841–843 (2014). 10.1007/s00125-013-3155-y [DOI] [PubMed] [Google Scholar]
- 17.Atkinson MA Pancreatic biopsies in type 1 diabetes: revisiting the myth of Pandora’s box. Diabetologia 57, 656–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henquin JC Glucose-induced insulin secretion in isolated human islets: Does it truly reflect β-cell function in vivo? Mol Metab 48, 101212 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Negi S et al. Analysis of beta-cell gene expression reveals inflammatory signaling and evidence of dedifferentiation following human islet isolation and culture. PLoS One. 7(1):e30415 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mawla AM & Huising MO Navigating the Depths and Avoiding the Shallows of Pancreatic Islet Cell Transcriptomes. Diabetes 68, 1380–1393 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barklin A Systemic inflammation in the brain-dead organ donor. Acta Anaesthesiol Scand 53, 425–35 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Boggi U et al. First world consensus conference on pancreas transplantation: Part I-Methods and results of literature search. Am J Transplant 21 Suppl 3, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berney T et al. Utilization of organs from donors after circulatory death for vascularized pancreas and islet of Langerhans transplantation: recommendations from an expert group. Transpl Int 29, 798–806 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Markmann JF et al. The use of non-heart-beating donors for isolated pancreatic islet transplantation. Transplantation 75, 1423–9 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Barovic M et al. Metabolically phenotyped pancreatectomized patients as living donors for the study of islets in health and diabetes. Mol Metab 27S, S1–S6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mezza T et al. Increased beta-Cell Workload Modulates Proinsulin-to-Insulin Ratio in Humans. Diabetes 67, 2389–2396 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Mezza T et al. Pancreaticoduodenectomy model demonstrates a fundamental role of dysfunctional beta cells in predicting diabetes. J Clin Invest 131(2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niwano F et al. Glucose Metabolism After Pancreatectomy: Opposite Extremes Between Pancreaticoduodenectomy and Distal Pancreatectomy. J Clin Endocrinol Metab 106, e2203–e2214 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y et al. Paraneoplastic beta Cell Dedifferentiation in Nondiabetic Patients with Pancreatic Cancer. J Clin Endocrinol Metab 105, (2020). dgz224. doi: 10.1210/clinem/dgz22 [DOI] [PubMed] [Google Scholar]
- 30.Panzer JK, Cohrs CM & Speier S Using Pancreas Tissue Slices for the Study of Islet Physiology. Methods Mol Biol 2128, 301–312 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Cefalo CMA, Mezza T, Giaccari A & Kulkarni RN A Systematic Comparison of Protocols for Recovery of High-Quality RNA from Human Islets Extracted by Laser Capture Microdissection. Biomolecules 11 (2021) doi: 10.3390/biom11050625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brereton MF, Vergari E, Zhang Q & Clark A Alpha-, Delta- and PP-cells: Are They the Architectural Cornerstones of Islet Structure and Coordination? J Histochem Cytochem 63, 575–91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahlqvist E et al. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol 6, 361–369 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Walker JT et al. The Human Islet: Mini-Organ With Mega-Impact. Endocr Rev 42:605–657 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manning Fox JE et al. Human islet function following 20 years of cryogenic biobanking. Diabetologia 58, 1503–12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henquin JC The challenge of correctly reporting hormones content and secretion in isolated human islets. Mol Metab 30, 230–239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verge CF et al. Combined use of autoantibodies (IA-2 autoantibody, GAD autoantibody, insulin autoantibody, cytoplasmic islet cell antibodies) in type 1 diabetes: Combinatorial Islet Autoantibody Workshop. Diabetes 47, 1857–66 (1998) [DOI] [PubMed] [Google Scholar]
- 38.Wilkinson MD et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3, 160018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dragan I, Sparsø T, Kuznetsov D, Slieker R & Ibberson M SwissKnife: An R package for federated data analysis. bioRxiv, 2020.11.17.386813 (2020). [Google Scholar]
- 40.Warnat-Herresthal S, et al. Swarm Learning for decentralized and confidential clinical machine learning. Nature. 594, 265–270 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner R et al. Pathophysiology-based subphenotyping of individuals at elevated risk for type 2 diabetes. Nat Med 27, 49–57 (2021). [DOI] [PubMed] [Google Scholar]


