Abstract
Immunosenescence contributes to systematic aging and plays a role in the pathogenesis of Alzheimer’s disease (AD). Therefore, the objective of this study was to investigate the potential of immune rejuvenation as a therapeutic strategy for AD. To achieve this, the immune systems of aged APP/PS1 mice were rejuvenated through young bone marrow transplantation (BMT). Single-cell RNA sequencing revealed that young BMT restored the expression of aging- and AD-related genes in multiple cell types within blood immune cells. The level of circulating senescence-associated secretory phenotype proteins was decreased following young BMT. Notably, young BMT resulted in a significant reduction in cerebral Aβ plaque burden, neuronal degeneration, neuroinflammation, and improvement of behavioral deficits in aged APP/PS1 mice. The ameliorated cerebral amyloidosis was associated with an enhanced Aβ clearance of peripheral monocytes. In conclusion, our study provides evidence that immune system rejuvenation represents a promising therapeutic approach for AD.
Young bone marrow transplantation rejuvenates the immune system and attenuates Alzheimer’s disease-like pathologies.
INTRODUCTION
Alzheimer’s disease (AD) is the most prevalent form of dementia (1). It has been found that approximately half of the AD risk genes (BIN1, TREM2, CD33, etc.) are involved in immune processes (2, 3), providing compelling evidence for a connection between immune function and AD. In recent years, numerous studies have emphasized the crucial role of immune cell dysfunction in the pathogenesis of AD (4). The involvement of the immune system in AD pathogenesis is multifaceted, encompassing immune cells in both the brain and periphery (4, 5).
The aged immune system experiences a decline in the production of immune cells, a reduction in immune repertoire diversity, and an increase in dysfunctional immune cells. These changes are collectively referred to as immunosenescence, which not only plays a causal role in driving systemic aging, including brain aging (6), but also contributes to an increased susceptibility to age-related diseases such as AD (7–9). Therefore, rejuvenating aged immune cells represents a potential therapeutic strategy for AD.
Hematopoietic stem cells (HSCs), which are the source of peripheral immune cells, undergo functional decline with aging (10). On the basis of this, we hypothesized that replenishing the bone marrow with young HSCs could be a viable approach to rejuvenating aged immune cells and might have the potential for AD intervention. In this study, we established a heterochronic bone marrow transplantation (BMT) model and observed that transplanting young bone marrow cells rejuvenated peripheral immune cells in aged APP/PS1 mice. Furthermore, this approach resulted in reduced central and peripheral Aβ levels, alleviated cerebral Aβ plaque burden, neuronal degeneration, neuroinflammation, and improvement in behavioral deficits. These findings suggest that immune rejuvenation through young BMT may serve as a novel intervention strategy for AD.
RESULTS
Aging led to changes in the cell composition of PBMCs
In this study, old APP/PS1 mice (9 months) were transplanted with BMCs from young APP/PS1 mice (2 months, YTg BMCs), and transplantation with BMCs from old APP/PS1 mice (9 months, OTg BMCs) served as a control (fig. S1A). The transplantation efficiency exceeded 90% at 3 months after BMT (fig. S1B), indicating successful transplantation.
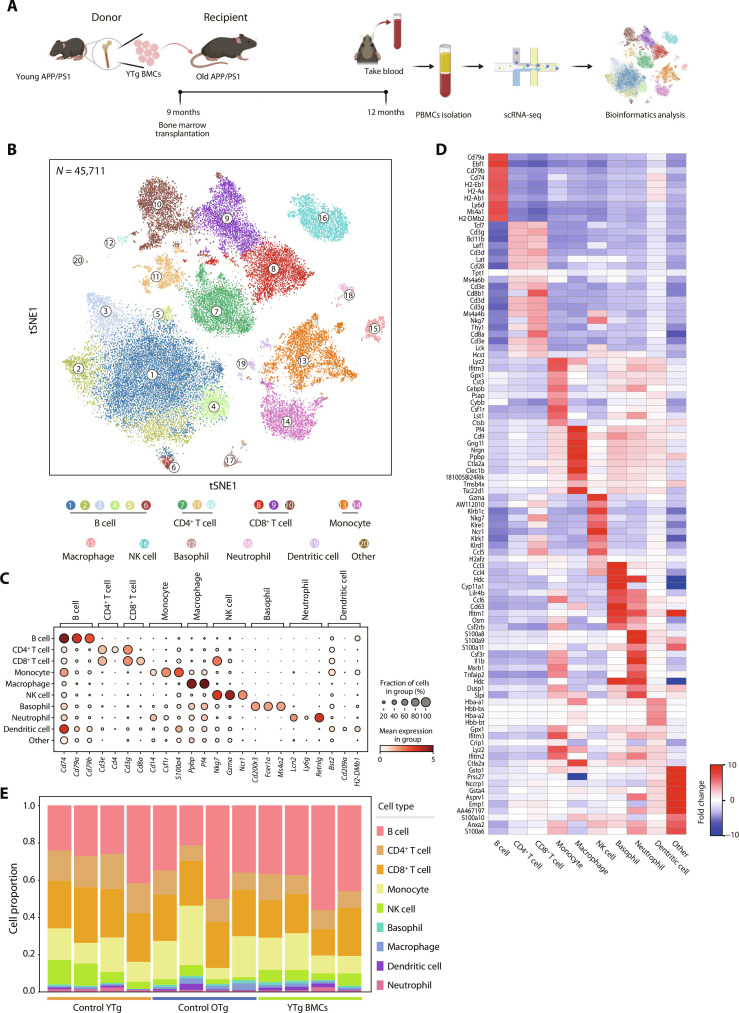
To characterize the gene expression alteration of peripheral immune cells at single-cell resolution, peripheral blood mononuclear cells (PBMCs) were collected, and a total of 45,711 high-quality mononuclear cells were acquired (Fig. 1, A and B). After marker gene annotation, all the major immune cell types were identified (Fig. 1C). The top 10 featured genes of each cell type were displayed in the heatmap (Fig. 1D).
Fig. 1. Construction of a PBMC single-cell transcriptomic atlas.
(A) Schematic diagram of the establishment of the BMT mouse model. (B) t-Distributed stochastic neighbor embedding (t-SNE) projection of 45,711 cells from PBMCs in the control YTg (n = 4), control OTg (n = 4), and YTg BMC groups (n = 4). A total of nine cell types, including B cells (clusters 1 to 6), CD4+ T cells (clusters 7, 11, and 12), CD8+ T cells (clusters 8 to 10), monocytes (clusters 13 and 14), macrophages (cluster 15), natural killer (NK) cells (cluster 16), basophils (cluster 17), neutrophils (cluster 18), and dendritic cells (cluster 19), were characterized. (C) Canonical cell type–specific markers define B cells, CD4+ T cells, CD8+ T cells, monocytes, basophils, neutrophils, NK cells, and macrophages. (D) Heatmap of the top 10 marker genes from each cell type. The color key indicates the fold change level. (E) Proportions of the main immune cell types in control YTg, control OTg, and YTg BMC mice. These proportions were calculated by dividing the number of cells of each type by the total number of cells in each sample from the processed single-cell sequencing data.
The effects of aging and young BMT on the composition of peripheral immune cells in old AD mice were first analyzed (Fig. 1E). The B cell and CD8+ T cell proportions remained stable during aging and after young BMT. The proportion of CD4+ T cells and natural killer (NK) cells significantly decreased from young to old mice and did not return to youthful levels post-BMT. These results were consistent with those from flow cytometry analysis (fig. S2, A and B). The monocytic proportion significantly increased in aged mice, and young BMT brought the monocytic proportion back to the youthful level.
Young BMT restored the expression of aging DEGs in various cell types of PBMCs
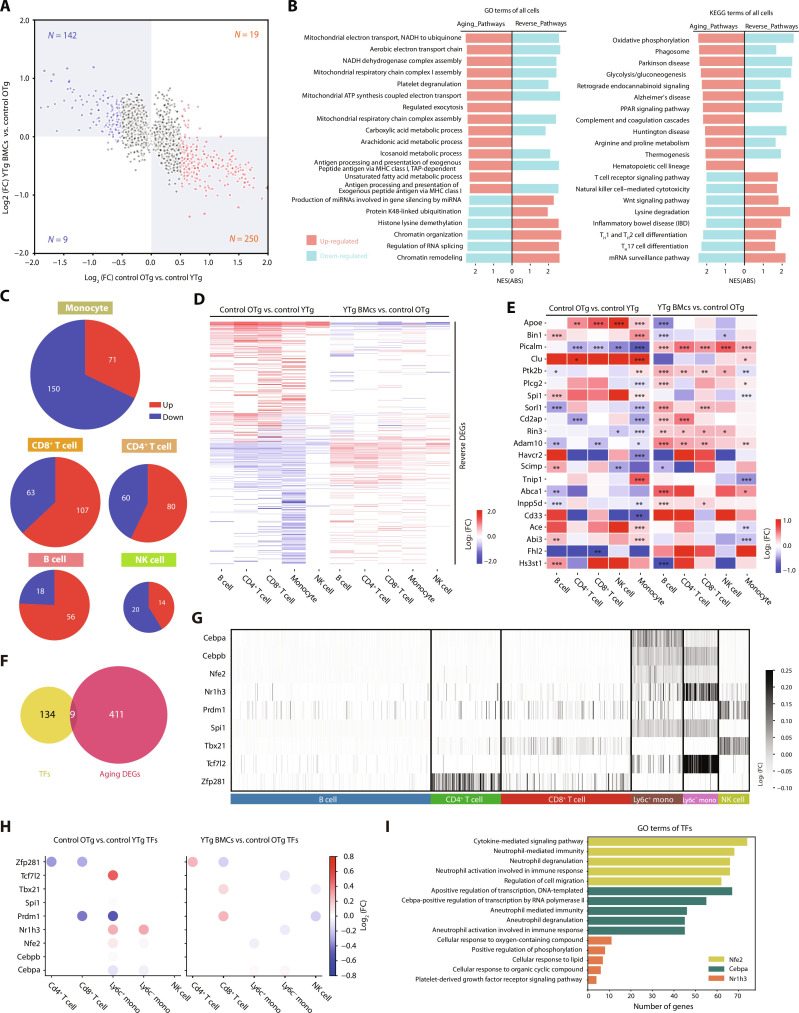
A total of 269 genes were up-regulated, and 151 genes were down-regulated in the PBMCs of old mice compared to those of young mice (Fig. 2A). These were collectively termed aging-related differentially expressed genes (aging DEGs). Approximately one-third of aging DEGs exhibited reversed expression patterns after young BMT, and we named these genes “reverse DEGs” (fig. S3A). To provide further insight into the underlying pathways modulated by aging and BMT in PBMCs, we analyzed functionally annotated aging and reverse DEGs. We found that up-regulated aging pathways were commonly associated with mitochondrial function (Fig. 2B), and mitochondrial dysfunction is one of the hallmarks of aging (11). Pathways related to neurodegenerative diseases, such as AD, were also up-regulated, indicating an association between immunosenescence with AD. By contrast, down-regulated aging pathways were commonly associated with epigenetic regulation and immune processes. These alterations were restored after young BMT (Fig. 2B).
Fig. 2. Young BMT restored the expression of aging DEGs in multiple cell types of PBMCs.
(A) Scatter plot of the general aging differentially expressed genes (DEGs; cutoff by P < 0.05, log2FC > 0.5) in PBMCs. (B) The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) terms enriched by the general aging and reverse DEGs. Red: up-regulated pathways; blue: down-regulated pathways. (C) Pie plots of the aging DEGs in monocytes, CD8+ T cells, CD4+ T cells, B cells, and NK cells. (D) Heatmap of the reversed DEGs in each cell type alongside the young BMT. The color key indicates the log2FC values. (E) The expression of differentially expressed AD risk genes in aging and BMT of B cells, CD4+ T cells, CD8+ T cells, NK cells, and monocytes. The color indicates the log2FC values. (F) Venn plot of the identified transcription factors (TFs) and aging DEGs in PBMCs. (G) Heatmap of the expression level of TFs in each cell type. The color key indicates the log2FC values. (H) Dot plots of the effects of aging and young BMT on TFs expression. The color key indicates the log2FC values. (I) Bar plots of the GO terms enriched by the target genes of Nfe2, Cepbp, and Nr1h3. The downstream genes of Nfe2, Cepbp, and Nr1h3 were obtained from the regulon analysis using SCENIC, and the genes identified through SCENIC analysis may not be DEGs. *P < 0.05; **P < 0.01; ***P < 0.001. The error bars are the SEMs. NES(ABS), absolute normalized enrichment score; FC, fold change.
Furthermore, we characterized the aging DEGs in each cell type. Among these cells, monocytes, CD4+ T, and CD8+ T cells exhibited the most pronounced changes (Fig. 2C). Approximately one-third of the aging DEGs exhibited reversed expression patterns after young BMT (Fig. 2D and fig. S3B). Similar results were observed by pseudobulk DEG analysis (fig. S4A). About 10 to 20% of aging DEGs were reversed by young BMT (fig. S4, B and C). We assessed how aging and young BMT affect the expression of epigenetic regulation-related genes across various blood cell types. The results showed that genes regulating DNA methylation, histone methylation, histone acetylation, and chromatin organization typically decreased with aging. Young BMT effectively counteracted this decline (figs. S3C and S4D).
In addition, we compared the expression of the top 45 AD risk genes (12) in the major immune cell types during aging and after young BMT (Fig. 2E and fig. S3D). Notably, all the top 10 AD risk genes displayed significant differential expression in monocytes with aging, and young BMT reversed these expression changes; among which, five risk genes in monocytes were verified by pseudobulk DEG analysis (fig. S4E).
The above findings demonstrate that the gene expression of major blood immune cells undergoes changes with aging, with the most notable changes occurring in monocytes, and that young BMT can mitigate these alterations.
Young BMT mitigated aging pathways in various cell types of PBMCs
We performed gene set enrichment analysis (GSEA) analysis on the transcriptome associated with aging and reversal in each cell type. In monocytes, aging pathways primarily linked to innate immunity, such as regulation of defense response, showed down-regulation (fig. S5A). In CD8+ T cells, mitochondrial function-related pathways were up-regulated, while cell homeostasis–related pathways were down-regulated during aging (fig. S5B). In CD4+ T cells, most aging pathways were up-regulated and associated with inflammation and AD (fig. S5C). The aging pathways were mostly up-regulated in B cells and were associated with mitochondrial function and AD (fig. S5D). Conversely, in NK cells, most aging pathways—tied to protein degradation and pathogen clearance—were down-regulated (fig. S5E). In addition, we observed that young BMT mitigated and restored most aging pathways.
Young BMT restored the transcriptional regulatory network of PBMCs
To elucidate the alterations in transcriptional regulatory networks during aging and after young BMT, single-cell regulatory network inference and clustering (SCENIC) analysis was used. This approach allowed us to predict core transcription factors (TFs) involved in regulating aging and reverse DEGs. We identified 143 TFs for further analysis. Among these, nine TFs exhibited dysregulation during aging (Fig. 2F), with predominant expression observed in monocytes (Fig. 2G). Notably, the expression levels of Nfe2, Cebpa, and Nr1h3, which are among the dysregulated TFs, were restored in monocytes upon exposure to young bone marrow (Fig. 2H). Downstream genes regulated by Nfe2, Cebpa, and Nr1h3 were found to be involved in innate immunity, specifically neutrophil-mediated immunity (Fig. 2I). Among these three TFs, the expression of Nr1h3 was also detected by pseudobulk DEG analysis (fig. S6, A to D). These results indicate that the gene expression changes in monocytes are the most notable during aging, which may be because the transcriptional regulatory network of monocytes is most affected by aging, and young BMT can restore this alternation.
Young BMT down-regulated plasma SASP proteins in aged APP/PS1 mice
The effects of aging and young BMT on the plasma proteome were investigated using mass spectrometry (fig. S7A and table S1). These proteins were mostly extracellular proteins, originating from various organismal systems, and participated in the cell community (fig. S7, B and C). Quantification analysis indicated that there were 23 up-regulated and 16 down-regulated proteins in the context of aging (fig. S7D) which were collectively referred to as aging-related differentially expressed proteins (aging DEPs). The up-regulated DEPs, such as CD5 antigen-like protein (Cd5l) (13), tubulin beta 2A chain (Tubb2a) (14), and galectin-3–binding protein (Lgals3 bp) (15), are involved in proinflammatory immune processes. The down-regulated DEPs, including leukemia inhibitory factor receptor (Lifr) (16), transketolase (Tkt) (17), and galectin-1 (Lgals1) (18), are important immune homeostasis regulators. These changes coincide with the dysregulation of immune function observed in aged mice. The above three up-regulated aging DEPs were all decreased after young BMT (fig. S7E). Functional enrichment analysis showed that the DEPs after young BMT were enriched in the immune process, such as the ErbB signaling pathway and the relaxin signaling pathway (fig. S7F). ErbB signaling pathway was reported to play a role in inhibiting pro-inflammatory macrophage activity (19), and relaxin was reported to be able to reverse inflammatory and immune signals (20).
Out of a formerly reported senescence-associated secretory phenotype (SASP) atlas (21), a total of 88 SASP proteins were detected in our dataset (table S2), with 33 of them increasing with aging (fig. S7G). More than half of the aging-related SASP proteins (such as Lgals3bp, Vim, and Mmp2) were reduced after young BMT (fig. S7G). Overall, young BMT was able to lower the plasma levels of aging-related proteins.
Young BMT restored the impaired Aβ phagocytosis capacity of blood monocytes
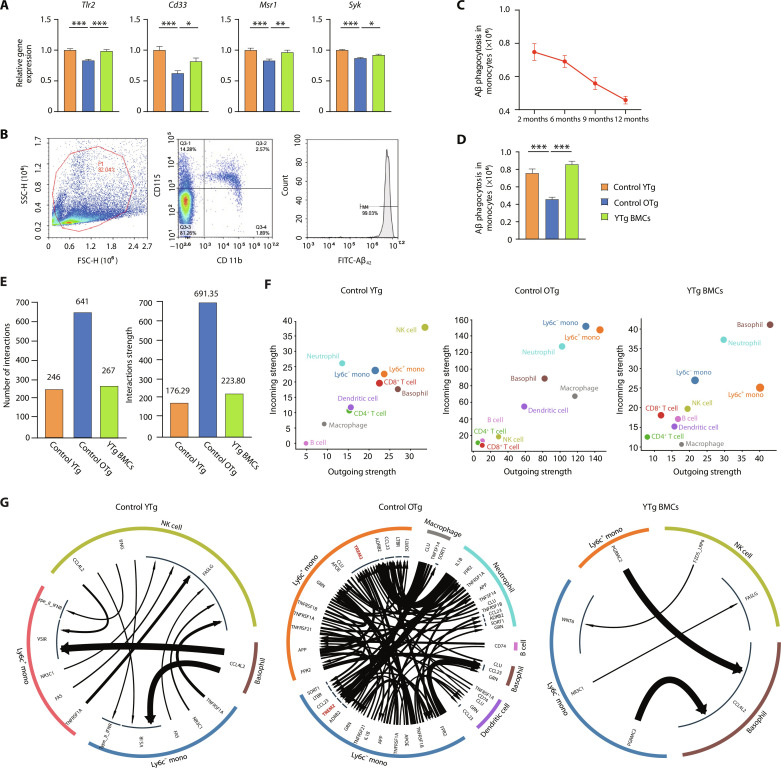
Blood monocytes clear circulating Aβ (22). The gene expression of key Aβ uptake receptors, such as Tlr2, Cd33, and Msr1, decreased in monocytes with aging, but young BMT rescued this decline (Fig. 3A). Syk expression, which is associated with microglial Aβ phagocytosis, was also reduced in aged monocytes and restored by young BMT (Fig. 3A).
Fig. 3. Young BMT rescued the impaired Aβ phagocytosis capacity of blood monocytes.
(A) Levels of expression of the Aβ uptake-related receptors Tlr2, Cd33, Msr1, and Syk. One-way analysis of variance (ANOVA). (B) In vitro analysis of Aβ42 phagocytosis by monocytes with flow cytometry. (C and D) Quantification of the levels of monocyte Aβ42 uptake. One-way ANOVA. (E) Bar plots of the number and strength of cell interactions in control YTg, control OTg, and YTg BMC mice. (F) Scatter plots of cell interaction strength in control YTg, control OTg, and YTg BMC mice. (G) Cell-to-cell interaction network between monocytes and the rest of the cell types. *P < 0.05; **P < 0.01; ***P < 0.001. The error bars are the SEMs.
Classic monocytes (Ly6c+) primarily mediate Aβ phagocytosis (fig. S8, A and B), and we found that they were more susceptible to aging (figs. S8C and S9A). The expression of genes involved in Aβ phagocytosis decreased with aging in Ly6c+ monocytes but not in Ly6c− monocytes (fig. S8D). Young BMT successfully reversed these changes in monocytes (fig. S8, C to E). An in vitro Aβ phagocytosis assay also indicated that young BMT rescued the impaired Aβ phagocytosis ability of senescent monocytes (Fig. 3, B to D). We analyzed the expression of genes involved in phagocytosis in monocytes, and the results showed that the expression of these genes decreased globally with aging (figs. S8F and S9B). We also performed a cell debris phagocytosis experiment, which showed that the cell debris phagocytosis of senescent monocytes also decreased (fig. S8G and table S3), suggesting that the reduced phagocytosis in senescent monocytes is not Aβ specific but rather overall reduced phagocytosis.
Moreover, we compared peripheral blood monocytes from patients with AD and aged-matched healthy controls (table S4) on senescent phenotypes, such as Aβ phagocytosis and SASP factor expression. The result showed that monocytes from patients with AD had impaired Aβ phagocytosis capacity (fig. S10, A to D) and higher SASP factor expression (fig. S10E). It indicated that monocytes in patients with AD were more senescent than in healthy controls.
Cell-to-cell communication, mediated by cytokines, is crucial for effective immune responses. Cell-to-cell communications were generally up-regulated during aging but restored by young BMT (Fig. 3E). Specifically, the aging-related gain of communication among monocytes (Lyc6+ monocytes and Lyc6− monocytes) and macrophages was markedly restored by young BMT (Fig. 3F), potentially rebalancing innate immune function. Impaired Aβ phagocytosis is linked to the loss of TREM2 function in monocytes (23). Communications between APOE/APP/CLU (ligand in macrophage/neutrophil/basophil/DC cell) and TREM2 (receptor in monocytes) were up-regulated in aging (Fig. 3G), which might be responsible for the impaired Aβ phagocytosis in senescent monocyte.
Young BMT reduced cerebral and plasma Aβ levels
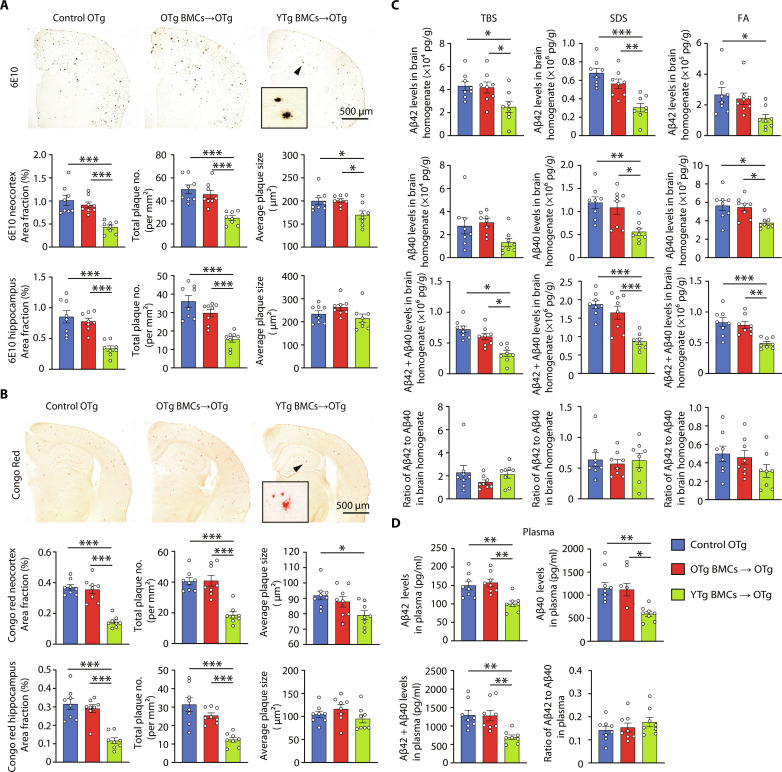
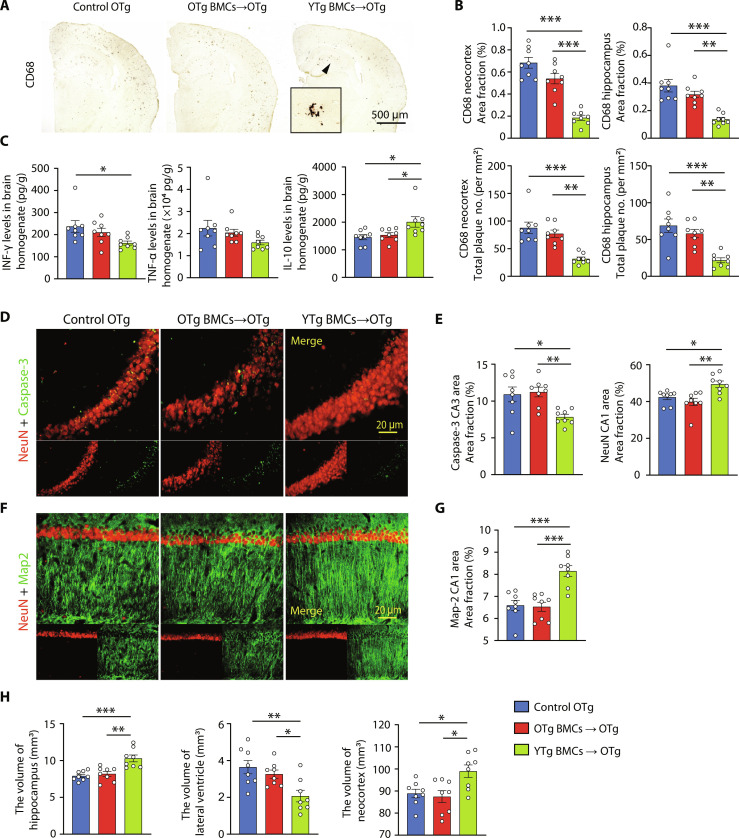
Bone marrow reconstitution using YTg BMCs resulted in a significant decrease in amyloid plaques in the neocortex and hippocampus (Fig. 4, A and B), whereas no such reduction was observed with OTg BMCs. In addition, the levels of Aβ40 and Aβ42 were markedly lower in the YTg BMC group than in the OTg BMC and control OTg groups (Fig. 4C). However, there were no discernible differences in the levels of APP itself or its metabolites [C-terminal fragment–β (CTF-β) and CTF-α], APP secretases [β-secretase 1 (BACE1), a disintegrin and metalloproteinase 10 (ADAM10), and Presenilin-1 (PS1)], Aβ degrading enzymes [insulin-degrading enzyme (IDE) and neprilysin (NEP)], and Aβ transport receptors [low-density lipoprotein receptor–related protein 1 (LRP-1) and receptor for advanced glycation end products (RAGE)] among the YTg BMC, OTg BMC, and control OTg groups (fig. S11). The number of microglia per plaque was also similar between YTg BMC and control OTg groups (fig. S12A). We performed bulk RNA sequencing (RNA-seq) to elucidate whether young BMT reinstalled phagocytosis gene expression in microglia (fig. S12B). Overall, we observed 150 up-regulated and 959 down-regulated genes when comparing YTg BMC and control OTg microglia (fig. S12C). Up-regulated genes mainly enriched in amino acid metabolism pathways, while the down-regulated genes enriched in inflammation pathways, such as mitogen-activated protein kinase signaling pathway, apoptosis, and tumor necrosis factor (TNF) signaling pathway (fig. S12D). We analyzed the expression of genes involved in phagocytosis in microglia, unlike in blood monocytes (37 phagocytosis-related DEGs after young BMT; fig. S8F), there were only seven DEGs when comparing YTg BMC and control OTg microglia, and half of these genes (three out of seven) were even down-regulated (fig. S12E). Microglia sustain an Aβ-responsive program widely known as disease-associated microglia (DAM), which has the potential to restrict Aβ deposition (24). The expression of DAM marker genes was not changed after young BMT (fig. S12F). Infiltrated monocytes constituted only 1% of CD11b+ cells in the brain (fig. S12, G to I), the role of infiltrated monocytes in Aβ clearance is also limited. Overall, these results suggested that the amelioration of cerebral amyloidosis following young BMT is rarely, if ever, attributed to the reduction in Aβ production or enhancement in Aβ degradation within the brain but may be more likely caused by the enhanced clearance of peripheral Aβ. The plasma levels of both Aβ species were significantly diminished in YTg BMC recipients (Fig. 4D). We also compared the levels of Aβ plaques in YTg BMC and 9-month-old APP/PS1 mice and found that the density of plaques in YTg BMC mice was lower (fig. S13, A and B), suggesting that young BMT may reverse AD pathology.
Fig. 4. Young BMT reduced Aβ levels in APP/PS1 mice.
(A and B) Immunostaining and quantification of Aβ plaques stained with 6E10 (total plaques) and Congo red (dense plaques) in the neocortices and hippocampus of APP/PS1 mice with or without BMT. One-way ANOVA. (C) Comparison of Aβ40 and Aβ42 levels, Aβ40/Aβ42 ratios in the tris-buffered saline (TBS; soluble extracellular Aβ), SDS (intracellular Aβ), and FA (dense plaques Aβ) fractions of brain homogenates among BMT and control mice. One-way ANOVA. (D) Comparison of Aβ40 and Aβ42 levels, Aβ40/Aβ42 ratio in plasma among BMT and control mice. One-way ANOVA. n = 8 per group. *P < 0.05; **P < 0.01; ***P < 0.001. The scale bar is 500 μm. The error bars are the SEMs. FA, formic acid.
Young BMT attenuated neuroinflammation and neuronal loss in APP/PS1 mice
Compared to the old control mice, recipients of YTg BMCs but not OTg BMCs showed lower levels of microglial activation (Fig. 5, A and B), as well as reduced levels of proinflammatory cytokines such as TNF-α and interferon-γ (IFN-γ). Conversely, they exhibited higher levels of the anti-inflammatory cytokine interleukin-10 (IL-10) (Fig. 5C) 3 months after BMT. This is consistent with the above RNA-seq data of microglia (fig. S12, C and D). The microgliosis level in YTg BMC mice was also lower than in 9-month-old APP/PS1 mice (fig. S13, C and D). In addition, when comparing the young BMT recipients with the OTg BMC and control OTg groups, the levels of neuronal apoptosis and degeneration were significantly decreased. This was evidenced by a decrease in caspase-3–positive neurons and an increase in Map2-positive dendrites in the hippocampus (Fig. 5, D to G). We also observed that atrophy occurred in the hippocampus and neocortex, accompanied by significant lateral ventricular enlargement in the control OTg and OTg BMC groups, and young BMT significantly alleviated this situation (Fig. 5H).
Fig. 5. Young BMT alleviated AD-related pathogenesis.
(A and B) Immunostaining and quantification of microglia stained with CD68 in the neocortex and hippocampus of APP/PS1 mice with or without BMT. One-way ANOVA. (C) Statistical analysis of the levels of inflammatory factors, including interferon-γ (IFN-γ), tumor necrosis factor–α (TNF-α), and interleukin-10 (IL-10), in brain homogenate. One-way ANOVA. (D to G) Representative images and quantification of neurons (NeuN, red) and dendrites (MAP2, green) in the hippocampal CA1 region and neural apoptosis (caspase-3, green) in the hippocampal CA3 region. One-way ANOVA. (H) Volumes of the hippocampus, lateral ventricle, and neocortex. One-way ANOVA. n = 8 per group; *P < 0.05; **P < 0.01; ***P < 0.001. The scale bar in (A) is 500 μm, and the scale bars in (D) and (F) are 20 μm. The error bars are the SEMs.
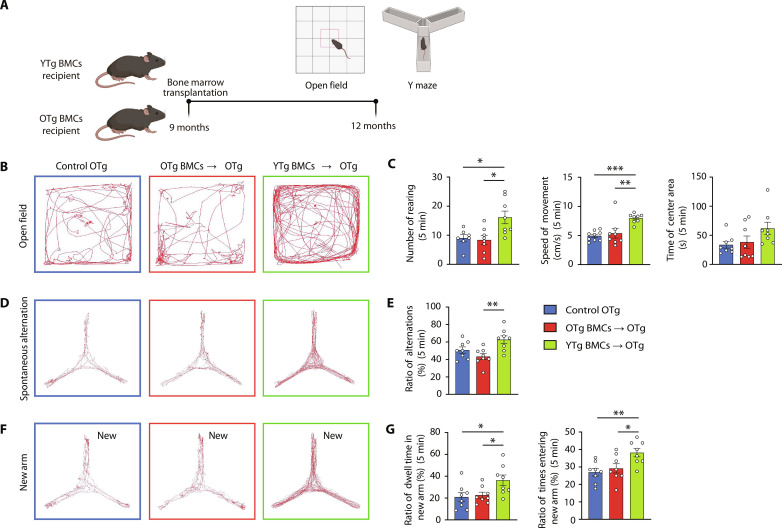
Young BMT rescued the cognitive impairment of APP/PS1 mice
Behavioral testing was conducted 3 months after BMT (Fig. 6A). In the open-field test, recipients of YTg BMCs, but not OTg BMCs, displayed greater activity than control OTg mice, indicating improved locomotion ability (Fig. 6, B and C). In addition, the YTg BMC group exhibited increased rearing behavior, suggesting an ameliorated anxiety state. In the spontaneous alternation maze, YTg BMC recipients exhibited a higher number of spontaneous alternations between the arms of the maze than OTg BMC recipients and control OTg mice (Fig. 6, D and E). In the new arm maze, YTg BMC recipients performed better, with a significant increase in the number of entries and the dwell time in the novel arm (Fig. 6, F and G). Both tests suggested that young BMT could enhance the spatial and working memory of AD mice. Overall, these findings demonstrated that young BMT effectively rescued cognitive deficits in aged APP/PS1 mice.
Fig. 6. Young BMT rescued the cognitive impairment of APP/PS1 mice.
(A) Schematic presentation of the experimental design used to investigate the effect of young BMT on cognition in APP/PS1 mice. (B and C) Representative images of the open-field test and statistics of the number of rearing events, average speed, and time spent in the center. One-way ANOVA. (D and E) Representative images of the spontaneous alteration test and statistics of the ratio of alternations. One-way ANOVA. (F and G) Representative images of the new arm test and statistics of the ratio of time and times entering the new arm. One-way ANOVA. n = 8 per group; *P < 0.05; **P < 0.01; ***P < 0.001. The error bars are the SEMs.
DISCUSSION
Aging represents the most significant risk factor for AD, and immunosenescence, a critical component of the aging process, has attracted considerable attention due to its potential impact on AD (25). Previous research has primarily focused on investigating the involvement of immune cells, particularly microglia, the resident macrophages of the CNS, in the pathogenesis of AD. Numerous studies have extensively examined and confirmed the characteristics of aging microglia (26), their role in the development of AD (27), and the effectiveness of interventions targeting aging microglia (28). However, it is important to recognize that AD is systemically influenced by both central and peripheral factors. Consequently, senescent peripheral immune cells also play an important role in the development of AD (29–31). To fully comprehend the occurrence of AD, it is imperative to further investigate the potential involvement of dysfunction in aging-related peripheral immune cells and determine whether they can serve as viable targets for intervention.
Although numerous studies have demonstrated significant alterations in the composition and gene expression profiles of peripheral immune cells in individuals with AD (4), the impact of aging on the functionality of peripheral immune cells and, consequently, the progression of AD remains unclear. Our findings revealed that aging induced changes in the gene expression in both innate and adaptive immune cells, aligned with the dysfunction of both innate and adaptive immune responses observed in aging animals or elderly individuals, such as diminished phagocytosis function of monocytes (22), impaired antiviral immunity of NK cells (32), elevated production of autoantibodies by B cells (9, 33), and expansion of cytotoxic T cells (34). Functional annotation analysis further demonstrated that these DEGs associated with aging were significantly enriched in pathways related to AD, implying the potential involvement of senescent peripheral immune cells in the development of AD.
In addition, our study revealed that monocytes may have a significant impact on the process of immunosenescence leading to AD. Among the DEGs associated with aging, senescent monocytes displayed the highest proportion of AD risk genes. Previous studies have demonstrated that impaired peripheral clearance of Aβ contributes to the development of AD (35). Monocytes, found in the bloodstream, have a physiological capacity for Aβ clearance, and aging-induced impairment of monocytic Aβ clearance can accelerate the occurrence of AD (22). The findings from this study suggest that the diminished monocytic Aβ clearance capacity is a consequence of the down-regulation of key receptors involved in Aβ phagocytosis within aging monocytes. Collectively, these pieces of evidence indicate that the senescence of peripheral immune cells plays a critical role in the pathogenesis of AD, and rejuvenating peripheral immune cells in aging individuals may represent a promising intervention strategy.
Blood immune cells originate from HSCs, and previous research has indicated that the functionality of HSCs is compromised during the aging process (36). This impairment leads to a decreased capacity for differentiation and the production of immune cells with reduced functionality (10). The senescence of HSCs serves as a source for the senescence observed in immune cells. Rejuvenation of peripheral immune cells can potentially be achieved through young BMT. Our findings demonstrate that young BMT can reverse the expression of approximately one-third of aging DEGs, alleviate aging-related pathways, restore altered cell-to-cell communication in aging PBMCs, rescue the dysfunction in monocyte-mediated Aβ clearance, and reduce the levels of blood SASP factors. It is also noteworthy that young BMT can significantly decrease both central and peripheral Aβ levels, ameliorate Aβ burden, mitigate neuroinflammation and neuronal apoptosis in the brain, and improve the cognitive function of AD mice. We investigated the effect of young BMT on several major Aβ clearance mechanisms, including microglial clearance, enzyme degradation, blood-brain barrier (BBB) transport, and blood monocytic clearance. Only blood monocytic Aβ clearance was improved after young BMT. Therefore, we speculate that the main mechanism for the amelioration of Aβ pathology by young BMT may be the enhanced phagocytic ability of young monocytes, thereby promoting the peripheral pathway for cerebral Aβ clearance. Moreover, adaptive immune functions were also disordered during aging which was reversed by young BMT, suggesting that the rejuvenation of the immune system may also contribute to Aβ clearance via an adaptive immune pathway. Previous studies have shown that the efflux of Aβ from the brain to the periphery is an important pathway for cerebral Aβ clearance (37). However, although no discernible difference in both the number and gene expression of Aβ-induced microglial activation was observed in our study, it cannot rule out the possibility that young BMT enhances the efficiency of microglial Aβ clearance mechanisms, enabling these cells to maintain effective Aβ phagocytosis even at reduced plaque levels. Moreover, BBB transporters LRP-1 and RAGE were not changed after young BMT in our study, but other transporters on BBB and nonreceptor-mediated transport capacity, as well as Aβ drainage through lymphatic vessels (38), were not examined in our present study. Therefore, the efflux of Aβ from the brain to the peripheral may be enhanced via other pathways. In addition, we believe that the neuroprotective effects after BMT are comprehensive, including the protective effect of the rejuvenated immune system and the neuroprotective substances produced by the young BMCs. A comprehensive understanding of the protective mechanism of BMT on AD requires future work to reveal.
Before our study, there were also reports of improving immune senescence through BMT, thereby improving AD pathology and cognitive impairment. For example, Das et al. (39) proved that young BMT in 18-month-old WT mice can preserve learning and memory in old mice. However, wild-type (WT) mice do not develop AD, so it cannot prove whether immune rejuvenation has an ameliorative effect on AD-specific pathology (such as Aβ deposition). There are some reports showing that young plasma can significantly improve AD pathology (40, 41). These studies revealed the effects of biofluid factors on aging and AD. Compared with them, an advance of our study is that we demonstrated the effects of blood cells, mainly immune cells, on aging and AD, suggesting that rejuvenating immune cells is a promising therapeutic strategy for AD. In addition, tau pathology, the other hallmark of AD, is more closely associated with cognitive impairment. Our previous study has shown that pathologic tau proteins can also efflux out of the brain to be cleared in the periphery, and the enhancement of peripheral clearance via peritoneal dialysis can ameliorate tau pathology in the brain of tau transgenic mice (42). Given that young BMT enhances the overall phagocytosis of monocytes, the intervention effects of young BMT in tau transgenic mice are worth exploring, which will provide opportunities for the treatment of other tauopathies. Future studies may focus on exploring other new strategies that can rejuvenate immune cells to advance the possibility of clinical translation.
MATERIALS AND METHODS
Animals
APPswe/PS1dE9 (APP/PS1) transgenic mice were obtained from Nanjing Junke Bioengineering Co., Ltd. (Nanjing, China). B6.SJL-Ptprca pepcb/BoyJ mice (Wt CD45.1) were obtained from the Peking University Health Science Center. Female C57BL/6 J (wild type, WT) mice were provided by the Animal Centre of Daping Hospital affiliated with the Third Military Medical University. We used female mice in this study to exclude the impact of sex on AD pathologies in the brain (43). APP/PS1 mice and B6. SJL-Ptprca pepcb/BoyJ mice share the same C57BL/6 J genetic background. The transgenic APP/PS1 mouse model harbors the human APP gene containing the Swedish mutant and human presenilin 1 (PS1) gene encoding the deleted exon 9 mutation under the control of the mouse PrP promoter, which is active predominantly in brain neurons, and develops amyloid plaques at 6 months of age (44). All experiments that involved mice were approved by the Laboratory Animal Welfare and Ethics Committee of the Third Military Medical University.
Establishment of the BMT mouse model
Nine-month-old AD mice were irradiated [8 gray (Gy) single dose at ~0.4 Gy/min from a gamma source], while the head was shielded without irradiation and then transplanted with BMCs from 2-month-old female APP/PS1 transgenic mice (YTg BMCs, n = 8) or BMCs from 9-month-old female APP/PS1 transgenic mice (OTg BMCs, n = 8). The control group was 9-month-old APP/PS1 transgenic mice without irradiation or transplantation (control OTg group, n = 8). BMT was performed following previously described procedures (45). Briefly, donor BMCs were harvested from both the tibias and femurs of mice with basic RPMI 1640 medium. Previous BMT studies have demonstrated that a minimum of 2.0 × 106 to 5.0 × 106 BMCs are generally injected to obtain functional immune cells after BMT (46). To ensure that most of the senescent immune cells are replaced with young immune cells, a total of 1 × 107 BMCs in 300 μl of basic RPMI 1640 medium were intravenously transplanted into irradiated recipients via the tail vein.
Behavior tests
Previous studies showed that peripheral lymphohematopoietic reconstitution of all cell lineages may become normal by day 21 after BMT (47), suggesting that BMT may begin to display anti-AD effects in host AD animals after peripheral lymphohematopoietic reconstitution which is approximately 21 to 30 days after BMT. At 3 months after BMT, all mice were subjected to open-field and Y-maze tests following previously described methods (30). Briefly, in the open-field test, the mice were placed in the center of the open-field apparatus for 5 min. Rearing was recorded for each mouse. The paths were tracked using a computer tracking system (LimeLight, ActiMetrics, Wilmette, IL, USA), and the distance traveled and speed were recorded and measured. In a spontaneous alternation test, the mice were allowed to move freely through a Y-maze during a 5-min session. Alternation was defined as successive entries into the three arms on overlapping triplet sets. The percentage of alternation was calculated as the total number of alternations × 100/(total number of arm entries − 2). A new arm exploration test was also performed in the Y-maze. One arm was blocked (defined as the new arm), and the mice were allowed to explore the other two arms for 5 min. After a 2-hour interval, the mice were allowed to freely explore all three arms for 5 min. The number of new arm entries and dwell time spent in the new arm were recorded.
Aβ phagocytosis assay
The assay was performed following previously described procedures (48). Briefly, PBMCs were isolated by density gradient centrifugation using Ficoll-Hypaque (TBD Science, Tianjin, China), and mononuclear sections were collected and resuspended in RPMI medium with 10% fetal calf serum and 1% penicillin/streptomycin and incubated with FITC-Aβ 1–42 (2 μg/ml) (GL Biochem, Shanghai, China) at 37°C in a 5% CO2 incubator for 1 hour. Then, the cell suspensions were washed and stained with the following fluorophore-labeled antibodies: APC anti-mouse CD11b and phycoerythrin PE anti-mouse CD115 (BD Biosciences, NJ, USA) for mice blood monocytes, and APC anti-human CD14 and PE anti-human CD16 (BD Biosciences, NJ, USA) for human blood monocytes. Following incubation, flow cytometry was performed on a NovoCyte Flow Cytometer (ACEA Biosciences, CA, USA) after appropriate compensation. The data were analyzed by NovoExpress software based on forwards and side scatter and the mean fluorescence intensity.
Cell debris phagocytosis assay
Blood monocytes were isolated according to a published method. Briefly, fresh blood was collected in EDTA tubes and left at room temperature for 1 hour. Then, the sample was centrifuged at 2000g for 10 min to obtain the lower layer blood cells. PBMCs were isolated by density gradient centrifugation using Ficoll-Hypaque (TBD Science, Tianjin, China), mononuclear sections were collected, washed with phosphate-buffered saline (PBS) three times, and then used for monocytes isolation using CD14 MicroBeads Human (Miltenyi Biotec, Germany) and MS sorting columns (Miltenyi Biotec, Germany). Isolated monocytes were resuspended in RPMI medium 1640 basic (1×) with 10% fetal calf serum and 1% penicillin/streptomycin and incubated at 37°C in a 5% CO2 incubator for 2 hours. Fluorescein-labeled (100 μl; Abs/Em 494/518 nm) Escherichia coli K-12 BioParticles (Invitrogen, USA) per 1 × 105 monocytes were added and incubated together for 2 hours. Last, the cells were washed and resuspended in Dulbecco's PBS (Thermo Fisher Scientific, USA), and images were taken by Primovert Microscope (Zeiss, Germany) with an excitation wavelength of 488 nm. Cell debris phagocytosis capacity was quantified by the proportion of GFP+ monocytes. The positive proportion of phagocytic debris in the monocytes was counted in ImageJ, and the differences between the groups were analyzed in GraphPad Prism.
Immunohistochemistry and immunofluorescence
The left hemi-brain of each mouse was sectioned coronally at 30 μm using a freezing sliding microtome. Five sections from each mouse, corresponding approximately to bregma coordinates +3.0, +1.3, −0.9, −2.1, and −3.0 mm respectively, were used for 6E10, Congo red staining; two sections −2.1 and −3.0 mm were used for CD68 staining. For immunofluorescence, two sections (bregma −2.1 mm for Map2 staining, bregma −3.0 mm for caspase-3 staining) from each mouse were used.
We conducted Aβ plaque staining with anti-6E10 antibody (1:500; Abcam) and Congo red (1:1000, Sigma-Aldrich) to detect the Aβ burden. Microglia were stained with CD68 (1:500, Abcam) and IBA-1 (1:1000, DAKO). Neurodegeneration was indicated by neuronal loss, neurite degeneration, and apoptosis of neurons. Neuronal loss and neurite degeneration in the CA1 region of the hippocampus were detected by double immunofluorescence staining for NeuN and microtubule-associated protein 2 (Map2) (NeuN: 1:500, Abcam; Map2: 1:2000, Abcam). Apoptosis of neurons in the CA3 region of the hippocampus was assessed by double immunofluorescence staining for NeuN and caspase-3 (NeuN: 1:500, Abcam; caspase-3: 1:500, Abcam).
Brain volume analysis
Every 10th coronal brain section (300 μm between sections) starting rostrally at bregma +2.1 mm to the dorsal end of the hippocampus at bregma −3.9 mm was mounted for each mouse. For the hippocampus, posterior lateral ventricle, and neocortex, quantification started from bregma −1.1 mm and ended at bregma −3.0 mm. The volume was calculated using the following formula: volume = (sum of area) × 0.3 mm.
Western blotting
Brain samples were homogenized in ice-cold radioimmunoprecipitation assay lysis buffer. Protein samples were loaded on a 4 to 20% SDS–polyacrylamide gel electrophoresis, and then transferred onto nitrocellulose (NC) membranes. After blocking with 5% fat-free milk, the NC membranes were incubated with the following primary antibodies overnight at 4°C: anti-APP C-terminal antibody (1:1000, Millipore) to detect CTF-α and CTF-β; anti-6E10 was used to detect Aβ, full-length APP (flAPP) (1:400, BioLegend); anti-RAGE (1:1000, Millipore); anti-LRP-1 (1:1000, Abcam); anti-IDE antibody (1:1000, Abcam); anti-NEP antibody (1:1000, Millipore); and anti-BACE1 antibody (1:1000, Abcam). The membranes were incubated with the corresponding IRDye 800 CW-conjugated secondary antibodies and scanned using an Odyssey fluorescent scanner. Relative band intensities were normalized to the band intensity of the internal reference protein for analysis.
Enzyme-linked immunosorbent assay
Proteins were extracted from brain tissues in tris-buffered saline (TBS), 2% SDS, and 70% formic acid (FA) to measure soluble and insoluble Aβ levels. The levels of human Aβ42 and Aβ40 in the TBS, SDS, and FA fractions were measured using enzyme-linked immunosorbent assay (ELISA) kits (Invitrogen). The levels of inflammatory factors in brain homogenates, including TNF-α, IFN-γ, and IL-10, were measured with ELISA kits (Raybiotech, USA).
Quantitative reverse-transcription PCR assays
CD14+ monocytes were directly sorted into 1 ml Trizol reagent (Thermo Fisher Scientific, USA). Total RNA was extracted by using Trizol reagent followed by quantification with NanoDrop ND-100 spectrophotometer (Thermo Fisher Scientific, USA). cDNA was synthesized by reverse transcription for qPCR with a HiScript II one-step quantitative reverse-transcription polymerase chain reaction (qRT-PCR) Kit (Takara Bio, Japan) on CFX96 real-time PCR system (Bio-Rad, USA). Glyceraldehyde-3-phosphate dehydrogenase was used as the internal control. The sequence of qRT-PCR primers used in this study is listed in table S5.
Bulk RNA-seq analysis of microglia
Mice were anesthetized using isoflurane (RWD, CN) and perfused via the left ventricle with ice-cold PBS without Ca2+ and Mg2+ for 6 min. Brains were removed and cut into two pieces along the midline. One hemisphere was minced with a surgical knife, and then transferred to ice-cold Hanks’ balanced salt solution (HBSS). Minced hemi-brains were enzymatically digested at 37°C for 15 min by using papain (Sangon Biotech, CN) according to the manufacturer’s protocol. The cell suspension was washed by ice-cold HBSS. The single-cell suspension was stained with anti-CD11b (BD Biosciences, USA) antibodies for 30 min at 4°C in the dark. CD11b+ microglia were then sorted by MA900 multi-application cell sorter (Sony, Japan). Total RNA was isolated using a TRIzol total RNA extraction kit (TIANGEN, CN). RNA quality was examined by 0.8% agarose gel electrophoresis and spectrophotometry. High-quality RNA with a 260/280 absorbance ratio of 1.8 to 2.2 was used for library construction and sequencing. Illumina library construction was performed according to the manufacturer’s instructions (Illumina, USA). Oligo-dT primers are used to transverse mRNA to obtain cDNA (APExBIO, USA). cDNA for the synthesis of the second chain of cDNA was amplified. cDNA products by AMPure XP system (Beckman Coulter, Beverly, USA) were purified. After library construction, library fragments were enriched by PCR amplification and selected according to a fragment size of 350 to 550 base pairs. The library was quality-assessed using an Agilent 2100 Bioanalyzer (Agilent, USA). The library was sequenced using the Illumina NovaSeq 6000 sequencing platform (Paired end150) to generate raw reads. Raw paired-end fastq reads were filtered by TrimGaloreto to discard the adapters and low-quality bases via calling the Cutadapt tool. The clean reads obtained were then aligned to the mm10 mouse genome using HISAT2, followed by reference genome-guided transcriptome assembly and gene expression quantification using StringTie. DEGs were identified by DEseq2 (for the sample with replications) with a cutoff value of log2|fold change (FC)| > 0.5 and adjusted P < 0.05. The clusterProfiler was used to perform functional enrichment analysis for the annotated significant DEGs, the potential genes in identified modules based on Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway categories. Terms with P < 0.05 were considered significant. GSEA was performed by the function in package clusterProfiler with a gene list sorted by log2FC.
LC–MS/MS analysis of the plasma proteome
Plasma samples were incubated with Bio-Rad Proteominer beads to remove high-abundance proteins. The processed sample was frozen into dry powder and then dissolved in dissolution buffer [8 M urea and 100 mM triethylammonium bicarbonate (TEAB) (pH 8.5)]. The solution was reduced with 10 mM dithiothreitol and subsequently alkylated with sufficient iodoacetamide. Each protein sample was taken, and the volume was made up to 100 μl with DB lysis buffer [8 M urea and 100 mM TEAB (pH 8.5)]. Trypsin and 100 mM TEAB buffer were added, and the sample was mixed and digested at 37°C for 4 hours. FA was mixed with the digested sample, and the mixture was adjusted to pH 3 and centrifuged at 12,000g for 5 min at room temperature. The supernatant was desalted and then fractionated on a Rigol L3000 HPLC system. A half sample containing 4 μg of fraction supernatant and 0.8 μl of iRT reagent was injected into the EASY-nLCTM 1200 UHPLC system (Thermo Fisher Scientific) coupled with an Orbitrap Q ExactiveTM series mass spectrometer (Thermo Fisher Scientific) operating in the data-independent acquisition (DIA) mode.
The resulting spectra from each fraction were searched separately by Proteome Discoverer 2.2 (PD 2.2, Thermo Fisher Scientific). The results of the search and identification by PD2.2 software were imported into Spectronaut (version 14.0, Biognosys) software to generate a library. The eligible peptides and product ions were selected from the spectrum by setting peptides and ion pair selection rules to generate a Target List. The DIA data were imported, and ion-pair chromatographic peaks were extracted according to the Target List. The ions were matched, and the peak area was calculated to achieve qualitative and quantitative peptides. iRT was added to the sample to correct the retention time, and the precursor ion q value cutoff was set to 0.01. The protein quantitation results were statistically analyzed by t test. The proteins whose quantitation was significantly different between the experimental and control groups (P < 0.05 and |log2FC| >0.5) were defined as DEPs.
Analyses of single-cell RNA sequencing
Quality control and normalization
Data preprocessing was performed by the Scanpy Python package v.1.8.1 (49). All cells were combined into a single dataset. We excluded cells with read counts less than 1000, gene counts less than 500, mitochondrial gene proportions greater than 10%, and cells identified as doublets. Doublets were detected using the Python package doublet detection v.4.2. Genes expressed in less than three cells were also discarded. Integration and normalization of all samples followed the standard workflow of Scanpy, with the batch effect corrected by sc.external.pp.bbknn (50).
Cell clustering and identification
Principal components analysis was performed on variable genes, and the top 40 principal components (PCs) were used in neighbor graph building. We confirmed that the top 40 PCs capture 99% of the variance. Then, we clustered the data using the Leiden method (51). The data were visualized using Uniform Manifold Approximation and Projection and t-distributed stochastic neighbor embedding projection. For each precluster, DEGs were detected using the Wilcoxon-rank sum test as implemented in the function rank_genes_groups in SCANPY. Cell types were assigned to each cluster using known marker genes.
Differential gene expression analysis
In our differential gene expression analysis, we used the rank_genes_groups function, which uses the Wilcoxon rank sum test, to sensitively detect DEGs at a single-cell level. For the five main cell types, genes with an adjusted P < 0.05 and an absolute log2FC > 0.5 were considered significantly different. On the basis of the DE genes, genes with different signs in the two comparisons were marked as reversed genes. To address the issue of potential false positives inherent in the Wilcoxon method, we also applied a pseudobulk approach to aggregating cell counts and performed additional validation using the DESeq2 package (52).
TF regulatory network analysis
TF regulatory network analysis was performed according to the SCENIC workflow (pySCENIC v.0.11.2) (53) with default parameters. Briefly, SCENIC includes three steps: First, GRNBoost is used to infer coexpression modules; then, the cisTarget database is used as a reference to filter directly regulated regulons; last, the activity scores of regulons are quantified by AUCell. Through SCENIC, we obtained 143 regulons for further analysis.
Functional annotation analysis
Pathway enrichment analysis was performed using the ‘GSEA’ function in the Python package GSEApy v.1.0.4 (https://github.com/zqfang/GSEApy), and enrichment analysis was conducted on the GO database, which provides functional annotations and biological process information for genes. For gene ranking in GSEA, we used the “signal-to-noise” algorithm. This algorithm ranks genes based on their differential expression between two phenotypic groups, considering the variability within each group. Higher rankings indicate greater differential expression. In terms of parameters, we conducted a permutation test with 1000 permutations to assess the statistical significance of gene set enrichment. The permutation test reshuffles the phenotype labels of the samples to generate a null distribution and determine the significance of the observed enrichment score. Pathways with FDR q < 0.05 were retained.
Cell-to-cell communication analysis
CellPhoneDB software v.2.1.7 (54) was used to perform cell-to-cell communication analysis for three groups (control YTg, control OTg, and YTg BMCs) separately. Ligand-receptor interactions with P < 0.05 were retained. The results were visualized using the R package iTALK (55).
Statistical analyses
The normality and homoscedasticity of the data were tested by the Kolmogorov-Smirnov test and Bartlett’s test, respectively. All data were checked for outliers. One-way analysis of variance (ANOVA) was used for the comparison of groups where applicable. All data are presented as the means ± SEM. Two-sided P < 0.05 was defined as statistically significant. All analyses were carried out using GraphPad Prism version 8.0 (GraphPad Software Inc., San Diego, CA, USA).
Acknowledgments
We thank all the members of Y.-J. Wang’s laboratory for the discussions and suggestions.
Funding: The study was supported by the National Natural Science Foundation of China (92249305 and 81930028 to Y.-J.W.) and the National Key Research and Development Program Foundation of China (2023YFC3605400 to Y.-J.W.).
Author contributions: Y.-J.W. and J.W. conceived and designed the project. P.-Y.S., J.L., J.-N.H., Q.J., Y.-J.J., H.-L.S., S.-H.C., J.-Y.X., Z.-Y.Y., and Z.-H.L. conducted animal and in vitro experiments. Y.-F.T., C.-R.T., G.-H.Z., A.-Y.S., Y.-H.L., and X.-L.B. analyzed data. Y.-J.W., J.W., P.-Y.S., and J.L. wrote and revised the manuscript. All authors discussed the results and commented on the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The scRNA-seq data that support the findings of this study are deposited in the Gene Expression Omnibus repository under accession number GSE235527. Source data are provided in this paper.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Tables S3 and S4
Legends for tables S1, S2, and S5
Other Supplementary Material for this : manuscript includes the following:
Tables S1, S2, and S5
REFERENCES AND NOTES
- 1.Huang Y., Mucke L., Alzheimer mechanisms and therapeutic strategies. Cell 148, 1204–1222 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sims R., Hill M., Williams J., The multiplex model of the genetics of Alzheimer's disease. Nat. Neurosci. 23, 311–322 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Bellenguez C., Küçükali F., Jansen I. E., Kleineidam L., Moreno-Grau S., Amin N., Naj A. C., Campos-Martin R., Grenier-Boley B., Andrade V., Holmans P. A., Boland A., Damotte V., van der Lee S. J., Costa M. R., Kuulasmaa T., Yang Q., de Rojas I., Bis J. C., Yaqub A., Prokic I., Chapuis J., Ahmad S., Giedraitis V., Aarsland D., Garcia-Gonzalez P., Abdelnour C., Alarcón-Martín E., Alcolea D., Alegret M., Alvarez I., Álvarez V., Armstrong N. J., Tsolaki A., Antúnez C., Appollonio I., Arcaro M., Archetti S., Pastor A. A., Arosio B., Athanasiu L., Bailly H., Banaj N., Baquero M., Barral S., Beiser A., Pastor A. B., Below J. E., Benchek P., Benussi L., Berr C., Besse C., Bessi V., Binetti G., Bizarro A., Blesa R., Boada M., Boerwinkle E., Borroni B., Boschi S., Bossù P., Bråthen G., Bressler J., Bresner C., Brodaty H., Brookes K. J., Brusco L. I., Buiza-Rueda D., Bûrger K., Burholt V., Bush W. S., Calero M., Cantwell L. B., Chene G., Chung J., Cuccaro M. L., Carracedo Á., Cecchetti R., Cervera-Carles L., Charbonnier C., Chen H. H., Chillotti C., Ciccone S., Claassen J., Clark C., Conti E., Corma-Gómez A., Costantini E., Custodero C., Daian D., Dalmasso M. C., Daniele A., Dardiotis E., Dartigues J. F., de Deyn P. P., de Paiva Lopes K., de Witte L. D., Debette S., Deckert J., Del Ser T., Denning N., DeStefano A., Dichgans M., Diehl-Schmid J., Diez-Fairen M., Rossi P. D., Djurovic S., Duron E., Düzel E., Dufouil C., Eiriksdottir G., Engelborghs S., Escott-Price V., Espinosa A., Ewers M., Faber K. M., Fabrizio T., Nielsen S. F., Fardo D. W., Farotti L., Fenoglio C., Fernández-Fuertes M., Ferrari R., Ferreira C. B., Ferri E., Fin B., Fischer P., Fladby T., Fließbach K., Fongang B., Fornage M., Fortea J., Foroud T. M., Fostinelli S., Fox N. C., Franco-Macías E., Bullido M. J., Frank-García A., Froelich L., Fulton-Howard B., Galimberti D., García-Alberca J. M., García-González P., Garcia-Madrona S., Garcia-Ribas G., Ghidoni R., Giegling I., Giorgio G., Goate A. M., Goldhardt O., Gomez-Fonseca D., González-Pérez A., Graff C., Grande G., Green E., Grimmer T., Grünblatt E., Grunin M., Gudnason V., Guetta-Baranes T., Haapasalo A., Hadjigeorgiou G., Haines J. L., Hamilton-Nelson K. L., Hampel H., Hanon O., Hardy J., Hartmann A. M., Hausner L., Harwood J., Heilmann-Heimbach S., Helisalmi S., Heneka M. T., Hernández I., Herrmann M. J., Hoffmann P., Holmes C., Holstege H., Vilas R. H., Hulsman M., Humphrey J., Biessels G. J., Jian X., Johansson C., Jun G. R., Kastumata Y., Kauwe J., Kehoe P. G., Kilander L., Ståhlbom A. K., Kivipelto M., Koivisto A., Kornhuber J., Kosmidis M. H., Kukull W. A., Kuksa P. P., Kunkle B. W., Kuzma A. B., Lage C., Laukka E. J., Launer L., Lauria A., Lee C. Y., Lehtisalo J., Lerch O., Lleó A., Longstreth W. Jr., Lopez O., de Munain A. L., Love S., Löwemark M., Luckcuck L., Lunetta K. L., Ma Y., Macías J., MacLeod C. A., Maier W., Mangialasche F., Spallazzi M., Marquié M., Marshall R., Martin E. R., Montes A. M., Rodríguez C. M., Masullo C., Mayeux R., Mead S., Mecocci P., Medina M., Meggy A., Mehrabian S., Mendoza S., Menéndez-González M., Mir P., Moebus S., Mol M., Molina-Porcel L., Montrreal L., Morelli L., Moreno F., Morgan K., Mosley T., Nöthen M. M., Muchnik C., Mukherjee S., Nacmias B., Ngandu T., Nicolas G., Nordestgaard B. G., Olaso R., Orellana A., Orsini M., Ortega G., Padovani A., Paolo C., Papenberg G., Parnetti L., Pasquier F., Pastor P., Peloso G., Pérez-Cordón A., Pérez-Tur J., Pericard P., Peters O., Pijnenburg Y. A. L., Pineda J. A., Piñol-Ripoll G., Pisanu C., Polak T., Popp J., Posthuma D., Priller J., Puerta R., Quenez O., Quintela I., Thomassen J. Q., Rábano A., Rainero I., Rajabli F., Ramakers I., Real L. M., Reinders M. J. T., Reitz C., Reyes-Dumeyer D., Ridge P., Riedel-Heller S., Riederer P., Roberto N., Rodriguez-Rodriguez E., Rongve A., Allende I. R., Rosende-Roca M., Royo J. L., Rubino E., Rujescu D., Sáez M. E., Sakka P., Saltvedt I., Sanabria Á., Sánchez-Arjona M. B., Sanchez-Garcia F., Juan P. S., Sánchez-Valle R., Sando S. B., Sarnowski C., Satizabal C. L., Scamosci M., Scarmeas N., Scarpini E., Scheltens P., Scherbaum N., Scherer M., Schmid M., Schneider A., Schott J. M., Selbæk G., Seripa D., Serrano M., Sha J., Shadrin A. A., Skrobot O., Slifer S., Snijders G. J. L., Soininen H., Solfrizzi V., Solomon A., Song Y., Sorbi S., Sotolongo-Grau O., Spalletta G., Spottke A., Squassina A., Stordal E., Tartan J. P., Tárraga L., Tesí N., Thalamuthu A., Thomas T., Tosto G., Traykov L., Tremolizzo L., Tybjærg-Hansen A., Uitterlinden A., Ullgren A., Ulstein I., Valero S., Valladares O., Broeckhoven C. V., Vance J., Vardarajan B. N., van der Lugt A., Dongen J. V., van Rooij J., van Swieten J., Vandenberghe R., Verhey F., Vidal J. S., Vogelgsang J., Vyhnalek M., Wagner M., Wallon D., Wang L. S., Wang R., Weinhold L., Wiltfang J., Windle G., Woods B., Yannakoulia M., Zare H., Zhao Y., Zhang X., Zhu C., Zulaica M., Farrer L. A., Psaty B. M., Ghanbari M., Raj T., Sachdev P., Mather K., Jessen F., Ikram M. A., de Mendonça A., Hort J., Tsolaki M., Pericak-Vance M. A., Amouyel P., Williams J., Frikke-Schmidt R., Clarimon J., Deleuze J. F., Rossi G., Seshadri S., Andreassen O. A., Ingelsson M., Hiltunen M., Sleegers K., Schellenberg G. D., van Duijn C. M., Sims R., van der Flier W. M., Ruiz A., Ramirez A., Lambert J. C., New insights into the genetic etiology of Alzheimer's disease and related dementias. Nat. Genet. 54, 412–436 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao W., Zheng H., Peripheral immune system in aging and Alzheimer's disease. Mol. Neurodegener. 13, 51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu H., Jia J., Single-cell RNA sequencing of peripheral blood reveals immune cell signatures in Alzheimer's disease. Front. Immunol. 12, 645666 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yousefzadeh M. J., Flores R. R., Zhu Y., Schmiechen Z. C., Brooks R. W., Trussoni C. E., Cui Y., Angelini L., Lee K. A., McGowan S. J., Burrack A. L., Wang D., Dong Q., Lu A., Sano T., O'Kelly R. D., McGuckian C. A., Kato J. I., Bank M. P., Wade E. A., Pillai S. P. S., Klug J., Ladiges W. C., Burd C. E., Lewis S. E., LaRusso N. F., Vo N. V., Wang Y., Kelley E. E., Huard J., Stromnes I. M., Robbins P. D., Niedernhofer L. J., An aged immune system drives senescence and ageing of solid organs. Nature 594, 100–105 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinti M., Appay V., Campisi J., Frasca D., Fülöp T., Sauce D., Larbi A., Weinberger B., Cossarizza A., Aging of the immune system: Focus on inflammation and vaccination. Eur. J. Immunol. 46, 2286–2301 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikolich-Žugich J., The twilight of immunity: Emerging concepts in aging of the immune system. Nat. Immunol. 19, 10–19 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Cancro M. P., Age-associated B cells. Annu. Rev. Immunol. 38, 315–340 (2020). [DOI] [PubMed] [Google Scholar]
- 10.de Haan G., Lazare S. S., Aging of hematopoietic stem cells. Blood 131, 479–487 (2018). [DOI] [PubMed] [Google Scholar]
- 11.López-Otín C., Blasco M. A., Partridge L., Serrano M., Kroemer G., Hallmarks of aging: An expanding universe. Cell 186, 243–278 (2023). [DOI] [PubMed] [Google Scholar]
- 12.Yang A. C., Vest R. T., Kern F., Lee D. P., Agam M., Maat C. A., Losada P. M., Chen M. B., Schaum N., Khoury N., Toland A., Calcuttawala K., Shin H., Pálovics R., Shin A., Wang E. Y., Luo J., Gate D., Schulz-Schaeffer W. J., Chu P., Siegenthaler J. A., McNerney M. W., Keller A., Wyss-Coray T., A human brain vascular atlas reveals diverse mediators of Alzheimer's risk. Nature 603, 885–892 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C., Yosef N., Gaublomme J., Wu C., Lee Y., Clish C. B., Kaminski J., Xiao S., Horste G. M. Z., Pawlak M., Kishi Y., Joller N., Karwacz K., Zhu C., Ordovas-Montanes M., Madi A., Wortman I., Miyazaki T., Sobel R. A., Park H., Regev A., Kuchroo V. K., CD5L/AIM regulates lipid biosynthesis and restrains TH17 cell pathogenicity. Cell 163, 1413–1427 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maillard C., Roux C. J., Charbit-Henrion F., Steffann J., Laquerriere A., Quazza F., Buisson N. B., Tubulin mutations in human neurodevelopmental disorders. Semin. Cell Dev. Biol. 137, 87–95 (2023). [DOI] [PubMed] [Google Scholar]
- 15.Li J., Zhao C., Li Y., Wen J., Wang S., Wang D., Dong H., Wang D., Zhao Y., Wang X., He X., Qin J., Osteosarcoma exocytosis of soluble LGALS3BP mediates macrophages toward a tumoricidal phenotype. Cancer Lett. 528, 1–15 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Pollen A. A., Nowakowski T. J., Chen J., Retallack H., Sandoval-Espinosa C., Nicholas C. R., Shuga J., Liu S. J., Oldham M. C., Diaz A., Lim D. A., Leyrat A. A., West J. A., Kriegstein A. R., Molecular identity of human outer radial glia during cortical development. Cell 163, 55–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q., Zhu F., Liu X., Lu Y., Yao K., Tian N., Tong L., Figge D. A., Wang X., Han Y., Li Y., Zhu Y., Hu L., Ji Y., Xu N., Li D., Gu X., Liang R., Gan G., Wu L., Zhang P., Xu T., Hu H., Hu Z., Xu H., Ye D., Yang H., Li B., Tong X., Non-oxidative pentose phosphate pathway controls regulatory T cell function by integrating metabolism and epigenetics. Nat. Metab. 4, 559–574 (2022). [DOI] [PubMed] [Google Scholar]
- 18.Cutine A. M., Bach C. A., Veigas F., Merlo J. P., Laporte L., Manselle Cocco M. N., Massaro M., Sarbia N., Perrotta R. M., Mahmoud Y. D., Rabinovich G. A., Tissue-specific control of galectin-1-driven circuits during inflammatory responses. Glycobiology 31, 891–907 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Schumacher M. A., Dennis I. C., Liu C. Y., Robinson C., Shang J., Bernard J. K., Washington M. K., Polk D. B., Frey M. R., NRG4-ErbB4 signaling represses proinflammatory macrophage activity. Am. J. Physiol. Gastrointest. Liver Physiol. 320, G990–G1001 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin B., Gabris-Weber B. A., Reddy R., Romero G., Chattopadhyay A., Salama G., Relaxin reverses inflammatory and immune signals in aged hearts. PLOS ONE 13, e0190935 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basisty N., Kale A., Jeon O. H., Kuehnemann C., Payne T., Rao C., Holtz A., Shah S., Sharma V., Ferrucci L., Campisi J., Schilling B., A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biol. 18, e3000599 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S. H., Tian D. Y., Shen Y. Y., Cheng Y., Fan D. Y., Sun H. L., He C. Y., Sun P. Y., Bu X. L., Zeng F., Liu J., Deng J., Xu Z. Q., Chen Y., Wang Y. J., Amyloid-beta uptake by blood monocytes is reduced with ageing and Alzheimer's disease. Transl. Psychiatry 10, 423 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleinberger G., Yamanishi Y., Suárez-Calvet M., Czirr E., Lohmann E., Cuyvers E., Struyfs H., Pettkus N., Wenninger-Weinzierl A., Mazaheri F., Tahirovic S., Lleó A., Alcolea D., Fortea J., Willem M., Lammich S., Molinuevo J. L., Sánchez-Valle R., Antonell A., Ramirez A., Heneka M. T., Sleegers K., van der Zee J., Martin J. J., Engelborghs S., Demirtas-Tatlidede A., Zetterberg H., Van Broeckhoven C., Gurvit H., Wyss-Coray T., Hardy J., Colonna M., Haass C., TREM2 mutations implicated in neurodegeneration impair cell surface transport and phagocytosis. Sci. Transl. Med. 6, 243ra86 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T. K., David E., Baruch K., Lara-Astaiso D., Toth B., Itzkovitz S., Colonna M., Schwartz M., Amit I., A unique microglia type associated with restricting development of Alzheimer's disease. Cell 169, 1276–1290.e17 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Prinz M., Priller J., The role of peripheral immune cells in the CNS in steady state and disease. Nat. Neurosci. 20, 136–144 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Srinivasan K., Friedman B. A., Etxeberria A., Huntley M. A., van der Brug M. P., Foreman O., Paw J. S., Modrusan Z., Beach T. G., Serrano G. E., Hansen D. V., Alzheimer's patient microglia exhibit enhanced aging and unique transcriptional activation. Cell Rep. 31, 107843 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leng F., Edison P., Neuroinflammation and microglial activation in Alzheimer disease: Where do we go from here? Nat. Rev. Neurol. 17, 157–172 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Zhang L., Wang Y., Liu T., Mao Y., Peng B., Novel microglia-based therapeutic approaches to neurodegenerative disorders. Neurosci. Bull. 39, 491–502 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng Y., Tian D. Y., Wang Y. J., Peripheral clearance of brain-derived Aβ in Alzheimer's disease: Pathophysiology and therapeutic perspectives. Transl. Neurodegener. 9, 16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin W. S., Shen L. L., Bu X. L., Zhang W. W., Chen S. H., Huang Z. L., Xiong J. X., Gao C. Y., Dong Z., He Y. N., Hu Z. A., Zhou H. D., Song W., Zhou X. F., Wang Y. Z., Wang Y. J., Peritoneal dialysis reduces amyloid-beta plasma levels in humans and attenuates Alzheimer-associated phenotypes in an APP/PS1 mouse model. Acta Neuropathol. 134, 207–220 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Tian D. Y., Cheng Y., Zhuang Z. Q., He C. Y., Pan Q. G., Tang M. Z., Hu X. L., Shen Y. Y., Wang Y. R., Chen S. H., Sun H. L., Sun P. Y., Yu Z. Y., Fan D. Y., Bu X. L., Tan C. R., Zeng G. H., Wang J., Zhao H. W., Wang Y. J., Physiological clearance of amyloid-beta by the kidney and its therapeutic potential for Alzheimer's disease. Mol. Psychiatry 26, 6074–6082 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Hazeldine J., Lord J. M., The impact of ageing on natural killer cell function and potential consequences for health in older adults. Ageing Res. Rev. 12, 1069–1078 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin J., Ibrahim S., Petersen F., Yu X., Autoimmunomic signatures of aging and age-related neurodegenerative diseases are associated with brain function and ribosomal proteins. Front. Aging Neurosci. 13, 679688 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hashimoto K., Kouno T., Ikawa T., Hayatsu N., Miyajima Y., Yabukami H., Terooatea T., Sasaki T., Suzuki T., Valentine M., Pascarella G., Okazaki Y., Suzuki H., Shin J. W., Minoda A., Taniuchi I., Okano H., Arai Y., Hirose N., Carninci P., Single-cell transcriptomics reveals expansion of cytotoxic CD4 T cells in supercentenarians. Proc. Natl. Acad. Sci. U.S.A. 116, 24242–24251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J., Gu B. J., Masters C. L., Wang Y. J., A systemic view of Alzheimer disease - insights from amyloid-β metabolism beyond the brain. Nat. Rev. Neurol. 13, 612–623 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Tabula Muris Consortium , A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583, 590–595 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiang Y., Bu X. L., Liu Y. H., Zhu C., Shen L. L., Jiao S. S., Zhu X. Y., Giunta B., Tan J., Song W. H., Zhou H. D., Zhou X. F., Wang Y. J., Physiological amyloid-beta clearance in the periphery and its therapeutic potential for Alzheimer's disease. Acta Neuropathol. 130, 487–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Da Mesquita S., Louveau A., Vaccari A., Smirnov I., Cornelison R. C., Kingsmore K. M., Contarino C., Onengut-Gumuscu S., Farber E., Raper D., Viar K. E., Powell R. D., Baker W., Dabhi N., Bai R., Cao R., Hu S., Rich S. S., Munson J. M., Lopes M. B., Overall C. C., Acton S. T., Kipnis J., Functional aspects of meningeal lymphatics in ageing and Alzheimer's disease. Nature 560, 185–191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Das M. M., Godoy M., Chen S., Moser V. A., Avalos P., Roxas K. M., Dang I., Yáñez A., Zhang W., Bresee C., Arditi M., Liu G. Y., Svendsen C. N., Goodridge H. S., Young bone marrow transplantation preserves learning and memory in old mice. Commun. Biol. 2, 73 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villeda S. A., Plambeck K. E., Middeldorp J., Castellano J. M., Mosher K. I., Luo J., Smith L. K., Bieri G., Lin K., Berdnik D., Wabl R., Udeochu J., Wheatley E. G., Zou B., Simmons D. A., Xie X. S., Longo F. M., Wyss-Coray T., Young blood reverses age-related impairments in cognitive function and synaptic plasticity in mice. Nat. Med. 20, 659–663 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Middeldorp J., Lehallier B., Villeda S. A., Miedema S. S., Evans E., Czirr E., Zhang H., Luo J., Stan T., Mosher K. I., Masliah E., Wyss-Coray T., Preclinical assessment of young blood plasma for Alzheimer disease. JAMA Neurol. 73, 1325–1333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J., Jin W. S., Bu X. L., Zeng F., Huang Z. L., Li W. W., Shen L. L., Zhuang Z. Q., Fang Y., Sun B. L., Zhu J., Yao X. Q., Zeng G. H., Dong Z. F., Yu J. T., Hu Z., Song W., Zhou H. D., Jiang J. X., Liu Y. H., Wang Y. J., Physiological clearance of tau in the periphery and its therapeutic potential for tauopathies. Acta Neuropathol. 136, 525–536 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Jiao S. S., Bu X. L., Liu Y. H., Zhu C., Wang Q. H., Shen L. L., Liu C. H., Wang Y. R., Yao X. Q., Wang Y. J., Sex dimorphism profile of Alzheimer's disease-type pathologies in an app/ps1 mouse model. Neurotox. Res. 29, 256–266 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Jankowsky J. L., Fadale D. J., Anderson J., Xu G. M., Gonzales V., Jenkins N. A., Copeland N. G., Lee M. K., Younkin L. H., Wagner S. L., Younkin S. G., Borchelt D. R., Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: Evidence for augmentation of a 42-specific gamma secretase. Hum. Mol. Genet. 13, 159–170 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Kemp K. C., Dey R., Verhagen J., Scolding N. J., Usowicz M. M., Wilkins A., Aberrant cerebellar Purkinje cell function repaired in vivo by fusion with infiltrating bone marrow-derived cells. Acta Neuropathol. 135, 907–921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duran-Struuck R., Dysko R. C., Principles of bone marrow transplantation (BMT): Providing optimal veterinary and husbandry care to irradiated mice in BMT studies. J. Am. Assoc. Lab. Anim. Sci. 48, 11–22 (2009). [PMC free article] [PubMed] [Google Scholar]
- 47.Ojielo C. I., Cooke K., Mancuso P., Standiford T. J., Olkiewicz K. M., Clouthier S., Corrion L., Ballinger M. N., Toews G. B., Paine R. III, Moore B. B., Defective phagocytosis and clearance of Pseudomonas aeruginosa in the lung following bone marrow transplantation. J. Immunol. 171, 4416–4424 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Chen S. H., He C. Y., Shen Y. Y., Zeng G. H., Tian D. Y., Cheng Y., Xu M. Y., Fan D. Y., Tan C. R., Shi A. Y., Bu X. L., Wang Y. J., Polysaccharide krestin prevents Alzheimer's disease-type pathology and cognitive deficits by enhancing monocyte amyloid-β processing. Neurosci. Bull. 38, 290–302 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolf F. A., Angerer P., Theis F. J., SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Polanski K., Young M. D., Miao Z., Meyer K. B., Teichmann S. A., Park J. E., BBKNN: Fast batch alignment of single cell transcriptomes. Bioinformatics 36, 964–965 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Traag V. A., Waltman L., van Eck N. J., From Louvain to Leiden: Guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van de Sande B., Flerin C., Davie K., De Waegeneer M., Hulselmans G., Aibar S., Seurinck R., Saelens W., Cannoodt R., Rouchon Q., Verbeiren T., De Maeyer D., Reumers J., Saeys Y., Aerts S., A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat. Protoc. 15, 2247–2276 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Efremova M., Vento-Tormo M., Teichmann S. A., Vento-Tormo R., CellPhoneDB: Inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Y. Wang, R. Wang, S. Zhang, S. Song, C. Jiang, G. Han, M. Wang, J. Ajani, A. Futreal, L. Wang, iTALK: An R package to characterize and illustrate intercellular communication. bioRxiv 507871v1 (2019). www.biorxiv.org/content/10.1101/507871v1.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Tables S3 and S4
Legends for tables S1, S2, and S5
Tables S1, S2, and S5