Abstract
Antigen 43 (Ag43) is a surface-displayed autotransporter protein of Escherichia coli. By virtue of its self-association characteristics, this protein is able to mediate autoaggregation and flocculation of E. coli cells in static cultures. Additionally, surface display of Ag43 is associated with a distinct frizzy colony morphology in E. coli. Here we show that Ag43 can be expressed in a functional form on the surface of the environmentally important Pseudomonas fluorescens strain SBW25 with ensuing cell aggregation and frizzy colony types. Using green fluorescence protein-tagged cells, we demonstrate that Ag43 can be used as a tool to provide interspecies cell aggregation between E. coli and P. fluorescens. Furthermore, Ag43 expression enhances biofilm formation in P. fluorescens to glass surfaces. The versatility of this protein was also reflected in Ag43 surface display in a variety of other gram-negative bacteria. Display of heterologous Ag43 in selected bacteria might offer opportunities for rational design of multispecies consortia where the concerted action of several bacterial species is required, e.g., waste treatment and degradation of pollutants.
In nature, many bacteria have the ability to form aggregates consisting of tight communities of cells. Such aggregating phenotypes often seem to be associated with particular colony morphology forms and environmental niche specificities, for example with surface or bottom dwelling in static liquid environments, as elegantly demonstrated in the case of Pseudomonas fluorescens (20). In spite of the prominent phenotypic characteristics connected with bacterial cell-cell aggregation, little is known about the underlying molecular mechanisms. Some Escherichia coli strains have the ability to autoaggregate, seen as characteristic flocculation and settling of cells from static liquid suspensions. This phenomenon was first described by Diderichsen (4), who defined a locus, flu, mapping at 43 min on the E. coli K-12 chromosome. The flu locus appeared to control several interesting phenotypes, i.e., in addition to autoaggregation, a characteristic frizzy colony morphology. In separate studies, it was found that the product of the flu locus was identical to an outer membrane protein termed antigen 43 (Ag43) (11, 18). Ag43 was reported to consist of two equimolar protein subunits, α and β, with apparent molecular masses of about 50 and 53 kDa, respectively (3, 17). The α subunit is attached to the cell surface via interaction with the β subunit, which is an integral outer membrane component. Only recently was Ag43 unambiguously identified as the product of the flu gene (8, 13). In fact, the flu gene encodes an open reading frame of 1,039 codons. The corresponding product is processed into a 987-amino-acid protein by removal of a signal peptide, and subsequent autocatalytic processing results in an N-terminal α subunit of 499 amino acids and a C-terminal β subunit of 488 amino acids (8). It was further established that autoaggregation of cells takes place through an Ag43-Ag43 interaction (8). Further studies revealed that the frizzy colony morphology form was due to the presence of Ag43 on virtually all cells of such colonies, whereas Ag43-negative cells gave rise to flat opaque colonies (9). The expression of Ag43 is phase variable, with cells shifting from Ag43+ to Ag43− and vice versa at about 10−3 per cell per generation. The phase variation results from the combined regulation of the flu gene by the Dam methylase and OxyR (7, 10, 13, 18, 24). Interestingly, the frizzy and opaque colony forms, respectively, can be correlated with niche specificity under static liquid growth conditions; progeny from frizzy colonies, predominantly Ag43+ cells, give rise to bottom dwellers, and progeny from opaque colonies, predominantly Ag43− cells, are pelagic (9).
Ag43 is a member of the family of autotransporter proteins (12). These are characterized by the fact that all information required for transport to the outer membrane and secretion through this cell envelope is contained within the protein itself (Fig. 1A). This prompted us to investigate whether the Ag43 protein of E. coli K-12 could be expressed in a functional form in other gram-negative bacteria.
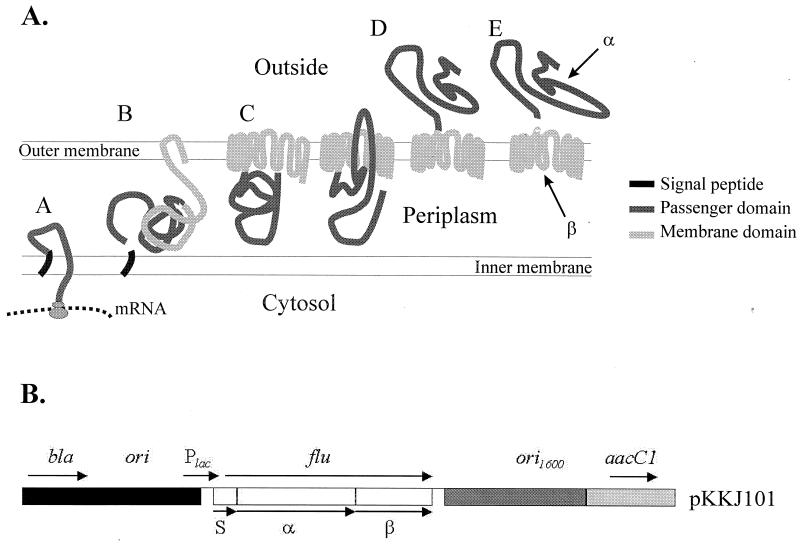
FIG. 1.
(A) Model of the biogenesis and processing of the Ag43 autotransporter protein. The signal peptide (black) enables the autotransporter to reach the periplasm (A). Subsequently, the β-membrane domain (light gray) starts inserting into the outer membrane (B) where it forms a pore (C) through which the α-passenger domain (dark gray) is translocated to the cell exterior (D). Once the α domain has reached the cell surface, it is processed by autocatalysis but remains associated with the cell through interaction with the β domain (E). (B) Structure of the plasmid pKKJ101 (not drawn to scale). Indicated are the β-lactamase gene (bla), E. coli origin of replication (ori) (black), the broad-host-range origin of replication (ori1600) (dark grey), the gentamycin resistance gene (aacC1) (light grey), the lac promoter, and the flu gene. The positions of the signal peptide, α domain, and β domain in the flu gene are indicated.
P. fluorescens strains are highly versatile microorganisms capable of establishing in a wide range of environments and capable of mineralizing a large spectrum of organic substances, including many pollutants (2, 21). These characteristics have spurred significant interest in such strains from the fields of microbial ecology and bioremediation. The well-characterized P. fluorescens reference strain SBW25 is an excellent plant colonizer and was originally isolated from sugar beets (19). This strain was recently characterized in depth with regard to colony morphology, colony morphology variation, and niche specificity in static liquid environments (20). These attributes make it a prime candidate for testing of transspecies Ag43 expression.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are described in Table 1. Cells were grown at 37 or 30°C (P. fluorescens and Klebsiella pneumonia, respectively) on solid or in liquid Luria-Bertani (LB) medium supplemented with the appropriate antibiotics unless otherwise stated. E. coli K-12 strain HEHA16 is a fim::kan Δflu derivative of BD1428 (leu lysA metE proB PurE thi trp str supE tonA). To delete the flu gene in strain BD1428, we constructed the deletion plasmid pHHA174. This plasmid consists of a 1.5-kb fragment from the region upstream of the flu gene (amplified with primers P1, 5′-ACGGCTGACTAAGCTTACTC, and P2, 5′-GGGCCTTGGATCCAGGGTGAATAAAAAAGCCGG) and a 1.5-kb fragment from the region downstream of the flu gene (amplified with primers P3, 5′-CGGCAGATCTCGAGGCGGATCTTCAACGCCACATTCG, and P4, 5′-CGGGAGATCTCTCGAGGTTCATGAAGATGGCGGTCCA) ligated into HindIII-BamHI-digested pMAK700oriT. This plasmid carries a temperature-sensitive origin of replication which permits it to replicate only at temperatures at or below 30°C. Plasmid pHHA174 was transformed into BD1428 and was allowed to grow overnight at 30°C on 17-μg/ml chloramphenicol (CAM) plates. Single colonies, after being restreaked at 30°C for 2 days, were streaked out on 17-μg/ml CAM plates and placed at 42°C to select for recA-mediated single-crossover events. Camr colonies were then grown in liquid LB medium containing 17 μg of CAM/ml at 42°C. Overnight cultures were diluted 1:1,000 in liquid LB medium without antibiotics, grown to an optical density at 600 nm (OD600) of 0.5, and then plated out on LB plates. Colonies from these plates were replica plated on 17-μg/ml CAM plates and LB plates, and chromosomal DNA from Cams colonies was tested by PCR employing primers P5, 5′-GGCGTCGACGACCGATTGAGGTTTCC, and P6, 5′-GCGCGGATCCGTTAAATCAAACCTCTTC. A colony which had a deletion in the flu gene was selected and named HEHA14.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| E. coli K-12 (BD1428) | 4 | |
| E. coli K-12 (HEHA16) | BD1428; fim::kan Δflu | This study |
| E. cloacae (SM825) | Fim+ | Institute strain collection |
| K. pneumonia (SM830) | Fim+ | Institute strain collection |
| P. fluorescens (SBW25) | 19 | |
| S. liquefaciens (MG1) | Fim+ | 6 |
| Plasmids | ||
| pUCP22 | E. coli-Pseudomonas shuttle vector; Apr; Gmr | 25 |
| pKKJ101 | flu gene in pUCP22 | This study |
| pKKJ128 | flu gene in pACYC184 | This study |
| pJBA25 | gfp gene in pUC18 | J. B. Andersen |
A fim deletion mutant of HEHA14 was constructed by use of the λpir-dependent plasmid pLBJ312 containing a truncated fim gene cluster with an npt gene (Kanr) inserted between truncated fimB and fimH. Insertion on the chromosome was done as described previously (22). A representative clone carrying the correct insertion was verified by PCR and Southern blotting and was designated HEHA16. This strain was deficient in Ag43-mediated cell aggregation and type 1 fimbria-mediated yeast agglutination.
DNA manipulations.
Isolation of plasmid DNA was carried out by using the QIAprep Spin Miniprep Kit (QIAGEN). Restriction endonucleases were used according to the manufacturer's specifications (Biolabs). Chromosomal DNA purification was carried out by using the GenomicPrep Cell and Tissue DNA isolation kit (Amersham Pharmacia Biotech Inc.). PCRs were carried out as previously described (23). Amplified products were sequenced to ensure fidelity of the PCR by using the ABI PRISM BigDye Terminator cycle sequencing ready reaction kit (PE Applied Biosystems). Samples were electrophoresed on a Perkin-Elmer ABI PRISM 310 Genetic Analyzer (PE Applied Biosystems) as described in the manufacturer's specifications.
Construction of flu-expressing shuttle vector.
The flu gene was amplified by PCR from E. coli K-12 chromosomal DNA by using the primers P7, 5′-CCCGCGGCCGCGATATCCTTTGTCAGTAACATGC, and P8, 5′-CCCGCGGCCGCGGATCCTGTGGCGTTGAAGATCCG. The resulting fragment was cut with EcoRV-BamHI and inserted directly into the EcoRV-BamHI site of the shuttle vector pUCP22 to produce plasmid pKKJ101. In this construct, expression of the flu gene is under transcriptional control of the lac promoter (Fig. 1B).
Additionally, the flu gene from pKKJ101 was isolated after digestion with HindIII-BamHI and inserted directly into the HindIII-BamHI site of pACYC184 to produce pKKJ128.
Colony morphology.
Colony morphology was assayed by employing a Carl Zeiss Axioplan epifluorescence microscope, and digital images were captured with a 12-bit cooled slow-scan charge-coupled device camera (KAF 1400 chip; Photometrics, Tucson, Ariz.) controlled by PMIS software (Photometrics).
Immunofluorescence microscopy.
Surface presentation of Ag43 was assessed by immunofluorescence microscopy as previously described (8).
Western immunoblotting.
Overnight cultures of the strains were harvested and adjusted to an OD600 of 2.5. After resuspension in 1× phosphate-buffered saline the cells were held at 60°C for 10 min, allowing detachment of the α subunit of Ag43 from the cells. The supernatant was transferred to new tubes and the α subunit was precipitated by 75% acetone. Samples were subjected to sodium dodecyl sulfate–15% polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride microporous membrane filters with a semidry blotting apparatus. Reconstituted dried skimmed milk (10% [wt/vol]) was used as the blocking reagent, and serum raised against the α subunit of Ag43 (8) was used as the primary serum. Peroxidase-conjugated anti-rabbit serum was used as the secondary antibody.
Autoaggregation assay.
Overnight cultures of the strains were adjusted to the same OD600, and 10-ml samples of each culture were placed in sterile 20-ml tubes. At the beginning of each experiment, all cultures were vigorously shaken for 10 s. Two 100-μl samples were taken from each tube, approximately 1 cm from the top, and transferred to two new tubes, each containing 1 ml of 0.9% NaCl. The OD600 was then measured.
Interspecies cell aggregation.
Plasmid pJBA25 (a kind gift from Jens Bo Andersen) encoding constitutive expression of the green fluorescence protein (GFP) was transformed into HEHA16 carrying plasmid pUCP22 or pKKJ101. GFP-expressing cells were mixed with equal amounts of Ag43-expressing cells from strains lacking GFP (either E. coli or P. fluorescens), and cell aggregation was observed by using a Carl Zeiss Axioplan epifluorescence microscope equipped with a filter for detecting GFP.
Detection of type 1 fimbriae.
The capacity of bacteria to express a d-mannose-binding phenotype was assayed by their ability to agglutinate yeast cells on glass slides. Aliquots of liquid cultures grown to an OD600 of 4.0 and 5% (wt/vol) suspensions of yeast cells were mixed, and the time until agglutination was measured.
Biofilm assay.
The ability of P. fluorescens expressing Ag43 to form a biofilm on glass surfaces was assessed by growing cells statically in LB medium. After 48 h, growth cultures were removed and the glass tubes were stained with 0.1% crystal violet as previously reported (5). Quantification of cells in biofilms was done by adding acetone-ethanol (80:20) and measuring the OD590 of dissolved crystal violet.
RESULTS
Colony morphology changes induced by Ag43 expression.
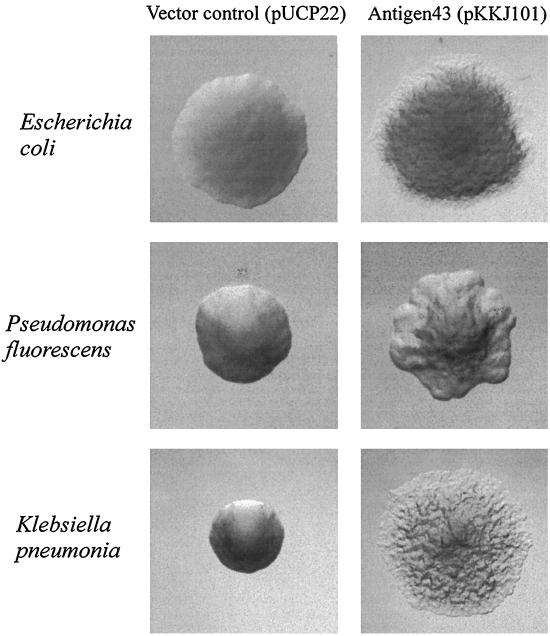
The E. coli K-12 reference strain BD1428 exhibits different colony morphotypes. We have previously demonstrated that phase-variable expression of Ag43 is directly involved in this phenomenon. In particular, cells expressing Ag43 form frizzy colonies, while Ag43− cells form colonies with a smooth surface (9). In order to investigate whether Ag43 could be expressed on the surface of P. fluorescens, the Ag43-encoding flu gene was inserted into the broad-host-range shuttle plasmid pUCP22 (25), resulting in plasmid pKKJ101 (Fig. 1B). When plasmid pKKJ101 was introduced into P. fluorescens SBW25, a distinct frizzy colony morphotype was noted, similar to that observed in Ag43+ E. coli (Fig. 2). In contrast, the vector control, pUCP22, gave rise to colonies which appeared to be identical to the parent strain (Fig. 2).
FIG. 2.
Colony morphology of cells examined by phase-contrast microscopy. Ag43-expressing strains containing pKKJ101 give rise to changes in colony morphology. Strains containing the control plasmid pUCP22 display the same morphotype as the parent strain. Colonies were grown overnight and photographed at the same magnification.
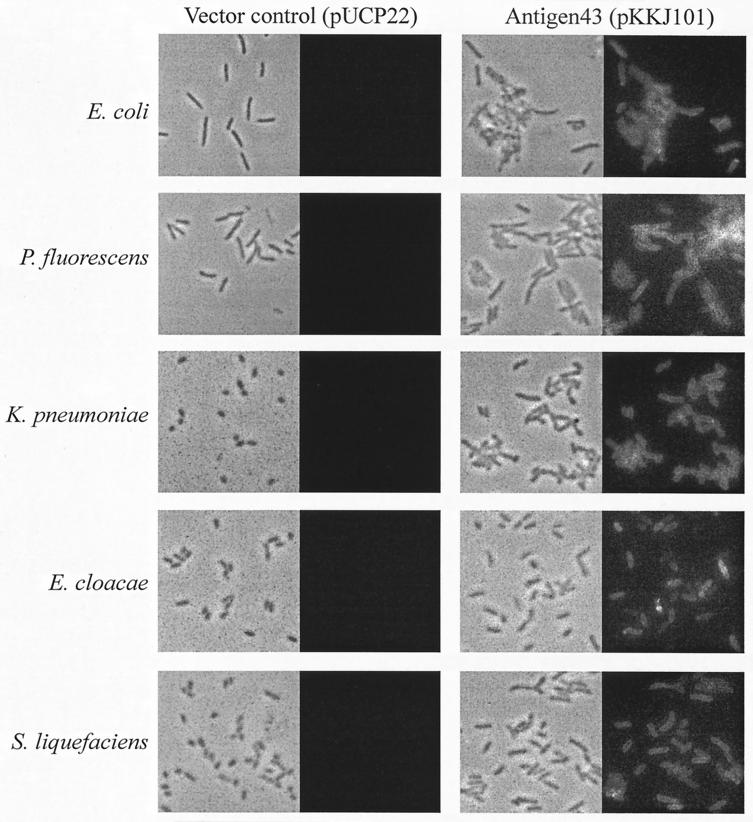
To investigate whether the frizzy colony morphology type was concomitant with cell surface display of Ag43, colonies were picked from plates and submitted to immunofluorescence microscopy with specific Ag43 antiserum. Virtually all cells from frizzy colonies of P. fluorescens harboring plasmid pKKJ101 reacted strongly, indicative of surface display of Ag43 (Fig. 3). These results suggest that despite the E. coli K-12 ancestry of Ag43, this protein was indeed translocated to the surface of P. fluorescens.
FIG. 3.
Phase-contrast microscopy (left panels on each side) and fluorescence microscopy (right panels on each side) of various gram-negative strains containing either the vector alone (pUCP22) or the Ag43-expressing plasmid pKKJ101, respectively. To detect the presence of Ag43 on the cell surface, a polyclonal rabbit serum raised against the α subunit of Ag43 was used, and this was detected by fluorescein isothiocyanate-labeled anti-rabbit serum.
Ag43-mediated autoaggregation of cells.
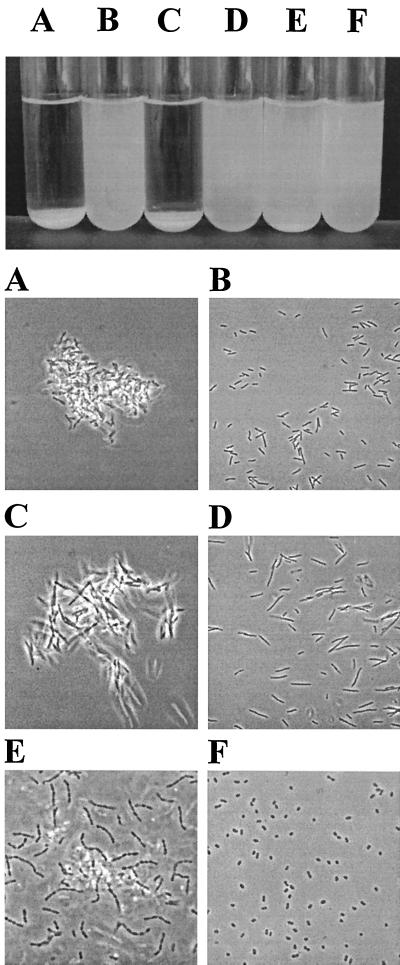
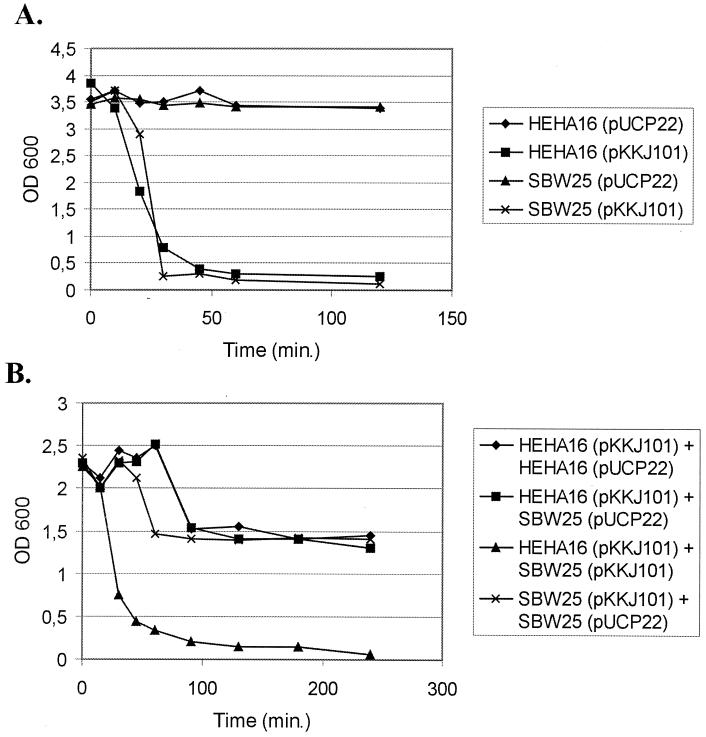
In E. coli, expression of Ag43 induces autoaggregation and settling of cells from standing liquid cultures. This phenomenon is mediated by Ag43-Ag43 interaction and follows first-order kinetics in a cell-density-dependent manner (8). When cells from shaken liquid overnight cultures of P. fluorescens harboring plasmid pKKJ101 were left standing, they were observed to autoaggregate, flocculate, and settle, forming a thick precipitate at the bottom of the tubes (Fig. 4). The kinetics of Ag43-mediated settling demonstrated by P. fluorescens expressing Ag43 was almost identical to that of Ag43+ E. coli (Fig. 5A). This indicates that Ag43 is presented on the surface of P. fluorescens in a fully functional form and imparts the same autoaggregating phenotype with ensuing cell settling as observed in E. coli. Furthermore, when P. fluorescens(pKKJ101) was submitted to mild heat treatment, a protein with the same apparent molecular mass as the α fragment of Ag43 was released, and this protein reacted with specific anti-Ag43 antiserum in Western immunoblotting (data not shown). This indicates that Ag43 is processed identically in P. fluorescens and in E. coli.
FIG. 4.
Settling (upper panel) and aggregation (lower panel) of cells from static liquid suspensions. (A) E. coli HEHA16(pKKJ101), (B) E. coli HEHA16(pUCP22), (C) P. fluorescens SBW25(pKKJ101), (D) P. fluorescens SBW25(pUCP22), (E) K. pneumoniae(pKKJ101), and (F) K. pneumoniae(pUCP22).
FIG. 5.
(A). Autoaggregation assay performed with monocultures of E. coli HEHA16 and P. fluorescens SBW25 containing pUCP22 (vector control) or pKKJ101 (Ag43-expressing plasmid), respectively. (B) Settling profiles for the following mixed cultures are shown: E. coli HEHA16(pKKJ101) + E. coli HEHA16(pUCP22), E. coli HEHA16(pKKJ101) + P. fluorescens SBW25(pUCP22), E. coli HEHA16(pKKJ101) + P. fluorescens SBW25(pKKJ101), and P. fluorescens SBW25(pKKJ101) + P. fluorescens SBW25(pUCP22).
Ag43 causes interspecies cell aggregation.
In order to examine whether the cell aggregation properties of Ag43 could work between cells of two different species, the settling profiles of mixed cultures were studied. This was done to investigate whether the aggregation was due to Ag43-Ag43 interaction or Ag43 interaction with another cell surface component. A mixed culture containing equal amounts of Ag43-expressing E. coli and P. fluorescens cells behaved exactly the same as monocultures of Ag43-expressing cells of either species, respectively, suggesting that Ag43-Ag43 interaction was responsible for interspecies cell aggregation (Fig. 5B). Furthermore, when Ag43-expressing cells were mixed with Ag43-negative cells, only half of the cells precipitated, as expected if interspecies autoaggregation was due to Ag43-Ag43 interaction (Fig. 5B).
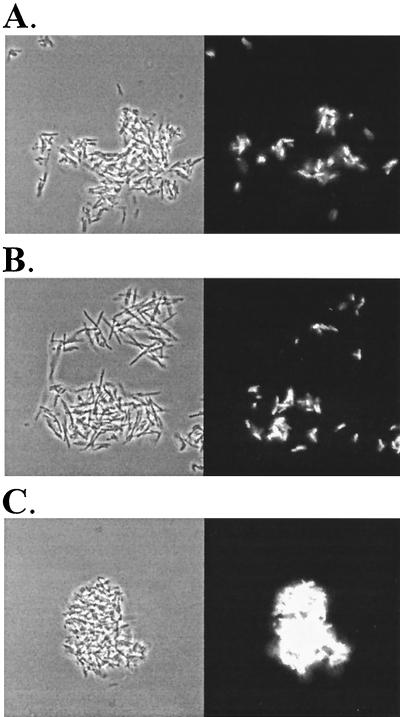
To expand on these findings, we introduced plasmid pJBA25, carrying the GFP gene (gfp) into E. coli cells containing the Ag43 expression plasmid pKKJ128. Plasmid pKKJ128 was constructed by transferring the flu gene from pKKJ101 into pACYC184 and used in order to ensure compatibility with plasmid pJBA25. No phenotypic difference was observed between cells harboring this plasmid and pKKJ101. By using a phase-contrast microscope equipped with a filter for detecting GFP, E. coli cells expressing both GFP and Ag43 could be detected visually. After mixing and subsequent settling of Ag43-expressing E. coli and P. fluorescens cells, aggregation between the two strains was observed (Fig. 6). This finding confirmed that interspecies cell aggregation is indeed due to Ag43-Ag43 interaction.
FIG. 6.
Phase-contrast microscopy showing aggregation of E. coli and P. fluorescens cells expressing Ag43 and/or GFP. E. coli HEHA16 (Ag43+, GFP+) mixed with (A) E. coli HEHA16 (Ag43+), (B) P. fluorescens SBW25 (Ag43+), or (C) E. coli HEHA16 (Ag43+, GFP+). Left panels, phase-contrast microscopy; right panels, visualization of cells expressing GFP.
Ag43 expression enhances biofilm formation.
P. fluorescens has previously been shown to form biofilms on abiotic surfaces (16). The autoaggregating phenotype of P. fluorescens cells expressing Ag43 prompted us to examine the effect of Ag43 expression with respect to biofilm formation on glass surfaces. Ag43+ P. fluorescens cells were grown statically for 48 h in glass tubes. Unbound cells were removed, the tubes were rinsed thoroughly with water, and bound cells were subsequently stained with 0.1% crystal violet. By using this procedure, cells attached to the glass surface could be visualized, as they were stained purple with crystal violet while the glass surface was not stained (data not shown). We found that Ag43 expression by P. fluorescens resulted in an enhanced ability to form biofilms on such surfaces.
Ag43 expression in other gram-negative organisms.
To examine the versatility of the Ag43 protein, we assessed its ability to be transported to and displayed on the surface of other gram-negative bacteria available from our strain collection. These were K. pneumoniae, Enterobacter cloacae, and Serratia liquefaciens. Transformation of K. pneumoniae with plasmid pKKJ101 resulted in a characteristic frizzy colony morphology type like the one seen in for E. coli and P. fluorescens (Fig. 2). Furthermore, virtually all cells from such colonies reacted strongly with specific Ag43 antiserum (Fig. 3). When overnight cultures of K. pneumoniae(pKKJ101) were inspected, only small precipitates were observed. In E. coli, we previously demonstrated that type 1 fimbriation physically blocks Ag43-mediated cell aggregation. Consequently, aliquots of K. pneumoniae (pKKJ101) cells were assayed for their ability to agglutinate yeast cells in a mannose-sensitive manner. A strong reaction indicative of type 1 fimbriation was observed. Like in E. coli, the expression of type 1 fimbriae in K. pneumoniae is phase variable. In line with this tenet, the fraction of the nonfimbriated cells in the population would be expected to undergo Ag43-mediated autoaggregation and settle. Cells from precipitates were carefully sampled and reincubated. After several rounds of enrichment, we obtained cultures in which most cells were seen to autoaggregate and settle (Fig. 4), suggesting that Ag43 is also fully functional in K. pneumoniae.
We also observed Ag43-induced colony morphology changes in S. liquefaciens (data not shown). While Ag43 could be detected on the surface of this strain (Fig. 3), it did not display the autoaggregation phenotype observed in E. coli and P. fluorescens. In contrast, E. cloacae did not exhibit any colony morphology changes after transformation with plasmid pKKJ101. The lack of a visible Ag43-induced change in colony morphotype in this organism could be due to several factors, including poor expression of the Ag43 protein, improper export to the surface, improper integration into the outer membrane, or interference or shielding by other surface structures. Therefore, to examine these possibilities, colonies were picked directly from plates and submitted to immunofluorescence microscopy with specific Ag43 antiserum. Both E. cloacae and S. liquefaciens reacted strongly, indicative of surface display of Ag43 (Fig. 3). Furthermore, when E. cloacae and S. liquefaciens cells containing either plasmid pKKJ101 or pUCP22 were assayed for mannose-sensitive yeast agglutination, they reacted strongly, indicative of type 1 fimbriation. This result suggests that type 1 fimbriae physically block Ag43-Ag43 interaction, as previously demonstrated in E. coli. Taken together, the data indicate that despite the E. coli K-12 origin of Ag43, this protein was indeed translocated to the surface of not only members of the Enterobacteriaceae, but also a more distantly related bacterium, viz. P. fluorescens.
DISCUSSION
The double-membrane system of gram-negative bacteria forms an efficient barrier which is instrumental in maintaining a stable milieu in the cytoplasm. Only a few low-molecular-weight substances are able to cross this barrier unassisted from the exterior. On the other hand, the gram-negative cell envelope also forms a formidable obstacle which proteins destined for the cell exterior have to cross. Gram-negative bacteria have developed various systems for protein transport to the cell exterior; four major secretion systems can be distinguished that are widely distributed in gram-negative bacteria (15). In contrast to the secretory systems that require a variety of auxiliary proteins, it seems that members of the autotransporter family contain all information required for export to the cell exterior within a single peptide chain. The N-terminal signal peptide directs translocation across the cytoplasmic membrane to the periplasm via the general secretory pathway. Subsequently, the β domain forms a β barrel structure in the outer membrane through which the α domain gains access to the cell exterior. Many autotransporters are processed, potentially by autocatalytic cleavage, between the α and β domains (12). Autotransporter proteins are functionally very diverse, some are adhesins, e.g., AIDA-I from certain clinical isolates (1), while others are proteases, e.g., the immunoglobulin A (IgA)-specific protease of Neisseriaceae (14). All available evidence suggests that Ag43 is a self-recognizing adhesin which is processed by autocatalytic action and thus exhibits the functional hallmarks of the autotransporter family. Ag43 shows 34% identity to the AIDA-I autotransporter (1). However, it exhibits only marginal homology to autotransporters from other bacteria. Furthermore, even though the overall architecture of the cell envelope is similar in gram-negative bacteria, large variations with regard to composition are found among different genera and strains. These aspects and the characteristic cell-aggregating phenotype of Ag43, not observed with other autotransporters, could indicate that Ag43 might be functionally restricted to E. coli. In order to test this tenet, the environmentally important, well-characterized P. fluorescens strain SBW25 was chosen to monitor transspecies expression and functionality of Ag43. The results of this study strongly suggest that Ag43 is indeed fully functional in this strain.
Previous experiments by us strongly indicated that Ag43-Ag43 interaction is responsible for the cell aggregation observed in Ag43-expressing E. coli (8). This prompted us to see whether Ag43 could be employed as a tool to induce interspecies cell aggregation of bacteria. Indeed, when GFP-labelled E. coli expressing Ag43 was mixed with Ag43-expressing P. fluorescens, interspecies cell aggregation was readily evident (Fig. 6). The use of Ag43 to confer interspecies cell aggregation represents a novel approach to engineer aggregation between different bacteria. This concept might have a number of applications, such as in rational design of selected consortia of bacteria with uses in waste treatment and mineralization of pollutants.
The apparent ease of transferring Ag43 from E. coli to P. fluorescens prompted us to investigate Ag43 expression in other gram-negative bacteria. Interestingly, in all strains tested, Ag43 was observed to be located on the surface, evidenced by immunofluorescence microscopy (Fig. 3). However, of these strains only K. pneumoniae and S. liquefaciens colonies exhibited the characteristic frizzy colony form upon transformation with plasmid pKKJ101 (Fig. 2). Also, the characteristic cell aggregation phenomenon was not observed in the tested bacteria and only weak aggregation and settling were observed in K. pneumoniae. This apparent lack of activity could be due to incorrect folding of Ag43; however, in a previous study we showed that fimbriation abolished Ag43-mediated autoaggregation in E. coli (8). In fact, the E. cloacae, S. liquefaciens, and K. pneumoniae strains tested readily agglutinated yeast cells in a mannose-sensitive manner, indicative of type 1 fimbriation. In the case of K. pneumoniae, a non- or scarcely fimbriated fraction of the population could be selected by subculturing of small primary precipitates, resulting in a subpopulation that turned out to contain copious amounts of aggregating cells (Fig. 4). Ag43-expressing K. pneumoniae was observed to form chains. We have no clear understanding of the background for this phenomenon. Ag43 seems to be evenly distributed on the bacterial surface (Fig. 3); however, one might invoke capsule-Ag43 interference and uneven capsule distribution as a tentative explanation.
E. coli is able to adjust to surface or bottom dwelling in static liquid environments through the production of different surface structures, viz., type 1 fimbriae and Ag43, respectively. These phenotypes correlate with distinct colony morphology types (9). A similar correlation between niche specificity in static liquid environments and colony morphotypes was also demonstrated for P. fluorescens, although the specific gene or gene products were not identified (20). The heterologous expression of Ag43 in P. fluorescens created a bottom-dwelling phenotype with enhanced ability to form biofilms on an inert surface (glass). Thus, it is possible that the colony morphotypes exhibited by P. fluorescens might result from the expression of similar types of proteins. This would suggest that one way bacteria can adapt to different environmental niches is by coordinated expression of specific surface features such as Ag43 that favors proliferation in a specific environment. Understanding such processes may have important implications for the prevention of the establishment of bacterial surface communities in environments where such are unwanted (e.g., biofouling) or, on the other hand, allow rational design of bacterial consortia where such are desired, for example, in the degradation of pollutants.
ACKNOWLEDGMENTS
This work was supported in part by the BIOPRO Center, part of the Danish National Strategic Environmental Research Program, and the Danish Medical Research Council (grant no. 9802358). Plasmid pJBA25 was kindly provided by Jens Bo Andersen, Technical University of Denmark.
REFERENCES
- 1.Benz I, Schmidt M A. Isolation and serologic characterization of AIDA-I, the adhesin mediating the diffuse adherence phenotype of the diarrhea-associated Escherichia coli strain 2787 (O126:H27) Infect Immun. 1992;60:13–18. doi: 10.1128/iai.60.1.13-18.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brazil G M, Kenefick L, Callanan M, Haro A, de Lorenzo V, Dowling D N, O'Gara F. Construction of a rhizosphere pseudomonad with potential to degrade polychlorinated biphenyls and detection of bhd gene expression in the rhizosphere. Appl Environ Microbiol. 1995;61:1946–1952. doi: 10.1128/aem.61.5.1946-1952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caffrey P, Owen P. Purification and N-terminal sequence of the α subunit of antigen 43, a unique protein complex associated with the outer membrane of Escherichia coli. J Bacteriol. 1989;171:3634–3640. doi: 10.1128/jb.171.7.3634-3640.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diderichsen B. flu, a metastable gene controlling surface properties of Escherichia coli. J Bacteriol. 1980;141:858–867. doi: 10.1128/jb.141.2.858-867.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Genevaux P, Muller S, Bauda P. A rapid screening procedure to identify mini-Tn10 insertion mutants of Escherichia coli K-12 with altered adhesion properties. FEMS Microbiol Lett. 1996;142:27–30. doi: 10.1111/j.1574-6968.1996.tb08402.x. [DOI] [PubMed] [Google Scholar]
- 6.Givskov M, Olsen L, Molin S. Cloning and expression in Escherichia coli of the gene for an extracellular phospholipase from Serratia liquefaciens. J Bacteriol. 1988;170:5855–5862. doi: 10.1128/jb.170.12.5855-5862.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haagmans W, van der Woude M. Phase variation of Ag43 in Escherichia coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression and transcription. Mol Microbiol. 2000;35:877–887. doi: 10.1046/j.1365-2958.2000.01762.x. [DOI] [PubMed] [Google Scholar]
- 8.Hasman H, Chakraborty T, Klemm P. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J Bacteriol. 1999;181:4834–4841. doi: 10.1128/jb.181.16.4834-4841.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasman H, Schembri M A, Klemm P. Antigen 43 and type 1 fimbriae determine colony morphology of Escherichia coli K-12. J Bacteriol. 2000;182:1089–1095. doi: 10.1128/jb.182.4.1089-1095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson I R, Meehan M, Owen P. A novel regulatory mechanism for a novel phase-variable outer membrane protein of Escherichia coli. Adv Exp Med Biol. 1997;412:349–355. doi: 10.1007/978-1-4899-1828-4_56. [DOI] [PubMed] [Google Scholar]
- 11.Henderson I R, Meehan M, Owen P. Antigen 43, a phase-variable bipartite outer membrane protein, determines colony morphology and autoaggregation in Escherichia coli K-12. FEMS Microbiol Lett. 1997;149:115–120. doi: 10.1111/j.1574-6968.1997.tb10317.x. [DOI] [PubMed] [Google Scholar]
- 12.Henderson I R, Navarro-Garcia F, Nataro J P. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 1998;6:370–378. doi: 10.1016/s0966-842x(98)01318-3. [DOI] [PubMed] [Google Scholar]
- 13.Henderson I R, Owen P. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and OxyR. J Bacteriol. 1999;181:2132–2141. doi: 10.1128/jb.181.7.2132-2141.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lomholt H, Poulsen K, Kilian M. Comparative characterization of the iga gene encoding IgA1 protease in Neisseria meningitidis, Neisseria gonorrhoeae and Haemophilus influenzae. Mol Microbiol. 1995;15:495–506. doi: 10.1111/j.1365-2958.1995.tb02263.x. [DOI] [PubMed] [Google Scholar]
- 15.Lory S. Secretion of proteins and assembly of bacterial surface organelles: shared pathways of extracellular protein targeting. Curr Opin Microbiol. 1998;1:27–35. doi: 10.1016/s1369-5274(98)80139-2. [DOI] [PubMed] [Google Scholar]
- 16.O'Toole G A, Kolter R. The initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 17.Owen P, Caffrey P, Josefsson L G. Identification and partial characterization of a novel bipartite protein antigen associated with the outer membrane of Escherichia coli. J Bacteriol. 1987;169:3770–3777. doi: 10.1128/jb.169.8.3770-3777.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Owen P, Meehan M, de Loughry-Doherty H, Henderson I. Phase-variable outer membrane proteins in Escherichia coli. FEMS Immunol Med Microbiol. 1996;16:63–76. doi: 10.1111/j.1574-695X.1996.tb00124.x. [DOI] [PubMed] [Google Scholar]
- 19.Rainey P B, Bailey M J. Physical and genetic map of the Pseudomonas fluorescens SBW25 chromosome. Mol Microbiol. 1996;19:521–533. doi: 10.1046/j.1365-2958.1996.391926.x. [DOI] [PubMed] [Google Scholar]
- 20.Rainey P B, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 21.Ramos J L, Diaz E, Dowling D, de Lorenzo V, Molin S, O'Gara F, Ramos C, Timmis K N. The behavior of bacteria designed for biodegradation. Bio/Technology. 1994;12:1349–1356. doi: 10.1038/nbt1294-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schembri M A, Pallesen L, Connell H, Hasty D L, Klemm P. Linker insertion analysis of the FimH adhesin of type 1 fimbriae in an Escherichia coli fimH-null background. FEMS Microbiol Lett. 1996;137:257–263. doi: 10.1111/j.1574-6968.1996.tb08115.x. [DOI] [PubMed] [Google Scholar]
- 23.Stentebjerg-Olesen B, Pallesen L, Jensen L B, Christiansen G, Klemm P. Authentic display of a cholera toxin epitope by chimeric type 1 fimbriae: effects of insert position and host background. Microbiology. 1997;143:2027–2038. doi: 10.1099/00221287-143-6-2027. [DOI] [PubMed] [Google Scholar]
- 24.Warne S R, Varley J M, Boulnois G J, Norton M G. Identification and characterization of a gene that controls colony morphology and auto-aggregation in Escherichia coli K12. J Gen Microbiol. 1990;136:455–462. doi: 10.1099/00221287-136-3-455. [DOI] [PubMed] [Google Scholar]
- 25.West S E, Schweizer H P, Dall C, Sample A K, Runyen-Janecky L J. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]