Abstract
Xanthomonas oryzae pv. oryzae causes bacterial leaf blight, a serious disease of rice. Spontaneous mutants which are deficient for virulence and extracellular polysaccharide (Eps) production accumulate in large numbers in stationary-phase cultures of this bacterium, a phenomenon which we have called stationary-phase variation. A clone (pSD1) carrying the Eps biosynthetic gene (gum) cluster of X. oryzae pv. oryzae restored Eps production and virulence to several spv (for stationary-phase variation) mutants. Data from localized recombination analysis, Southern hybridization, PCR amplification, and sequence analysis showed that the mutations are due to insertion of either one of two novel endogenous insertion sequence (IS) elements, namely, ISXo1 and ISXo2, into gumM, the last gene of the gum gene cluster. The results of Southern analysis indicate the presence of multiple copies of both IS elements in the genome of X. oryzae pv. oryzae. These results demonstrate the role of IS elements in stationary-phase variation in X. oryzae pv. oryzae.
In their natural environment, bacteria often encounter nutritionally limited conditions which resemble the stationary-phase conditions of laboratory cultures. Gram-positive bacteria respond to the adverse conditions by forming resistant spores. Gram-negative bacteria exhibit reduced growth, and several genes that confer resistance to stress during starvation are transcriptionally induced in the stationary phase. Much of the transcriptional regulation in the stationary phase is mediated by rpoS, a gene that encodes the stationary-phase sigma factor (ςS) of RNA polymerase (14, 21). Several Escherichia coli mutants that have a competitive advantage in stationary-phase cultures have been shown to carry mutations in the rpoS gene (38).
Spontaneous and reversible phenotypic variations mediated by various DNA rearrangements, such as insertions, DNA duplications, inversions, deletions, and frameshift mutations, are strategies adopted by bacteria to cope with adverse environmental conditions (7). A few examples are intragenic duplications in a regulatory gene in the mushroom pathogen Pseudomonas tolaasii (11), large chromosomal duplications that promote growth during carbon starvation in Salmonella enterica serovar Typhimurium (34), insertions in Shigella flexneri (24), frameshift mutations in Bordetella pertussis (35), and flagellar-phase variation mediated by DNA inversion in serovar Typhimurium (39). In the well-studied plant-pathogenic bacterium Ralstonia solanacearum, phenotypic changes were shown to be due to either mutations in the phcA regulatory gene (2), overproduction of a negative regulatory element (16), or excision of an episome from the bacterial chromosome (26).
Xanthomonas oryzae pv. oryzae causes bacterial leaf blight, a serious disease of rice. Like other xanthomonads, this bacterium produces copious amounts of an extracellular polysaccharide (Eps). The Eps produced by a related bacterium, Xanthomonas campestris pv. campestris, is the industrially important xanthan gum, which is comprised of a repeating pentamer unit made up of two subunits of glucose, two of mannose, and one of glucuronic acid, along with certain modifications like acetylation (4). A cluster of X. campestris pv. campestris genes (gumA through gumM) spanning 16 kb has been shown to encode enzymes involved in Eps biosynthesis (3, 17). A cosmid clone carrying the X. oryzae pv. oryzae homologue of the X. campestris pv. campestris gum cluster was isolated using transposon tagging methods (5).
The spontaneous loss of virulence associated with reduced levels of Eps production has been reported for long-term stabs and aging liquid cultures of X. oryzae pv. oryzae (9, 30). These mutants were shown to accumulate in large numbers in stationary-phase cultures but not in exponentially growing cultures (29). There was at least a 5,000-fold increase in the frequency of the mutants 10 days after day 1 in the stationary phase, with a large number of the cells comprised of these mutants. This phenomenon was referred to as stationary-phase variation. Independently isolated spv (for stationary-phase variation) mutants were reported to show phenotypic variation in the degree of loss of virulence, stability of the mutant phenotype, ability to outcompete the wild-type strain in stationary-phase cocultures, and sensitivity to hydrogen peroxide (29).
We are interested in determining the nature of mutations in the various spv strains and to understand why the mutants accumulate in stationary-phase cultures of X. oryzae pv. oryzae. In this study, we report that in some of the spv strains, the mutant phenotype is due to insertion of either one of two novel endogenous insertion sequence (IS) elements into the gum cluster of X. oryzae pv. oryzae.
MATERIALS AND METHODS
Bacterial strains, media, and antibiotics.
The bacterial strains and plasmids used in this study are listed in Table 1. The wild-type strain of X. oryzae pv. oryzae (BXO1) was obtained from the Directorate of Rice Research, Hyderabad, India, and the spontaneous rifampin-resistant strain BXO112 was derived from it in our laboratory. Several independent Eps- and virulence-deficient mutants (spv mutants) were isolated from stationary-phase cultures of the BXO112 strain (29). The Rifr marker was useful for counterselection of E. coli donors in conjugal matings. All strains of X. oryzae pv. oryzae were grown at 28°C for 48 h in peptone-sucrose (PS) medium or PS agar (PSA) when maintained on plates (36). E. coli strains were grown in Luria broth medium at 37°C (23). The antibiotics kanamycin and rifampin were added wherever required (50 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| X. oryzae pv. oryzae | ||
| BXO1 | Natural isolate (Chinsuria, West Bengal, India) | Lab collection |
| BXO112 | rif-1; Rifr derived from BXO1 | 29 |
| BXO113 | spv-2 rif-1 | 29 |
| BXO114 | spv-3 rif-1 | 29 |
| BXO115 | spv-4 rif-1 | 29 |
| BXO117 | spv-5 rif-1 | 29 |
| BXO119 | spv-6 rif-1 | 29 |
| BXO121 | spv-7 rif-1 | 29 |
| BXO123 | spv-8 rif-1 | 29 |
| BXO125 | spv-9 rif-1 | 29 |
| BXO127 | spv-10 rif-1 | 29 |
| BXO148 | rif-1; Eps+ Vir+ recombinant of BXO125 | This study |
| BXO149 | rif-1; Eps+ Vir+ recombinant of BXO125 | This study |
| BXO150 | rif-1; Eps+ Vir+ recombinant of BXO125 | This study |
| BXO151 | BXO125/pSD2 | This study |
| BXO152 | BXO125/pSD1 | This study |
| E. coli | ||
| DH5α | F′ end A1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA relA1 φ80dlacZ ΔM15 Δ(lacZYA-argF)U169 | Lab collection |
| S17-1 | RP4-2-TC::Mu-Km::Tn7 pro hsdR recA | 33 |
| Plasmids | ||
| pSD1 | pUFR034 + 36 kb of BXO1 DNA containing the gum gene cluster | 5 |
| pSD2 | pUFR034 + 14.2 kb of BXO1 DNA containing part of the gum gene cluster | 5 |
| pRR7 | pBKS + the 4.5-kb BamHI fragment from pSD2 | This study |
Estimation of Eps.
Eps was isolated from colonies grown on PSA for 8 to 10 days at 28°C by precipitation with acetone, as described by Hancock and Poxton (12). It was quantitated by colorimetric estimation of pentoses and hexoses (6) using d-glucose as the standard; the amount of Eps was calculated in milligrams per 1010 cells of X. oryzae pv. oryzae.
Virulence assays.
Cultures of X. oryzae pv. oryzae that were grown to saturation, pelleted, and resuspended in sterile water to about 109 cells/ml were used as the inoculum. Forty-day-old greenhouse-grown plants of the susceptible rice cultivar Taichung Native-1 (TN-1) were inoculated by the leaf clip method of inoculation (18). Symptoms were measured as lesion lengths at regular intervals.
Conjugation and complementation of spv mutants.
The cosmid clones containing the gum genes of X. oryzae pv. oryzae were transferred from E. coli to BXO112 using biparental mating procedures (5, 15). Cultures of BXO112 were concentrated to 1012 cells/ml, and 200 μl was spotted onto Hybond N+ membranes placed on nutrient agar plates and then mixed with 10 μl of the donor E. coli strain, S17-1 (109 cells/ml), carrying either pSD1 or pSD2. The plates were incubated for 48 h at 28°C before the cells were washed into sterile distilled water and plated on selection medium containing PSA with rifampin and kanamycin.
Isolation of genomic DNA and Southern blot analysis.
Genomic DNA was isolated according to the protocol of Leach et al. (20). Restriction digestions were done using enzymes obtained from New England Biolabs (Beverly, Mass.), as per the manufacturer's instructions. The digested DNA was separated on agarose gels, denatured, neutralized, and vacuum transferred to a Hybond N+ membrane as described by Sambrook et al. (32). Probes were labeled with [α-32P]dATP using a random prime labeling kit (BRIT, Mumbai, India). Prehybridization, hybridization, and autoradiography were performed as described by Yashitola et al. (37).
PCR assays.
Six forward (F1 to F6) and six reverse (R1 to R6) primers with sequences specific to gum genes were used in various combinations to PCR amplify regions from genomic DNA of the wild-type and spv strains of X. oryzae pv. oryzae. The sequences of the primers can be obtained from us on request. Genomic DNA (100 ng) was used as the template in 25-μl reaction volumes containing 1× PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2, and 0.01% gelatin) from Perkin-Elmer (Foster City, Calif.); dATP, dGTP, dTTP, and dCTP (all 0.2 mM; Pharmacia Biotech, Uppsala, Sweden); 10 pmol of each primer; and 2 U of Taq polymerase. PCRs were carried out in a PTC-200 Peltier thermal cycler (MJ Research Inc., Watertown, Mass.), with denaturation at 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 5 min, and a final extension at 72°C for 5 min. PCR-amplified products were analyzed in 0.7% agarose gels stained with ethidium bromide (10 mg/ml) and visualized under UV.
Sequence analysis and homology search.
The PCR-amplified products were purified from agarose gels using the QIAEX kit (Qiagen Inc., Chatsworth, Calif.) according to the supplier's instructions. The purified DNA samples were sequenced using the same set of primers as were used for their amplification. The 4.5-kb BamHI DNA fragment cloned into pBluescript (pRR7) was sequenced using vector-specific primers. The internal sequences of the IS elements and the pRR7 clone were obtained by primer walking. Sequencing reactions, polyacrylamide gel electrophoresis, and sequence output processing were performed on an automated sequencing unit (ABI Prism 377; Perkin-Elmer) according to the manufacturer's instructions. Computer-based sequence homology searches were performed using the BLAST algorithm (1), available on the World Wide Web.
Isolation of spv mutants.
A culture of BXO112 was grown by inoculating a single colony in 20 ml of PS medium for 48 h at 28°C. The culture was then placed on top of the laboratory bench, and bacterial cells were plated at appropriate dilutions at various time intervals to yield single colonies on PSA plates. The mutants were scored visually for altered colony morphology (i.e., Eps deficiency).
Nucleotide sequence accession numbers.
The nucleotide sequences of the gumI and gumJ genes and the gumK, gumL, and gumM genes have been deposited in GenBank under accession no. AF231923 and AF231924, respectively. The nucleotide sequences of ISXo1 and ISXo2 have been deposited under accession no. AF225214 and AF225215, respectively.
RESULTS
The phenotype of several spv strains is due to mutations in the gum locus.
In a previous study, inactivation of gum genes by transposon insertion was shown to affect Eps production and virulence in X. oryzae pv. oryzae (5) and the transposon tag was used to isolate cosmid clones carrying the X. oryzae pv. oryzae homologue of the X. campestris pv. campestris gum cluster. One clone, pSD1, carried a 36-kb insert containing six EcoRI fragments of 1.2, 5.2, 5.5, 6.5, 7.8, and 10 kb, whereas a second clone, pSD2, had three fragments of 1.2, 5.2, and 7.8 kb. Only the pSD1 clone complemented the Eps and virulence deficiency of a gumGXo::Tn5 mutant. To determine whether the functional gum gene cluster could restore a similar phenotype to the spv mutants, these two clones were transferred in biparental matings to nine independently isolated spv mutants. The Eps-deficient phenotype of four spv mutants (BXO119, -121, -123, and -125) was complemented by pSD1 as determined by the appearance of mucoid colonies and quantification of Eps. The wild-type strain produced ∼110 mg of glucose/1010 cells, whereas the mutants produced only ∼10 mg for the same number of cells. The pSD1 clone restored wild-type levels of Eps production to these four spv mutants. The mucoid phenotype depended on the continued presence of the plasmid, since the exconjugants grown in the absence of the drug marker, viz., kanamycin, showed Eps− phenotypes and kanamycin sensitivity due to the loss of the transforming plasmid. Introduction of the pSD2 clone did not restore Eps production to the four mutants, as judged by colony morphology and further confirmed by quantification of Eps production for two of the strains (data not shown). Neither pSD1 nor pSD2 restored Eps production to the remaining five spv mutants (BXO113, -114, -115, -117, and -127), as judged by colony morphology.
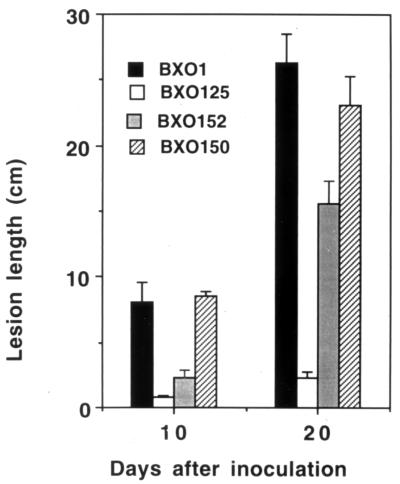
Pathogenicity tests on rice cultivar TN-1 revealed that the spv mutants which were restored for mucoidy by complementation with pSD1 also regained virulence, though the lesion lengths were not as extensive as those caused by the wild-type strain (Fig. 1). Although only the data for BXO125 are presented, similar results were obtained with BXO119, -121, and -123. While the lesions caused by the wild-type strain covered the entire length of the leaf (∼30 cm) within 20 days of inoculation and the mutant strain was restricted to 4 cm, the exconjugants showed lesion lengths of ∼15 cm (about 40 to 50% of that caused by the wild type), possibly due to the progressive loss of recombinant pSD1. Introduction of the pSD2 clone did not restore virulence to these spv mutants (data not shown). The virulence-deficient phenotype of the remaining five spv mutants (BXO113, -114, -115, -117, and -127) was not complemented by either the pSD1 or pSD2 clone (data not shown).
FIG. 1.
Virulence of X. oryzae pv. oryzae strains on rice. Inoculations were performed on 40 one-day-old plants of rice cultivar TN-1 as described in Materials and Methods. The data are the averages of measurements from 10 leaves that had been inoculated with particular strains: BXO1 (wild type), BXO125 (spv mutant), BXO152 (BXO125/pSD1), and BXO150 (an Eps+ Vir+ recombinant obtained in a BXO125/pSD2 background). Error bars indicate standard deviations. Similar results were obtained in independent experiments.
However, it was observed that approximately 3 out of every 10 leaves inoculated with transconjugants of BXO119, -121, -123, and -125 carrying pSD2 showed extended lesions. Bacteria isolated from these leaves were mucoid and kanamycin sensitive and appeared to be recombinants that had regained wild-type characteristics. In an attempt to isolate the putative recombinants from laboratory cultures, the exconjugants were also dilution plated in the laboratory on PSA. Mucoid revertants were obtained at a frequency of 1 in 1,000 for spv mutants BXO119 and -125 carrying pSD2 as the noncomplementing clone. These mucoid colonies were patched simultaneously on plates containing PSA and PSA with kanamycin. The majority of the colonies were found to be kanamycin sensitive, indicating the loss of the transforming plasmid. No such revertants were obtained when the spv strains alone or containing only the vector were used to inoculate rice plants (none in 30 inoculated leaves) or plated on PSA (none out of ∼10,000 colonies), indicating a reversion frequency of less than 1 in 104. These observations indicate that the wild-type revertants were obtained by homologous recombination of DNA between the plasmid and the bacterial chromosome, accompanied by the subsequent loss of plasmid in the kanamycin-sensitive colonies. Pathogenicity tests revealed that virulence was restored to these recombinants, as the lesion lengths on rice cultivar TN-1 were comparable to those caused by the wild-type strain (represented by BXO150 in Fig. 1). The complementation and localized recombination data suggest that mutations in the gum locus are the cause of the mutant phenotype in four out of nine spv strains.
RFLP indicates a DNA rearrangement at the gum locus.
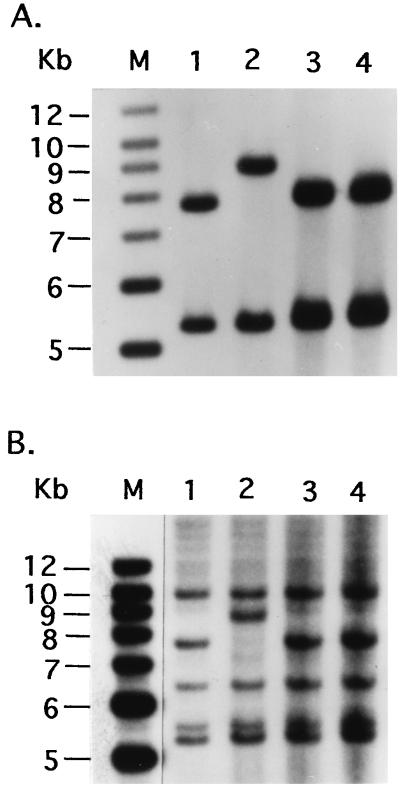
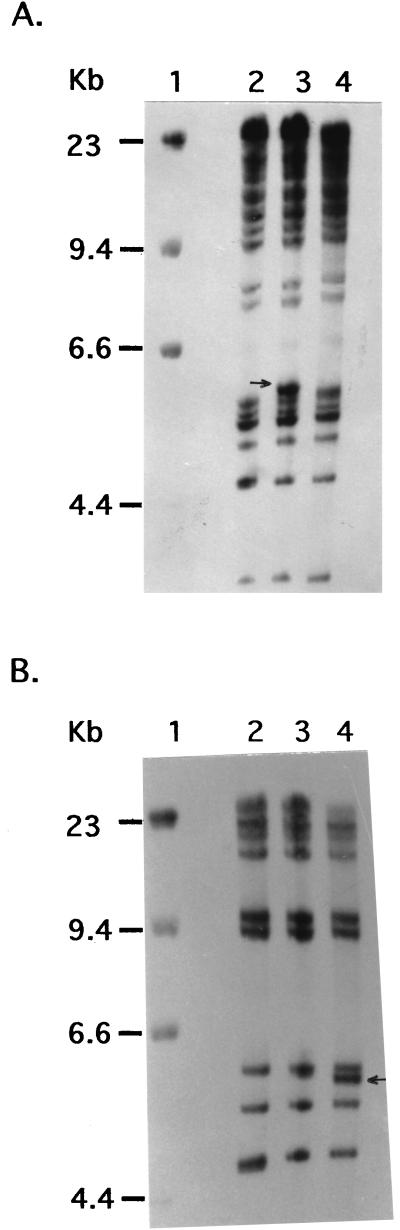
In order to determine the nature of mutations in the gum region of the spv strains which are complemented by the pSD1 clone, the total genomic DNA from the wild-type and mutant strains was each digested with EcoRI and analyzed by Southern hybridization with pSD2 (Fig. 2A) and pSD1 (Fig. 2B) clones. The results revealed the presence of a restriction fragment length polymorphism (RFLP) in the four spv strains (BXO119, -121, -123, and -125) in comparison to the wild-type strain. The spv strains were characterized by a 9-kb EcoRI fragment instead of the 7.8-kb band observed in the wild-type strain, while the other bands hybridizing to the two clones were unaltered in all the strains (representative data for BXO125 are shown in Fig. 2). Southern analysis of three independently isolated recombinants from each of the exconjugants between BXO123/pSD2 and BXO125/pSD2 showed that the altered 9-kb fragment characteristic of the mutant strains was restored to the original 7.8-kb fragment in all the recombinant strains that were examined (data not shown). The data therefore indicate that a DNA rearrangement of the gum cluster occurred in several spv mutants of X. oryzae pv. oryzae. This DNA rearrangement was not observed in two spv strains (BXO113 and -114) that were not complemented by pSD1 (Fig. 2).
FIG. 2.
DNA rearrangement at the gum locus in several spv strains. Southern analyses of EcoRI-digested genomic DNA were performed using α-32P-labeled probes as described in Materials and Methods. pSD2 (A) and pSD1 (B) were used as probes. Lanes: M, ladder marker DNA from Stratagene; 1, BXO1 (wild type); 2, BXO125 (spv mutant); 3 and 4, BXO113 and BXO114 (both are spv mutants which cannot be complemented by pSD1). The 7.8-kb fragment is present as a 9-kb fragment in BXO125.
PCR amplification of mutant alleles from spv strains.
Southern analysis of BamHI-digested genomic DNA from the wild type and the four spv strains (BXO119, -121, -123, and -125) probed with pSD2 revealed a 4.5-kb fragment in the wild-type strain which was altered by 1.2 kb to yield a 5.7-kb fragment in the four mutants (data not shown). The 4.5-kb BamHI fragment was subcloned from pSD2 into pBluescript (pRR7), and sequence analysis using vector-specific primers indicated homology to the gumI gene of X. campestris pv. campestris at one end and to the gumM gene at the other end (Fig. 3A). The sequence of most of the pRR7 clone was then obtained by primer walking. A sequence of 1,637 bp from the gumI end showed homology to the gumI and gumJ genes of X. campestris pv. campestris (85 and 82% identity to gumI and gumJ, respectively). A 2,181-bp sequence from the gumM end showed homology to the gumK, gumL, and gumM genes from X. campestris pv. campestris (∼83% identity at the nucleotide level). A contiguous stretch of sequence spanning an ∼700-bp interval between the 3′ end of gumJ and the start of the gumK gene of X. oryzae pv. oryzae remains to be sequenced. The gene order in the gum locus is gumA through gumM in X. campestris pv. campestris (3) and appears to be so also in X. oryzae pv. oryzae (at least from gumG through gumM based on earlier [5] and our present work).
FIG. 3.
(A) The gumI-to-gumM region of the gum locus, included in the pRR7 clone. The locations where PCR primers were designed are indicated. F1 to F6 are forward primers, whereas R1 to R6 are reverse primers. (B) The gumM region of the gum locus. Sites of insertions of the IS elements are shown. ▿, sites of ISXo1 insertion in BXO121, -123, and -125. ISXo1 is inserted in the same site in BXO121 and -125. ▾, site of ISXo2 insertion in BXO119. Note that the putative transposase genes of ISXo1 and ISXo2 insertions are of a transcriptional orientation (←) opposite to that of the gumM gene (→). (C) Schematic representation of the organization of ISXo1 and ISXo2 elements. ▧, the ORF encoding putative transposase; ░⃞, transposon-specific flanking sequences; ■, terminal inverted repeats. ATG is the start codon; TAA and TGA are the stop codons.
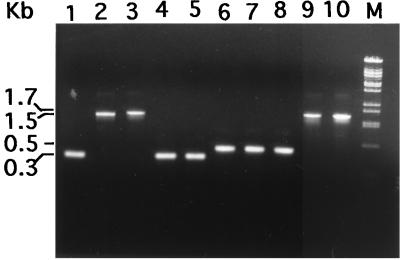
For use in PCRs, six pairs of forward as well as reverse primers were then designed, the locations of which are shown in Fig. 3A. PCRs were then performed to identify the region which shows polymorphism in the spv strains. One set of primers (F6 and R6) amplified a 300-bp fragment in BXO112, -121, and -125 and a 1.5-kb fragment in two of the spv strains, BXO119 and -123 (Fig. 4, lanes 1 through 5). Another set of primers (F5 and R5) amplified a 500-bp fragment in BXO112, -119, and -123 compared to a 1.7-kb fragment in the other two spv strains, BXO121 and -125 (Fig. 4, lanes 6 through 10). In both cases the altered fragment showed an increase of ∼1.2 kb over the normal allele, suggesting a DNA rearrangement in that region.
FIG. 4.
PCR amplification of mutant alleles from spv strains. Agarose gel electrophoresis of PCR products was performed by using the F6-R6 (lanes 1 to 5) and F5-R5 (lanes 6 to 10) sets of gum-specific primers with genomic DNA from X. oryzae pv. oryzae strains. Lanes: 1 and 6, BXO112; 2 and 7, BXO119; 3 and 8, BXO123; 4 and 9, BXO121; 5 and 10, BXO125. The molecular size marker (lane M) is a BstEII digest of lambda DNA.
The DNA rearrangement is due to IS element insertion into the gumM gene of X. oryzae pv. oryzae.
The 300-bp fragment amplified in the wild-type strain (BXO112) by the F6-R6 primer combination showed homology to the gumM gene, whereas the 500-bp fragment amplified by the F5 and R5 primers showed homology to the gumL and gumM genes of X. campestris pv. campestris. The fragments amplified from the spv strains were sequenced with the aid of the primers used for PCR as well as with internal primers. Two distinct and novel IS elements, designated ISXo1 and ISXo2, were found to be inserted in different regions of the gumM gene (Fig. 3B) in the four spv strains. ISXo1 was inserted at an identical site in gumM in the independently isolated mutants BXO121 and BXO125. The BXO123 strain had an ISXo1 insertion at another location in the gumM gene. The BXO119 strain had an insertion of ISXo2 at a third site within the gumM gene. The ISXo1 sequence was 1,156 bp long, with a 16-bp inverted repeat (Fig. 3C), and caused a 4-bp target site duplication. There was one open reading frame (ORF) with the potential to encode a 322-amino-acid-long polypeptide, which showed over 88% identity with the gene for the putative transposase of IS1646, an IS element from X. campestris pv. vesicatoria (19). The ∼150-bp flanking region on either side of the ORF, however, did not show homology to any known sequence and was unique to ISXo1. The ISXo2 element was 1,070 bp long, with a 24-bp terminal inverted repeat (Fig. 3C), and caused an 8-bp target site duplication. The element has the potential to encode a polypeptide of 320 amino acids which exhibits ∼46% similarity with the putative transposase of IH4, an IS element from Halobacterium halobium (28). The termination codon of the ISXo2 ORF extended into the right inverted terminal repeat. The putative transposase genes of ISXo1 and ISXo2 insertions in gumM were of a transcriptional orientation opposite to that of the gumM gene (Fig. 3B).
ISXo1 and ISXo2 are present in multiple copies within the genome of X. oryzae pv. oryzae.
Genomic DNA from each of the strains BXO112, -119, -121, -123, and -125 that had been digested with BamHI was probed after Southern blotting, with the PCR-amplified product containing either ISXo1 or ISXo2. Sequence analysis had indicated that neither of these elements is cleaved by BamHI. Southern analysis showed the presence of multiple copies of each of the elements in the genome of X. oryzae pv. oryzae (Fig. 5). The number of copies of ISXo1 was >20, whereas the number of copies of ISXo2 was 9 in BXO112. As expected, an additional 5.7-kb fragment hybridized to ISXo1 in BXO121, -123, and -125 (representative data shown for BXO125 in Fig. 5A) and to ISXo2 in BXO119 (Fig. 5B).
FIG. 5.
Distribution of ISXo1 and ISXo2 in the X. oryzae pv. oryzae genome. Southern analyses of BamHI-digested genomic DNA were performed using α-32P-labeled probes ISXo1 (A) and ISXo2 (B). Lanes: 1, HindIII-digested lambda DNA marker; 2, BXO1; 3, BXO125; 4, BXO119. An additional 5.7-kb band (indicated by arrows) was detected in BXO125 with the ISXo1 probe and in BXO119 with the ISXo2 probe.
spv mutants from a single stationary-phase culture of X. oryzae pv. oryzae are genetically heterogeneous.
The data presented so far indicate that independently isolated spv mutants are heterogeneous in nature. To determine whether spv mutants from a single stationary-phase culture show similar variation, several mutants were isolated from a single colony of BXO112 as described in Materials and Methods. As observed earlier (29), the frequency of mutants in the culture increased from undetectable levels (<1 in 105 cells) after 1 day to ∼5 × 108 cells after 10 days in the stationary phase. The pSD1 clone was transferred in biparental matings from E. coli strain S17-1 to 13 such spv mutants, and only 5 of them were complemented by pSD1 for Eps production and virulence (data not shown). This suggests that a single stationary-phase culture of X. oryzae pv. oryzae can have genetically heterogeneous spv mutants.
DISCUSSION
In prolonged stationary-phase cultures of X. oryzae pv. oryzae grown in the laboratory, spontaneous Eps- and virulence-deficient mutants (referred to as spv strains) accumulated in large numbers (29). This study shows that in several of these spv strains, the mutant phenotype is due to transposition of either one of two endogenous IS elements into gumM, the last gene of the Eps biosynthetic gene cluster. Four different IS elements (IS1112, IS1113, TNX6, and TNX7) have been previously reported for X. oryzae pv. oryzae (20, 27). All four elements are present in multiple copies within the X. oryzae pv. oryzae genome. However, when used as probes, they give DNA fingerprinting patterns in the BXO1 strain (37; J. Yashitola and R. V. Sonti, unpublished data) distinctly different from those of either ISXo1 or ISXo2, indicating that the latter two elements are different from the previously described IS elements. The sequences of IS1112 and IS1113 are also different from those of ISXo1 and ISXo2 (M. Ryba-White and J. E. Leach, unpublished results). TNX6 and TNX7 have not yet been sequenced.
IS elements have been shown to be ubiquitously distributed within bacterial genomes (22). The data presented in this paper demonstrate that mutations with clearly defined phenotypes caused by IS element transposition can accumulate in large numbers in stationary-phase cultures. IS element transposition has been demonstrated within bacterial cells recovered from stabs that have been stored for several decades (25). These elements have also been shown to play a role in adaptive mutation of the bgl operon of E. coli (10). These results suggest that IS elements may play prominent roles in the adaptation of X. oryzae pv. oryzae and possibly other bacteria to life in the stationary phase.
The results of this study indicate that the spv mutants of X. oryzae pv. oryzae are heterogeneous in nature, and even in a single stationary-phase culture, at least two different classes of spv mutants are found to coexist. Finkle and Kolter (8) have also found different morphotypes coexisting in populations of surviving cells in prolonged stationary-phase cultures of E. coli. An intriguing feature in the case of X. oryzae pv. oryzae is that the vast majority of spv mutants (including the four characterized in this study) do not appear to have a discernible growth advantage in stationary-phase cocultures with the wild-type strain (29). Yet in these same cocultures, new spv mutants that are derived from the wild-type strain were found to accumulate in large numbers. Further experimentation is required to determine if the increase in frequency of the spv strains is due either to a selection for preexisting mutants or to the generation of new mutants.
The spontaneous insertion of IS1-like elements into the virF gene of an invasion plasmid in S. flexneri has been found to be associated with the stability of the plasmid outside the host, whereas its precise excision allowed the pathogen to revert to the invasive form inside the host (24). Under natural conditions, X. oryzae pv. oryzae has been reported to survive on dried plant parts and seeds for extended periods of likely starvation (31). Mutants that are similar to the spv strains might accumulate under such conditions. This variability may not be a dead end for the pathogen, if the IS elements can be excised at reasonable frequencies and permit selection for the virulent form under conditions favorable for disease development.
In summary, this study shows that in four of the nine spv mutants tested, the spontaneous loss of virulence and Eps production was due to transposition of one of the two endogenous IS elements, ISXo1 or ISXo2, in the gum locus. The other spv mutants do not show any RFLP in the gum locus containing the biosynthetic genes, nor are they complemented by the pSD1 clone. It would be interesting to see if these strains show an RFLP when probed with the IS elements. At least three different non-gum loci have been shown to be involved in the production of Eps precursor molecules (UDP-glucose, UDP-mannose, UDP-glucosamine, and GDP-mannose) in X. campestris pv. campestris (13). It is possible that either the X. oryzae pv. oryzae homologues of these genes or putative Eps regulatory loci are affected in some of the spv mutants. The nature of the mutations in these mutants is being investigated.
ACKNOWLEDGMENTS
We thank Suvendra Kumar Ray and J. Gowrishankar for helpful discussion and Marietta Ryba-White and Jan E. Leach for communicating unpublished data. We acknowledge the help of Meher Sultana in oligonucleotide synthesis and that of N. Nagesh in automated DNA sequencing.
This work was funded in part by a grant to R.V.S. from the Department of Biotechnology, Government of India.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Brumbley S M, Denny T P. Cloning of wild-type Pseudomonas solanacearum phcA, a gene that when mutated alters expression of multiple traits that contribute to virulence. J Bacteriol. 1990;172:5677–5685. doi: 10.1128/jb.172.10.5677-5685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capage, M. A., D. H. Doherty, M. Betlach, and R. W. Vanderslice. September 1996. Recombinant DNA mediated production of xanthan gum. U.S. patent 5,559,015.
- 4.Coplin D L, Cook D. Molecular genetics of extracellular polysaccharide biosynthesis in vascular phytopathogenic bacteria. Mol Plant-Microbe Interact. 1990;3:271–279. doi: 10.1094/mpmi-3-271. [DOI] [PubMed] [Google Scholar]
- 5.Dharmapuri S, Sonti R V. A transposon insertion in the gumG homologue of Xanthomonas oryzaepv. oryzae causes loss of extracellular polysaccharide production and virulence. FEMS Microbiol Lett. 1999;179:53–59. doi: 10.1111/j.1574-6968.1999.tb08707.x. [DOI] [PubMed] [Google Scholar]
- 6.DuBois M, Gilles K A, Hamilton J K, Rebers P A, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- 7.Dybvig K. DNA rearrangements and phenotypic switching in prokaryotes. Mol Microbiol. 1993;10:465–471. doi: 10.1111/j.1365-2958.1993.tb00919.x. [DOI] [PubMed] [Google Scholar]
- 8.Finkle S E, Kolter R. Evolution of microbial diversity during prolonged starvation. Proc Natl Acad Sci USA. 1999;96:4023–4027. doi: 10.1073/pnas.96.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goto M. Interrelationship between colony type, phage susceptibility and virulence in Xanthomonas oryzae. J Appl Bacteriol. 1972;35:505–515. doi: 10.1111/j.1365-2672.1972.tb03729.x. [DOI] [PubMed] [Google Scholar]
- 10.Hall B G. Activation of the bgloperon by adaptive mutation. Mol Biol Evol. 1998;15:1–5. doi: 10.1093/oxfordjournals.molbev.a025842. [DOI] [PubMed] [Google Scholar]
- 11.Han B, Pain A, Johnstone K. Spontaneous duplication of a 661 bp element within a two-component sensor regulator gene causes phenotypic switching in colonies of Pseudomonas tolaasii, cause of brown blotch disease of mushrooms. Mol Microbiol. 1997;25:211–218. doi: 10.1046/j.1365-2958.1997.4411811.x. [DOI] [PubMed] [Google Scholar]
- 12.Hancock I, Poxton I. Isolation of exopolysaccharides. In: Hancock I, Poxton I, editors. Bacterial cell surface techniques. Chichester, United Kingdom: Wiley Interscience; 1988. pp. 121–125. [Google Scholar]
- 13.Harding N E, Raffo S, Raimondi A, Cleary J M, Ielpi L. Identification, genetic and biochemical analysis of genes involved in sugar nucleotide precursors of xanthan gum. J Gen Microbiol. 1993;139:447–457. doi: 10.1099/00221287-139-3-447. [DOI] [PubMed] [Google Scholar]
- 14.Hengge-Aronis R. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell. 1993;72:165–168. doi: 10.1016/0092-8674(93)90655-a. [DOI] [PubMed] [Google Scholar]
- 15.Hopkins C M, White F F, Choi S H, Guo A, Leach J E. Identification of a family of avirulence genes from Xanthomonas oryzae. Mol Plant-Microbe Interact. 1992;5:451–459. doi: 10.1094/mpmi-5-451. [DOI] [PubMed] [Google Scholar]
- 16.Huang Y, Sequeira L. Identification of a locus that regulates multiple functions in Pseudomonas solanacearum. J Bacteriol. 1990;172:4728–4731. doi: 10.1128/jb.172.8.4728-4731.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katzen F, Ferreiro D U, Oddo C G, Ielmini M V, Becker A, Pühler A, Ielpi L. Xanthomonas campestris pv. campestris gummutants: effects on xanthan biosynthesis and plant virulence. J Bacteriol. 1998;180:1607–1617. doi: 10.1128/jb.180.7.1607-1617.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kauffman H E, Reddy A P K, Hsieh S P Y, Merca S D. An improved technique for evaluation of resistance of rice varieties to Xanthomonas oryzae. Plant Dis Rep. 1973;57:537–541. [Google Scholar]
- 19.Kousik C S, Ritchie D F. Identification of an insertion sequence (IS1646) from Xanthomonas campestris pv. vesicatoriaand its distribution among Xanthomonads. Phytopathology. 1998;88:S49. [Google Scholar]
- 20.Leach J E, White F F, Rhoads M L, Leung H. A repetitive DNA sequence differentiates Xanthomonas campestris pv. oryzae from other pathovars of X. campestris. Mol Plant-Microbe Interact. 1990;3:238–246. [Google Scholar]
- 21.Loewen P C, Hengge-Aronis R. The role of the sigma factor ςS (katF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 22.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller J H. A short course in bacterial genetics: a laboratory manual for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 24.Mills J A, Venkatesan M M, Baron L S, Buysse M. Spontaneous insertion of an IS1-like element into the virF gene is responsible for avirulence in opaque colonial variants of Shigella flexneri2a. Infect Immun. 1992;60:175–182. doi: 10.1128/iai.60.1.175-182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naas T, Blot M, Fitch W M, Arber W. Insertion sequence-related genetic variation in resting Escherichia coliK-12. Genetics. 1994;136:721–730. doi: 10.1093/genetics/136.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Negishi J, Yamada T, Shiraishi T, Oku H, Tanaka H. Pseudomonas solanacearum: plasmid pJTPS1 mediates a shift from the pathogenic to nonpathogenic phenotype. Mol Plant-Microbe Interact. 1993;6:203–209. [Google Scholar]
- 27.Nelson R J, Baraoidan M R, Vera Cruz C M, Yap I V, Leach J E, Mew T W, Leung H. Relationship between phylogeny and pathotype for the bacterial blight pathogen of rice. Appl Environ Microbiol. 1994;60:3275–3283. doi: 10.1128/aem.60.9.3275-3283.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng W-L, Kothakota S, DasSarma S. Structure of the gas vesicle plasmid in Halobacterium halobium: inversion isomers, inverted repeats, and insertion sequences. J Bacteriol. 1991;173:1958–1964. doi: 10.1128/jb.173.6.1958-1964.1991. . (Erratum, 173:3933.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajeshwari R, Yashitola J, Reddy A P K, Sonti R V. Characteristics of stationary-phase variation affecting virulence in Xanthomonas oryzae pv. oryzae. Can J Microbiol. 1997;43:862–867. [Google Scholar]
- 30.Reddy A P K, Kauffman H E. Loss of virulence associated with aging of Xanthomonas oryzaecultures. Indian Phytopathol. 1977;30:106–111. [Google Scholar]
- 31.Reddy R, Shang-zhi Y. Proceedings of the International Workshop on Bacterial Blight of Rice. Manila, the Philippines: International Rice Research Institute; 1988. Survival of Xanthomonas campestris pv. oryzae, the causal organism of bacterial blight of rice; pp. 65–77. [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivogenetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 34.Sonti R V, Roth J R. Role of gene duplications in the adaptation of Salmonella typhimuriumto growth on limiting carbon sources. Genetics. 1989;123:19–28. doi: 10.1093/genetics/123.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stibitz S, Aaronson W, Monack D, Falkow S. Phase variation in Bordetella pertussisby frameshift mutation in a gene for a novel two-component system. Nature. 1989;338:266–269. doi: 10.1038/338266a0. [DOI] [PubMed] [Google Scholar]
- 36.Tsuchiya K, Mew T W, Wakimoto S. Bacteriological and pathological characteristics of wild types and induced mutants of Xanthomonas campestris pv. oryzae. Phytopathology. 1982;72:43–46. [Google Scholar]
- 37.Yashitola J, Krishnaveni D, Reddy A P K, Sonti R V. Genetic diversity within the population of Xanthomonas oryzae pv. oryzaein India. Phytopathology. 1997;87:760–765. doi: 10.1094/PHYTO.1997.87.7.760. [DOI] [PubMed] [Google Scholar]
- 38.Zambrano M M, Siegele D A, Almiron M, Tormo A, Kolter R. Microbial competition: Escherichia colimutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 39.Zieg J, Silverman M, Hilman M, Simon M. Recombinational switch for gene expression. Science. 1977;196:170–172. doi: 10.1126/science.322276. [DOI] [PubMed] [Google Scholar]