Abstract
A type III secretion-translocation system allows Yersinia adhering at the surface of animal cells to deliver a cocktail of effector Yops (YopH, -O, -P, -E, -M, and -T) into the cytosol of these cells. Residues or codons 1 to 77 contain all the information required for the complete delivery of YopE into the target cell (release from the bacterium and translocation across the eukaryotic cell membrane). Residues or codons 1 to 15 are sufficient for release from the wild-type bacterium under Ca2+-chelating conditions but not for delivery into target cells. Residues 15 to 50 comprise the binding domain for SycE, a chaperone specific for YopE that is necessary for release and translocation of full-length YopE. To understand the role of this chaperone, we studied the delivery of YopE-Cya reporter proteins and YopE deletants by polymutant Yersinia devoid of most of the Yop effectors (ΔHOPEM and ΔTHE strains). We first tested YopE-Cya hybrid proteins and YopE proteins deleted of the SycE-binding site. In contrast to wild-type strains, these mutants delivered YopE15-Cya as efficiently as YopE130-Cya. They were also able to deliver YopEΔ17–77. SycE was dispensable for these deliveries. These results show that residues or codons 1 to 15 are sufficient for delivery into eukaryotic cells and that there is no specific translocation signal in Yops. However, the fact that the SycE-binding site and SycE were necessary for delivery of YopE by wild-type Yersinia suggests that they could introduce hierarchy among the effectors to be delivered. We then tested a YopE-Cya hybrid and YopE proteins deleted of amino acids 2 to 15 but containing the SycE-binding domain. These constructs were neither released in vitro upon Ca2+ chelation nor delivered into cells by wild-type or polymutant bacteria, casting doubts on the hypothesis that SycE could be a secretion pilot. Finally, it appeared that residues 50 to 77 are inhibitory to YopE release and that binding of SycE overcomes this inhibitory effect. Removal of this domain allowed in vitro release and delivery in cells in the absence as well as in the presence of SycE.
The three Yersinia species that are pathogenic to humans (Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica) all share the ability to deliver toxins, called YopE, YopH, YopM, YopT, YopO/YpkA, and YopP/YopJ, into eukaryotic host cells (8). These toxic Yop effectors induce a range of modifications to the normal processes of eukaryotic cells. For example, YopE has a GTPase-activating protein activity which downregulates Rho activity and leads to actin filament disruption and inhibition of phagocytosis by macrophages (21, 22, 28, 31). Together with their complex type III Ysc machinery for export and translocation, the Yops are encoded by a 70-kb virulence plasmid (8). Similar to structures observed in other bacteria endowed with type III secretion, the Yop secretion apparatus—the injectisome—is thought to form a “syringe” directly projecting through the bacterial membranes with a “needle” that connects to the translocation apparatus in the eukaryotic cell membrane (8, 10a). Secretion and translocation of the Yop effectors are normally triggered by contact with a eukaryotic cell. However, secretion can be artificially induced by chelating Ca2+ ions, which leads to a massive release of Yops into the culture supernatant.
A secretion signal for the Yops is located at the 5′ end of the gene (16, 29). It has been proposed that this signal could be in the mRNA, so that the Yops are cotranslationally secreted from the bacteria (1, 2). In the case of YopE, the first 15 codons or amino acids constitute this 5′ secretion signal. In addition, efficient secretion of some Yops requires the assistance of individual cytosolic chaperones, called Sycs (32, 33). These chaperones are small acidic proteins that possess a leucine repeat in their C-terminal moiety. SycE, the chaperone of YopE, binds amino acids 15 to 50 (the chaperone binding domain) of YopE (27, 35), and it prevents the intrabacterial degradation of this Yop (5, 10). The chaperone binding domain is not required for secretion of YopE fusion proteins by the 5′ secretion signal (5, 27, 28). Moreover, in the absence of this chaperone binding domain, SycE becomes dispensable for secretion of YopE, suggesting that it is the presence of the chaperone binding domain that creates the need for the chaperone (35). However, data have been presented to show that hybrid YopE-neomycin phosphotransferase (designated YopE-Npt) proteins lacking the first 5′ secretion signal are still secreted by the Ysc apparatus, suggesting that YopE could contain a second secretion signal (5). This proposed second secretion signal is localized to the site of the chaperone binding domain and, correspondingly, it is only operational in the presence of SycE (5).
Translocation of the effector Yops across the eukaryotic cell membrane was shown by several laboratories (4, 12, 20, 23) to be dependent on YopB and YopD, two other proteins exported by the bacterium, but this view has recently been questioned (15). Translocation of effector Yops can be demonstrated by several methods. A classical approach makes use of a calmodulin-dependent adenylate cyclase (Cya) reporter strategy (29). Translocation of Yop effectors can also be demonstrated by fractionation of the infected cell culture or by indirect immunofluorescence and confocal scanning laser microscopy (23). Demonstration of the translocation turned out to be more difficult with some Yops than with others, and it has been observed that translocation can be improved if expression of the other Yop effectors is abolished (4, 11). This could be due to a decrease in competition between the different Yop effectors for the secretion and translocation machineries. Strains of Y. enterocolitica that carry multiple yop mutations are thus sensitive tools for studying the translocation of Yop effectors.
It has been shown previously that a Cya reporter protein fused to just the first 15 amino acids of YopE (YopE15-Cya) can be released by wild-type (wt) bacteria upon Ca2+ chelation; however, this fusion protein is not delivered into eukaryotic cells. Indeed, at least the first 50 amino acids are required for the reporter protein to be translocated into eukaryotic cells by wt bacteria (27, 28). Therefore, amino acids 15 to 50, which are the residues that bind the SycE chaperone and which constitute the proposed second secretion signal, were thought to be a translocation domain (27, 28), although they are not sufficient, in the absence of the 5′ secretion signal, to direct delivery of YopE by wt bacteria into eukaryotic cells (14).
In this study, we investigated the requirement for the two proposed secretion signals for delivery of YopE into eukaryotic cells. We confirmed that SycE and residues 15 to 50 of YopE are required for delivery of YopE by wt bacteria, but we observed that they are dispensable for delivery by a multimutant strain. This suggests that SycE could be a hierarchy factor for YopE delivery. Moreover, we identified a secretion-inhibitory domain between residues 50 and 77.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Parental wt strain Y. enterocolitica MRS40(pYV40) is an ampicillin-sensitive derivative of serotype O:9 clinical isolate E40(pYV40) (25, 29). Escherichia coli LK111, XL-1 Blue, and BL21(DE3) were used for plasmid construction and protein expression. E. coli SM10λpir+ was used to conjugate plasmids into Y. enterocolitica. The full list of plasmids used in this study is given in Table 1. Bacterial strains were routinely grown in tryptic soy broth and plated on tryptic soy agar. For in vitro induction of the yop genes, Y. enterocolitica was grown in brain heart infusion (BHI), supplemented with 20 mM sodium oxalate, 20 mM MgCl2, and 0.4% (wt/vol) glucose (BHI-Ox). Yop induction under minimal-medium conditions was performed as described previously (5). Selective agents were used at the following concentrations: ampicillin, 200 μg · ml−1; chloramphenicol, 10 μg · ml−1; nalidixic acid, 35 μg · ml−1; streptomycin, 100 μg · ml−1; sucrose, 5% (wt/vol); and arsenite, 0.4 mM.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant description | Reference or source |
|---|---|---|
| pYV plasmids | ||
| pYV40 | Wild type | 26 |
| pAB4052 | ΔYopE yopE21 | 17 |
| pAB409 | ΔHOPEMYopB | 3 |
| yopHΔ1–352 yopOΔ65–558 yopP23 yopE21 yopM23 yopBΔ189–217 | ||
| pABL403 | ΔHOPEM yopHΔ1–352 yopOΔ65–558 yopP23 yopE21 yopM23 | 3 |
| pAPB4054 | ΔSycE | This work |
| sycE54 | ||
| pAPB4055 | ΔHOPEMSycE | This work |
| yopHΔ1–352 yopOΔ65–558 yopP23 yopE21 yopM23 sycE54 | This work | |
| pAPBD416 | ΔTHEB | This work |
| yopT135 yopHΔ1–352 yopE21 yopBΔ189–217 | This work | |
| pIM426 | ΔTHE yopT135 yopHΔ1–352 yopE21 | 4a |
| pMSK49 | ΔHOPEMYopD yopHΔ1–352 yopOΔ65–558 yopP23 yopE21 yopM23 yopDΔ121–165 | 18 |
| pMSK50 | ΔHOPEMYscN yopHΔ1–352 yopOΔ65–558 yopP23 yopE21 yopM23 yscNΔ169–177 | Sory and Cornelis, unpublished |
| pMSL41 | ΔYscN yscNΔ169–177 | 28 |
| Other plasmids | ||
| pAPB24 | pQE-30 containing his-sycE under PT5 promoter control | This work |
| pAPB26 | pCNR26 containing yopE under yopE promoter control | 4a |
| pAPB35 | pCNR26 containing yopEΔ2–15 under yopE promoter control | This work |
| pAPB36 | pCNR26 containing yopE+1[2–15] under yopE promoter control | This work |
| pAPB37 | pCNR26 containing yopEΔ2–77 under yopE promoter control | This work |
| pAPBD16 | pBluescript KS− containing yopE15-cya plus sycE | This work |
| pAPBD17 | pBluescript KS(−) containing yopE15-cya plus sycE54 | This work |
| pAPBD18 | pTM100 containing yopE15-cya and sycE54 | This work |
| pAPBG30 | pCNR26 containing yopEΔ17–49 under yopE promoter control | This work |
| pAPBL34 | pCNR26 containing yopEΔ17–77 under yopE promoter control | This work |
| pAPBL38 | pBluescript KS(−) containing yopE130-cya plus sycE54 | This work |
| pAPBL40 | pBluescript KS(−) containing yopE130-cya plus sycE | This work |
| pAPBL42 | pBluescript KS(−) containing yopE130(Δ2–15)-cya and sycE54 | This work |
| pAPBL43 | pBluescript KS(−) containing yopE130(Δ2–15)-cya and sycE | This work |
| pAPBL44 | pBluescript KS(−) containing yopE130[+1(2–15)]-cya and sycE54 | This work |
| pAPBL45 | pBluescript KS(−) containing yopE130[+1(2–15)]-cya and sycE | This work |
| pAPBL47 | pTM100 containing yopE130[+1(2–15)]-cya and sycE54 | This work |
| pAPBL48 | pTM100 containing yopE130[+1(2–15)]-cya and sycE | This work |
| pAPBL49 | pTM100 containing yopE130(Δ2–15)-cya and sycE54 | This work |
| pAPBL50 | pTM100 containing yopE130(Δ2–15)-cya and sycE | This work |
| pBC18R | Ampr, PlacZ, expression vector | 7 |
| pBC19R | Ampr, PlacZ, expression vector | 7 |
| pBC5 | pBC19R containing yopQ ylpa′ yopP′ yopO | 7 |
| pBluescript | Ampr, lacZ′, cloning vector | Stratagene |
| pCNR26 | Ampr, PyopE, expression vector | 25 |
| pIL14 | pCNR26 containing yopEΔ50–77 under yopE promoter control | This work |
| pIM153 | pBC18R containing yopM under yopM and lac promoter control in HincII fragment | Iriarte and Cornelis, unpublished |
| pKNG101 | Smr, sacBR, suicide vector | 13 |
| pMS111 | pTM100 containing yopE130-cya and sycE | 29 |
| pMSK13 | pCNR26 containing yopP under yopE promoter control | 17 |
| pMSL30 | pTM100 containing yopE130-cya and sycE54 | 35 |
| pMSL56 | pTM100 containing yopE2-cya | Sory and Cornelis, unpublished |
| pMSLE15 | pTM100 containing yopE15-cya and sycE | 28 |
| pPW54 | pKNG101 containing sycE54 | 33 |
| pQE-30 | Ampr, PT5, His tag expression vector | Qiagen |
| pSW6 | pKNG101 containing yscNΔ169–177 | 34 |
| pTM100 | Cmr, medium-copy-number cloning vector | 16 |
| pTM163 | pBC18R containing yopH under yopH and lac promoter control | 16 |
| pYOB2 | pCNR26 containing yopO under yopE promoter control | Geuijen and Cornelis |
Molecular biology techniques.
Molecular biology techniques were essentially performed as previously described (24). All chemicals were obtained from Sigma unless stated otherwise. Yops were precipitated from culture supernatants by ammonium sulfate (0.5 g · ml−1) (9), analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and, where appropriate, transferred to nitrocellulose membranes. Immunoblots were developed with secondary antibody conjugated to horseradish peroxidase (HRP) and Supersignal (NEN) as a chemiluminescent substrate. For detection of Cya fusion proteins on nitrocellulose membranes, biotinylated calmodulin (Calbiochem) was used according to the supplier's instructions, except that streptavidin-biotinylated HRP complex (Amersham Life Science) was substituted for streptavidin-alkaline phosphatase, and Supersignal was used as the chemiluminescent substrate.
Mutator plasmids were introduced into Y. enterocolitica strains by conjugation from E. coli SM10λpir+. After allelic exchange, the mutations were confirmed by PCR analysis or sequencing and, where appropriate, by Yop induction and SDS-PAGE analysis of the secreted proteins or by Western blot analysis of the bacterial proteins.
Plasmid constructions. (i) pAPBD18.
The 2.4-kb BamHI-ClaI fragments of pMSLE15 (containing yopE15-cya and sycE) were cloned into the corresponding sites of pBluescript KS(−) to create pAPBD16. pAPBD16 was digested with EcoRI, the ends were filled in with Klenow, and the DNA was religated to create pAPBD17. The 2.4-kb BamHI-ClaI fragments of pAPBD17 (containing yopE15-cya and sycE54) were cloned into the corresponding sites of pTM100 to create pAPBD18.
(ii) pAPBL49, pAPBL50, pAPBL47, and pAPBL48.
The 2.5-kb BamHI-ClaI fragments of pMS111 (containing yopE130-cya and sycE) and of pMSL30 (containing yopE130-cya and sycE54) were cloned into the corresponding sites of pBluescript KS(−) to create pAPBL40 and pAPBL38, respectively. pAPBL40 and pAPBL38 were then mutagenized with oligonucleotide MIPA 679 (GGGAATAAATAGTCATGTCAGTGTCAGGATCTAG), which is identical to nucleotides −14 to 3 and 46 to 62 of yopE, to produce pAPBL43 and pAPBL42, respectively, which contain yopE130(Δ2–15). pAPBL40 and pAPBL38 were then mutagenized with oligonucleotide MIPA 680 (TAAATAGTCATGGAAAATA TCATCATTTATTTCTACATCACTGCCCCTGCCGGGAGCTCAGTGTCA GGA), which introduces a G at position +4 and in which GAGC replaces CA at +44 to 45 to produce pAPBL45 and pAPBL44, respectively, containing yopE130[+1(2–15)] (replacement sites are indicated by boldface type). The 2.5-kb BamHI-ClaI fragments of pAPBL42, pAPBL43, pAPBL44, and pAPBL45 were cloned into the corresponding sites of pTM100 to produce pAPBL49, pAPBL50, pAPBL47, and pAPBL48, respectively.
(iii) pYOB2.
The 2.2-kb yopO gene was amplified by PCR with oligonucleotides MIPA 471 (GCATGAACATATGGGAACTA) and MIPA 473 (TATATCAAATGCATGGCTTAGGG) using pBC5 as a template. After digestion with NdeI (underlined) and NsiI (underlined), the DNA fragment was cloned into the NdeI and PstI sites of pCNR26 to give plasmid pYOB2. This construction places the second start codon of yopO at the NdeI site.
(iv) pAPBG30, pAPBL34, pAPB35, pAPB36, pAPB37, and pIL14.
Plasmid pAPB26 was mutagenized with (i) oligonucleotide MIPA 635 (CTGGAACCCTGAGGTGATGCCGGCAG), which is complementary to nucleotides 37 to 48 and 148 to 161 of the yopE gene, to produce plasmid pAPBG30 which encodes YopEΔ17–49; (ii) oligonucleotide MIPA 671 (GCTCCCCTCCGATGATGCCGGCAG), which is complementary to nucleotides 37 to 48 and 235 to 246 of the yopE gene, to produce plasmid pAPBL34 which encodes YopEΔ17–77; (iii) oligonucleotide MIPA 677 (CTAGATCCTGACACTGACATATGTATTTCCTCCTT), which is complementary to nucleotides −15 to 3 and 46 to 62 of the yopE gene to produce plasmid pAPB35 which encodes YopEΔ2–15; (iv) oligonucleotide MIPA 678 (TCCTGACACTGAGCTCCCGGCAGGGGCAGTGATGTAGAAATAAATGATGATATTTTCCATATGTATTTC), which adds a G at the +4 position and in which GAGC replaces CA at positions +44 and 45 of the yopE gene to produce plasmid pAPB36 which encodes YopE+1(2–15) (noncoding strand; replacement sites shown in boldface); (v) oligonucleotide MIPA 681 (GCTCCCCTCCGACATATGTATTTCCTCCTT), which is complementary to nucleotides −15 to 3 and 232 to 243 of the yopE gene to produce plasmid pAPB37 which encodes YopEΔ2–77; and (vi) oligonucleotide MIPA 892 (GCTCCCCTCCGAGCTTTCAGTGCG), which is complementary to nucleotides 135 to 147 and 234 to 246 of the yopE gene to produce plasmid pIL14 which encodes YopEΔ50–77.
(v) pAPB24.
SycE was amplified by PCR with oligonucleotides MIPA 599 (CGGGATCCTATTCATTTGAACAAGCTA), which is complementary to nucleotides 4 to 22 of sycE, and MIPA 521 (CTCAAGCTTCTACTCAACTAAATGACCG), which is identical to nucleotides 379 to 393 of sycE and four additional bases with pAPBD16 as a template (introduced restriction sites are underlined). The 400-bp product was digested with BamHI and HindIII and cloned into the corresponding sites of pQE-30 to create pAPB24.
Yop translocation assay.
The PU5-1.8 mouse monocyte/macrophage cell line (ATCC TIB-61) used in these studies was grown in RPMI 1640 medium (Gibco BRL) supplemented with 10% (wt/vol) fetal bovine serum, 2 mM l-glutamine, and streptomycin, 100 μg · ml−1. Translocation assays were carried out essentially as described by Sory and Cornelis (29). Cells were seeded into 24-well tissue culture plates at a density of 5 × 105 cells per ml of medium per well and allowed to adhere for 20 h. Before infection with Y. enterocolitica, cells were washed and covered with RPMI 1640 supplemented only with 2 mM l-glutamine. Cytochalasin D was added 30 min before infection, at a final concentration of 5 μg ml−1 (stock solution, 2 mg ml−1 in dimethyl sulfoxide). Cytochalasin D is not toxic to Y. enterocolitica at this concentration (29). A freshly isolated transconjugant colony of Y. enterocolitica was cultured overnight at 22°C and diluted the next day to an optical density at 600 nm of 0.2 in 5 ml of BHI medium. After being grown with shaking at 22°C for 2 h, bacteria were washed and suspended in saline. Samples of 100 μl, containing about 107 bacteria (multiplicity of infection, 20:1), were added to the monolayer, and the infected cultures were incubated at 37°C for 2 h in a 6% CO2 atmosphere. Cells were washed and then lysed under denaturing conditions (100°C for 5 min in 50 mM HCl and 0.1% [wt/vol] Triton X-100). The lysate was neutralized by NaOH, and cyclic AMP (cAMP) was extracted with ethanol. After centrifugation, the supernatant was dried, and cAMP was assayed by an enzyme immunoassay (Biotrak Amersham). All experiments were performed three times.
Cytotoxicity assay.
The HeLa human epithelial cell line (ATCC CCL-2) used in these studies was grown in RPMI 1640 supplemented with 10% (wt/vol) fetal bovine serum, 2 mM l-glutamine, and streptomycin (100 μg · ml−1) and prepared similarly to the PU5-1.8 macrophages as described above except that the HeLa cells were seeded at a density of 7 × 104 cells · ml−1. Bacteria were pregrown as described above, and cells were infected with Y. enterocolitica at a multiplicity of infection of 70. Two to three hours after infection, the morphology of the cells was observed by phase-contrast microscopy. The cells became rounded as a result of cytotoxicity.
Staining of actin filaments with phalloidin.
Rat I fibroblasts grown on coverslips were infected with the different strains. After 2.5 h of infection, the cells were fixed in 2% (wt/vol) paraformaldehyde for 20 min. After being washed with phosphate-buffered saline (PBS) (136 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, 1.8 mM KH2PO4 [pH 7.4]), membranes were permeabilized with 0.5% (wt/vol) Triton X-100 in PBS for 10 min. Cells were then incubated for 40 min at 37°C with fluorescein isothiocyanate-conjugated phalloidin. Samples were mounted on Mowiol and examined by fluorescence microscopy.
SycE-binding assays.
Native SycE was produced and purified as described in reference 33. His6-SycE was produced in E. coli XL-1 Blue(pAPB24) and purified on a His-Trap column by elution with 300 mM imidazole according to the manufacturer's instructions (Pharmacia Biotech). Total cell proteins of Y. enterocolitica were separated by SDS-PAGE and transferred to nitrocellulose membranes. After blockage in PBST plus BSA (PBS plus 0.1% Tween 20 plus 0.5% bovine serum albumin [BSA]), the membrane was incubated with His6-SycE (0.5 μg · ml−1) in PBST plus BSA for 2 h at room temperature. Bound SycE or His6-SycE was revealed with anti-SycE or anti-His antibody (Pharmacia Biotech), respectively, followed by HRP-conjugated secondary antibody and chemiluminescence detection.
RESULTS
Translocation of YopE15-Cya into eukaryotic cells by Yop effector polymutant Y. enterocolitica.
It has been previously demonstrated that wt bacteria deliver YopE130-Cya, but not YopE15-Cya, into eukaryotic cells, suggesting that the 5′ secretion signal is not sufficient for YopE translocation (28). We repeated these experiments using ΔHOPEM polymutant bacteria that lack the YopH, YopO, YopP, YopE, and YopM effectors (Table 1). Delivery of YopE15-Cya (encoded by plasmid pMSLE15 [8]) into the PU5-1.8 macrophage-monocyte cell line by ΔHOPEM and by wt Y. enterocolitica was compared. As a control, translocation of YopE130-Cya (encoded by plasmid pMS111 [29]) by the same bacteria was also monitored. In agreement with previously published results (28), wt bacteria delivered YopE130-Cya, but not YopE15-Cya, into eukaryotic cells. In contrast, ΔHOPEM bacteria delivered YopE15-Cya just as efficiently as YopE130-Cya (Table 2). Likewise, YopE20-Cya, YopE24-Cya, and YopE30-Cya (28) were delivered into eukaryotic cells by ΔHOPEM bacteria (data not shown). To confirm that delivery of YopE15-Cya into eukaryotic cells by ΔHOPEM bacteria was due to the type III secretion-translocation system, the delivery of YopE15-Cya by ΔHOPEMYscN bacteria (secretion deficient) and ΔHOPEMYopB and ΔHOPEMYopD bacteria (both translocation deficient) was tested (Table 2). YopE15-Cya was not translocated by these strains, confirming that YopE15-Cya was indeed delivered into eukaryotic cells by the type III injectisome (Table 2). To assess the necessity for the 5′ secretion signal, delivery of Cya fused to the first two amino acids of YopE (encoded by plasmid pMSL56) (Table 1) into eukaryotic cells was measured. This fusion protein was not delivered into macrophages by either wt (0.1 ± 0.1 nmole of cAMP/mg) or ΔHOPEM (0.1 ± 0.1 nmole of cAMP/mg) bacteria, showing that a Yop secretion signal is required for delivery of a protein by ΔHOPEM Y. enterocolitica. These results show that the translocation system of ΔHOPEM bacteria is still specific for the Yops. In conclusion, translocation of YopE-Cya hybrids is possible without the previously described translocation domain, which comprises amino acids 15 to 50, but not without the 5′ secretion signal (residues or codons 1 to 15 and upstream RNA).
TABLE 2.
Translocation of YopE15-Cya and YopE130-Cya into PU5-1.8 macrophages by Y. enterocolitica
| Y. enterocolitica strain | Intracellular cAMP accumulation (nmol of cAMP/mg)a in:
|
|
|---|---|---|
| YopE15-Cya | YopE130-Cya | |
| wt | 0.4 ± 0.2 | 22.8 ± 4.3 |
| ΔHOPEM | 14.3 ± 2.3 | 18.1 ± 3.0 |
| ΔHOPEMYopB | 0.2 ± 0.1 | 0.1 ± 0.1 |
| ΔHOPEMYopD | 0.2 ± 0.1 | 0.1 ± 0.1 |
| ΔHOPEMYscN | 0.1 ± 0.1 | 0.1 ± 0.1 |
| ΔSycE | 0.8b | 3.8 ± 2.2 |
| ΔHOPEMSycE | 16.5 ± 2.7 | 3.2 ± 0.4 |
Mean ± SD from three independent experiments.
Mean of two independent experiments.
Production and secretion levels of YopE15-Cya by wt and ΔHOPEM bacteria.
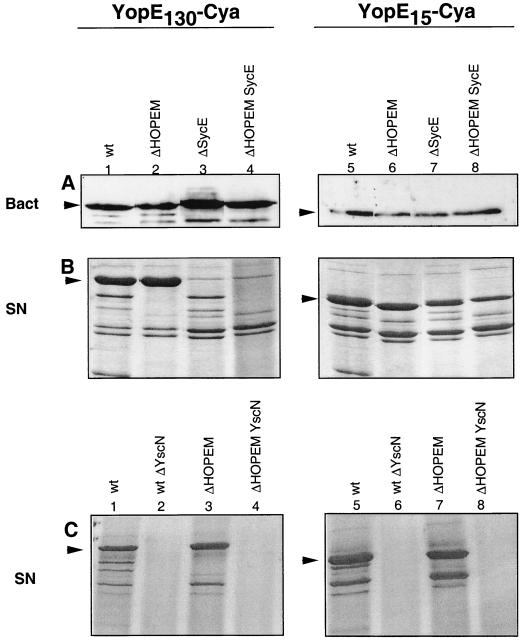
It was next investigated whether the delivery of YopE15-Cya by ΔHOPEM, but not wt Y. enterocolitica, could result from differences between strains in the production and secretion of this hybrid protein. Protein levels were analyzed following growth of the bacteria in Ca2+-chelating conditions, which induce Yop production and release. No differences were seen between the two strains in the levels of YopE15-Cya associated with the bacteria or released into the extracellular medium (Fig. 1A and Fig. 1B, lanes 5 and 6). Secretion of YopE15-Cya by both strains was strictly dependent on the Ysc system, since no secretion was observed in a yscN background (Fig. 1C). Although Yop release upon Ca2+ chelation may not necessarily reflect exactly what occurs upon contact of Y. enterocolitica with eukaryotic cells, these data do show that synthesis of the two fusion proteins and their passage through the bacterial membranes was equally efficient in the two strains and equally dependent on Ysc. The only difference between the two strains with regard to YopE15-Cya was thus the level of translocation of this protein into eukaryotic cells (Table 2). This suggests that the presence of additional Yops in the wild type directly reduces translocation of YopE15-Cya and that in order to enter into eukaryotic cells the Yops must thus compete with one another for passage through the secretion-translocation machinery.
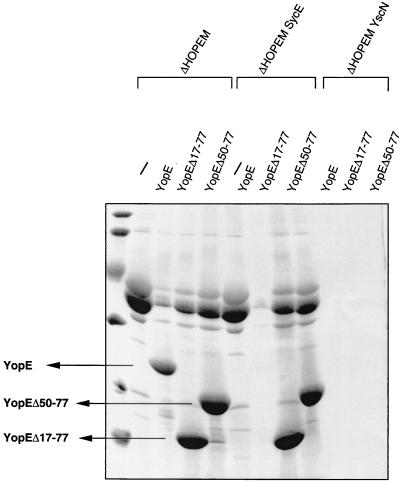
FIG. 1.
Ca2+ chelation-triggered release of YopE130-Cya and YopE15-Cya by wt and ΔHOPEM Y. enterocolitica. The role of Ysc and SycE is shown. (A) Immunoblot probed with calmodulin-biotin and streptavidin-HRP to detect bacteria-associated YopE130-Cya or YopE15-Cya (Bact). A total of 1.5 × 108 Y. enterocolitica bacteria grown under BHI-Ox conditions were loaded in each lane. (B and C) Coomassie blue-stained SDS-PAGE gel of proteins secreted (SN) by various Y. enterocolitica encoding YopE130-Cya or YopE15-Cya and grown under BHI-Ox conditions. Arrowheads indicate YopE130-Cya and YopE15-Cya. The hybrid proteins were encoded by pMS111 (YopE130-Cya plus SycE), pMSL3O (YopE130-Cya only), pMSLE15 (YopE15-Cya plus SycE) and pAPB18 (YopE15-Cya alone). The host strains were MRS40(pYV40) (wt), MRS40(pABL403) (ΔHOPEM), MRS40(pAPB4054) (ΔSycE), MRS40(pAPB4055) (ΔHOPEM SycE), MRS40(pMSL41) (wt ΔYscN), and MRS40(pMSK50) (ΔHOPEM YscN). In each lane, the proteins released by 1.5 × 109 bacteria were loaded. Arrowheads point to YopE130-Cya or YopE15-Cya.
Influence of SycE on secretion and translocation of YopE15-Cya.
In order to investigate the requirement for SycE for translocation of YopE15-Cya and YopE130-Cya into eukaryotic cells, an sycE mutation was introduced into the wt and ΔHOPEM strains (Table 1). The experiment was carried out with plasmids pMS111 and pMSLE15, which encode SycE along with YopE130-Cya and YopE15-Cya, respectively, and with plasmids pMSL30 and pAPBD18, which encode only the YopE-Cya fusion proteins (Table 1). The presence or absence of SycE did not affect the steady-state levels of YopE130-Cya or YopE15-Cya associated with the wt or ΔHOPEM bacteria when grown under BHI-Ox conditions (Fig. 1A). However, SycE was required for efficient secretion and translocation of YopE130-Cya into eukaryotic cells not only by wild-type but also by ΔHOPEM bacteria (Fig. 1B, compare lanes 1 and 3 and lanes 2 and 4; Table 2). In contrast, the presence or absence of SycE did not influence secretion or translocation of YopE15-Cya into eukaryotic cells by ΔHOPEM bacteria (Fig. 1B, compare lanes 5 and 7 and lanes 6 and 8; Table 2). Thus, efficient delivery of YopE15-Cya by ΔHOPEM bacteria occurred in the absence of SycE. We conclude from this that SycE is only required for efficient secretion and subsequent translocation when its binding domain is present. However, when the chaperone binding domain is present, the chaperone is required, irrespective of the presence of other effectors.
Translocation into eukaryotic cells of YopE lacking the SycE chaperone binding domain.
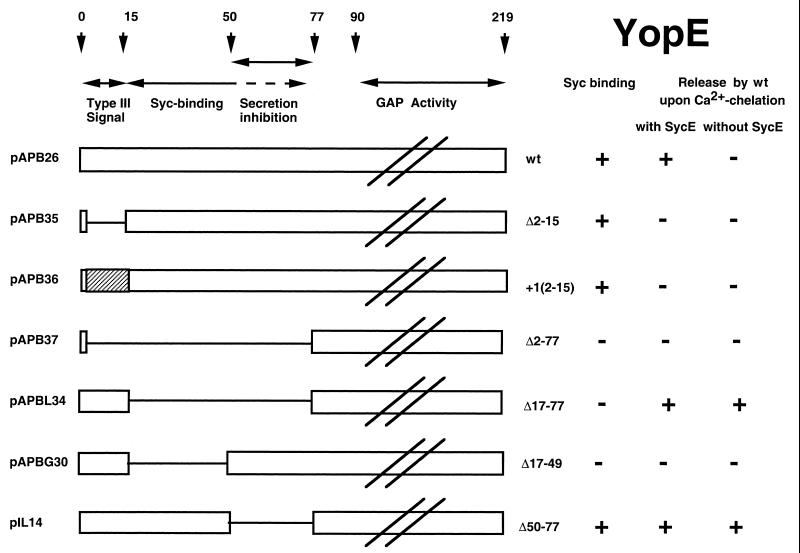
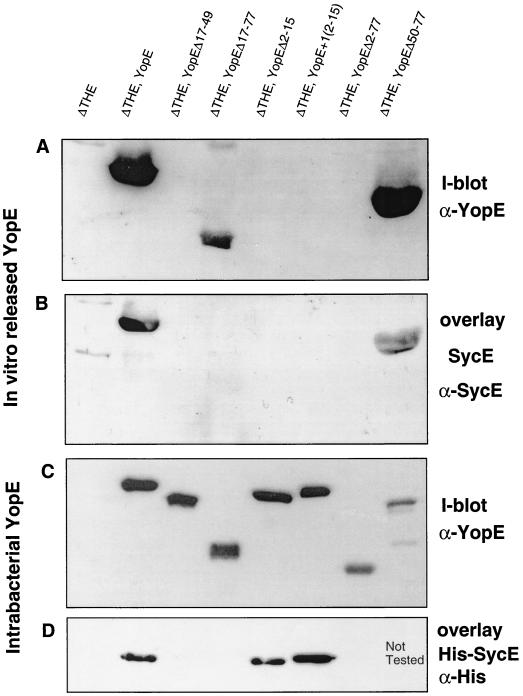
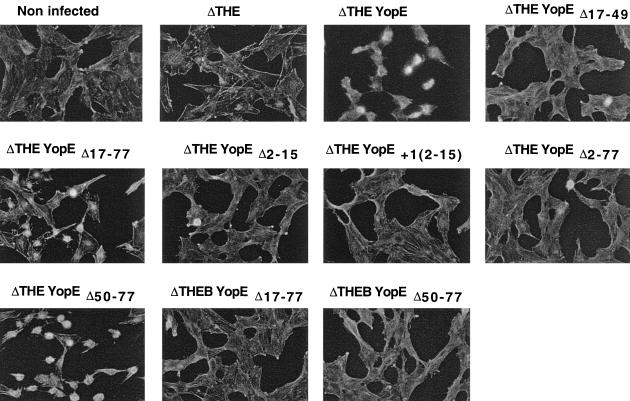
In order to confirm that codons or amino acids 1 to 15 of YopE are sufficient to translocate YopE into eukaryotic cells, we removed the chaperone binding domain (YopEΔ17–77) from YopE (Fig. 2), and we checked this removal by a SycE overlay experiment (33). Purified SycE or His6-SycE bound YopE but failed to bind YopEΔ17–77, verifying that the chaperone binding domain had been deleted from the latter protein (Fig. 3B and D). ΔHOPEM bacteria could not be used for cytotoxicity experiments because they still produce the YopT cytotoxin (8). We thus turned to ΔTHE bacteria (Table 1), which do not induce any morphological changes in eukaryotic cells (Fig. 4). YopEΔ17–77 was produced and released by ΔTHE bacteria (Fig. 3A and 3C), and release of YopEΔ17–77 did not occur in an yscN background (Fig. 5), confirming that this release was type III dependent. Delivery was then assayed by monitoring the rounding up of HeLa epithelial cells and by staining the actin of Rat-I cells. ΔTHE Y. enterocolitica strains producing YopE or YopEΔ17–77 were cytotoxic for HeLa epithelial cells (results not shown) and Rat I fibroblasts (Fig. 4), while ΔTHEB bacteria producing YopEΔ17–77 were not cytotoxic, indicating that translocation of YopEΔ17–77 was YopB dependent. This result confirmed that the first 16 amino acids of YopE are sufficient for delivery into eukaryotic cells and that the chaperone binding domain is not required.
FIG. 2.
Schematic representation of the YopE proteins used in this work.
FIG. 3.
SycE-binding and in vitro release of mutated YopE proteins. ΔTHE Y. enterocolitica [MRS40(pIM426)] producing YopE (pAPB26), YopEΔ17–49 (pAPBG30), YopEΔ17–77 (pAPBL34), YopEΔ2–15 (pAPB35), YopE+1[2–15] (pAPB36), YopEΔ2–77 (pAPB37), or YopEΔ50–77 (pIL14) were used in these experiments. (A) Immunoblot with anti-YopE antibodies to detect YopE proteins released by Y. enterocolitica incubated under BHI-Ox conditions. In each lane, the proteins released by 1.25 × 109 bacteria were loaded. (B) Overlay experiment with purified SycE protein. SycE bound to released YopE was detected with anti-SycE antibodies. The same gel described in the legend to panel A was used. (C) Immunoblot with anti-YopE antibodies to detect bacteria-associated YopE proteins (BHI-Ox conditions). A total of 1.5 × 108 bacteria were loaded in each lane. (D) Overlay experiment with purified His6-SycE protein. His6-SycE protein bound to bacteria-associated YopE was detected with anti-His antibodies. The same gel as that described in the legend to panel C was used. I-blot, immunoblot.
FIG. 4.
Cytotoxicity of Y. enterocolitica producing deleted YopE proteins. Rat I cells were infected with ΔTHE Y. enterocolitica [MRS40(pIM426)] producing WT YopE (pAPB26), YopEΔ17–49 (pAPBG30), YopEΔ17–77 (pAPBL34), YopEΔ2–15 (pAPB35), YopE+1[2–15] (pAPB36), YopEΔ2–77 (pAPB37), or YopEΔ50–77 (pIL14). As a control, Rat I cells were also infected with ΔTHEB Y. enterocolitica [MRS40(pAPBD4016)] producing YopEΔ17–77 (pAPBL34) or YopEΔ50–77 (pIL14). Actin was stained with fluorescent phalloidin. Cytotoxicity of YopE, YopEΔ17–77, or YopEΔ50–77 is manifested by rounding up of the cells. Note that YopEΔ50–77 is particularly active.
FIG. 5.
Ca2+ chelation-induced release of YopEΔ50–77. The role of Ysc and SycE is shown. Coomassie blue-stained SDS-PAGE analysis of the proteins released by ΔHOPEM [MRS40(pABL403)], ΔHOPEMSycE [MRS40(pAPB4055)], and ΔHOPEMYscN [MRS40(pMSK50)] producing no protein (/), wt YopE (pAPB26), YopEΔ17–77 (pAPBL34), or YopEΔ50–77 (pIL14) was carried out. In each lane, the proteins released by 1.25 × 109 bacteria were loaded.
Competition could play an important role in determining the level of translocation.
YopE15-Cya was delivered into eukaryotic cells by ΔHOPEM bacteria, but not by wt bacteria, suggesting that competition between the Yops is an important determinant for secretion and translocation and that the chaperone binding domain plays a significant role with regards to this competition. To investigate this theory, the ability of ΔHOPEM bacteria to deliver YopE15-Cya (encoded by pMSLE15) into eukaryotic cells when overproducing another Yop effector in trans was tested. Therefore, the translocation of YopE15-Cya into eukaryotic cells by ΔHOPEM bacteria overproducing YopH, YopO, YopP, YopE, or YopM was measured. ΔHOPEM(pMSLE15)(pBC18R) served as a vector control. In each case, the rate of translocation of YopE15-Cya into eukaryotic cells was lower than that of ΔHOPEM bacteria not overexpressing one of these Yop effectors in trans (Table 3). In contrast, translocation into eukaryotic cells of YopE130-Cya by ΔHOPEM was unaffected by overproducing another Yop in trans. The strongest effect on delivery of YopE15-Cya was observed with YopE and YopH (Table 3). As a control, we checked the profile of proteins released by these strains upon Ca2+ chelation. This control (data not shown) confirmed the overproduction of the Yops encoded in trans. Unfortunately, it also showed a concomitant reduction in the release of the Cya reporter and of the translocators LcrV, YopB, and YopD, indicating that the previous results must be interpreted with caution. To circumvent these difficulties, presumably linked to titration, we tested whether YopE15-Cya could be delivered into cells by Y. enterocolitica bacteria missing only YopE (ΔYopE strain, plasmid pAB4052). Delivery by the ΔYopE strain led to the synthesis of 2.7 ± 0.6 nmole of cAMP/mg of protein, while delivery by the wt strain led only to the synthesis of 0.4 ± 0.2 nmole of cAMP/mg of protein. Thus, lack of YopE alone significantly increased delivery of YopE15-Cya into eukaryotic cells. These results are consistent with the idea that amino acids 15 to 50 promote translocation of YopE by wt bacteria by assisting YopE to compete with other Yops for the secretion-translocation apparatus. If this was so, one would expect that YopE deprived of its chaperone binding domain (YopEΔ17–77) would not compete with YopE15-Cya for delivery into eukaryotic cells. We thus overproduced YopEΔ17–77 in trans, and we monitored translocation of YopE15-Cya. As expected, overproduction of YopEΔ17–77 did not inhibit translocation of YopE15-Cya (Table 3). Thus, amino acids 15 to 50 of YopE, in conjunction with SycE, seem to give YopE a competitive advantage over the other Yops for the secretion-translocation process.
TABLE 3.
Translocation of YopE15-Cya and YopE130-Cya into PU5-1.8 macrophages by HOPEM strain overexpressing other Yop effector
| Cya hybrid fusion | Yop overexpressed in trans (Yop expression plasmid) | Intracellular cAMP accumulation (nmole of cAMP/mg)a |
|---|---|---|
| YopE15-Cya | ||
| YopH (pTM163) | 0.3 ± 0.3 | |
| YopO (pYOB2) | 0.4 ± 0.3 | |
| YopP (pMSK13) | 1.6 ± 0.5 | |
| YopE (pAPB26) | 0.3 ± 0.1 | |
| YopM (pIM153) | 3.0 ± 0.3 | |
| None (pBC18R) | 14.0b | |
| YopEΔ17–77 (pAPBL34) | 9.5 ± 3.3 | |
| YopE130-Cya | ||
| None | 22.8 ± 4.3 | |
| YopO (pYOB2) | 25.0b | |
| YopE (pAPB26) | 15.1b |
Mean ± SD from three independent experiments.
Mean of two independent experiments.
Role of proposed second secretion signal in translocation.
Since the first secretion signal (amino acids or codons 1 to 15) was found to be sufficient for translocation into eukaryotic cells, we wondered whether the second secretion signal (amino acids 15 to 100) proposed by Cheng et al. (5) would also be sufficient to direct translocation into eukaryotic cells by the Yop effector multimutant strain ΔHOPEM. This second signal was previously shown to be insufficient for delivery into eukaryotic cells by wt bacteria (14). Therefore, three plasmids were constructed encoding YopE proteins lacking the first secretion signal (amino acids or codons 2 to 15). Plasmid pAPB35 encodes YopEΔ2–15. Plasmid pAPB36 encodes YopE(+1[2–15]) in which amino acids 2 to 15 have been shifted out of frame by the addition of 1 bp after the ATG and by compensatory changes before codon 16. A similar construct has previously been shown to have an inactive first secretion signal and to be secreted by the proposed second secretion signal (5). As well, plasmid pAPB37 encodes YopEΔ2–77. The three constructs were checked first for their capacity to bind His6-SycE in an overlay assay. As expected, YopEΔ2–77 did not bind SycE, while YopEΔ2–15 and YopE(+1[2–15]) were recognized by the chaperone (Fig. 3D). Each of the three proteins was produced by ΔTHE bacteria, but no secretion when grown in BHI-Ox medium could be detected (Fig. 3A). This result was expected for YopEΔ2–77, since it lacks both the first 5′ signal and the proposed second secretion signal, but not for the two others. Surprised by the inability of amino acids 15 to 50 (the proposed second secretion signal) to promote secretion of YopEΔ2–15 or YopE(+1[2–15]), the secretion of these proteins was tested under the same minimal-medium conditions as those used by Cheng et al. (5). Under these conditions, the proteins were produced but not secreted by ΔTHE bacteria (Fig. 6). In accordance with their non-secretion phenotype, neither ΔTHE encoding YopEΔ2–15, ΔTHE encoding YopE(+1[2–15]), nor ΔTHE encoding YopEΔ2–77 was cytotoxic for HeLa cells (data not shown) and Rat I cells (Fig. 4).
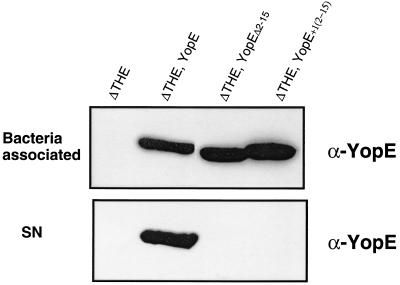
FIG. 6.
Lack of detectable release in the absence of residues or codons 1 to 15. ΔTHE Y. enterocolitica [MRS40(pIM426)] bacteria producing YopE (pAPB26), YopEΔ2–15 (pAPB35), and YopE+1[2–15] (pAPB36) were incubated in minimal medium. (Top) Immunoblot with anti-YopE antibodies to detect bacterium-associated YopE proteins. In each lane 2 × 108 bacteria were loaded. (Bottom) Immunoblot with anti-YopE antibodies to detect secreted YopE proteins. In each lane, the proteins released by 2 × 109 bacteria were loaded.
In addition, plasmids encoding YopE130-Cya reporter proteins lacking the first 5′ signal sequence were constructed. Plasmid pAPBL50 encodes YopE130(Δ2–15)-Cya and plasmid pAPBL48 encodes YopE130(+1[2–15])-Cya. These proteins were not translocated into eukaryotic cells by either of these strains of Y. enterocolitica (intracellular cAMP concentration, 0.1 ± 0.1 ng of cAMP/mg). From the experiments with modified full-length YopE and YopE130-Cya, we conclude that under our experimental conditions, the proposed second secretion signal is not functional and that the only functional secretion signal for YopE is contained within amino acids or codons 1 to 15.
A secretion-inhibitory sequence localized between residues 50 and 77.
While constructing plasmids encoding YopE deleted of its SycE-binding site, we constructed pAPBG30, which encodes YopEΔ17–49 (Table 1). Like YopEΔ17–77, YopEΔ17–49 did not bind SycE in an overlay experiment, since they both lack the chaperone binding domain at amino acids 15 to 50 (Fig. 3D). Unlike YopEΔ17–77, which was efficiently secreted by Y. enterocolitica, YopEΔ17–49 was neither secreted (Fig. 3A) nor delivered into HeLa (data not shown) and Rat-I cells (Fig. 4), even though it was well produced (Fig. 3C). This suggested that the portion of YopE between amino acids 49 and 77 inhibits YopE secretion and that binding of SycE overcomes this inhibition.
To check this hypothesis, we removed residues 50 to 77 from YopE, and we monitored in vitro release of YopE in the presence and in the absence of SycE. As expected, it was released equally as well as YopEΔ17–77, and this release was independent of SycE. This contrasted with wt YopE, which was only released in the presence of SycE (Fig. 5). Thus, amino acids 50 to 77 of YopE inhibit secretion of YopE in the absence of SycE. Interestingly, although this construct does not need SycE for secretion, it still binds SycE. Thus the secretion-inhibitory domain is distinct from the minimal SycE-binding domain, although this secretion-inhibitory domain must be somehow covered by SycE.
DISCUSSION
In this paper, we have analyzed the N-terminal domain of Y. enterocolitica YopE in order to clarify its roles in the in vitro release of YopE and its delivery into eukaryotic cells.
The results, summarized in Fig. 7, confirm previous data in showing that residues 1 to 50 of YopE are required for delivery of YopE into eukaryotic cells by wt Y. enterocolitica (27, 28). However, the current results also show that delivery of YopE by Yop effector multimutant bacteria does not require amino acids 15 to 50 but rather that the secretion signal encompassing amino acids or codons 1 to 15 is sufficient. This implies that the chaperone binding domain does not need to interact with the Yop translocators for Yop effector translocation. In addition, this suggests that any protein that can be released by the Ysc secretion machinery also has the capacity to be delivered into eukaryotic cells. This conclusion hence implies a continuity between the secretion and translocation apparatuses, so that a Yop can pass through the secretion channel, syringe and needle, and then directly through the translocation apparatus into the target cell.
FIG. 7.
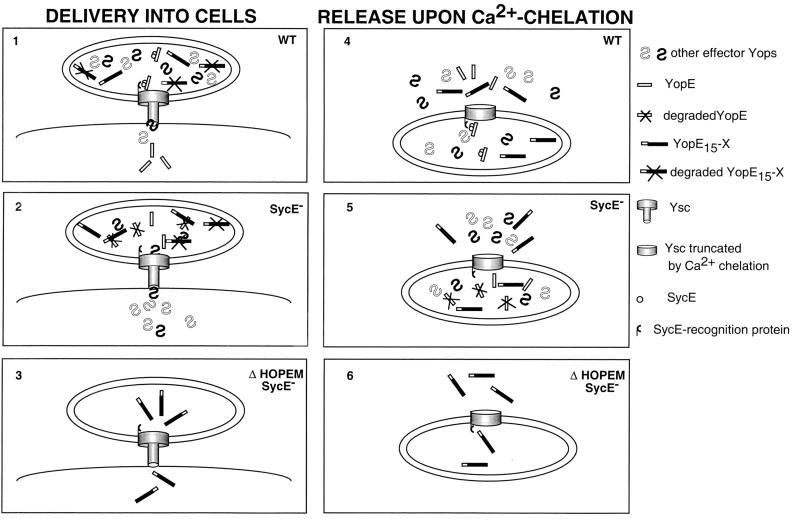
Schematic representation of the role of SycE binding to amino acids 15 to 50 of YopE in YopE delivery into eukaryotic cells and release under low-Ca2+ conditions. Y. enterocolitica bacteria are shown attached at the surface of a eukaryotic cell (panels 1, 2, and 3) or incubated under low-Ca2+ conditions (panels 4, 5, and 6). Three strains are presented: wt bacteria (panels 1 and 4), sycE mutant bacteria (2 and 5), and ΔHOPEM sycE bacteria (3 and 6). The wt bacteria synthesize full-length YopE, a YopE15-X hybrid protein, other effector Yops, and the SycE chaperone. Binding of SycE to amino acids 15 to 50 allows YopE to be delivered into cells (panel 1) or released under low-Ca2+ conditions (panel 4). YopE15-X, containing the N-terminal 5′ secretion signal but lacking the chaperone binding site, is prevented from entering eukaryotic cells (panel 1) but is nevertheless released under low-Ca2+ conditions (4). We hypothesize that competition is stronger for delivery into cells (small channel) than for release under low-Ca2+ conditions (large channel). In sycE mutant bacteria, the lack of SycE does not affect the pathway followed by YopE15-X (panels 2 and 5). However, full-length YopE is neither delivered into cells nor released under low-Ca2+ conditions. Removal of the domain encompassing amino acids 50 to 77 (not shown in this figure) allows YopE to be released independently of SycE. We conclude that this domain is inhibitory for release and that this inhibition is prevented by SycE. In ΔHOPEM sycE strains (panels 3 and 6), YopE15-X is not only released under low-Ca2+ conditions but also delivered into cells. This indicates that the N-terminal 5′ secretion signal is sufficient for delivery into cells. YopE and YopE15-X are partially degraded when blocked inside bacteria. This representation is based on the results presented in this paper and on previous results which are cited in the text.
The requirement for amino acids 15 to 50 for translocation of YopE into eukaryotic cells by wt Y. enterocolitica, but not by Yop effector multimutant bacteria, implies that these amino acids give YopE a competitive advantage over the other Yops for the Ysc secretion-translocation apparatus. Due to the competition, only the Yops that are avidly recognized by the Ysc apparatus would be successfully delivered inside eukaryotic cells. Competition between the Yops could determine the order of precedence of Yop entry into eukaryotic cells and/or the relative quantities of each Yop delivered inside a cell.
However, if there is continuity between secretion and translocation, how could one explain that domains 15 to 50 of YopE are required for translocation by wt bacteria but not for release of YopE under Ca2+-chelating conditions? This could be due to differences in the structure of the Ysc apparatus when opening is caused by Ca2+ chelation and when opening is triggered by contact with eukaryotic cells. It is possible that Ca2+ chelation shears the external part of the Ysc apparatus, resulting in a secretion channel (syringe) on the surface of the bacteria with an inner diameter that is much wider than that of the channel (needle) bridging the bacteria and the eukaryotic cell (Fig. 7). In support of this hypothesis, Ca2+ chelation leads to the release of some external parts of the Ysc apparatus, such as YscP (19, 30). Thus, passage through the secretion channel under Ca2+-chelating conditions would be far more abundant and far more permissive than upon bacteria-eukaryotic cell interaction.
In agreement with the observations of Lee et al. (14), domain encompassing amino acids 15 to 50 was not sufficient to direct YopE to the eukaryotic cytosol (14). However, unlike previous data (5), release of YopE to the extracellular milieu by this domain could not be detected, despite the use of various gene constructions, protein systems, and growth conditions. Although the same +1 frame-shift mutation of codons 2 to 15 was used here as that employed by Cheng et al. (5), in the present work the mutation was inserted in yopE and yopE130-cya, while Cheng et al. (5, 14) tested yopE-npt hybrids. This difference in protein backbone may explain the disparity of our results. In conclusion, the domain encompassing amino acids 15 to 50 is a secretion-translocation enhancer signal that is required for efficient delivery of YopE into eukaryotic cells by wt Y. enterocolitica, but it can not be considered as a physiological secretion signal.
Our results indicate that SycE plays a role as a factor introducing a hierarchical order in effector delivery, by abetting YopE to compete with the other Yops. This role should not be considered as exclusive, as SycE is required in addition when YopE contains amino acids 50 to 77. Indeed, the presence of this domain creates a need for the chaperone. This fits with older observations that bacteria missing SycE are unable to efficiently release or deliver full-length YopE or YopE130-Cya but are able to secrete YopE40-Cya (35). According to our previous observations, we suggested that it was the Syc-binding domain (residues 15 to 50) that created the need for the chaperone. The more refined present observations indicate that the secretion-inhibitory domain is localized immediately downstream of the minimal domain needed for Syc binding. Although residues 50 to 77 are neither sufficient nor necessary for SycE binding, they are likely to be covered by SycE. The determination of the three-dimensional structure of the YopE-SycE complex will clarify this.
The reason why residues 50 to 77 of YopE interfere with secretion of YopE is not clear. These amino acids could interfere with secretion through the Ysc machinery and/or they could affect the stability or solubility of YopE. Recently, Cheng et al. (6) have shown that SycE fused to glutathione S-transferase was unable to complement ΔSycE bacteria for delivery of YopE into eukaryotic cells, even though the SycE hybrid protein bound YopE in the bacterial cytosol and stabilized this Yop (6). These experiments support the results presented here, as they show that in addition to stabilizing YopE in the bacterial cytosol, SycE is also required for efficient Yop translocation by wt bacteria. It seems that glutathione S-transferase–SycE fusion proteins do not have this secondary function. In conclusion, the data presented in this paper present a more-complete picture of the functions of the N-terminal domains of YopE for secretion and translocation of this protein. Amino acids or codons 1 to 15 (secretion domain) are sufficient and absolutely necessary to direct translocation of YopE into eukaryotic cells by Yop effector multimutant Y. enterocolitica. Amino acids 15 to 50 bind the SycE chaperone and aid YopE to compete with the other Yops for entry into eukaryotic cells via the secretion-translocation machinery. Finally, amino acids 49 to 77 are inhibitory to YopE secretion, and this inhibition is overcome by binding of SycE to amino acids 15 to 50. Future crystallography studies of YopE alone and in complex with SycE will be very beneficial to the further studies of these domains, as would detailed studies on the other Yop-Syc interactions. It will be of great interest to investigate whether these other combinations have properties similar to those of YopE and SycE described here.
ACKNOWLEDGMENTS
We thank D. Desnoeck for excellent technical assistance and N. Grosdent for assistance with plasmid constructions. We also thank S. Tötemeyer, C. Geuijen, S. Bleves, N. Sauvonnet, and I. Stainier for discussions and a critical reading of the manuscript. In addition, we are grateful to Cecile Geuijen for plasmid pYOB2, Marie-Paule Sory and Corinne Kerbouch for plasmid pMSL56 and strain MRS40(pMSK50), and Maite Iriarte for pIM153.
A.P.B. was the recipient of an H. and A. Brenninkmeijer ICP fellowship and also received funding from EU TMR Programme Research Network contract FMRX-CT98-0164. This work was supported by the Belgian Fonds National de la Recherche Scientifique Médicale (Convention 3.4595.97), the Direction générale de la Recherche Scientifique-Communauté Française de Belgique (Action de Recherche Concertée 94/99-172) and the Interuniversity Poles of Attraction Program—Belgian State, Prime Minister's Office, Federal Office for Scientific, Technical and Cultural affairs (PAI 4/03).
REFERENCES
- 1.Anderson D M, Schneewind O. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science. 1997;278:1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- 2.Anderson D M, Schneewind O. Yersinia enterocolitica type III secretion: an mRNA signal that couples translation and secretion of YopQ. Mol Microbiol. 1999;31:1139–1148. doi: 10.1046/j.1365-2958.1999.01254.x. [DOI] [PubMed] [Google Scholar]
- 3.Boland A, Cornelis G R. Role of YopP in suppression of tumor necrosis factor alpha release by macrophages during Yersinia infection. Infect Immun. 1998;66:1878–1884. doi: 10.1128/iai.66.5.1878-1884.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boland A, Sory M-P, Iriarte M, Kerbourch C, Wattiau P, Cornelis G R. Status of YopM and YopN in the Yersinia Yop virulon: YopM of Y. enterocolitica is internalized inside the cytosol of PU5-1.8 macrophages by the YopB, D, N delivery apparatus. EMBO J. 1996;15:5191–5201. [PMC free article] [PubMed] [Google Scholar]
- 4a.Boyd, A. P., N. Grosdent, S. Tötemeyer, C. Geuijen, S. Bleves, M. Iriarte, I. Lambermont, J.-N. Octave, and G. R. Gornelis.Yersinia enterocolitica can deliver Yop proteins into a wide range of cell types: development of a delivery system for heterologous proteins. Eur. J. Cell Biol., in press. [DOI] [PubMed]
- 5.Cheng L W, Anderson D M, Schneewind O. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol Microbiol. 1997;24:757–765. doi: 10.1046/j.1365-2958.1997.3831750.x. [DOI] [PubMed] [Google Scholar]
- 6.Cheng L W, Schneewind O. Yersinia enterocolitica type III secretion. On the role of SycE in targeting YopE into HeLa cells. J Biol Chem. 1999;274:22102–22108. doi: 10.1074/jbc.274.31.22102. [DOI] [PubMed] [Google Scholar]
- 7.China B, Michiels T, Cornelis G R. The pYV plasmid of Yersinia encodes a lipoprotein, YlpA, related to TraT. Mol Microbiol. 1990;4:1585–1593. doi: 10.1111/j.1365-2958.1990.tb02070.x. [DOI] [PubMed] [Google Scholar]
- 8.Cornelis G R, Boland A, Boyd A P, Geuijen C, Iriarte M, Neyt C, Sory M-P, Stainier I. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev. 1998;62:1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelis G R, Vanooteghem J C, Sluiters C. Transcription of the yop regulon from Y. enterocolitica requires trans acting pYV and chromosomal genes. Microb Pathog. 1987;2:367–379. doi: 10.1016/0882-4010(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 10.Frithz-Lindsten E, Rosqvist R, Johansson L, Forsberg A. The chaperone-like protein YerA of Yersinia pseudotuberculosis stabilizes YopE in the cytoplasm but is dispensible for targeting to the secretion loci. Mol Microbiol. 1995;16:635–647. doi: 10.1111/j.1365-2958.1995.tb02426.x. [DOI] [PubMed] [Google Scholar]
- 10a.Galan J E, Collmer A. Type III secretion machines: bacterial devices for protein delivery into host cells. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 11.Håkansson S, Galyov E E, Rosqvist R, Wolf-Watz H. The Yersinia YpkA Ser/Thr kinase is translocated and subsequently targeted to the inner surface of the HeLa cell plasma membrane. Mol Microbiol. 1996;20:593–603. doi: 10.1046/j.1365-2958.1996.5251051.x. [DOI] [PubMed] [Google Scholar]
- 12.Håkansson S, Schesser K, Persson C, Galyov E E, Rosqvist R, Homble F, Wolf-Watz H. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 1996;15:5812–5823. [PMC free article] [PubMed] [Google Scholar]
- 13.Kaniga K, Delor I, Cornelis G R. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 14.Lee V T, Anderson D M, Schneewind O. Targeting of Yersinia Yop proteins into the cytosol of HeLa cells: one-step translocation of YopE across bacterial and eukaryotic membranes is dependent on SycE chaperone. Mol Microbiol. 1998;28:593–601. doi: 10.1046/j.1365-2958.1998.00822.x. [DOI] [PubMed] [Google Scholar]
- 15.Lee V T, Schneewind O. Type III machines of pathogenic yersiniae secrete virulence factors into the extracellular milieu. Mol Microbiol. 1999;31:1619–1629. doi: 10.1046/j.1365-2958.1999.01270.x. [DOI] [PubMed] [Google Scholar]
- 16.Michiels T, Cornelis G R. Secretion of hybrid proteins by the Yersinia Yop export system. J Bacteriol. 1991;173:1677–1685. doi: 10.1128/jb.173.5.1677-1685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mills S D, Boland A, Sory M-P, Van Der Smissen P, Kerbourch C, Finlay B B, Cornelis G R. YopP, a novel Yersinia effector protein, is delivered into macrophages to induce apoptosis. Proc Natl Acad Sci USA. 1997;94:12638–12643. doi: 10.1073/pnas.94.23.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neyt C, Cornelis G R. Insertion of a Yop translocation pore into the macrophage plasma membrane: requirement for YopB and YopD, but not LcrG. Mol Microbiol. 1999;33:971–981. doi: 10.1046/j.1365-2958.1999.01537.x. [DOI] [PubMed] [Google Scholar]
- 19.Payne P L, Straley S C. YscP of Yersinia pestis is a secreted component of the Yop secretion system. J Bacteriol. 1999;181:2852–2862. doi: 10.1128/jb.181.9.2852-2862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Persson C, Nordfelth R, Holmström A, Håkansson S, Rosqvist R, Wolf-Watz H. Cell-surface-bound Yersinia translocate the protein tyrosine phosphatase YopH by a polarized mechanism into the target cell. Mol Microbiol. 1995;18:135–150. doi: 10.1111/j.1365-2958.1995.mmi_18010135.x. [DOI] [PubMed] [Google Scholar]
- 21.Rosqvist R, Forsberg A, Rimpiläinen M, Bergman T, Wolf-Watz H. The cytotoxic protein YopE of Yersinia obstructs the primary host defence. Mol Microbiol. 1990;4:657–667. doi: 10.1111/j.1365-2958.1990.tb00635.x. [DOI] [PubMed] [Google Scholar]
- 22.Rosqvist R, Forsberg A, Wolf-Watz H. Intracellular targeting of the Yersinia YopE cytotoxin in mammalian cells induces actin microfilament disruption. Infect Immun. 1991;59:4562–4569. doi: 10.1128/iai.59.12.4562-4569.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosqvist R, Magnusson K E, Wolf-Watz H. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 1994;13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Sarker M R, Neyt C, Stainier I, Cornelis G R. The Yersinia Yop virulon: LcrV is required for extrusion of the translocators YopB and YopD. J Bacteriol. 1998;180:1207–1214. doi: 10.1128/jb.180.5.1207-1214.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarker M R, Sory M-P, Boyd A P, Iriarte M, Cornelis G R. LcrG controls internalization of Yersinia Yop effector proteins into eukaryotic cells. Infect Immun. 1998;66:2976–2979. doi: 10.1128/iai.66.6.2976-2979.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schesser K, Frithz-Lindsten E, Wolf-Watz H. Delineation and mutational analysis of the Yersinia pseudotuberculosis YopE domains which mediate translocation across bacterial and eukaryotic cellular membranes. J Bacteriol. 1996;178:7227–7233. doi: 10.1128/jb.178.24.7227-7233.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sory M-P, Boland A, Lambermont I, Cornelis G R. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc Natl Acad Sci USA. 1995;92:11998–12002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sory M-P, Cornelis G R. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol Microbiol. 1994;14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 30.Stainier, I., S. Bleves, C. Josenhans, L. Karmani, C. Kerbourch, I. Lambermont, S. Tötemeyer, A. Boyd, and G. R. Cornelis. YscP, a Yersinia protein required for Yop secretion that is surface exposed, and released in low Ca2+. Mol. Microbiol., in press. [DOI] [PubMed]
- 31.Von Pawel-Rammingen U, Telepnev M V, Schmidt G, Aktories K, Wolf-Watz H, Rosqvist R. GAP activity of the Yersinia YopE cytotoxin specifically targets the Rho pathway: a mechanism for disruption of action microfilament structure. Mol Microbiol. 2000;36:737–748. doi: 10.1046/j.1365-2958.2000.01898.x. [DOI] [PubMed] [Google Scholar]
- 32.Wattiau P, Bernier B, Deslee P, Michiels T, Cornelis G R. Individual chaperones required for Yop secretion by Yersinia. Proc Natl Acad Sci USA. 1994;91:10493–10497. doi: 10.1073/pnas.91.22.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wattiau P, Cornelis G R. SycE, a chaperone-like protein of Yersinia enterocolitica involved in the secretion of YopE. Mol Microbiol. 1993;8:123–131. doi: 10.1111/j.1365-2958.1993.tb01209.x. [DOI] [PubMed] [Google Scholar]
- 34.Woestyn S, Allaoui A, Wattiau P, Cornelis G R. YscN, the putative energizer of the Yersinia Yop secretion machinery. J Bacteriol. 1994;176:1561–1569. doi: 10.1128/jb.176.6.1561-1569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woestyn S, Sory M-P, Boland A, Lequenne O, Cornelis G R. The cytosolic SycE and SycH chaperones of Yersinia protect the region of YopE and YopH involved in translocation across eukaryotic cell membranes. Mol Microbiol. 1996;20:1261–1271. doi: 10.1111/j.1365-2958.1996.tb02645.x. [DOI] [PubMed] [Google Scholar]