Abstract
Penicillin-induced killing and murein hydrolase activity in Staphylococcus aureus are dependent on a variety of regulatory elements, including the LytSR two-component regulatory system and the virulence factor regulators Agr and Sar. The LytSR effects on these processes can be explained, in part, by the recent finding that a LytSR-regulated operon, designated lrgAB, affects murein hydrolase activity and penicillin tolerance. To examine the regulation of lrgAB expression in greater detail, we performed Northern blot and promoter fusion analyses. Both methods revealed that Agr and Sar, like LytSR, positively regulate lrgAB expression. A mutation in the agr locus reduced lrgAB expression approximately sixfold, while the sar mutation reduced lrgAB expression to undetectable levels. cis-acting regulatory elements involved in lrgAB expression were identified by fusing various fragments of the lrgAB promoter region to the xylE reporter gene and integrating these constructs into the chromosome. Catechol 2,3-dioxygenase assays identified DNA sequences, including an inverted repeat and intrinsic bend sites, that contribute to maximal lrgAB expression. Confirmation of the importance of the inverted repeat was achieved by demonstrating that multiple copies of the inverted repeat reduced lrgAB promoter activity, presumably by titrating out a positive regulatory factor. The results of this study demonstrate that lrgAB expression responds to a variety of positive regulatory factors and suggest that specific DNA topology requirements are important for optimal expression.
Peptidoglycan or murein hydrolases are a family of enzymes that catalyze the cleavage of specific structural components of the bacterial cell wall. These enzymes are involved in several important physiological processes, including peptidoglycan turnover and recycling, cell wall expansion during bacterial growth, septation, and daughter cell separation (13, 32). Due to the potential of these enzymes to compromise cell wall integrity, leading to cell lysis (autolysis), murein hydrolase activity must be carefully regulated. This regulation includes transcriptional control, blocked access to the specific substrate, inhibition by choline or teichoic acid, and substrate modification (13, 32).
The Staphylococcus aureus lytS and lytR genes, whose products are members of the two-component regulator family of proteins, are involved in the control of murein hydrolase activity (3). A lytS mutant phenotypically displays altered murein hydrolase activity and an increased rate of penicillin- and Triton X-100-induced lysis (3, 11). Immediately downstream of lytS and lytR are two genes, lrgA and lrgB, whose transcription is dependent upon lytS and lytR. LrgA and LrgB show no sequence similarity to known murein hydrolases. Instead, LrgA has structural characteristics in common with the bacteriophage-encoded holin proteins involved in murein hydrolase export (4, 11). Recent data generated in our laboratory indicate that the LrgA and LrgB gene products inhibit extracellular murein hydrolase activity and increase tolerance to penicillin (11). Based on these data, it was proposed that the function of LrgA, and possibly LrgB, is analogous to that of an antiholin, i.e., blocking murein hydrolase access to the substrate peptidoglycan (1).
In another study, we have shown that the staphylococcal virulence factor regulators Agr and Sar affect the rate of penicillin-induced lysis and killing (7). Mutations within the agr and sar genes result in decreased and increased rates of penicillin-induced lysis, respectively, possibly as a result of the different effects that these mutations have on murein hydrolase activity. In contrast, both mutant strains exhibited increased sensitivity to the bactericidal effects of penicillin, including the agr mutant, which exhibited reduced penicillin-induced lysis. These data demonstrate that, in addition to controlling virulence factor expression, Agr and Sar affect murein hydrolase activity and penicillin tolerance.
In the present study, we have extended our analysis of lrgAB regulation. We show that Agr and Sar, in addition to the LytSR regulatory system, positively regulate lrgAB expression. We also identified and characterized lrgAB cis-acting elements, including a region of intrinsic DNA bending (curvature), which contribute to maximal lrgAB expression. These data suggest that the effects of Agr and Sar on penicillin tolerance involve the regulation of lrgAB expression.
MATERIALS AND METHODS
Strains and growth conditions.
S. aureus strains (Table 1) were routinely cultivated in tryptic soy broth (Difco Laboratories, Detroit, Mich.) or NZY broth (3% casein enzymatic hydrolysate [Sigma Chemical Co., St. Louis, Mo.], 1% yeast extract [Fisher Scientific, Fair Lawn, N.J.] adjusted to pH 7.5). Escherichia coli DH5α (14) was grown in Luria-Bertani medium (Fisher Scientific). Liquid cultures were grown with shaking at 250 rpm and 37°C. The antibiotics used were purchased from either Sigma Chemical Co. or Fisher Scientific and used at the following concentrations: ampicillin, 50 μg/ml; chloramphenicol, 5 μg/ml; tetracycline, 3 μg/ml; kanamycin, 50 μg/ml; spectinomycin, 50 μg/ml.
TABLE 1.
Plasmids and bacterial strains used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| S. aureus strains | ||
| RN4220 | Highly transformable strain | 18 |
| RN6390 | Wild-type S. aureus laboratory strain | 22 |
| RN6911 | agr null mutant; Tcr | 16 |
| 8325-4 | Wild-type S. aureus laboratory strain | 22 |
| KB300 | lytS mutant; Emr | 3 |
| ALC488 | sar mutant; Emr | |
| RN6390Δagr-1 | agrACDB and partial RNAIII deletion | Gift from John Iandolo |
| KB700 | RN6390 geh::pDF17 | This study |
| KB701 | RN6390 geh::pDF32 | This study |
| KB705 | RN6390 geh::pDF34 | This study |
| KB706 | RN6390 geh::pDF35 | This study |
| KB707 | RN6390 geh::pDF36 | This study |
| KB708 | RN6390 Δagr-1 geh::pDF17 | This study |
| KB709 | ALC488 geh::pDF17 | This study |
| CYL316 | RN4220 containing plasmid pYL112Δ19 expressing int gene | 18 |
| Plasmids | ||
| pCR100 | pUC18 derivative containing xylE gene | Gift from Ron Yasbin |
| pLC4 | E. coli-S. aureus shuttle vector containing xylE gene | 25 |
| pCL84 | S. aureus integration vector containing L54a phage attP site | 18 |
| pDF4 | pLC4 containing AB396 lrgAB promoter fragment | This study |
| pDF16 | pCL84 derivative containing xylE reporter | This study |
| pDF17 | pDF16 containing AB396 fragment fused to xylE | This study |
| pDF32 | pDF16 containing AB396ΔIR fused to xylE | This study |
| pDF34 | pDF16 containing Δ42 fragment fused to xylE | This study |
| pDF35 | pDF16 containing Δ85 fragment fused to xylE | This study |
| pDF36 | pDF16 containing Δ147 fragment fused to xylE | This study |
| pMK4 | High-copy E. coli-S. aureus shuttle plasmid | 33 |
| pDF28 | pMK4 containing AB396 | This study |
| pDF29 | pMK4 containing AB396ΔIR | This study |
DNA manipulations.
S. aureus genomic DNA was isolated using the method of Dyer and Iandolo (6). Plasmid DNA purification was performed using the Wizard Plus kits from Promega, Inc. (Madison, Wis.). The restriction enzymes and T4 DNA ligase used in this study were purchased from GIBCO-BRL (Gaithersburg, Md.). Preparation and transformation of E. coli DH5α were accomplished as described by Inoue et al. (14). Electroporation of DNA into S. aureus was carried out using the procedure of Kraemer and Iandolo (17). S. aureus φ11-mediated transduction was performed using the method of Shafer and Iandolo (30).
Northern blot analysis.
S. aureus RNA was isolated using the procedure described by Hart et al. (12). A lrgA-specific probe was PCR amplified using the primers 5′-CCAGCACACTTTTTTCACC-3′ and 5′-GGTGCTTGGCTAATGACACC-3′, producing a 260-bp fragment. The PCR products were gel purified and radiolabeled with [α-32P]ATP by the random priming method as described previously (28). Northern blot analysis was performed as described by Sambrook et al. (28).
lrgAB promoter fusion construction.
To construct a single-copy reporter gene vector, the xylE reporter gene encoding the catechol 2,3-dioxygenase enzyme was PCR amplified from the plasmid pCR100. This was achieved using the M13 reverse primer as the 3′ primer and a 5′ primer designated EX (5′-CCTGAAT TCATGACTCGAGAAGAGGTGACGTCATGAACAAAGGTGTAATGCG ACC-3′) containing EcoRI and XhoI restriction sites (underlined). The amplified product was digested with EcoRI and BamHI and directionally ligated into the integration vector pCL84 (18). The resulting plasmid, designated pDF16, contained a promoterless xylE gene downstream of two EcoRI and XhoI restriction sites available for cloning. To examine specific sequences involved in lrgAB expression, PCR amplification was employed to generate consecutively smaller lrgAB promoter fragments. Four different 5′ oligomers that anneal to selected regions upstream of the lrgA gene (5′-CCTGTTGAAATTGAATTCAAAATTCACATGTTAAAGC-3′, 5′-AATGGTGTCAGAATTCAAGTTGGACGTCCA-3′, 5′-TACTTTAACAGGAATTCTTTTTTTTATGC-3′, and 5′-TTGTATTCGAATTCAAATCACGCAAATCG-3′) were used in separate reactions with the same 3′ oligomer (5′-CCGGAAGCTTGTGCTGGTTTTGATGCGTC-3′). All four 5′ oligomers generate an EcoRI restriction site (underlined), while the 3′ oligomer produces a HindIII site beginning at position +60 with respect to the transcription start site. The PCRs produced products designated AB396, Δ147, Δ85 and Δ42, respectively, where the names refer to the 5′ base present in each fragment with respect to the transcription start site of the lrgA gene (see Fig. 1). These fragments were digested with EcoRI and HindIII and directionally cloned into the polylinker of pBluescriptSK(+) (Stratagene), transformed into DH5α, and sequenced using the method developed by Sanger et al. (29). Each fragment was liberated from pBluescriptSK(+) by sequential restriction digestion using EcoRI and XhoI and ligated in front of the xylE gene in pDF16, producing plasmids pDF17, pDF36, pDF35, and pDF34, respectively (Table 1). These plasmids (along with pDF16) were electroporated into S. aureus CYL316, where they specifically integrated into the geh locus encoding lipase (18). The integrated loci were transferred into RN6390 by φ11-mediated transduction, producing strains designated KB700, KB707, KB706, and KB705 that were used for reporter gene assays (Table 1). Proper integration of the promoter fusion constructs was confirmed by Southern blot analysis and/or PCR analysis using the 5′ primer employed for the production of the promoter fragment along with a second oligomer (5′-GTTCTGCACCTTTACGTTG-3′) that is complementary to a DNA sequence downstream of the attB integration site within the geh locus (19).
FIG. 1.
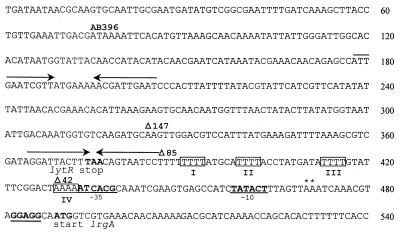
Nucleotide sequence of the S. aureus lrgAB promoter region. The potential ribosome binding sites and the −10 and −35 promoter elements are underlined and in bold. The asterisks mark the two transcription start sites identified previously (4). The translational stop site for the lytR gene and the translational start site for the lrgA structural gene are in bold. The four boxed sequences highlight AT tracts, and inverted repeat sequences are indicated by arrows.
Reporter gene assays.
Assays of catechol 2,3-dioxygenase activity were performed by growing S. aureus strains containing the reporter gene in 20 ml of NZY medium for 12 h (stationary phase). A 10-ml sample was removed, and the cells were pelleted by centrifugation at 4,000 × g. The cell pellet was washed with 10 ml of TES buffer (50 mM Tris-HCl [pH 7.5], 10 mM EDTA, 30 mM NaCl) and pelleted a second time. The cell pellet was resuspended in 2 ml of lysis buffer (100 mM potassium acetate, 10% acetone, 20 mM EDTA), and cells were lysed by incubation at 37°C in the presence of lysostaphin (AMBI Inc., Tarrytown, N.Y.) at 50 μg/ml. The cell lysate was homogenized by repeated passage through an 18-gauge needle, and then the cellular debris was removed by centrifugation at 27,000 × g. The clarified lysate was assayed for catechol 2,3-dioxygenase activity as described by Zukowski et al. (37). Specific activity units are defined as milliunits of the product generated divided by the total cellular protein used in each reaction mixture (1 mU corresponds to the formation of 1 nmol of 2-hydroxymuconic semialdehyde per min at 37°C). Total cellular protein concentration was determined using a Bradford protein assay kit (Bio-Rad, Richmond, Calif.) with bovine serum albumin as the standard.
Two-dimensional polyacrylamide gel electrophoresis.
A two-dimensional electrophoresis assay to assess the presence of intrinsic DNA bending was performed as described by Rohde et al. (26). Briefly, the far left lane(s) of a 5% nondenaturing polyacrylamide gel was loaded with 1 μg of a 1-kb DNA ladder (GIBCO-BRL) along with a mixture of EcoRI- and HindIII-digested plasmid clones that liberated different fragments originating from the lrgAB promoter region. The fragments were separated in the first dimension at 65°C for 30 min at 100 V. The gel was then rotated 90°, and the fragments were separated in the second dimension by electrophoresis at 4°C for 5 h at 80 V. The DNA fragments were stained with ethidium bromide and then visualized with UV light.
Site-directed mutagenesis.
A PCR-based strategy described by Chen and Przybyla (5) was used to generate site-directed changes in the lrgAB promoter region. The 3′ primers 5′-TAGGTAAAATGCATAAGCTAAAAAAAAGGATTACTGTTAAAG-3′, 5′-TAGGTAAAATGCAGCTCTAGACATAAAAAAAAGGATTACTGTTAAAG-3′, and 5′-GGTAAATGCATAAAAAAAATCGACGCTTTAAAATC-3′ were designed to generate lrgAB promoter fragments containing 5- and 10-bp insertions and the inverted repeat deletion, respectively (the 5- and 10-bp insertions and the point of deletion in the primers are underlined). These primers were used in PCRs along with a common 5′ primer, 5′-CCTGTTGAAATTGAATTCAAAATTCACATGTTAAAGC-3′. The PCR fragments generated were used in a second round of amplification along with the downstream 3′ primer 5′-CCGGAAGCTTGTGCTGGTTTTGATGCGTC-3′, which amplifies the entire lrgAB promoter region.
RESULTS
Regulatory elements involved in lrgAB expression.
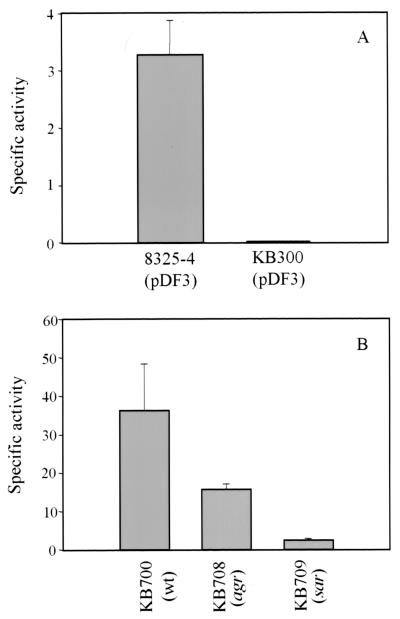
In previous studies, it was shown that a lytS mutation resulted in altered murein hydrolase activity, increased Triton X-100- and penicillin-induced lysis, and undetectable lrgAB transcription (3, 4, 11). Given that the lrgAB operon is located immediately downstream of the lytSR locus (Fig. 1), the possibility that the lytS mutation has a polar effect on lrgAB could not be ruled out. Thus, to examine the effects of LytSR on lrgAB expression in greater detail, an lrgAB-xylE promoter fusion construct was introduced into the lytS mutant KB300 (3) and the parental strain 8325-4, which were assayed for catechol 2,3-dioxygenase activity (Fig. 2A). The cloned lrgAB promoter region produced 3.27 ± 0.60 U of catechol 2,3-dioxygenase specific activity in 8325-4. In contrast, KB300 produced undetectable levels of catechol 2,3-dioxygenase activity. These data demonstrate that the gene products produced by the lytSR operon activate lrgAB transcription in trans, either directly or indirectly.
FIG. 2.
Catechol 2,3-dioxygenase assays. (A) Reporter gene assays of 8325-4 (wild-type [wt]) and KB300 (lytSR mutant) strains containing the plasmid reporter gene pDF3 (Table 1). (B) Enzyme activity detected in the virulence regulator mutants using the integrated reporter gene system with the full-length promoter (AB396). RN6390, wild type; RN6390Δagr-1, agr mutant; ALC488, sar mutant. Control strain RN6390, containing promoterless reporter gene plasmid pDF16, did not produce detectable enzyme activity. The specific activities shown are averages of three independent experiments reported in milliunits (1 mU equals 1 nmol of 2-hydroxymuconic semialdehyde min−1 mg of total protein−1).
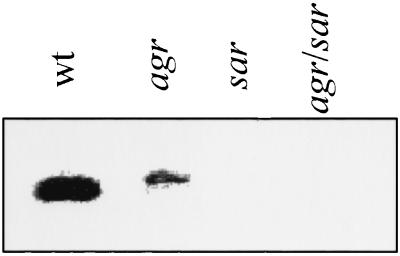
In addition to the LytSR regulatory system, we have also shown that the S. aureus Agr and Sar virulence regulators affect murein hydrolase activity and the rates of penicillin-induced lysis and killing of S. aureus cells (7). In light of the recent finding that the lrgAB operon increases tolerance to penicillin and inhibits murein hydrolase activity (11), it is possible that Agr and Sar influence these processes via lrgAB. To test this hypothesis, Northern blot analyses were used to compare lrgAB mRNA levels in agr and sar mutants and the parental wild-type strain. Using a lrgAB-specific probe, the level of lrgAB transcription was monitored in late-exponential-phase cells, when the lrgAB operon is maximally expressed (11). As shown in Fig. 3, the agr mutant RN6911 produced a greater-than-sixfold decrease in the lrgAB transcript compared to the wild-type strain. The sar and agr sar mutant strains produced undetectable levels of the lrgAB transcript. Thus, Agr and Sar, in addition to LytSR, are involved in the regulation of lrgAB expression.
FIG. 3.
Northern blot analysis. Total RNA was isolated from S. aureus strains RN6390 (wild type [wt]), RN6911 (agr), ALC136 (sar), and ALC135 (agr sar), and 10-μg samples were separated in a 1% formaldehyde gel. Hybridization was performed using an lrgA-specific probe.
To further examine the agr and sar effects on lrgAB expression, we generated a promoter fusion vector, pDF16, that takes advantage of the pCL84 plasmid integration system (18). This allowed the integration of lrgAB promoter fusions, in single copy, into the bacteriophage L54a attB site within the staphylococcal chromosome, thus giving a better representation of promoter activity compared to plasmid-based reporter gene systems. Plasmid pDF17 (containing a 456-bp lrgAB promoter fragment designated AB396; Fig. 1) was integrated into the chromosome of S. aureus CYL316 and then transduced into wild-type strain RN6390, agr mutant strain RN6390Δagr-1, and sar mutant strain ALC488, producing strains KB700, KB708, and KB709, respectively. As shown in Fig. 2B, the agr mutant displayed a greater-than-twofold reduction in reporter gene activity (15.59 ± 1.41 U of specific activity) compared to RN6390 (36.21 ± 12.16 U of specific activity). The sar mutant exhibited a 15-fold decrease in reporter gene activity (2.12 ± 0.75 U of specific activity) compared to RN6390. The results of these promoter fusion studies are consistent with those of the above-described Northern analysis, suggesting that the AB396 promoter fragment contains all of the sequences necessary for normal lrgAB regulation.
Identification of the lrgAB promoter cis-acting elements.
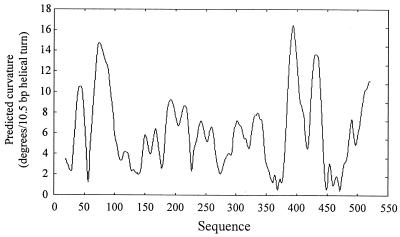
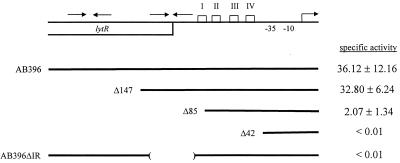
Inspection of the lrgAB promoter region (Fig. 1) revealed the presence of two inverted repeat sequences, centered at nucleotides (nt) 192 and 375, that are potential binding sites for regulatory proteins. Furthermore, four homopolymeric AT tracts (Fig. 1) are located immediately upstream of the −35 and −10 hexamers. Koo et al. (15) determined that homopolymeric AT tracts at least 4 bp long positioned in phase with the helical screw contribute to overall DNA bending. As shown in Fig. 1, AT tracts I and II are in phase with the helical turn and are thus a potential site containing intrinsic DNA curvature. Tracts III and IV are also nearly in phase and may also contain intrinsic curvature. In support of this, the Bend-It software created by Gabrielian et al. (8) to predict DNA curvature predicts that both of these sites contain intrinsic DNA curvature (Fig. 4). To examine the potential role of all these sequences in lrgAB regulation, three consecutively smaller deletions (designated Δ147, Δ85, and Δ42; Fig. 5) of the lrgAB promoter region were made, cloned in front of the xylE gene in pDF16, and inserted into the RN6390 chromosome. The Δ147 construct, which excluded the inverted repeat sequence centered at nt 192, produced 32.80 ± 6.24 U of enzyme specific activity. This level of activity is similar to that produced by the fusion strain containing AB396 (36.12 ± 12.16 U of specific activity), suggesting that the sequences upstream of nt 324 (including the inverted repeat centered at nt 192) do not affect lrgAB expression under these conditions. However, the Δ85 construct, which excludes the inverted repeat sequence centered on nt 375, produced 15-fold less enzyme activity (2.3 ± 1.8 U of specific activity) compared to AB396, indicating that the sequence between nt 324 and 386 (including the inverted repeat sequence) is required for optimal lrgAB expression. The Δ42 construct, which eliminates the potential intrinsic bend sites, produced undetectable levels of reporter gene activity. Since the removal of the sequences spanning the inverted repeat sequence centered on nt 375 significantly decreased reporter gene activity, a full-length promoter fusion construct in which this inverted repeat sequence was specifically deleted was generated and designated AB396ΔIR. The AB396ΔIR fragment was ligated into pDF16 and integrated into the RN6390 chromosome, producing strain KB701. Reporter gene activity was undetectable in this strain. These experiments show that cis-acting regulatory elements controlling lrgAB expression, including the inverted repeat centered at position 375, are contained within approximately 134 nt upstream of the transcription start site.
FIG. 4.
DNA curvature prediction. Predicted bending or curvature of the lrgAB promoter region as determined by the Bend-It program constructed by Gabrielian et al. (8) (http://www2.icgeb.trieste.it/∼dna/cgi-bin/BendIt2) using a 20-bp window.
FIG. 5.
Deletion analysis. Promoter fusion analysis of four consecutively smaller fragments of the lrgAB promoter and the internal deletion fragment of the inverted repeat centered on nt −96 analyzed using the integrated reporter gene construct. The specific activity values shown are averages of at least three independent experiments and are in milliunits (1 mU equals 1 nmol of 2-hydroxymuconic semialdehyde min−1 mg of total protein−1).
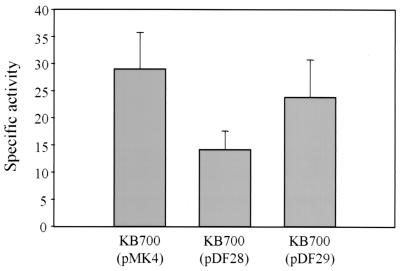
Several studies have used multiple copies of operator sites to titrate host-encoded trans-acting proteins (23, 27). We took a similar approach to further establish the role of the inverted repeat in the activation of lrgAB expression. Specifically, we subcloned the AB396 fragment and the identical fragment containing a specific deletion of the inverted repeat region, AB396ΔIR, into high-copy-number plasmid pMK4, producing plasmids pDF28 and pDF29, respectively. These plasmids were introduced into RN6390 containing the integrated reporter gene construct, producing strain KB700. As shown in Fig. 6, the introduction of pDF28 resulted in a twofold decrease in catechol 2,3-dioxygenase activity (14.17 ± 3.40 U of specific activity) compared to the same strain with pMK4 (28.94 ± 6.80 U of specific activity). In contrast, the strain containing pDF29 produced reporter gene activity (23.81 ± 6.93 U of specific activity) similar to that observed with the pMK4-containing strain.
FIG. 6.
Operator titration experiment using strain KB700 containing high-copy-number plasmids pMK4, pDF28, and pDF29. The specific activities shown are in milliunits (1 mU equals 1 nmol of 2-hydroxymuconic semialdehyde min−1 mg of total protein−1) and are averages of three independent experiments.
Two-dimensional gel electrophoresis.
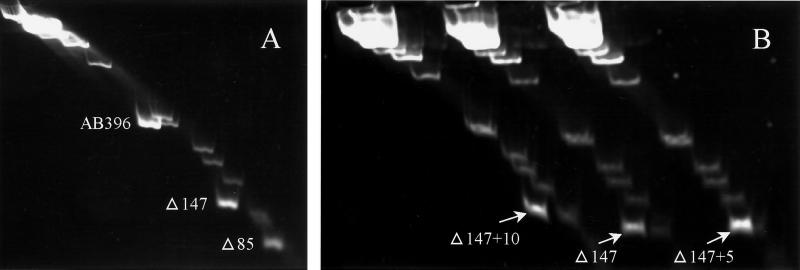
As described above, the Δ42 construct, which lacks the potential bend sites upstream of the −10 and −35 elements, produced no detectable reporter gene activity, indicating that these sequences affect lrgAB expression. To test if this promoter region contains intrinsic DNA bending, a two-dimensional gel electrophoresis assay was performed (Fig. 7A). First, the AB396, Δ147, and Δ85 promoter fragments were mixed with a 1-kb ladder and separated in a 5% polyacrylamide gel at 65°C. In this dimension, the rate of DNA migration was a function of size alone. Next, the gel was rotated 90° and the DNA fragments were separated at 4°C. During electrophoresis in this dimension, the migration rate of the DNA fragments is a function of size and secondary structure (intrinsic bending). As shown in Fig. 7A, significant retardation of the migration rate in the second dimension of the AB396 and Δ147 fragments and, to a lesser extent, the Δ85 fragment was observed. These data indicate that the lrgAB promoter region does contain intrinsic DNA bending, as predicted by DNA sequencing and computer analysis. Furthermore, the increased rate of migration of the Δ85 fragment compared to the AB396 and Δ147 fragments suggests that the bend is centered near the 5′ end of Δ85, which encompasses the AT tract sites.
FIG. 7.
Two-dimensional gel electrophoresis assays. (A) lrgAB promoter region fragments AB396, Δ147, and Δ85. (B) The Δ147 fragment and Δ147 fragments containing the 5- and 10-bp insertions between AT tracts I and II. Fragments are labeled Δ147, Δ147+5, and Δ147+10, respectively.
To confirm that localized bending was associated with AT tracts I and II, we generated fragments with 5- and 10-bp insertions between these AT tracts within the Δ147 fragment (Fig. 1). The 5-bp insertion represents the addition of a half-helical turn, while the 10-bp insertion adds a full turn. As shown in Fig. 7B, the 10-bp insertion had a retarded mobility rate similar to that of the wild-type sequence, indicating that it contains intrinsic bends. In contrast, the insertion of 5 bp displayed an increased rate of migration compared to the wild-type sequence, similar to the 1-kb ladder DNA, suggesting that the fragment had reduced intrinsic bending. These results strongly support the hypothesis that the AT tracts confer intrinsic bending and indicate that the altered migration rate due to bending is phase dependent and positioned near the 5′ end of the Δ85 fragment. Both the 5- and 10-bp insertions in the AB396 fragment abolish lrgAB promoter activity when these promoter alterations are fused with xylE (data not shown).
DISCUSSION
In a previous study, the global virulence regulator loci agr and sar were shown to affect murein hydrolase activity and tolerance to penicillin-induced lysis (7). Both the agr and sar mutant strains were found to be more sensitive to penicillin-induced lysis and killing compared to the parental strain (7). A more recent study demonstrated that the lrgAB gene products function to decrease extracellular murein hydrolase activity and increase tolerance to penicillin (11). Based on these findings, we hypothesized that the virulence regulators Agr and Sar may affect the expression of the lrgAB operon. Indeed, the results of promoter fusion (Fig. 2B) and Northern analyses (Fig. 3) demonstrate that this is the case. The agr mutation decreased the level of lrgAB expression detected approximately two- to sixfold, while the sar mutation reduced lrgAB expression to nearly undetectable levels. These data suggest that SarA controls lrgAB expression through an Agr-independent pathway. Furthermore, results from our laboratory also indicate that the Agr and Sar effects on lrgAB expression are mediated through a LytSR-independent pathway (unpublished data).
Inspection of the lrgAB promoter region revealed several potential cis-acting regulatory sequences, including two inverted repeat sequences and several AT tracts phased with the DNA helical turn positioned upstream of the −35 and −10 promoter elements. To determine if these sequences are involved in lrgAB expression, we constructed a set of nested deletions spanning the lrgAB promoter region and fused each fragment to the xylE reporter gene. These constructs were integrated into the S. aureus chromosome, and reporter gene activity was assayed. This analysis revealed that the inverted repeat sequence centered on nt 375 and the AT tracts present upstream of the −35 hexamer are important for maximal lrgAB expression. To examine the inverted repeat sequence in greater detail, we produced a promoter fragment (AB396ΔIR) that was identical to AB396 except for a specific deletion of this inverted repeat sequence. This fragment failed to produce detectable enzyme activity using the integrated reporter gene system.
To examine the function of the inverted repeat sequence further, we performed a titration experiment by subcloning the AB396 and AB396ΔIR fragments into a high-copy-number plasmid and introducing them into an S. aureus strain containing the integrated AB396 reporter gene fusion (KB700). Multiple copies of the full-length promoter sequence significantly decreased the level of chromosomally encoded reporter gene activity. These data suggest that the inverted repeat is a specific binding site for an activator protein, although plasmid copy number differences could also impact these results. However, this construct did not completely abolish detectable reporter gene activity. One explanation for this observation is that the promoter topology within the plasmid may not accurately reflect the DNA structure of the promoter region as it exists within the chromosome. Thus, the promoter sequence located within the chromosome may display higher affinity and outcompete the plasmid-encoded promoter sequence for trans-acting factors required for lrgAB expression. The 10-fold increase in promoter activity obtained using the integrated reporter gene construct (Fig. 2) supports this idea.
The results of two-dimensional gel electrophoresis analysis show that the lrgAB promoter also contains a region of intrinsic DNA curvature. Using the deletion fragments generated for the promoter fusion analysis, intrinsic DNA curvature was localized between the inverted repeat sequence centered on nt 375 and the −35 hexamer. This is based on the observation that the mobility of the Δ85 fragment was not affected by low temperature as much as the other promoter fragments were. It has been shown that when a bend occurs at the center of a DNA molecule it will affect the shape of that molecule to a greater degree than if the bend occurs near an end (9). The results of the two-dimensional gel assay (Fig. 7A) indicate that the intrinsic bend site is located near the end of the Δ85 fragment, near AT tracts I and II. To confirm this, we created 5- and 10-bp insertions between AT tracts I and II in the Δ147 fragment. The 5-bp insertion (which inserts half of a helical turn) would be predicted to disrupt the overall curvature of this region, producing a zigzag structure and increasing the migration rate in the second dimension compared to the wild-type fragment. As shown in Fig. 7B, this is precisely what was observed. On the other hand, the 10-bp insertion (which inserts a full helical turn) was predicted to create a deeper curve, reducing the migration rate compared to the wild-type fragment. Again, the results shown in Fig. 7B show this to be the case. When these 5- and 10-bp insertions were introduced into full-length promoter fragment AB396 and analyzed using the chromosomal reporter gene construct, no detectable enzyme activity was produced (data not shown). These data argue against an enhancer loop model of lrgAB activation in which the placement of the operator and the promoter on the same face of the helix is more important than the spacing between these cis-acting elements. On the other hand, the data suggest that the geometry and/or the position of the inverted repeat relative to the promoter is critical for lrgAB expression to occur. Alternatively, the insertions may disrupt primary structure elements necessary for binding of an activator protein.
Intrinsic DNA curvature has been implicated in the activation of transcription by promoting the juxtaposition of DNA sequences near the terminal loop of a superhelical domain (35). Yang et al. (36) suggested that the introduction of a curved DNA sequence brings about changes in the overall shape of a supercoiled polymer. These examples raise the possibility that intrinsic DNA bending influences the supercoiled topology within promoter regions. Furthermore, there are several cases in which intrinsic DNA bending and/or protein-induced bending upstream of promoters affect transcription initiation (24). Thus, it is clear that DNA bending, supercoiling, and transcription factor binding can work either in concert or independently to control gene expression.
In S. aureus, the expression of the eta gene has been shown to be enhanced by DNA relaxation (31). More recently, Morfeldt et al. (20) hypothesized that SarA binding to the agr promoter region affects the localized superhelicity of this promoter. Interestingly, SarA exhibits moderate sequence similarity to Shigella flexneri VirF (21), which has been shown to activate virB transcription by binding upstream in a DNA topology-dependent manner (34). Based on this observation, along with the results of the current study, it is possible that SarA recognizes and binds distinct topological features in the lrgAB promoter region that may lead to the generation of an active transcription complex. Recently, it has also been shown that SarA specifically binds upstream of the collagen adhesion gene (cna), controlling its expression independently of Agr (2, 10). When applying the Bend-It software to examine whether this and the agr P3 promoter also contained DNA curvature, we found that both analyses predicted DNA curvature within these promoter regions (data not shown). Thus, similar topological features may be required for the activation of other SarA-regulated promoters.
ACKNOWLEDGMENTS
This work was funded by NIH grant R29-AI38901 and NSF-Idaho EPSCoR grant EPS-9720634.
We thank Scott A. Minnich for careful review of the manuscript.
REFERENCES
- 1.Bayles K W. The bactericidal action of penicillin: new clues to an unsolved mystery. Trends Microbiol. 2000;8:280–283. doi: 10.1016/s0966-842x(00)01762-5. [DOI] [PubMed] [Google Scholar]
- 2.Blevins J S, Gillaspy A F, Rechtin T M, Hurlburt B K, Smeltzer M S. The Staphylococcal accessory regulator (sar) represses transcription of the Staphylococcus aureus collagen adhesin gene (cna) in an agr-independent manner. Mol Microbiol. 1999;33:317–326. doi: 10.1046/j.1365-2958.1999.01475.x. [DOI] [PubMed] [Google Scholar]
- 3.Brunskill E W, Bayles K W. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J Bacteriol. 1996;178:611–618. doi: 10.1128/jb.178.3.611-618.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunskill E W, Bayles K W. Identification of LytSR-regulated genes from Staphylococcus aureus. J Bacteriol. 1996;178:5810–5812. doi: 10.1128/jb.178.19.5810-5812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen B, Przybyla A E. An efficient site-directed mutagenesis method based on PCR. BioTechniques. 1994;17:657–659. [PubMed] [Google Scholar]
- 6.Dyer D W, Iandolo J J. Rapid isolation of DNA from Staphylococcus aureus. Appl Environ Microbiol. 1983;46:283–285. doi: 10.1128/aem.46.1.283-285.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujimoto D F, Bayles K W. Opposing roles of the Staphylococcus aureus virulence regulators, Agr and Sar, in Triton X-100- and penicillin-induced autolysis. J Bacteriol. 1998;180:3724–3726. doi: 10.1128/jb.180.14.3724-3726.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gabrielian A, Vlahovicek K, Pongor S. Distribution of sequence-dependent curvature in genomic DNA sequences. FEBS Lett. 1997;406:69–74. doi: 10.1016/s0014-5793(97)00236-6. [DOI] [PubMed] [Google Scholar]
- 9.Garabedian M J, LaBaer J, Liu W, Thomas J R. Analysis of protein-DNA interactions. In: Hames D B, Higgins S J, editors. Gene transcription: a practical approach. New York, N.Y: Oxford University Press; 1993. pp. 243–293. [Google Scholar]
- 10.Gillaspy A F, Lee C Y, Sau S, Cheung A L, Smeltzer M S. Factors affecting the collagen binding capacity of Staphylococcus aureus. Infect Immun. 1998;66:3170–3178. doi: 10.1128/iai.66.7.3170-3178.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groicher K H, Firek B A, Fujimoto D F, Bayles K W. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J Bacteriol. 2000;182:1794–1801. doi: 10.1128/jb.182.7.1794-1801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hart M E, Smeltzer M S, Iandolo J J. The extracellular protein regulator (xpr) affects exoprotein and agr mRNA levels in Staphylococcus aureus. J Bacteriol. 1993;175:7875–7879. doi: 10.1128/jb.175.24.7875-7879.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holtje J-V, Tuomanen E I. The murein hydrolases of Escherichia coli: properties, functions and impact on the course of infections in vivo. J Gen Microbiol. 1991;137:441–454. doi: 10.1099/00221287-137-3-441. [DOI] [PubMed] [Google Scholar]
- 14.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 15.Koo H S, Wu H M, Crothers D M. DNA bending at adenine/thymine tracts. Nature. 1986;320:501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- 16.Kornblum J, Kreiswirth B N, Projan S J, Ross H, Novick R P. Agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers, Inc.; 1990. pp. 373–402. [Google Scholar]
- 17.Kraemer G R, Iandolo J J. High-frequency transformation of Staphylococcus aureus by electroporation. Curr Microbiol. 1990;21:373–376. [Google Scholar]
- 18.Lee C Y, Buranen S L, Ye Z. Construction of single-copy integration vector for Staphylococcus aureus. Gene. 1991;103:101–105. doi: 10.1016/0378-1119(91)90399-v. [DOI] [PubMed] [Google Scholar]
- 19.Lee C Y, Iandolo J J. Lysogenic conversion of staphylococcal lipase is caused by insertion of the bacteriophage L54a genome into the lipase structural gene. J Bacteriol. 1986;166:385–391. doi: 10.1128/jb.166.2.385-391.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morfeldt E, Tegmark K, Arvidson S. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol Microbiol. 1996;21:1227–1237. doi: 10.1046/j.1365-2958.1996.751447.x. [DOI] [PubMed] [Google Scholar]
- 21.Novick R P. Pathogenicity factors and their regulation. In: Fischetti V A, Novick R P, Ferretti J J, Portnoy D A, Rood J I, editors. Gram-positive pathogens. Washington, D.C.: ASM Press; 2000. pp. 392–407. [Google Scholar]
- 22.Novick R P, Ross H F, Projan S J, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osuna R, Schwacha A, Bender R A. Identification of the hutUH operator (hutUo) from Klebsiella aerogenes by DNA deletion analysis. J Bacteriol. 1994;176:5525–5529. doi: 10.1128/jb.176.17.5525-5529.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez-Martin J, Rojo F, de Lorenzo V. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol Rev. 1994;58:268–290. doi: 10.1128/mr.58.2.268-290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ray C, Hay R E, Carter H L, Moran C P., Jr Mutations that affect utilization of a promoter in stationary-phase Bacillus subtilis. J Bacteriol. 1985;163:610–614. doi: 10.1128/jb.163.2.610-614.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohde J R, Luan X S, Rohde H, Fox J M, Minnich S A. The Yersinia enterocolitica pYV virulence plasmid contains multiple intrinsic DNA bends which melt at 37°C. J Bacteriol. 1999;181:4198–4204. doi: 10.1128/jb.181.14.4198-4204.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saint-Girons I, Fritz H J, Shaw C, Tillmann E, Starlinger P. Integration specificity of an artificial kanamycin transposon constructed by the in vitro insertion of an internal Tn5 fragment into IS2. Mol Gen Genet. 1981;183:45–50. doi: 10.1007/BF00270136. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shafer M W, Iandolo J J. Genetics of staphylococcal enterotoxin B in methicillin-resistant isolates of Staphylococcus aureus. Infect Immun. 1979;25:902–911. doi: 10.1128/iai.25.3.902-911.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheehan B J, Foster T J, Dorman C J, Park S, Stewart G S A B. Osmotic and growth-phase dependent regulation of the eta gene of Staphylococcus aureus: a role for DNA supercoiling. Mol Gen Genet. 1992;232:49–57. doi: 10.1007/BF00299136. [DOI] [PubMed] [Google Scholar]
- 32.Shockman G D, Holtje J-V. Microbial peptidoglycan (murein) hydrolases. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Vol. 27. Amsterdam, The Netherlands: Elsevier Science B.V.; 1994. pp. 131–166. [Google Scholar]
- 33.Sullivan M A, Yasbin R E, Young F E. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene. 1984;29:21–26. doi: 10.1016/0378-1119(84)90161-6. [DOI] [PubMed] [Google Scholar]
- 34.Tobe T, Yoshikawa M, Sasakawa C. Thermoregulation of virB transcription in Shigella flexneri by sensing of changes in local DNA superhelicity. J Bacteriol. 1995;177:1094–1097. doi: 10.1128/jb.177.4.1094-1097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsen H, Levene S D. Supercoiling-dependent flexibility of adenosine-tract-containing DNA detected by a topological method. Proc Natl Acad Sci USA. 1997;94:2817–2822. doi: 10.1073/pnas.94.7.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Y, Westcott T P, Pedersen S C, Tobias I, Olson W K. Effects of localized bending on DNA supercoiling. Trends Biochem Sci. 1995;20:313–319. doi: 10.1016/s0968-0004(00)89058-1. [DOI] [PubMed] [Google Scholar]
- 37.Zukowski M M, Gaffney D F, Speck D, Kauffmann M, Findeli A, Wisecup A, Lecocq J P. Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned Pseudomonas gene. Proc Natl Acad Sci USA. 1983;80:1101–1105. doi: 10.1073/pnas.80.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]