Abstract
Transcription of algD, encoding GDP-mannose dehydrogenase, the key enzyme in the alginate biosynthetic pathway, is highly regulated in Azotobacter vinelandii. We describe here the characterization of a Tn5 insertion mutant (AC28) which shows a higher level of expression of an algD::lacZ fusion. AC28 cells were morphologically abnormal and unable to encyst. The cloning and nucleotide sequencing of the Tn5-disrupted locus in AC28 revealed an operon homologous to the Escherichia coli ampDE operon. Tn5 was located within the ampD gene, encoding a cytosolic N-acetyl-anhydromuramyl-l-alanine amidase that participates in the intracellular recycling of peptidoglycan fragments. The ampE gene encodes a transmembrane protein, but the function of the protein is not known. We constructed strains carrying ampD or ampE mutations and one with an ampDE deletion. The strain with a deletion of the ampDE operon showed a phenotype similar to that of mutant AC28. The present work demonstrates that both alginate production and bacterial encystment are greatly influenced by the bacterial ability to recycle its cell wall.
Azotobacter vinelandii is a soil bacterium that undergoes a process of cellular differentiation to form metabolically dormant cysts resistant to desiccation (for a review, see reference 38). Wild-type strains of A. vinelandii produce the extracellular polysaccharide alginate, a linear copolymer of d-mannuronic acid and its C-5 epimer l-guluronic acid. Alginate is essential for the encystment process, since it is a component of the intine and exine layers of the cysts (31) and since nonmucoid strains fail to form cysts (5, 26, 29).
The pathway for alginate biosynthesis in A. vinelandii has been elucidated (36); fructose-6-P is converted by four enzymatic reactions to GDP-mannuronic acid, which is the substrate of the polymerase. The resultant polymannuronic acid is secreted and modified by an O-acetylase and several extracellular C-5 epimerases to give the final product, alginate (9, 36, 37).
The algD gene codes for GDP-mannose dehydrogenase, which converts GDP-mannose, a metabolite used for the synthesis of different saccharides, to GDP-mannuronic acid. algD is located in a biosynthetic gene cluster which contains most of the genes that code for the enzymes involved in the synthesis and modification of alginate (5, 23, 26, 27, 37, 42). A correlation between alginate production and algD transcription was found for three A. vinelandii strains producing different alginate levels (5).
In Pseudomonas aeruginosa, the ςE factor AlgU (also known as AlgT) controls transcription of the alginate biosynthetic operon and is negatively regulated by MucA, an inner membrane protein that has been shown to act as an antisigma factor, and MucB, a periplasmic protein proposed to sense denatured proteins in the periplasm (45). In A. vinelandii the algUmucABCD genes have been characterized and have been shown to regulate alginate production (25, 29). Transcription of the A. vinelandii algD gene is initiated from three sites (5, 29, 30); initiation from one of them, p2, requires AlgU (5, 29). Transcription of algD is also under the control of the two-component regulatory system GacSA (6).
The bacterial cell wall is composed of a heteropolymer known as murein or peptidoglycan. The peptidoglycan consists of glycan chains of alternating units of N-acetylglucosamine and N-acetylmuramic acid that are frequently cross-linked by short peptides to each other (34). Escherichia coli and presumably most other gram-negative bacteria degrade up to 50% of their murein per generation; however, most of the liberated murein fragments (1-6-anhydro-N-acetylmuramyl [MurNAc]-l-Ala-d-Glu-mA2pm) are transported from the periplasm into the cytoplasm to reutilize the tripeptide l-Ala-d-Glu-A2pm to form new murein (12, 13, 32).
In enterobacteria the ampD and ampE genes form an operon (15, 22). The ampE gene encodes an inner membrane protein with an ATP-binding motif whose function is unknown (15, 22). The ampD gene encodes a cytosolic N-acetylmuramyl-l-alanine amidase essential for murein tripeptide recycling. AmpD is specific for anhydro-MurNAc-tripeptides (anhydro-MurNAc-l-Ala-d-Glu-mA2pm) (17); thus, it distinguishes the newly synthesized peptidoglycan precursors which lack the anhydrous bond from the peptidoglycan-derived anhydromuropeptides transported back into the cell for recycling (17).
In many gram-negative bacteria, the inducible β-lactamase gene ampC is transcriptionally controlled by a regulator encoded by ampR, which belongs to the LysR family of transcriptional regulators (21). Mutations in ampD result in constitutive expression of AmpC β-lactamase even in the absence of β-lactam antibiotics. AmpR is inhibited in the presence of the main murein precursor UDP-MurNac-pentapeptide, and this inhibition is reversed by anhydro-MurNac-tripeptide (18). In ampD mutants, the substrate (anhydro-MurNac-tripeptide) accumulates and acts as an intracellular positive effector for ampC expression (16). The presence of ampD, even in bacteria lacking an inducible ampC β-lactamase gene, suggests that cell wall recycling may have a signaling role in other cellular processes (33). In agreement with this suggestion, in Bacillus subtilis, transport of wall-derived peptides into the cell has been proposed to have a signaling role in the initiation of sporulation (35).
In this study, we describe the isolation and characterization of a Tn5 insertion mutation which increases transcription of algD and, consequently, alginate production in A. vinelandii. This was accomplished by isolating a mutant derivative of strain WI12 carrying an algD::lacZ fusion that showed a deep blue color on plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The Tn5 insertion was shown to interrupt an operon homologous to E. coli ampDE. The results presented here indicate that A. vinelandii ampD and ampE gene products contribute to normal cell morphology and suggest that cell wall recycling has a signaling role for the control of alginate synthesis and cyst formation in A. vinelandii.
MATERIALS AND METHODS
Microbiological procedures.
Bacterial strains and plasmids used are listed in Table 1. Media and growth conditions were as follows. A. vinelandii was grown at 30°C in Burk's nitrogen-free salts supplemented with 2% sucrose (BS) (19). E. coli DH5α was grown on Luria-Bertani (LB) medium (28) at 37°C. Antibiotic concentrations used for A. vinelandii and E. coli, respectively, were as follows: tetracycline, 20 and 20 μg/ml; kanamycin, 2 μg/ml (not used for E. coli); ampicillin, 100 μg/ml (not used for A. vinelandii); nalidixic acid, 20 and 10 μg/ml; spectinomycin, 100 and 100 μg/ml; streptomycin, 2 and 20 μg/ml; gentamicin, 1.5 and 10 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| A. vinelandii | ||
| ATCC 9046 | Highly mucoid | ATCC |
| AEIV | Mucoid | Svein Valla |
| WI12 | algD::lacZ derivative of ATCC 9046, Kmr | 5 |
| AC28 | ampD::Tn5 derivative of WI12, Tcr | This work |
| ATD1 | ATCC 9046 with a nonpolar ampD::Gm mutation | This work |
| ATE1 | ATCC 9046 with an ampE::Sp mutation | This work |
| ATDE1 | ATCC 9046 with a ΔampDE::Gm mutation | This work |
| AED1 | AEIV with a nonpolar ampD::Gm mutation | This work |
| AEE1 | AEIV with an ampE::Sp mutation | This work |
| AEDE1 | AEIV with a ΔampDE::Gm mutation | This work |
| AE28 | AEIV carrying an ampD::Tn5 insertion, Tcr | This work |
| WID1 | WI12 with a nonpolar ampD::Gm mutation | This work |
| WIE1 | WI12 with an ampE::Sp mutation | This work |
| WIDE1 | WI12 with a ΔampDE::Gm mutation | This work |
| E. coli | ||
| DH5α | supE44 ΔlacU169 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 14 |
| Plasmids | ||
| pCP13 | RK2-derived cosmid vector; Tcr Kmr | 7 |
| pBluescript SK+ | Plasmid used for subcloning DNA; Apr | Stratagene |
| pKT230 | Broad-host-range vector; Smr Kmr | 2 |
| pUT-mini-Tn5luxAB | Suicide vector for mutagenesis with mini-Tn5luxAB; able to replicate only in strains expressing λ pir protein; Tcr | 8 |
| pRK2013 | ColE1-tra (RK2); Kmr | 11 |
| pHP45Ω-Sp | Plasmid used to provide an Ω Spr cassette | 10 |
| pBSL141 | Plasmid used to provide a Gmr cassette | 1 |
| pK28 | pBluescript SK+ with a 3.0-kb KpnI fragment containing the site of transposition in AC28; Tcr | This work |
| pSM194 | Cosmid containing 25 kb of A. vinelandii DNA including ampDE operon derived from pCP13; Tcr | This work |
| pS194.5 | pBluescript SK+ with a 2.0-kb SmaI fragment containing ampDE operon | This work |
| pS195 | pS194.5 derivative containing ampD 5′ end | This work |
| pS196 | pS194.5 derivative containing ampD gene and ampE 5′ end | This work |
| pS196.2 | pS194.5 derivative containing ampE 5′ end | This work |
| pSD1 | pS196 derivative containing ampD::Gm mutation | This work |
| pSE1 | pS196.2 derivative containing ampE::Sp mutation | This work |
| pSDE1 | pS194.5 derivative containing ΔampDE::Gm mutation | This work |
| pKT194.5 | pKT230 derivative carrying ampDE operon; Smr | This work |
Triparental and biparental matings were carried out as previously reported (19). A. vinelandii transformation was carried out as described by Bali et al. (3).
β-Galactosidase activity was measured as reported by Miller (28); 1 U corresponds to 1 nmol of o-nitrophenyl-β-d-galactoside hydrolyzed per min per μg of protein. Protein was determined by the Lowry method (24). All measurements were done in triplicate.
Alginate production was determined as previously described (26), and all determinations were done in triplicate.
Encystment was determined as previously described (5) by measuring the percentage of cells resistant to desiccation after induction with n-butanol.
Transposon mutagenesis.
Random transposon mutagenesis of WI12 was carried out as described previously with a pUT derivative containing the mini-Tn5-luxAB transposon (8). Matings were done on Burk's sucrose (BS)-LB medium plates overnight at 30°C. The cells were resuspended in 10 mM MgSO4 and plated on BSTcNal plates, and 200 WI12 Tcr derivatives were isolated from each mating.
Cloning of the AC28 transposon insertion.
Southern blot analysis was used to determine suitable restriction fragments on which the transposon-inactivated locus in strain AC28 was to be recovered. We identified a 3-kb KpnI fragment which contained the Tcr cassette derived from the transposon (data not shown). KpnI-digested genomic DNA from AC28 was then size fractionated by agarose gel electrophoresis, purified from the gel, and ligated into pBluescript KS+ (Stratagene), resulting in plasmid pK28. Ligations were transformed into E. coli DH5α, and transformed cells were plated on LB medium containing tetracycline.
Nucleic acid procedures.
RNA and DNA isolation, cloning, Southern blotting, and nick translation procedures were carried out as described previously (39). Plasmids pS194.5, pS195, pS196, and pS196.2 (Fig. 1; Table 1) were used to determine the nucleotide sequence reported in this study. DNA sequencing was done with the Taq FS DNA polymerase and fluorescent dideoxy terminators using a cycle sequencing method. The precise site of transposition in AC28 was determined by nucleotide sequencing of pK28 across the transposon insertion junction. Primer extension of algD was carried out as previously described (5) using a primer extension kit (Amersham) as instructed by the manufacturer.
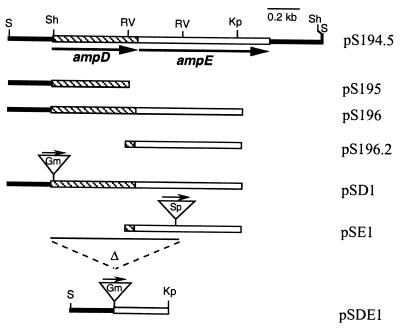
FIG. 1.
Physical map of the A. vinelandii ampDE region and the plasmids constructed in this study. Arrows, direction of transcription; triangles, antibiotic resistance cassettes. Abbreviations: Kp, KpnI; RV, EcoRV; S, SmaI; Sh, SphI.
Construction of plasmid pKT194.5 carrying the A. vinelandii ampDE operon.
Cosmid clone pSM194, derived from an A. vinelandii genomic library, was shown to contain the ampDE operon in a 2-kb SmaI fragment. This fragment was cloned into the pBluescript KS+ vector to yield plasmid pS194.5 (Fig. 1). The ampDE operon was released from this construction as a BamHI-HindIII fragment and cloned into the pKT230 vector, previously digested with BamHI and HindIII endonucleases, producing plasmid pKT194.5.
Construction of ampD::Gm, ampE::Sp, and ΔampDE::Gm mutants.
Plasmid pS196 was used to introduce into the unique SphI site of ampD a 0.8-kb SmaI fragment with a gentamicin resistance (Gmr) cassette (1), and plasmid pSD1 with the Gmr cassette ligated in the same orientation as that for the ampD transcription was isolated (Fig. 1), generating an ampD::Gm nonpolar mutation. Plasmid pS196.2 contains a unique EcoRV site within ampE. Therefore, in order to construct an ampE::Sp mutation, plasmid pS196.2 was cleaved with EcoRV and the 2-kb SmaI fragment containing an Ω spectinomycin resistance cassette (10) was inserted to create pSE1. To generate an ampDE::Gm deletion mutant, plasmid pS196 was cleaved with SphI and EcoRV (releasing the entire ampD gene as well as the 5′ end of ampE), blunt ended, and ligated to a 0.8-kb SmaI fragment containing a gentamicin resistance cassette, resulting in pSDE1. Plasmids pSD1, pSE1, and pSDE1 (Fig. 1), which were unable to replicate in A. vinelandii, were used to introduce the ampD::Gm, ampE::Sp, and ΔampDE::Gm mutations into strains ATCC 9046, AEIV, and WI12. Transformants were selected using the corresponding antibiotic and were confirmed by Southern blot analysis to carry the desired mutations (data not shown).
Microscopic analysis.
A. vinelandii cells, grown for 48 h on BS plates, were resuspended, mounted between two coverslips, and visualized under a microscope (Eclipse 330; Nikon) with a ×60 air lens (numerical aperture, 0.7) (Nikon Inc., Melville, N.Y.). Cell morphology was observed using differential interference contrast microscopy, and image acquisition was made by using a Sensys chip-cooled device (CCD) camera (Photometrics). The CCD camera was driven with PMIS processing software (Photometrics), and acquired images were processed using Adobe Photoshop (version 3.0).
Nucleotide sequence accession number.
The A. vinelandii ampDE operon sequence reported here has been assigned GenBank accession no. AF237388.
RESULTS
Isolation of strain AC28.
In A. vinelandii the synthesis of alginate correlates well with the transcription of the gene algD (5). We hypothesized that mutations in genes whose products regulate transcription of algD can be identified by a loss or increase in the β-galactosidase activity of a strain carrying an algD::lacZ transcriptional fusion such as WI12. Random Tn5 mutagenesis of strain WI12 was carried out as described in Materials and Methods. Tcr derivatives (3,200) were isolated and screened for a deep blue color on BS plates containing X-Gal. Five derivatives that were bluer than WI12 were identified (data not shown). Derivative AC28, which showed the deepest blue phenotype, was chosen for further analysis.
Cloning and DNA sequence.
The sequence contiguous to the Tn5 insertion in AC28 was cloned to produce plasmid pK28. The DNA sequence of pK28 revealed homology to those of the P. aeruginosa and E. coli ampD genes. Southern blot analysis with pK28 as the probe led to the identification, in an A. vinelandii genomic library, of cosmid pSM194 carrying a 2-kb SmaI fragment with the ampD wild-type sequence. Sequence analysis of this DNA fragment revealed two open reading frames, one encoding a 187-amino-acid polypeptide (AmpD) and the second encoding a 280-amino-acid protein (AmpE). Potential Shine-Dalgarno sequences (AGGAG and GGGAG) are present upstream of both the ampD and ampE start codons. As in other bacteria, such as E. coli and P. aeruginosa, the ampE start codon overlaps the ampD TGA termination codon, suggesting that, as in P. aeruginosa and E. coli, these two genes form an operon (15, 20, 22). The predicted A. vinelandii AmpD and AmpE proteins exhibited 70 and 57% identity, respectively, to their homologues in P. aeruginosa.
β-Galactosidase activity of strain AC28.
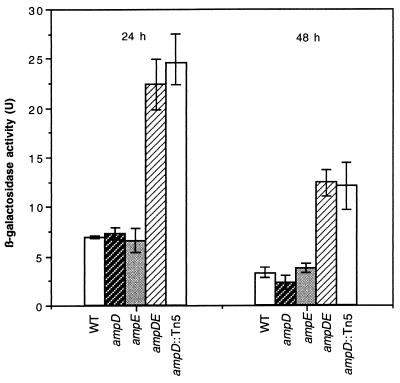
To confirm and evaluate the effect of the ampD::Tn5 mutation on the transcription of algD, we compared the β-galactosidase activity of strain WI12 with that of strain AC28 during growth in BS. As shown in Fig. 2, transcription of algD in AC28, measured as β-galactosidase activity, was threefold higher than that in strain WI12.
FIG. 2.
algD transcription measured as β-galactosidase activity in strains WI12 (wild type [WT]), WID1 (ampD), WIE1 (ampE), WIDE1 (ampDE), and AC28 (ampD::Tn5). Cells were grown on BS solid medium and were harvested at the indicated times.
Transcription of algD in ampD and ampE mutants.
Since ampD and ampE genes seem to be arranged as an operon transcribed from a promoter located upstream ampD, it seemed likely that the ampD::Tn5 mutation in AC28 was polar on ampE expression. To investigate whether polarity on ampE was contributing to the increase in algD transcription, we constructed WI12 derivatives carrying a nonpolar ampD::Gm mutation (WID1) and an ampE::Sp mutation (WIE1) and strain WIDE1 with an ampDE deletion. Similar to strain AC28, strain WIDE1 showed a deep blue color on X-Gal plates and had increased levels of β-galactosidase activity. In contrast, the ampD and ampE single mutations did not affect the transcription of algD (Fig. 2). These results indicate a cooperative effect of both mutations on algD transcription.
Production of alginate.
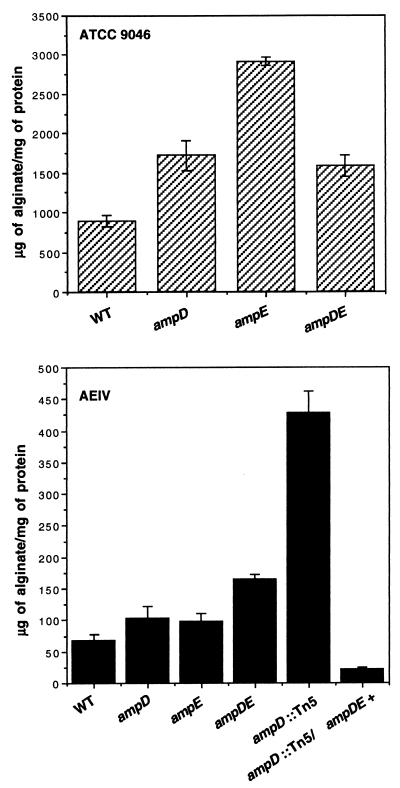
In strain WI12 alginate production is reduced 75% compared to that in its parental strain, ATCC 9046; this reduction is likely due to the fact that the lacZ insertion is located on the 3′ end of the algD gene, resulting in a GDP-mannose dehydrogenase with a reduced activity (5). To determine if the increased AC28 β-galactosidase activity shown above correlated with the production of alginate, ATCC 9046 and AEIV derivatives carrying the ampD::Gm, ampE::Sp, and ΔampDE::Gm mutations were constructed. The ampD::Tn5 insertion from AC28 was transferred by transformation to strain AEIV, thus creating an AE28 mutant. The resulting strains were found to produce more alginate than their parental strains (Fig. 3); however, the extent of such an increase depended on the strain tested. Several attempts were made to transfer the ampD::Tn5 mutation to the highly mucoid strain ATCC 9046 with no success. The AE28 strain exhibited a higher alginate production level than strain AEDE1, which carries a nonpolar ΔampDE::Gm mutation, whereas AE28 carries a polar Tn5 mutation; therefore, polarity on transcription of a gene downstream of ampE also affecting alginate production could explain this difference. Plasmid pKT194.5, which carries a copy of the A. vinelandii ampDE operon, was transferred to strain AE28. As shown in Fig. 3, strain AE28/pKT194.5 produced alginate levels similar to that produced by wild-type AEIV, indicating that the high level of alginate production observed in strain AE28 was due to the absence of ampDE gene products.
FIG. 3.
Alginate production in ampD, ampE, ΔampDE, and ampD::Tn5 derivatives of strains ATCC 9046 (top) and AEIV (bottom). Cells were grown on solid BS medium for 48 h. Alginate was separated from the cells and measured as described in Materials and Methods. WT, wild type.
The ampD::Tn5 mutation increases transcription of algD from its three promoters.
As in strain ATCC 9046, transcription of algD in strain WI12 is initiated from three sites (Fig. 4). We tested whether the increase in algD transcription in strain AC28 was caused by specifically increasing initiation from one of these promoters. Primer extension analysis using RNA isolated from strains WI12 and AC28 was carried out. As shown in Fig. 4, the ampD::Tn5 mutation increased transcription of algD from its three promoters.
FIG. 4.
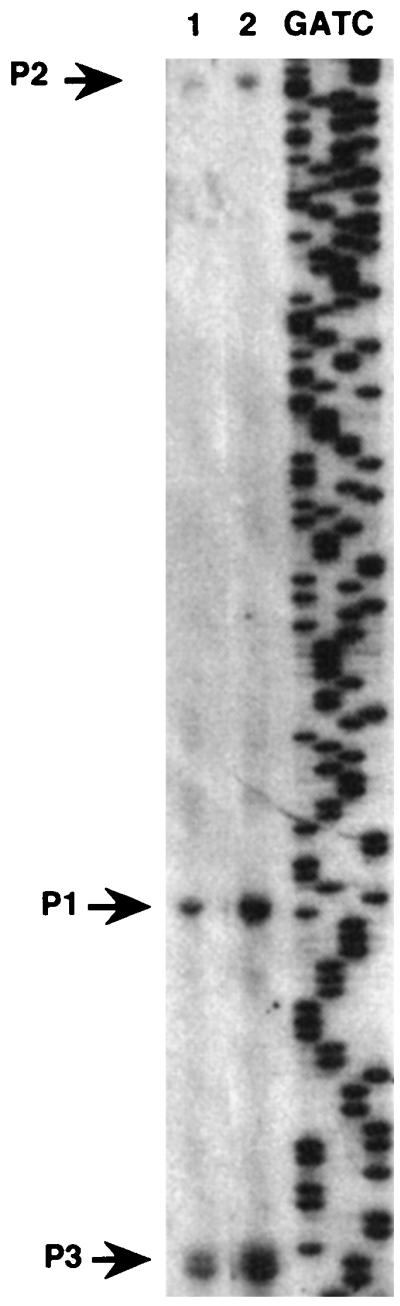
Primer extension analysis of algD transcription in strains WI12 (lane 1) and AC28 (lane 2). RNA (50 μg) isolated from bacterial cultures grown for 48 h in Burk's medium supplemented with 2.0% sucrose was used. Arrows, transcription initiation sites from the three algD promoters.
Effect of ampD and ampE mutations on the morphology of A. vinelandii.
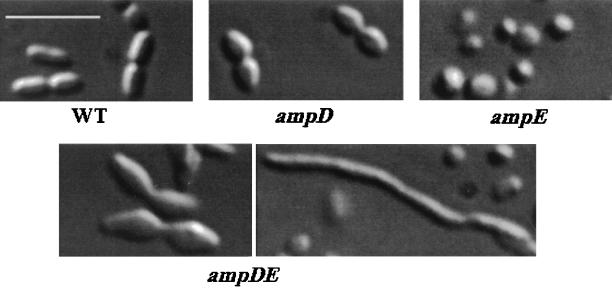
The bacterial cell wall determines the shape of the cell (34). As ampD mutants are impaired in peptidoglycan recycling, the effect of the ampD and ampE mutations on cell shape was investigated. Microscopic examination of strains WID1, WIE1, WIDE1, and AC28 grown on BS plates was carried out. As shown in Fig. 5, the parental WI12 strain cells had a normal rod shape and were 3 to 5 μm long. The nonpolar ampD mutation produced cells similar to those found in WI12, whereas the ampE mutation produced spherical cells. Cultures derived from AC28 and WIDE1 mutants showed a heterogeneous cell morphology; they produced filamented and spherical cells. In addition they produced cells whose poles have an arrow shape, and the septs between dividing cells are abnormal, suggesting a failure to produce a normal septum (Fig. 5). The effect of the ampDE mutations on cell morphology was less dramatic when the mutants were grown on liquid medium (data not shown).
FIG. 5.
Effect of mutations in the ampDE operon on cell morphology in A. vinelandii. All photographs were captured with a ×60 air lens. Bar, 10 μm. WT, wild type.
Effect of the ampDE mutation on encystment.
Transport of wall-derived peptides into the cell has been proposed to have a signaling role in the initiation of sporulation in B. subtilis (35). We tested the effect of the ampDE mutation on encystment. The formation of desiccation-resistant cells in WI12 and AC28 cultures was measured. Strain WI12 has a frequency of cyst formation of about 5.0%, while mutant AC28 has a frequency of less than 10−6. After induction, vegetative cells divide to give small rounded cells that eventually become mature cysts. As expected, WI12 cells induced for encystment on n-butanol plates produced small rounded cells; in contrast, AC28 cells induced for encystment had the abnormal morphology found in vegetative cultures of this mutant (data not shown).
DISCUSSION
The aim of this study was to identify genes whose products participate in the control of alginate production. The isolation of mutants in which algD transcription was increased led us to the identification of the A. vinelandii ampD and ampE genes. In P. aeruginosa and other gram-negative bacteria, the ampDE operon participates in a pathway for recycling the cell wall. This recycling pathway was proposed to regulate other functions besides β-lactamase induction (33, 34).
This study presents evidence suggesting that, in A. vinelandii, peptidoglycan recycling participates in the regulation of alginate production. Similar to the effect of the ampDE mutation on transcription of ampC, inactivation of the ampDE operon in A. vinelandii resulted in an increase in algD transcription accompanied by an increase in alginate production. Neither ampD nor ampE single mutations seemed to affect algD transcription although they resulted in an increased alginate production. Similarly, in an algR mutant, alginate production was reduced 50% despite the fact that algD transcription was not affected (30). This suggests the presence of another regulated point, besides the algD promoter, which also determines the extent of alginate production. The ampD and ampE mutations could affect this regulation point, resulting in higher alginate levels.
In A. vinelandii the increase in algD expression observed in the ampDE mutants might be caused by accumulation of the AmpD substrate. We propose that inactivation of ampE also contributes to the accumulation of the anhydromuropeptide in the cytoplasm. This hypothesis implies that AmpE also participates in the peptidoglycan recycling pathway. The fact that ampE and ampD genes constitute an operon supports this hypothesis. AmpE might be involved in regulating the extent of cell wall breakdown, slowing down the lytic process by controlling the activity of lytic transglycosylases. In E. coli three lytic transglycosylases and two endopeptidases are present in the periplasm or bound to the cytoplasmic membrane with the active sites oriented toward the periplasm (40). If AmpE was involved in cell wall breakdown, lysis of the cell wall would increase in the absence of AmpE, with the subsequent increase of anhydromuropeptides in the cytoplasm, which in the absence of AmpD accumulate and serve as a regulatory signal.
The ampDE mutation increased the transcription of algD from its three initiation sites. We have recently reported that the two-component global regulatory system GacSA also controls algD transcription from its three sites (6); thus, it is possible that the GacSA system participates in the signal transduction pathway turned on in the ampDE mutants. Accumulation of murein precursors in the A. vinelandii ampDE mutants might increase algD transcription directly or indirectly by activating an AmpR-like regulator.
This study shows that mutations in the ampDE operon also affected cell shape. We found that the ampE mutation produced rounded cells. In E. coli, strains with mutations in rodA-pbpA or the mre region (41, 43) are defective in the elongation process and grow as spherical cells, suggesting that in the ampE mutant cell elongation may be affected. The ampDE mutations produced, in addition to rounded cells, filamented cells of different lengths, a phenotype similar to those of E. coli pbpB mutants. In E. coli, PBP3, the product of gene pbpB, is involved in septum formation (4). Temperature-sensitive pbpB mutants stop dividing at high temperature and form long filaments (44). Nothing is known about the regulatory mechanisms controlling cell wall elongation and septum formation; however, it was suggested that changes in the cytoplasmic concentration of anhydromuropeptides could play a role (33). In this context, the morphological aberrations observed in the double mutant ampDE could be due to the presence of high levels of anhydromuropeptide which, in turn, deregulates the events of cell wall elongation and septum formation. Since the cell wall determines the shape of a cell (34), this result indicates that cell wall recycling has important implications in determining cell shape.
Upon induction of encystment, A. vinelandii vegetative cells divide to give two small rounded cells, which are later surrounded by an inner coat, the intine, and a thick laminated exine (38). Alginate is a component of the intine and exine layers. A. vinelandii algU or algK mutants are unable to produce alginate and therefore to form a mature cyst; upon encystment, these mutants form rounded cells lacking the intine and exine layers (26, 29). A mutation in algR did not impair alginate production but caused a cyst-defective phenotype, producing cysts lacking the intine layer (30). The ampDE mutant reported here is impaired in encystment. Upon conditions for induction of encystment, cells of this mutant remained as vegetative cells. Thus, encystment seems to be blocked at the first step. This result suggests that the level of muropeptides in the cytoplasm could be a signal necessary for A. vinelandii cells to undergo the differentiation process.
Finally, alginate synthesis also occurs independently of encystment; this polysaccharide is normally produced by vegetative cells not undergoing this differentiation process. This indicates that these two cellular processes can be separated and is in agreement with the finding that the ampDE mutation enhanced alginate synthesis but abolished encystment.
ACKNOWLEDGMENTS
This work was supported by grant 27767-N from CONACyT to G.E. The microscopic study was carried out with an equipment acquired with grants 27698-N and 27640-N from CONACyT and grant IN212298 from DGAPA to F. Sánchez and C. Quinto. L.C. was supported by installation grant for young scientists I29972-N from CONACyT.
We thank J. Guzmán for technical support and O. Geiger and M. Villanueva for reviewing the manuscript.
REFERENCES
- 1.Alexeyev M F, Shokolenko I, Croughan T P. Improved antibiotic-resistance gene cassettes and omega elements for Escherichia coli vector construction and in vitro deletion/insertion mutagenesis. Gene. 1995;160:63–67. doi: 10.1016/0378-1119(95)00108-i. [DOI] [PubMed] [Google Scholar]
- 2.Bagdasarian M, Lurz L, Ruckert B, Franklin F C, Bagdasarian M M, Frey J, Timmis K N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF 1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 3.Bali A, Blanco G, Hill S, Kennedy C. Excretion of ammonium by a nifL mutant of Azotobacter vinelandii fixing nitrogen. Appl Environ Microbiol. 1992;58:1711–1718. doi: 10.1128/aem.58.5.1711-1718.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Botta G A, Park J T. Evidence for involvement of penicillin-binding protein 3 in murein synthesis during septation but not during cell elongation. J Bacteriol. 1981;145:333–340. doi: 10.1128/jb.145.1.333-340.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campos M E, Martínez-Salazar J M, Lloret L, Nuñez C, Espín G, Soberón-Chávez G. Cloning and characterization of the gene coding for GDP-mannose dehydrogenase (algD) from Azotobacter vinelandii. J Bacteriol. 1996;178:1793–1799. doi: 10.1128/jb.178.7.1793-1799.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castañeda M, Guzmán J, Moreno S, Espín G. GacS sensor kinase regulates alginate and poly-β-hydroxybutyrate production in Azotobacter vinelandii. J Bacteriol. 2000;182:2624–2628. doi: 10.1128/jb.182.9.2624-2628.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darzins A, Chakrabarty A M. Cloning of genes controlling alginate biosynthesis from a mucoid cystic fibrosis isolate of Pseudomonas aeruginosa. J Bacteriol. 1984;159:9–19. doi: 10.1128/jb.159.1.9-18.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lorenzo V, Herrero M, Jakubzik V, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ertesvag H, Hoidal H K, Hals I K, Rian A, Doseth B, Valla S. A family of modular type mannuronan C-5-epimerase genes controls alginate structure in Azotobacter vinelandii. Mol Microbiol. 1995;16:719–731. doi: 10.1111/j.1365-2958.1995.tb02433.x. [DOI] [PubMed] [Google Scholar]
- 10.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 11.Figurski D, Helinski R D. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodell E W. Recycling of murein by Escherichia coli. J Bacteriol. 1985;163:305–310. doi: 10.1128/jb.163.1.305-310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodell E W, Schwarz U. Release of cell wall peptides into culture medium by exponentially growing Escherichia coli. J Bacteriol. 1985;162:391–397. doi: 10.1128/jb.162.1.391-397.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanahan D. Studies on transformation of E. coli. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 15.Honoré N, Nicolas M H, Cole S T. Regulation of enterobacterial cephalosporinase production: the role of a membrane bound sensory transducer. Mol Microbiol. 1989;3:1121–1130. doi: 10.1111/j.1365-2958.1989.tb00262.x. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs C, Huang L, Bartowsky E, Normark S, Park J T. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for β-lactamase induction. EMBO J. 1994;13:4684–4694. doi: 10.1002/j.1460-2075.1994.tb06792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs C, Joris B, Jamin M, Klarsov K, Beeumen J V, Mengin-Lecreulx D, van Heijenoort J, Park J T, Normark S, Frère J M. AmpD, essential for both β-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-l-alanine amidase. Mol Microbiol. 1995;15:553–559. doi: 10.1111/j.1365-2958.1995.tb02268.x. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs C, Frère J M, Normark S. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible β-lactam resistance in gram-negative bacteria. Cell. 1997;88:823–832. doi: 10.1016/s0092-8674(00)81928-5. [DOI] [PubMed] [Google Scholar]
- 19.Kennedy C, Gamal R, Hummprey R, Ramos J, Brigle K, Dean D. The nifH, nifM, and nifN genes of Azotobacter vinelandii: characterization by Tn5 mutagenesis and isolation from pLARF1 gene bank. Mol Gen Genet. 1986;205:318–325. [Google Scholar]
- 20.Langaee T Y, Dargis M, Huletasy A. An ampD gene in Pseudomonas aeruginosa encodes a negative regulator of AmpC β-lactamase expression. Antimicrob Agents Chemother. 1998;42:3296–3300. doi: 10.1128/aac.42.12.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindberg F, Lindquist S, Normark S. Inactivation of the ampD gene causes semiconstitutive overproduction of the inducible Citrobacter freundii β-lactamase. J Bacteriol. 1987;169:1923–1928. doi: 10.1128/jb.169.5.1923-1928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindquist S, Galleni M, Lindberg F, Normark S. Signalling proteins in enterobacterial AmpC β-lactamase regulation. Mol Microbiol. 1989;3:1091–1102. doi: 10.1111/j.1365-2958.1989.tb00259.x. [DOI] [PubMed] [Google Scholar]
- 23.Lloret L, Barreto R, León R, Moreno S, Martínez-Salazar J, Espín G, Soberón-Chávez G. Genetic analysis of the transcriptional arrangement of Azotobacter vinelandii alginate biosynthetic genes: identification of two independent promoters. Mol Microbiol. 1996;21:449–457. doi: 10.1111/j.1365-2958.1996.tb02554.x. [DOI] [PubMed] [Google Scholar]
- 24.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 25.Martínez-Salazar J M, Moreno S, Nájera R, Boucher C, Espín G, Soberón-Chávez G, Deretic V. Characterization of the genes coding for the putative sigma factor AlgU and its negative regulators MucA, MucB, MucC, and MucD in Azotobacter vinelandii and evaluation of their role in alginate biosynthesis. J Bacteriol. 1996;178:1800–1808. doi: 10.1128/jb.178.7.1800-1808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mejía-Ruíz H, Moreno S, Guzmán J, Nájera R, León R, Soberón-Chávez G, Espín G. Isolation and characterization of an Azotobacter vinelandii algK mutant. FEMS Microbiol Lett. 1997;156:101–106. doi: 10.1111/j.1574-6968.1997.tb12712.x. [DOI] [PubMed] [Google Scholar]
- 27.Mejía-Ruíz H, Guzmán J, Moreno S, Soberón-Chávez G, Espín G. The Azotobacter vinelandii alg8 and alg44 genes are essential for alginate synthesis and can be transcribed from an algD-independent promoter. Gene. 1997;199:271–277. doi: 10.1016/s0378-1119(97)00380-6. [DOI] [PubMed] [Google Scholar]
- 28.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. pp. 431–435. [Google Scholar]
- 29.Moreno S, Guzmán J, Nájera R, Soberón-Chávez G, Espín G. Role of the alternative ςE factor AlgU in encystment of Azotobacter vinelandii. J Bacteriol. 1998;180:2766–2769. doi: 10.1128/jb.180.10.2766-2769.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Núñez C, Moreno S, Soberón-Chávez G, Espín G. The Azotobacter vinelandii response regulator AlgR is essential for cyst formation. J Bacteriol. 1999;181:141–148. doi: 10.1128/jb.181.1.141-148.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Page W J, Sadoff H L. Relationship between calcium and uronic acids in the encystment of Azotobacter vinelandii. J Bacteriol. 1975;122:145–151. doi: 10.1128/jb.122.1.145-151.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park J T. Turnover and recycling of the murein sacculus in oligopeptide permease-negative strains of Escherichia coli: indirect evidence for an alternative permease system and monolayered sacculus. J Bacteriol. 1993;175:7–11. doi: 10.1128/jb.175.1.7-11.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park J T. Why does Escherichia coli recycle its wall peptides? Mol Microbiol. 1995;17:421–426. doi: 10.1111/j.1365-2958.1995.mmi_17030421.x. [DOI] [PubMed] [Google Scholar]
- 34.Park J T. The murein sacculus. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1996. pp. 48–57. [Google Scholar]
- 35.Perego M, Higgins C F, Pearce S R, Gallagher M P, Hoch J A. The oligopeptide transport system of Bacillus subtillis plays a role in the initiation of sporulation. Mol Microbiol. 1991;5:173–185. doi: 10.1111/j.1365-2958.1991.tb01838.x. [DOI] [PubMed] [Google Scholar]
- 36.Pindar D F, Bucke C. The biosynthesis of alginic acid by Azotobacter vinelandii. Biochem J. 1975;152:617–622. doi: 10.1042/bj1520617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rehm H A, Ertesvag H, Valla S. A new Azotobacter vinelandii mannuronan C-5 epimerase gene (algG) is part of an alg gene cluster physically organized in a manner similar to that in Pseudomonas aeruginosa. J Bacteriol. 1996;178:5884–5889. doi: 10.1128/jb.178.20.5884-5889.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sadoff H L. Encystment and germination in Azotobacter vinelandii. Bacteriol Rev. 1975;39:516–539. doi: 10.1128/br.39.4.516-539.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 40.Shockman G D, Höltje J V. Bacterial cell wall. New Compr Biochem. 1994;27:131–166. [Google Scholar]
- 41.Spratt B G, Boyd A, Stoker N. Defective and plaque forming lambda transducing bacteriophage carrying penicillin-binding protein-cell shape genes: genetic and physical mapping and identification of gene products from the lip-dacA-rodA-pbpA-leuS region of Escherichia coli chromosome. J Bacteriol. 1980;143:569–581. doi: 10.1128/jb.143.2.569-581.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vazquez A, Moreno S, Guzmán J, Alvarado A, Espín G. Transcriptional organization of the Azotobacter vinelandii algGXLVIFA genes: characterization of algF mutants. Gene. 1999;232:217–222. doi: 10.1016/s0378-1119(99)00119-5. [DOI] [PubMed] [Google Scholar]
- 43.Wachi M, Doi M, Tamaki S, Park W, Nakajima-Iijima S, Matsuhashi M. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J Bacteriol. 1987;169:4935–4940. doi: 10.1128/jb.169.11.4935-4940.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker J, Kovarik A, Allan J, Gustafson R. Regulation of bacterial cell division: temperature-sensitive mutants of Escherichia coli that are defective in septum formation. J Bacteriol. 1975;123:693–703. doi: 10.1128/jb.123.2.693-703.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie Z-D, Hershberger C D, Shankar S, Ye R W, Chakrabarty A M. Sigma factor–anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J Bacteriol. 1996;178:4990–4996. doi: 10.1128/jb.178.16.4990-4996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]