Abstract
The β-N-acetylglucosaminidase of Escherichia coli was found to have a novel specificity and to be encoded by a gene (nagZ) that maps at 25.1 min. It corresponds to an open reading frame, ycfO, whose predicted amino acid sequence is 57% identical to that of Vibrio furnissii ExoII. NagZ hydrolyzes the β-1,4 glycosidic bond between N-acetylglucosamine and anhydro-N-acetylmuramic acid in cell wall degradation products following their importation into the cell during the process for recycling cell wall muropeptides. From amino acid sequence comparisons, the novel β-N-acetylglucosaminidase appears to be conserved in all 12 gram-negative bacteria whose complete or partial genome sequence data are available.
β-N-Acetylglucosaminidase in Escherichia coli K-12 was first described by Yem and Wu in 1976 (18, 19). It was shown to be a cytoplasmic enzyme active against both p-nitrophenyl-β-N-acetyl-d-glucosaminide and a muropeptide released by lysozyme from E. coli cell wall murein (peptidoglycan). However, based on indirect evidence, it was clear that the enzyme would also be active against anhydro-muropeptides (6). These muropeptides contain N-acetylglucosamine (GlcNAc) linked β-1,4 to anhydro-N-acetylmuramyl peptides (aMurNAc-peptides) (15). aMurNAc, which possesses a 1,6 anhydro bond, is formed by the lytic transglycosylases of E. coli that digest murein during normal growth as the initial step of the murein tripeptide recycling pathway. The murein tripeptide recycling pathway is a major metabolic pathway of E. coli (3) in which, during each generation of growth, about 40% of the cell wall murein is broken down into anhydro-muropeptides (3, 6). The anhydro-muropeptides are then transported into the cytoplasm via AmpG permease (6) and are rapidly degraded by the combined actions of β-N-acetylglucosaminidase (NagZ), anhydro-N-acetylmuramyl-l-alanine amidase (AmpD) (5, 7), and an ld-carboxypeptidase (LdcA) (17) to release GlcNAc, aMurNAc, d-alanine, and the murein tripeptide (l-alanyl-γ-d-glutamyl-meso-diaminopimelic acid). The tripeptide is then linked to UDP-MurNAc by the murein peptide ligase, Mpl (10), and efficiently recycled to form murein de novo.
Previous results imply the participation of a β-N-acetylglucosaminidase in recycling (6), and the present results identify NagZ as the enzyme involved. In this work, we also identify the gene encoding β-N-acetylglucosaminidase (nagZ), characterize a null mutation and a mutation in the structural gene, and report initial observations on the specificity of NagZ.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The E. coli K-12 strains and plasmids used in this work are listed in Table 1. Bacteria were grown aerobically at 37°C in L broth, which is LB broth (12) modified to contain only 5 g of NaCl per liter. Ampicillin (100 μg/ml), kanamycin (25 μg/ml), and chloramphenicol (10 μg/ml) were used as required.
TABLE 1.
E. coli K-12 strains and plasmids used in this work
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| E. coli strain | ||
| TP71 | F−lysA opp araD139 rpsL150 relA1 deoC1 ptsF25 flbB5301 rbsR Δ(argF-lac) | 6 |
| TP73 | F− ΔampDE lysA opp araD139 rpsL150 relA1 deoC1 ptsF25 flbB5301 rbsR Δ(argF-lac) | 6 |
| TP73B | TP73 nagB::Kan | J. Park |
| TP75 | TP71 nagZ1 | This work |
| JM109 | F−traD36 lacIq Δ(lacZ)M15 proA+B+/e14− (McrA−) Δ(lac-proAB) thi gyrA96 (Nalr) endA1 hsdR17(rK− mK+) relA1 supE44 | New England Biolabs |
| BL21(DE3) | F−ompT [lon] hsdSB(rB− mB−; E. coli B strain) with DE3, a λ prophage carrying the T7 RNA polymerase gene | New England Biolabs |
| DY330 | W3110 Δ(lac)169 gal490* λ[cI857 Δ(cro-bio)] | D. Court |
| TP76 | DY330 nagZ::Cm | This work |
| TP77 | TP71 nagZ::Cm | P1(TP76) × TP71 |
| TP78 | TP73 nagZ::Cm | P1(TP76) × TP73 |
| TP78B | TP73B nagZ::Cm | P1(TP76) × TP73B |
| Plasmids | ||
| pGem-T | Cloning vector, Ampr | Promega |
| pKM1 | pGem-T carrying nagZ | This work |
| pKM3 | 178-bp NruI-ClaI fragment removed from pKM1, blunt ended, and replaced with a blunt 1,413-bp fragment containing the chloramphenicol acetyltransferase gene; Cmr | This work |
| pExoII | Plasmid expressing ExoII, Ampr | S. Roseman (2) |
| pEtslt70 | Plasmid expressing slt70, Ampr | A. Dijkstra |
| pACYC184 | Cloning vector, Cmr | New England Biolabs |
Isolation of a β-N-acetylglucosaminidase-deficient mutant.
E. coli TP71 cells were treated with nitrosoguanidine (10, 20, or 40 μg/ml in 0.1 M citrate buffer [pH 5.5]) for 30 min at 37°C and then washed with saline and grown overnight in L broth. Dilutions of overnight cultures were plated on L agar, and individual colonies were screened for NagZ activity. One clone, TP75, which had greatly reduced activity was chosen for further studies.
NagZ assay.
The qualitative assay for β-N-acetylglucosaminidase was carried out by incubating the cells from one colony or from 1 ml of overnight culture with 0.8 ml of 1 mM p-nitrophenyl-β-N-acetyl-d-glucosaminide in 50 mM Tris-HCl (pH 7.4). After 3 or 4 h of incubation at 37°C, the reaction was stopped by the addition of 0.2 ml of 1.25 M K2CO3. A reduction in the amount of yellow p-nitrophenol present in the supernatant following centrifugation of the incubation mixture relative to that of the control indicated a loss of enzyme activity. Alternatively, the cells were incubated at 37°C with 0.2 ml of 0.25 mM 4-methylumbelliferyl-β-N-acetyl-d-glucosaminide in 10 mM sodium phosphate buffer (pH 6.8). After 4 or more h incubation, a decrease in the amount of purple fluorescence observed under UV light relative to the control indicated a loss of enzyme activity. For quantitative assay, the method employing p-nitrophenyl-β-N-acetyl-d-glucosaminide was used with cells equivalent to 1 ml of culture having an optical density at 600 nm of 2. The reaction was stopped after 2 h, and the optical density of the cell-free reaction mixture was determined at 420 nm. When radioactive substrates were used, substrates and products were separated by high-pressure liquid chromatography (HPLC) as described below, and the radioactivity present in the different peaks was determined with a Beckman LS7500 liquid scintillation system.
Mapping.
Mapping was done by transduction with P1vir (12), using a set of strains containing Tn5 kanamycin resistance (Kanr) elements at known map positions (1, 16).
HPLC analysis.
HPLC was performed with Rainin Rabbit HP pumps and mixer equipment (Rainin Instrument Co., Woburn, Mass.) by two different methods. In method 1, the column used was a LiChrosphere RP-18 column (250 by 4 mm, 3-μm particle size; E. Merck). At a flow rate of 0.5 ml/min, isocratic elution with 50 mM sodium phosphate (pH 4.31) for 20 min was followed by a linear gradient of 0 to 35% 75 mM sodium phosphate (pH 4.95) in 15% methanol over 40 min and then isocratically for 60 min. In method 2, the sample was adjusted to pH ∼2.5 with trifluoroacetic acid, adsorbed on a 150- by 4.6-mm X-Terra RP-18 column (Waters, Milford, Mass.), and eluted at 0.5 ml/min with 0.05% trifluoroacetic acid for 5 min, followed by a gradient from 0.05% trifluoroacetic acid to 10% of acetonitrile containing 0.035% trifluoroacetic acid over a period of 50 minutes.
Isolation of radioactive NagZ substrates.
E. coli TP78B (nagZ::Cm nagB::Kan ΔampDE) was labeled for about five generations during growth in L broth supplemented with 0.25 strength M-9 salts (14), 1 mM MgCl2, and 1 μCi of d-[6-3H(N)]glucosamine (21.6 Ci/mmol; NEN Life Science Products, Boston, Mass.) per ml. Well-washed sacculi were recovered after the cells had been boiled in 4% sodium dodecyl sulfate for 30 min. Mixed muropeptide monomers and dimers containing 3H-labeled GlcNAc-β-1,4-MurNAc were obtained by digestion of murein sacculi with Chalaropsis muramidase. These muropeptides contained the native muramic acid present in murein.
To obtain mixed muropeptide monomers and dimers containing anhydro-muramic acid in the disaccharide (GlcNAc-β-1,4-aMurNAc), a portion of the labeled sacculi was digested with a partially purified preparation of soluble lytic transglycosylase (Slt). Slt was obtained from E. coli BL21(DE3)/pEtSlt70, carrying a plasmid that overexpresses slt from a T7 promoter when T7 RNA polymerase of the host is induced with isopropyl-β-d-thiogalactopyranoside (IPTG) (A. Dijkstra, personal communication). Cells were grown at 37°C from a 1% inoculum in L broth with vigorous aeration. The inoculum was grown overnight from a single colony. All cultures contained 100 μg of ampicillin per ml. After addition of 1 mM IPTG in early log phase, growth was continued for approximately 4 h. Cells were harvested in the cold when the optical density at 600 nm reached the range of 0.8 to 1 and were washed with buffer A (10 mM sodium phosphate [pH 7.0], containing 0.02% sodium azide and 0.1 mM dithioerythritol). The cell pellet was resuspended in the same buffer (0.25 g of cells [wet weight]/ml of buffer) and opened by sonication (Sonicator 150; T.S. Ultrasons, Annemasse, France). A 50-ml aliquot of cell suspension containing DNase I (30 μg/ml) and RNase (10 μg/ml) was treated for 4 min under cooling. After centrifugation (20,000 × g, 1 h), the supernatant was used as an enzyme source.
Enzyme was purified by chromatography on Blue Sepharose CL-6B (8). The cell extract was dialyzed against buffer A for 20 h and passed through a column (38 by 1.6 cm) of Blue Sepharose (Pharmacia). After washing with 200 ml of buffer A, the column was eluted with 120 ml of 0.07 M sodium chloride in buffer A followed by a linear salt gradient (120 ml of 0.07 M sodium chloride and 120 ml of 0.6 M sodium chloride, both in buffer A). The elution velocity was 8.25 ml/h, and fractions of 5.5 ml were collected. The transglycosylase activity was assayed by release of radioactivity from 3H-labeled sacculi. The enzyme-containing fractions, which eluted in fractions 95 to 103, were combined and dialyzed against buffer A for 20 h.
Radioactive GlcNAc-β-1,4-aMurNAc-tripeptide was isolated from E. coli TP78B (nagZ::Cm ΔampDE) labeled as described above. The washed cells were extracted with water at 95°C for 5 min; the extract was concentrated and fractionated by HPLC using the acetonitrile gradient method. The principal fraction, which eluted after 50 min, was neutralized with NH3, concentrated, and stored frozen. Its identity was confirmed by mass spectrometry.
The radioactive disaccharide, GlcNAc-β-1,4-aMurNAc, was obtained by treatment of the disaccharide-tripeptide with 3 μg of AmpD amidase in 10 mM phosphate buffer (pH 7.0) for 3 h at 37°C followed by purification by the same HPLC method.
Cloning the gene for nagZ.
A search of GenBank revealed an open reading frame, ycfO, at approximately 25.1 min whose hypothetical protein had 57% sequence identity to ExoII, a novel β-N-acetylglucosaminidase of Vibrio furnissii (2). To clone the ycfO gene, which may code for NagZ, a forward primer (5′-TGGCTGCTGATGCTCAAA), starting 124 nucleotides upstream of the start codon, and a reverse primer (5′-AATCATCGCTTCCTCACA), starting 28 nucleotides downstream of the stop codon, were used to amplify the hypothetical nagZ gene from E. coli TP71 cells by PCR. The amplified DNA was ligated directly into a TA cloning vector (pGem-T; Promega, Madison, Wis.), and the resulting plasmid, pKM1, was transformed into competent E. coli JM109. Sequencing of the gene confirmed its exact identity with the open reading frame designated ycfO. To determine the orientation of the gene in the plasmid, the plasmid was digested with restriction enzymes AatII and ClaI and analyzed by electrophoresis on a 1% agarose gel. The presence of an 893-bp fragment, rather than a 325-bp fragment, proved that nagZ was present in a clockwise orientation relative to the T7 promoter.
Construction of a nagZ null mutant.
Plasmid pKM1, which contains the nagZ gene on a 1,177-bp PCR product ligated to the pGem-T vector, was cut with restriction enzymes NruI and ClaI to remove a 178-bp fragment from nagZ. The linearized plasmid was blunted by filling with the aid of the Klenow fragment of DNA polymerase I and then dephosphorylated with calf intestinal alkaline phosphatase. A blunt 1,413-bp fragment, containing the chloramphenicol acetyltransferase gene, was isolated from pACYC184 that had been cut with restriction enzyme BsaAI. The fragments of interest were separated by electrophoresis on agarose gels and purified using a Qiagen gel extraction kit. The chloramphenicol acetyltransferase gene was ligated to the linearized plasmid from which a 178-bp fragment of nagZ had been removed. The ligation mixture was transformed into competent JM109 cells; following 3 h of incubation to allow expression, the cells were plated on L agar containing 15 μg of chloramphenicol per ml. Chloramphenicol-resistant (Cmr) colonies were purified, and plasmids were isolated. Analysis of the fragments released from each plasmid by EcoRI identified plasmids that contained fragments of 3,018, 2,102, and 310 bp as predicted for the desired construct. One such plasmid was saved and named pKM3. To construct a null mutation of nagZ, the procedure described by Yu et al. (20), which utilizes the RED system of phage lambda, was used. pKM3 was cut with restriction enzymes PstI and SphI for which restriction sites are present in the multiple cloning sites of pGem-T on opposite sides of the nagZ::Cm insert but are absent in the insert. Then 100 ng of DNA from the DNA digest were introduced into strain DY330 by electroporation following induction of the RED system present in DY330 by incubation of mid-log-phase cells for 15 min at 42°C. After growth for 1.5 h at 30°C to allow expression, the cells were plated at 30°C on L agar containing 10 μg of chloramphenicol per ml. Among 24 Cmr colonies purified, six were sensitive to ampicillin. When tested for NagZ activity, all six were found to lack NagZ activity. The null mutant of DY330 was designated TP76.
Other methods.
DNA manipulations, transductions, and transformations were performed as described elsewhere (12, 14). Mass spectrometry was performed at the Tufts Protein Chemistry Facility utilizing a PE Biosystems Voyager Maldi mass spectrometer. For genome database searches, the GenBank site as well as the preliminary sequence data available from The Institute for Genomic Research website (http://www.tigr.org) were searched.
RESULTS
Map position of the presumed structural gene for NagZ in mutant TP75.
Hrebenda had reported that the gene for E. coli β-N-acetylglucosaminidase cotransduced with trp at a low frequency (4) such that one would expect the gene to be located roughly 1.5 min away from 28.3 min, the position of trp in edition 9 of the E. coli linkage map (1). However, we were unable to obtain cotransduction of nagZ with trp. Instead, when kanamycin resistance was transduced into TP75 from strains carrying Kanr markers at known locations (16), only those Kanr markers 3 or 4 min counterclockwise from trp cotransduced with nagZ. nagZ cotransduced with Kanr markers at 24.6 min (9 of 30), 25.5 min (10 of 30), and 26.6 min (only 1 of 30), indicating that the gene is located at about 25 min.
Proof that a gene located at 25.1 min is the structural gene for β-N-acetylglucosaminidase.
As described in Materials and Methods, we cloned an open reading frame, ycfO, which has 57% identity to a novel β-N-acetylglucosaminidase present in V. furnissii (2) and is located at 25.1 min in the E. coli genome. This plasmid, pKM1, was transformed into TP75 (nagZ1). The transformant, TP75/pKM1, had more than four times the NagZ activity of the wild type, whereas TP75 had less than 5% of the wild-type NagZ activity. Since ycfO is an ortholog of the structural gene for a known β-N-acetylglucosaminidase, the open reading frame ycfO is the structural gene for NagZ.
Mutation site in the structural gene for NagZ in strain TP75.
The nucleotide sequence of the mutated gene was determined on a PCR product obtained using the aforementioned primers and TP75 cells as the source of chromosomal DNA. The sequencing revealed a point mutation, 66 nucleotides upstream from the expected stop codon, which converted a G to an A, resulting in creation of an opal stop codon, TGA. Thus, the mutant protein should lack the C-terminal 22 amino acids present in wild-type NagZ.
The lack of NagZ causes accumulation of amino sugar-containing compounds in the cytoplasm.
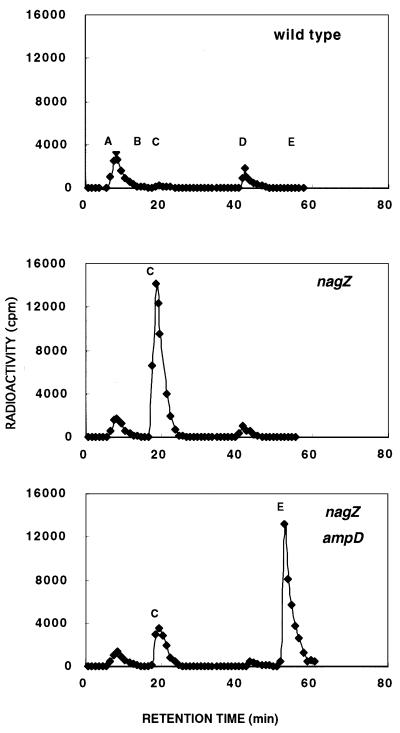
The nagZ::Cm strain, TP77, had no detectable activity against p-nitrophenyl-β-GlcNAc under our assay conditions (data not shown). TP77 (nagZ::Cm), TP78 (nagZ::Cm ΔampDE), and the parent TP71 were each grown from a 0.5% inoculum for approximately five generations in 8 ml of L broth supplemented with 2 mM MgCl2 and 1 μCi of [3H]glucosamine/ml. The cells were collected by centrifugation, washed once with 15 ml of 0.9% NaCl, and suspended in 2 ml of water. The suspensions were heated at 95°C for 5 min to obtain hot water extracts. Extract equivalent to 2 ml of each culture was analyzed by HPLC using the acetonitrile gradient method. The results (Fig. 1) show that the wild-type strain contains two principal peaks. Peak A contains UDP-GlcNAc, GlcNAc, and glucosamine; peak D contains UDP-MurNAc-pentapeptide. A trace amount of the free disaccharide, GlcNAc-β-1,4-aMurNAc (peak C), and an even smaller amount of free aMurNAc (peak B) are present. In contrast with the wild-type strain, the nagZ mutant contains a huge amount of the disaccharide (peak C), and the double mutant, lacking both NagZ and AmpD, accumulates large amounts of the disaccharide-tripeptide (peak E) and also contains a substantial amount of free disaccharide (Fig. 1). The large accumulation of disaccharide by the nagZ null mutant demonstrates the need for NagZ in order to dispose of the GlcNAc and aMurNAc produced by the recycling pathway.
FIG. 1.
HPLC analysis of the cytoplasmic pools of E. coli TP71 (wild type), TP77 (nagZ), and TP78 (nagZ ampD). The hot water extract from 2 ml of culture grown in the presence of 1 μCi of [3H]glucosamine per ml for approximately five generations was analyzed by HPLC (method 2). Identities of fractions A to E: A, a mixture containing glucosamine and GlcNAc and their phosphorylated derivatives and UDP-GlcNAc; B, aMurNAc; C, GlcNAc-aMurNAc; D, UDP-MurNAc-pentapeptide; E, GlcNAc-aMurNAc-tripeptide.
Enzyme activity.
As already noted, NagZ is very similar in amino acid sequence to ExoII of V. furnissii. ExoII was shown to have an unusual substrate specificity (2). It had negligible activity against N,N′-diacetylchitobiose but rapidly cleaved p-nitrophenyl-β-GlcNAc and slowly cleaved p-nitrophenyl-β-N-acetylgalactosaminide (GalNAc). Since NagZ has high sequence identity with ExoII, it seemed likely that their substrate profiles would be similar. As shown in Table 2, this proved to be the case. Under our assay conditions, neither enzyme has activity against N,N′-diacetylchitobiose, and the activity against p-nitrophenyl-β-GlcNAc is about 30 times greater than that against p-nitrophenyl-β-GalNAc. These initial tests of enzyme specificity were done using whole cells of TP75 (nagZ1), carrying a plasmid expressing either nagZ+ or exoII+.
TABLE 2.
Comparison of the substrate specificities of NagZ and ExoII
| Substrate | GlcNAc released (nmol/h)a
|
|
|---|---|---|
| NagZ | Exo II | |
| N,N′-Diacetylchitobiose | 0.0 | 0.0 |
| p-Nitrophenyl-β-N-acetyl-d-glucosaminide (1) | 25 | 240 |
| p-Nitrophenyl-β-N-acetyl-d-galactosaminide (2) | 0.8 | 6.9 |
| Activity ratio, (1)/(2) | 31 | 35 |
The source of enzyme was toluene-treated intact cells from 1 ml of overnight culture of TP75 containing plasmids expressing the relevant gene. The specific activities of the two enzymes should not be compared since the relative expression of the β-N-acetylglucosaminidase genes from the two plasmids is unknown.
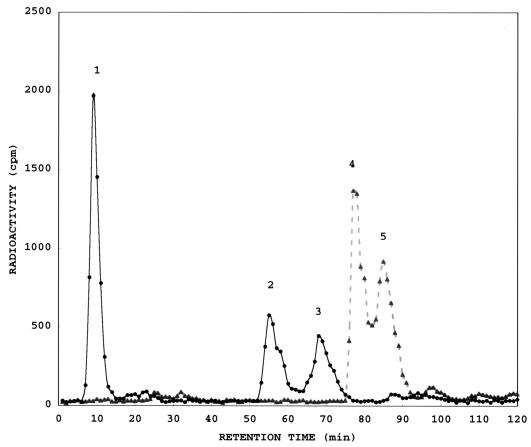
Figure 2 illustrates that NagZ completely cleaves GlcNAc from anhydro-muropeptides. GlcNAc-aMurNAc-tripeptide (peak 4) and GlcNAc-aMurNAc-tetrapeptide (l-Ala-γ-d-Glu-meso-Dap-d-Ala) (peak 5) were quantitatively converted to the aMurNAc-tripeptide (peak 2) and aMurNAc-tetrapeptide (peak 3), respectively, with the release of GlcNAc (peak 1). Table 3 summarizes experiments which demonstrate that NagZ cleaves muropeptides almost as efficiently as anhydro-muropeptides and that NagZ readily hydrolyzes the free anhydro-disaccharide (GlcNAc-aMurNAc).
FIG. 2.
HPLC analysis of a mixture of anhydro-muropeptides before (▴) and after (●) digestion by NagZ. A mixture of anhydro-muropeptides, all containing [3H]GlcNAc and [3H]aMurNAc, was digested with toluene-treated cells from approximately 2 ml of overnight culture of E. coli TP78 (nagZ::Cm ampD)/pKM1 (nagZ+). TP78 cells without the plasmid lacked NagZ activity. Identities of fractions 1 to 5 (confirmed with authentic reference compounds): 1, GlcNAc; 2, aMurNAc-tripeptide; 3, aMurNAc-tetrapeptide; 4, GlcNAc-aMurNAc-tripeptide; 5, GlcNAc-aMurNAc-tetrapeptide. HPLC method 1 was used.
TABLE 3.
Activity of NagZ against muropeptides from E. coli
| Enzyme | Substrate | Time of incubation | % Hydrolyzed |
|---|---|---|---|
| TP78 cells, 50 μl | GlcNAc-aMurNAc-tripeptideb | 2 h | 0 |
| NagZa | |||
| 50 μl | GlcNAc-aMurNAc-tripeptideb | 2 h | 100 |
| GlcNAc-aMurNAc-tripeptideb | 1 h | 90 | |
| GlcNAc-aMurNAc-tripeptideb | 15 min | 35 | |
| 100 μl | GlcNAc-aMurNAc | 2 h | 100 |
| 165 μl | Anhydro-muropeptidesc | 3 h | 100 |
| 50 μl | Anhydro-muropeptidesc | 30 min | 64 |
| 200 μl | Muropeptidesd | 3 h | 100 |
| 50 μl | Muropeptidesd | 30 min | 75 |
Cells from 3 ml of an overnight culture of TP78 (nagZ::Cm)/pKM1 (nagZ+) were suspended in 0.2 ml of 50 mM Tris (pH 7.5) and vortexed with 8 μl of toluene. The amount of cell suspension used is indicated. Digests were analyzed by HPLC (method 1).
Approximately 4 μg of material with 131,000 cpm of total radioactivity.
Anhydro-muropeptides released from [3H]glucosamine-labeled TP73B sacculi by digestion with a crude preparation of soluble lytic transglycosylase from E. coli BL21(DE3)/pEtslt70.
Muropeptides released from [3H]glucosamine-labeled TP73B sacculi by digestion with Chalaropsis muramidase.
Prevalence of NagZ.
From a search of the sequences of unfinished as well as complete microbial genomes, possible orthologs with an amino acid sequence similar to that of NagZ were found to be present in the 12 gram-negative genera currently being sequenced (Table 4).
TABLE 4.
Possible orthologs of proteins involved in murein recycling in E. coli
| Bacterium | % Identitya
|
|||
|---|---|---|---|---|
| NagZ | AmpD amidase | AmpG permease | Mpl ligase | |
| Salmonella typhi | 91 | 81 | 90 | 96 |
| Yersinia pestis | 73 | 48 | 73 | 83 |
| Klebsiella pneumoniae | 65 | 72 | 90 | |
| Haemophilus influenzae | 52 | 40 | 38 | 68 |
| Pasteurella multicida | 55 | 41 | 37 | 66 |
| Vibrio cholerae | 56 | 37 | 32 | 69 |
| Pseudomonas aeruginosa | 52 | 42 | 32 | 59 |
| Bordetella pertussis | 48 | 45 | 59 | 63 |
| Shewanella putrefaciens | 51 | 38 | 37 | 64 |
| Neisseria meningitidis | 45 | 37 | 40 | 58 |
| Actinobacillus actinomycecomitans | 53 | 40 | 64 | |
| Thiobacillus ferrooxidans | 45 | 40 | 29 | 57 |
Boldface numbers indicate contigs of 45 to 90% of full length; all others are greater than 90% of full length.
DISCUSSION
We have shown that nagZ, the gene for the β-N-acetylglucosaminidase of E. coli, is the open reading frame, ycfO, present at 25.1 min on the E. coli map. TP75, a mutant with greatly reduced NagZ activity, facilitated mapping. TP75 was shown to have a single base pair change in the nagZ gene that produced a new opal stop codon 22 codons upstream of the normal stop codon. This mutant has less than 5% of the wild-type NagZ activity. A null mutant was constructed and shown to completely lack NagZ activity. Hence, NagZ is the only β-N-acetylglucosaminidase expressed in E. coli. That the null mutant grew normally and had normal morphology proved that NagZ is not essential when E. coli is grown in rich medium.
The amino acid sequence of NagZ is 57% identical to the sequence of ExoII of V. furnissii. Not surprisingly, NagZ and ExoII have very similar substrate specificities (Table 2). Figure 2 demonstrates that NagZ completely cleaves GlcNAc from anhydro-muropeptides. Surprisingly, NagZ also readily cleaves GlcNAc linked β-1,4 to MurNAc-peptides (Table 3). This is in contrast to AmpD anhydro-N-acetylmuramyl-l-alanine amidase, a cytoplasmic enzyme that cleaves anhydro-muropeptides to release murein tripeptide for recycling. The AmpD amidase is 10,000 times more active against the anhydro-MurNAc-l-alanine bond compared to the MurNAc-l-alanine bond (7). Presumably the AmpD amidase evolved to attack the amide bond linking l-alanine to anhydro-MurNAc without cleaving the MurNAc-l-alanine bond present in the UDP-MurNAc-pentapeptide precursor required for cell wall synthesis since this would be fatal.
A similar argument could be made that NagZ should not cleave the β-1,4 bond between GlcNAc and MurNAc that is present in lipid 2. During synthesis of murein, GlcNAc becomes linked β-1,4 to MurNAc-pentapeptide at the lipid intermediate stage, thereby converting lipid 1 to lipid 2 (9). Since lipid 2 is an essential murein biosynthetic intermediate, it would appear either that lipid 2 is not a substrate for NagZ or that as soon as it is formed, the disaccharide-pentapeptide of lipid 2 is protected from NagZ by its lipid environment or by immediate transfer to the outer surface of the membrane.
In the absence of NagZ, E. coli accumulates large amounts of the disaccharide, GlcNAc-aMurNAc, in its cytoplasm (Fig. 1). This reflects the high level of activity of the murein tripeptide recycling pathway and indicates that the cell cannot utilize or dispose of the amino sugars in the absence of NagZ. Figure 1 also shows that when both NagZ and AmpD are absent, E. coli accumulates GlcNAc-β-1,4-aMurNAc-tripeptide. Interestingly, a significant amount of the free disaccharide also accumulates, suggesting that E. coli has another amidase, in addition to AmpD, that cleaves the aMurNAc-l-alanine bond.
As noted earlier, possible orthologs of NagZ are present in the 12 gram-negative bacteria thus far partially or completely sequenced. This strongly suggests that the novel β-N-acetylglucosaminidase may be present in most gram-negative bacteria. Table 4 illustrates how closely the percent identities of E. coli NagZ, Mpl UDP-N-acetylmuramate:l-alanyl-γ-d-glutamyl-meso-diaminopimelate ligase, AmpD anhydro-N-acetylmuramyl-l-alanine amidase, and AmpG permease are to their respective orthologs in these gram-negative bacteria. These data suggest that, indeed, murein recycling is widespread among gram-negative bacteria. Though NagZ β-N-acetylglucosaminidase is not obviously needed for recycling of murein tripeptide, it nevertheless appears to have been conserved in gram-negative bacteria and to be an integral component of the murein tripeptide recycling pathway.
ACKNOWLEDGMENTS
We thank Saul Roseman for the plasmid expressing exoII and Amoud Dijkstra for the plasmid expressing slt70 and for authentic reference muropeptides.
This work was supported in part by Public Health Service grant GM51610 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Berlyn M K B, Low K B, Rudd K E. Linkage map of Escherichia coli K-12, edition 9. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: American Society for Microbiology; 1996. pp. 1715–1902. [Google Scholar]
- 2.Chitlaru E, Roseman S. Molecular cloning and characterization of a novel β-N-acetyl-d-glucosaminidase from Vibrio furnissii. J Biol Chem. 1996;271:33433–33439. doi: 10.1074/jbc.271.52.33433. [DOI] [PubMed] [Google Scholar]
- 3.Goodell E W. Recycling of murein by Escherichia coli. J Bacteriol. 1985;163:305–310. doi: 10.1128/jb.163.1.305-310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hrebenda J. Mutants of Escherichia coli with altered level of β-N-acetylglucosaminidase activities. Acta Microbiol Pol. 1979;28:53–62. [PubMed] [Google Scholar]
- 5.Holtje J-V, Kopp U, Ursinus A, Wiedemann B. The negative regulator of β-lactamase induction AmpD is a N-acetyl-anhydromuramyl-l-alanine amidase. FEMS Microbiol Lett. 1994;122:159–164. doi: 10.1111/j.1574-6968.1994.tb07159.x. [DOI] [PubMed] [Google Scholar]
- 6.Jacobs C, Huang L-J, Bartowsky E, Normark S, Park J T. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for β-lactamase induction. EMBO J. 1994;13:4684–4694. doi: 10.1002/j.1460-2075.1994.tb06792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobs C, Joris B, Jamin M, Klarsov K, van Beemen J, Mengin-Lecreulx D, van Heijenoort J, Park J T, Normark S, Frere J-M. AmpD, essential for both β-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-l-alanine amidase. Mol Microbiol. 1995;15:553–559. doi: 10.1111/j.1365-2958.1995.tb02268.x. [DOI] [PubMed] [Google Scholar]
- 8.Kusser W, Schwarz U. Escherichia coli murein transglycosylase. Eur J Biochem. 1980;103:277–281. doi: 10.1111/j.1432-1033.1980.tb04312.x. [DOI] [PubMed] [Google Scholar]
- 9.Mengin-Lecreulx D, Texier L, Rousseau M, van Heijenoort J. The murG gene of Escherichia coli codes for the UDP-N-acetylglucosamine:N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase involved in the membrane steps of peptidoglycan synthesis. J Bacteriol. 1991;173:4625–4636. doi: 10.1128/jb.173.15.4625-4636.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mengin-Lecreulx D, van Heijenoort J, Park J T. Identification of the mpl gene encoding UDP-N-acetylmuramate:l-alanyl-γ-d-glutamyl-meso-diaminopimelate ligase in Escherichia coli and its role in recycling of cell wall peptidoglycan. J Bacteriol. 1996;178:5347–5352. doi: 10.1128/jb.178.18.5347-5352.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 12.Miller J H. A short course in bacterial genetics, a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 268–274. [Google Scholar]
- 13.Park J T, Raychaudhuri D, Li H, Normark S, Mengin-Lecreulx D. MppA, a periplasmic binding protein essential for import of the bacterial cell wall peptide l-alanyl-γ-d-glutamyl-meso-diaminopimelate. J Bacteriol. 1998;180:1215–1223. doi: 10.1128/jb.180.5.1215-1223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 15.Shockman G D, Höltje J-V. Microbial peptidoglycan (murein) hydrolases. In: Ghuysen J M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science B.V.; 1994. pp. 131–166. [Google Scholar]
- 16.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Templin M F, Ursinus A, Holtje J-V. A defect in cell wall recycling triggers autolysis during the stationary growth phase of Escherichia coli. EMBO J. 1999;18:4108–4117. doi: 10.1093/emboj/18.15.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yem D W, Wu H C. Purification and properties of β-N-acetylglucosaminidase from Escherichia coli. J Bacteriol. 1976;125:324–331. doi: 10.1128/jb.125.1.324-331.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yem D W, Wu H C. Isolation of Escherichia coli K-12 mutants with altered levels of β-N-acetylglucosaminidase. J Bacteriol. 1976;125:372–373. doi: 10.1128/jb.125.1.372-373.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu D, Ellis H M, Lee E-C, Jenkins N A, Copeland N G, Court D L. An efficient recombination system for chromosome engineering in E. coli. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]