Abstract
As part of a Brucella abortus 2308 genome project carried out in our laboratory, we identified, cloned, and sequenced a genomic DNA fragment containing a locus (virB) highly homologous to bacterial type IV secretion systems. The B. abortus virB locus is a collinear arrangement of 13 open reading frames (ORFs). Between virB1 and virB2 and downstream of ORF12, two degenerated, palindromic repeat sequences characteristic of Brucella intergenic regions were found. Gene reporter studies demonstrated that the B. abortus virB locus constitutes an operon transcribed from virB1 which is turned on during the stationary phase of growth. A B. abortus polar virB1 mutant failed to replicate in HeLa cells, indicating that the virB operon plays a critical role in intracellular multiplication. Mutants with polar and nonpolar mutations introduced in virB10 showed different behaviors in mice and in the HeLa cell infection assay, suggesting that virB10 per se is necessary for the correct function of this type IV secretion apparatus. Mouse infection assays demonstrated that the virB operon constitutes a major determinant of B. abortus virulence. It is suggested that putative effector molecules secreted by this type IV secretion system determine routing of B. abortus to an endoplasmic reticulum-related replication compartment.
Brucella spp. are facultative intracellular gram-negative bacteria that are pathogenic for many mammalian species including humans, causing a chronic infectious disease known as brucellosis, a major zoonosis in several countries (6). In humans, brucellosis is a serious debilitating disease characterized by diverse pathological manifestations like undulant fever, osteoarticular complications, endocarditis, and several neurological disorders. In domestic animals like cattle, goats, and sheep, the outstanding manifestation of the pathology is abortion in pregnant females and sterility of males due to colonization of placenta, fetal tissues, and sexual organs (15).
Brucella spp. belong, like Agrobacterium spp., Rhizobium spp., and Rickettsia spp., to the alpha-2 subgroup of the Proteobacteria (14). Most genera of this group are characterized by their ability to interact pericellularly or intracellularly with eukaryotic cells either as pathogens or as endosymbionts.
In Brucella spp., virulence is associated with their capacity to multiply inside the host cell. In view of recent data reported by Pizarro-Cerda et al. (19), it is clear that intracellular survival and multiplication of Brucella depend on effectively avoiding the fusion of the phagosome-containing bacteria with the lysosome and replication in an endoplasmic reticulum-like vesicle. Genes that allow Brucella spp. to invade and reach the appropriate intracellular replication niche remain to be identified.
Recently, operons coding for export mechanisms specializing in transfer of a variety of multimolecular complexes across the bacterial membrane to the extracellular space or into other cells have been described. These complexes, named type IV secretion systems, are present in Bordetella pertussis (ptl genes) (11, 29), Agrobacterium tumefaciens (virB genes) (13, 27), Escherichia coli (tra genes) (20, 30), Legionella pneumophila (dot-icm genes and lvh genes) (3, 23, 25), and Helicobacter pylori (cag genes) (7). The paradigmatic example of type IV secretion machinery is the virB operon of the phytopathogen A. tumefaciens, which comprises a collinear arrangement of 11 genes coding for a pilus-like structure necessary for transfer of T-DNA (transfer DNA) from the bacterium to the plant cell. The Agrobacterium virB operon shares high sequence similarities with E. coli tra genes, which code for the conjugative pilus and other components necessary for transfer of DNA from one bacterium to another (4). In B. pertussis, the ptl operon codes for the apparatus that allows secretion of pertussis toxin. The H. pylori cag pathogenicity island is composed of 31 genes, six of them displaying homologies with others bacterial type IV secretion systems. The dot-icm virulence genes of the intracellular pathogen L. pneumophila encode a type IV secretion system that controls the intracellular trafficking of the bacteria. dot and icm mutants reside in phagosomes that rapidly fuse with lysosomes, resulting in a decrease in intracellular survival.
During the course of our work, it was reported that the virB region is essential for intracellular replication of Brucella suis 1330 in an in vitro infection model (17). Here, we describe the entire Brucella abortus virB region coding for a type IV secretion apparatus and demonstrate that it is a stationary-phase-induced operon that plays a critical role in virulence in vivo and intracellular multiplication within nonprofessional phagocytes.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
Bacterial strains and plasmids used in this work are listed in Table 1. Brucella strains were grown at 37°C on a rotary shaker (200 rpm) for 24 h in tryptic soy broth (TSB). E. coli strains were grown at 37°C on a rotary shaker (200 rpm) overnight in Luria-Bertani broth. When necessary, the following antibiotics were added to the indicated final concentrations: kanamycin (50 μg/ml), gentamicin (2.5 μg/ml), tetracycline (2 μg/ml), and ampicillin (50 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| Strains | ||
| Brucella abortus | ||
| 2308 | Wild type, smooth, virulent, Nalr | Laboratory stock |
| S19 | Smooth, vaccine strain, Nalr | Laboratory stock |
| virB1::Kan | 2308 Nalr Kanr, polar mutant of virB1 | This work |
| virB10::Kan | 2308 Nalr Kanr, polar mutant of virB10 | This work |
| virB10::Gm | 2308 Nalr Gmr, nonpolar mutant of virB10 | This work |
| virB10::lacZ-Gm | 2308 Nalr Gmr, chromosomal transcriptional fusion virB10-lacZ | This work |
| virB1::Kan virB10::lacZ-Gm | 2308 Nalr Kanr Gmr, polar mutant of virB1, chromosomal transcriptional fusion virB10-lacZ | This work |
| virB1::Kan(pVK8.3) | B. abortus virB1::Kan harboring the plasmid pVK8.3 | This work |
| virB10::Kan(pVK8.3) | B. abortus virB10::Kan harboring the plasmid pVK8.3 | This work |
| virB10::Gm(pBBR-virB10) | B. abortus virB10::Gm harboring the plasmid pBBR2-virB10 | This work |
| virB1::Kan(pBBR4-virB1) | B. abortus virB1::Kan harboring the plasmid pBBR4-virB1 | This work |
| Brucella suis | ||
| 1330 | Wild type, smooth, virulent | Laboratory stock |
| Brucella ovis | ||
| REO198 | Wild type, rough, virulent | Laboratory stock |
| Plasmids | ||
| pBBR1MCS-2 | Broad-host-range cloning vector, Kanr | 12 |
| pBBR1MCS-4 | Broad-host-range cloning vector, Ampr | 12 |
| pB2A3 | 1.5-kbp fragment with high homology to the virB9-virB10 genes of Agrobacterium tumefaciens cloned into pBluescript SK | 28 |
| pGEM-T-virB1 | 881-bp PCR product containing the B. abortus 2308 virB1 gene cloned into pGEM-T | This work |
| pGEM-T-virB1::Kan | 1.3-kbp BamHI fragment containing a kanamycin resistance cassette with transcriptional terminator cloned into the pGEM-T-virB1 plasmid | This work |
| pVK8.3 | 20-kb BamHI genomic DNA fragment of B. abortus 2308 containing the virB operon cloned into pVK102 | This work |
| pBBR2-virB10 | 1.2-kb EcoRI fragment containing the B. abortus virB10 gene cloned into pBBR1MCS-2 | This work |
| pBBR4-virB1 | 0.88-kb EcoRI fragment containing the B. abortus virB1 gene cloned into pBBR1MCS-4 | This work |
Cloning of virB region.
In order to clone the virB region of B. abortus 2308, plasmid pB2A3, containing a 1.5-kb DNA fragment with high homology to the virB9-virB10 genes of A. tumefaciens, was obtained from the B. abortus genome project (28) (accession no. AQ752936). A pVK102 cosmid library of B. abortus 2308 (10) was screened using pB2A3 as a probe. Three reactive cosmids, pVK8.3, pVK8.25, and pVK8.38, with different restriction enzyme patterns, were isolated. Southern blot analysis was carried out on the three cosmids digested with EcoRI, HindIII, and AvaII as described previously (22), using pB2A3 as a probe. Cosmid pVK8.3, containing the smallest DNA insert (20 kb), was chosen for genetic complementation and sequencing analysis.
Nucleotide sequencing and database search.
Based on the recently published B. suis 1330 virB sequence (17), a set of 44 primers were designed to obtain 22 PCR overlapping DNA fragments covering the entire virB region (average length, 530 bp). Every PCR fragment was subsequently cloned into pGEM-T-Easy vector (Promega Corp.). Eight independent clones of each fragment were sequenced. Sequencing reactions were performed on each template with T7 or SP6 primers by using the BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems) and were analyzed in an ABI Prism 377 sequencer (Applied Biosystems). Sequences were assembled and edited using the Phred-Phrap-Consed System (courtesy of P. Green, D. Gordon, and B. Erwing, University of Washington, Seattle) running in a LINUX environment. The sequences were compared against the National Center for Biotechnology Information nonredundant protein database by using the programs BLASTx or BLASTp on the BLAST network service at the National Center for Biotechnology Information (1).
Mutagenesis of virB1 and virB10 genes.
A PCR product of 881 bp containing the virB1 gene was amplified using primers 5′-CGCTGATCTATAATTAAGGCTA-3′ and 5′-TGCGACTGCCTCCTATCGTC-3′ and ligated to pGEM-T-Easy (Promega Corp.) to generate pGEM-T-virB1. The resulting plasmid was linearized with BamHI and ligated to a 1.3-kb BamHI fragment containing a kanamycin resistance cassette (18) to generate the plasmid pGEM-T-virB1::Kan. This plasmid was electroporated into B. abortus 2308 where it is incapable of autonomous replication, and double-homologous recombinant events (Kanr Amps) were selected using kanamycin resistance. PCR and Southern blot analysis showed that the wild-type virB1 gene had been replaced by the disrupted one. This procedure generated a B. abortus virB1::Kan polar mutant. To obtain polar and nonpolar mutations on the virB10 gene, two kinds of targeted insertional mutants were constructed. Plasmid pB2A3 was linearized with the restriction enzyme NruI and ligated with a 1.3-kb HincII fragment encoding a kanamycin resistance cassette or with a 0.7-kb SmaI fragment encoding a gentamicin resistance cassette devoid of transcriptional terminator sequences (J. Ugalde, unpublished data). The resulting plasmid pB2A3::Kan or pB2A3::Gm was electroporated into B. abortus 2308. pB2A3 is a derivative of pBluescript SK (Stratagene, La Jolla, Calif.) that does not replicate in Brucella spp. Accordingly, colonies that were Kanr Amps or Gmr Amps were selected as possible double-homologous recombinants. PCR and Southern blot analysis confirmed that the wild-type virB10 gene was replaced. This procedure generated polar mutant B. abortus virB10::Kan and nonpolar mutant B. abortus virB10::Gm.
Construction of pBBR-virB10 complementing plasmids.
PCR was carried out with primers 5′-GGGAATTCGTCAGGCACAATAAAGTCAC-3′ and 5′-CCACAGTGCCAGGCGTCAAG-3′ to amplify a 1,244-bp DNA fragment containing the complete (from the ATG1 to the stop codon) B. abortus 2308 virB10 gene. This PCR product was cloned into pGEM-T-Easy, generating plasmid pGEM-T-virB10. A 1.2-kb EcoRI fragment was excised from pGEM-T-virB10 and ligated to the EcoRI site of pBBR1MCS-2 or pBBR1MCS-4 (12) under the lacZ promoter. The resulting plasmid, named pBBR2-virB10 or pBBR4-virB10, respectively, was conjugated into the B. abortus virB10::Gm nonpolar mutant or the B. abortus virB10::Km polar mutant, respectively, by triparental mating.
Construction of plasmid pBBR4-virB1.
PCR was carried out with primers 5′-CGCTGATCTATAATTAAGGCTAT-3′ and 5′-TGCGACTGCCTCCTATCGTC-3′ to amplify an 881-bp DNA fragment containing the complete B. abortus 2308 virB1 gene. This PCR product was cloned into pGEM-T-Easy, generating plasmid pGEM-T-virB1. An 0.88-kb EcoRI fragment was excised from pGEM-T-virB1 and ligated to the EcoRI site of pBBR1MCS-4. The resulting plasmid, named pBBR4-virB1, was conjugated into the B. abortus virB1::Kan polar mutant by biparental mating.
Complementation of B. abortus virB1::Kan and virB10::Kan polar mutants.
Cosmid pVK8.3 was conjugated into B. abortus virB1::Kan or B. abortus virB10::Kan polar mutants by biparental mating as described previously (5).
Construction of lacZ-Gm transcriptional fusion.
Plasmid pAB2001 (2) was digested with HindIII, producing a 4.5-kb fragment containing a lacZ-Gmr promoter-probe cassette. This DNA fragment was blunt ended with T4 DNA polymerase (New England Biolabs) and ligated into the NruI site of pB2A3. The resulting plasmid, pB2A3::lacZ-Gmr (which does not replicate in Brucella spp.), was electroporated into B. abortus 2308 (5). Gmr Amps colonies were selected as possible double-homologous recombinants by plating them on TSB agar containing gentamicin (2.5 μg/ml) or ampicillin (100 μg/ml). PCR and Southern blot analysis confirmed correct recombination events. This procedure generated strain B. abortus virB10::lacZ-Gm, which has a transcriptional fusion of a promoterless lacZ gene to virB10.
Construction of the B. abortus virB1::Kan virB10::lacZ-Gm double mutant strain.
Plasmid pGEM-T-virB1::Kan was electroporated into the B. abortus virB10::lacZ-Gm strain. Bacteria were plated on TSB agar containing gentamicin (2.5 μg/ml) and kanamycin (50 μg/ml). Gmr Kanr Amps colonies were selected as possible double-homologous recombinants, and gene replacement was confirmed by PCR analysis.
β-Galactosidase activity assays.
β-Galactosidase assays were carried out with whole cells as described elsewhere (22) with the following modification: the reaction mixture was centrifuged before the determination of A420. β-Galactosidase activity was expressed as A420/volume × A600.
Cell infection assays.
Log-phase growing cultures of B. abortus were prepared as described previously (19). HeLa cells seeded in 24-well plates (105 cells per well) were inoculated with 1 ml of minimal essential medium (Gibco, Paisley, Scotland) supplemented with 5% fetal calf serum and 2 mM glutamine (cell culture medium) containing 5 × 107 CFU of bacteria, without antibiotics. In order to ensure close contact between cells and bacteria, multiwell plates were centrifuged for 10 min at 141 × g at room temperature and placed in a 5% CO2 atmosphere at 37°C. After 1 h, wells were washed five times with phosphate-buffered saline (PBS) (pH 7.4) and further incubated with cell culture medium containing 100 μg of gentamicin per ml and 50 μg of streptomycin per ml to eliminate remaining extracellular brucellae. At different times, the number of intracellular viable B. abortus bacteria was determined as follows: cells were washed four times with PBS and treated for 10 min with 1 ml of 0.1% Triton X-100 in deionized sterile water, and lysates were serially diluted in PBS and plated on tryptic soy agar with the appropriate antibiotic to determine CFU.
Experimental infection of mice.
The infection assay was carried out as described previously (5). Briefly, 60-day-old female BALB/c mice were injected intraperitoneally with 104 CFU of B. abortus 2308 wild-type or mutant strains. At 15 days postinfection (p.i.), mice were sacrificed, and spleens were removed, weighed, and homogenized in 1 ml of 150 mM NaCl. Tissue homogenates were serially diluted and plated in duplicate on tryptic soy agar with the appropriate antibiotic. CFU were counted after 3 to 4 days of incubation at 37°C.
Nucleotide sequence accession number.
The DNA sequence of the complete B. abortus 2308 virB operon was deposited in GenBank under accession no. AF226278.
RESULTS
Identification of a homologue to the A. tumefaciens virB operon in B. abortus 2308.
A partial genome project carried out in our laboratory led us to identify a clone, named pB2A3, consisting of a 1.5-kb DNA genomic fragment of B. abortus 2308 containing sequences highly similar to A. tumefaciens virB9 and virB10 genes and to ptlF and ptlG of B. pertussis (28). Plasmid pB2A3 was used as a probe to screen a pVK102 B. abortus 2308 genomic library. Cosmid pVK8.3 was selected, and the complete DNA sequence was obtained (see Materials and Methods).
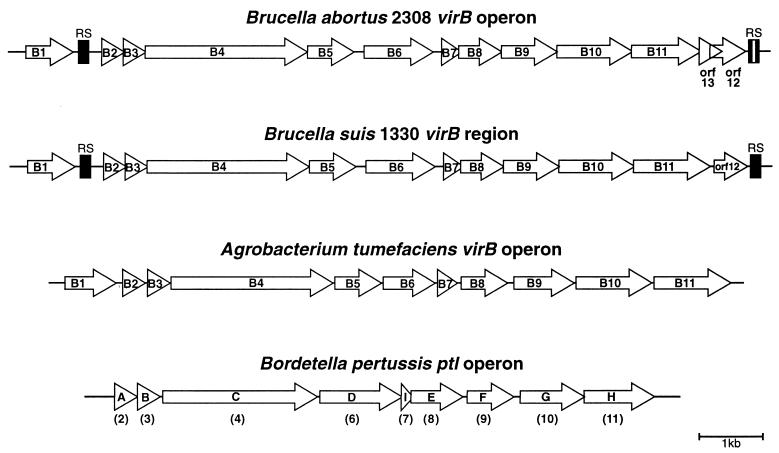
Analysis of the sequence revealed the presence of a collinear array of 13 open reading frames (ORFs) with the same orientation, all of them having putative ribosome binding sites. These ORFs are organized in a manner very similar to that of the recently published B. suis 1330 virB region (17) and of other operons coding for type IV secretion systems such as the A. tumefaciens virB and the B. pertussis ptl operons (Fig. 1). Comparison of this operon with the recently described B. suis 1330 virB region revealed that the two sequences are 99.7% identical. However, a few differences were observed. B. abortus 2308 has a 9-bp in-frame deletion at position 8897 on the virB10 homologue gene and a 1-bp insertion at position 10739 on the virB11 homologue gene. This insertion resulted in a VirB11 predicted protein of 363 amino acids (aa) that differs from the previously reported B. suis 1330 VirB11, which is 397 aa long. The VirB11 homologues of B. abortus 2308 and B. suis 1330 are 99% identical in the first 309 aa, but the sequences diverge at the C-terminal portion of the predicted proteins due to the 1-bp insertion present in the B. abortus 2308 virB11 gene. In order to assess whether this difference is due to divergence between the two Brucella species or to a mistake in the sequencing process, we amplified, cloned, and subsequently sequenced the corresponding virB11 3′ region of B. abortus S-19, B. suis 1330, and the most divergent member of the group, Brucella ovis REO198. We found that all sequences analyzed code for predicted products of 363 aa identical to that of B. abortus 2308 (data not shown). Thus, the difference was due to a misreading in the recently published B. suis 1330 virB11 sequence.
FIG. 1.
Schematic representation of B. abortus 2308 virB operon and comparison with B. suis 1330 virB, A. tumefaciens virB, and B. pertussis ptl operons. The white bar denotes the 18-bp inversion of the B. abortus 2308 RS with respect to the B. suis 1330 virB operon. The homologues between ptl and virB are shown in parentheses.
Another important difference is the presence of an additional overlapping ORF between VirB11 and ORF12 of B. abortus. This ORF, named ORF13, encodes a hypothetical 83-aa-long protein and has a conserved ribosome binding site sequence, and, as in other ORFs of the B. abortus virB operon, its ATG codon overlaps with the stop codon of the preceding ORF. ORF13 lacks similarity with any protein sequence deposited in GenBank databases.
It is interesting to note that B. abortus has two degenerated palindromic repeat sequences (RS in Fig. 1), one located between the VirB1 and VirB2 homologues and the other one downstream of ORF12. These RS are dispersed over the genome of all Brucella species and possibly behave as hot spots for insertion of IS711 (9). The presence of these flanking RS suggests that the Brucella VirB operon could have been acquired by horizontal transfer, although the region has a GC content similar to that of the rest of the genome. These RS have an organization similar to that found in the recently described B. suis virB region, except for an 18-bp inversion of the region flanked by the inverted repeats located at the end of the B. abortus operon (Fig. 1).
virB genes of B. abortus 2308 constitute an operon that is turned on in the stationary phase.
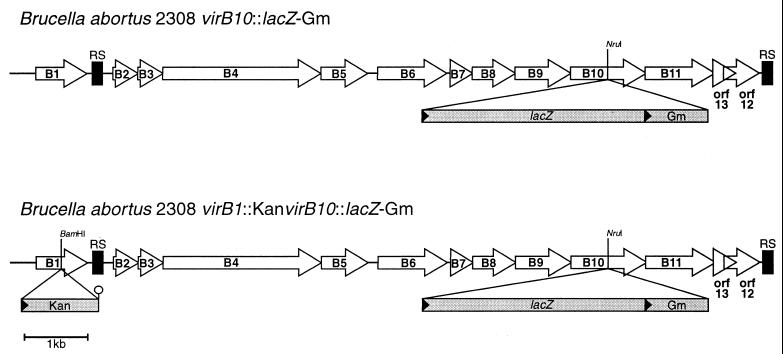
To study how these genes are expressed in B. abortus, we constructed two kinds of transcriptional fusions. First, a promotorless lacZ-Gm cassette was ligated at the NruI site on the coding sequence of the virB10 gene. This procedure generated strain B. abortus 2308 virB10::lacZ-Gm. Second, to determine if Brucella virB genes function as an operon starting at virB1, a polar insertional virB1 mutant was constructed on a B. abortus virB10::lacZ-Gm strain to generate the double mutant strain B. abortus virB1::Km virB10::lacZ-Gm (Fig. 2).
FIG. 2.
Schematic representation of the mutations introduced into the virB operon. The restriction sites where the cassettes were introduced are indicated. The hairpin indicates the transcriptional terminator of the kanamycin resistance cassette.
Both strains were used to measure β-galactosidase activity during growth. Figure 3A shows that, after 20 h of growth, β-galactosidase activity starts to increase, reaching a 30-fold induction after 36 h of growth (optical density at 600 nm of 2.6 [Fig. 3B]), indicating that the B. abortus virB region is turned on at the beginning of the stationary phase. However, β-galactosidase was not induced when a polar mutation was introduced in the virB1 gene (Fig. 3A). This result indicates that the B. abortus virB region behaves as an operon transcribed from virB1.
FIG. 3.
virB operon transcriptional induction. (A) Expression of the virB10::lacZ fusions of B. abortus 2308 virB10::lacZ-Gm (●) and B. abortus 2308 virB1::Kan virB10::lacZ-Gm (○). β-Galactosidase activity was expressed as A420/volume × A600. (B) Growth curve of B. abortus 2308 virB10::lacZ-Gm and B. abortus 2308 virB1::Kan virB10::lacZ-Gm strains. Symbols are the same as in panel A. OD600, optical density at 600 nm.
The virB operon is essential for intracellular replication of B. abortus.
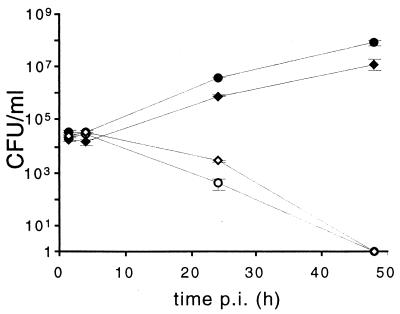
The effect of the virB operon on intracellular replication in HeLa cells was studied with a virB1 polar mutant (B. abortus virB1::Kan) generated as described in Materials and Methods. Figure 4 shows that at 4 h p.i. the number of intracellular B. abortus virB1::Kan mutants was the same as that of the wild-type parental strain. However, at longer times p.i., the CFU of the mutant decreased and no bacteria were recovered after 48 h. It can be observed that the number of wild-type intracellular bacteria grew exponentially during this period, reaching 7.4 × 107 CFU/ml at 48 h p.i. These results indicate that the B. abortus virB operon is essential for intracellular survival and is not involved with the invasion process.
FIG. 4.
Intracellular replication of parental B. abortus 2308 and virB1 polar mutant strain in HeLa cells. Monolayers of HeLa cells were inoculated with 5 × 107 CFU of bacteria. After 1 h of incubation at 37°C, cells were washed as described in the text and streptomycin and gentamicin were added. CFU were determined at the indicated times. Each determination is the average of two independent experiments performed in duplicate. In all cases, the standard error of each mean was less than 5%. ●, B. abortus 2308; ○, B. abortus 2308 virB1::Kan; ⧫, B. abortus 2308 virB1::Km(pVK8.3); ◊, B. abortus 2308 virB1::Km(pBBR4-virB1).
Plasmid pVK8.3, containing the entire virB operon introduced into B. abortus virB1::Kan, restored intracellular multiplication of the mutant to wild-type levels. Plasmid pBBR4-virB1, which contains only the complete virB1 gene under the lacZ promoter, failed to complement the polar virB1 mutation. This result confirms the polar nature of the virB1::Kan mutant.
Role of virB10 in B. abortus intracellular multiplication.
The A. tumefaciens virB operon is the best-studied type IV secretion system. Structure-function studies have been made with the T-complex transport machinery, assigning a possible role in transmembrane transport to most of its components. VirB10 is a member of this transport apparatus that has homologues in all type IV secretions systems described so far (4, 7). VirB10 is an inner membrane-bound protein that has a C-terminal periplasmic domain, as shown by PhoA fusion experiments (8).
To test the role of the B. abortus virB10 gene in intracellular replication, two targeted insertion mutants were constructed. B. abortus virB10::Kan has an insertion of a polar kanamycin resistance cassette interrupting the virB10 sequence at an NruI restriction site, and the B. abortus virB10::Gm mutant has an insertion of a nonpolar gentamicin resistance cassette interrupting the virB10 sequence at the same restriction site.
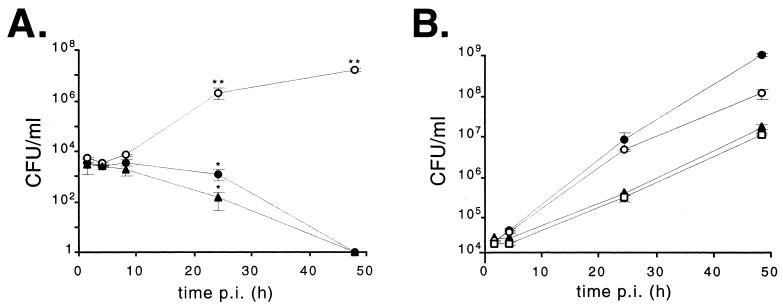
The intracellular behavior of both mutants was studied with HeLa cells. As shown in Fig. 5A, during the first 8 h p.i. the numbers of intracellular bacteria for both mutants were similar to that of the parental wild-type strain. At 24 h p.i., both polar and nonpolar virB10 mutants were affected in their ability to survive inside HeLa cells. However, the number of B. abortus virB10::Gm mutants remained significantly higher than the number of polar mutants. After 48 h p.i., no viable virB10 mutants were recovered, while the wild-type strain grew exponentially, reaching 1.38 × 107 CFU/ml. No host cell damage was observed with the wild type or the mutants during the course of the experiment. Thus, the lack of recovery of viable Brucella bacteria was the result of effective control of intracellular survival rather than the result of cell damage.
FIG. 5.
Intracellular replication of parental B. abortus 2308, virB10 mutants, and virB10 complemented mutant strains in HeLa cells. (A) B. abortus 2308 (○), B. abortus virB10::Kan (▴), and B. abortus virB10::Gm (●). (B) B. abortus 2308 (○), B. abortus 2308 virB10::Kan(pVK8.3) (▴), B. abortus 2308 virB10::Gm(pBBR-virB10) (●), and B. abortus 2308 virB10::Kan(pBBR-virB10) (□). Each value is the mean ± standard deviation of two independent experiments performed in duplicate. ∗, P < 0.05; ∗∗, P < 0.01 (compared with strain 2308). Statistical analysis was performed with a t test.
Plasmid pBBR2-virB10 containing the complete virB10 gene under the lacZ promoter was used to complement the nonpolar B. abortus virB10::Gm mutant strain. The B. abortus virB10::Kan polar mutant was complemented either with cosmid pVK8.3 containing the entire virB operon or with pBBR4-virB10. In every case, the ability of mutants to replicate within HeLa cells was restored, the bacteria reaching numbers similar to those of the wild-type strain (Fig. 5B). These results indicate that virB10 per se is essential for B. abortus intracellular multiplication within nonprofessional phagocytes and that virB11-ORF12-ORF13 are not required for intracellular survival.
Since many virB genes of A. tumefaciens and B. abortus, as well as those in other multicomponent secretion systems, overlap, it has been proposed that they are probably translationally coupled, suggesting that stoichiometry and order of expression may be important for the biogenesis of the secretion apparatus (4, 16). The fact that plasmid pBBR2-virB10 restored the intracellular replication of the B. abortus virB10::Gm mutant at levels similar to those of the wild type suggests that this hypothesis may not be applicable to the B. abortus virB operon.
virB10 is essential for virulence in mice.
The effect of B. abortus virB10 on virulence was studied in a mouse infection assay. Equal numbers of bacteria of the viable B. abortus 2308 wild-type strain or the virB10 polar and nonpolar insertion mutants were injected intraperitoneally into 90-day-old female BALB/c mice. Mice were sacrificed 15 days p.i., spleens were removed, and the number of Brucella bacteria recovered from spleens was estimated as described in Materials and Methods. As shown in Fig. 6, 4 × 105 viable bacteria were recovered from spleens of mice receiving the wild-type B. abortus 2308 whereas no viable bacteria were obtained from spleens of mice inoculated with the B. abortus virB10::Kan polar mutant. The number of bacteria recovered from spleens of mice inoculated with the virB10 nonpolar mutant was significantly lower than the number recovered from mice inoculated with the wild type (3.5 × 102 CFU/spleen) but significantly higher than CFU recovered from mice inoculated with the B. abortus virB10::Kan polar mutant (<10 CFU/spleen). B. abortus virB10::Kan harboring pVK8.3 and B. abortus virB10::Gm harboring pBBR2-virB10 recovered virulence, reaching viable counts similar to those of the wild-type parental strain.
FIG. 6.
Persistence of B. abortus 2308 and B. abortus virB10 mutants in spleens of infected mice. Mice were inoculated intraperitoneally with 104 CFU of wild-type B. abortus 2308, the virB10 mutant strains, or the virB10 complemented mutant strains as indicated. Numbers of bacteria in spleens were determined at 15 days p.i. Values are expressed as means ± standard errors of the means (n = 5). ∗, P < 0.05 (compared with the control group). Statistical analysis was performed with a t test.
These results indicate that virB10 and downstream sequences (virB11-ORF12-ORF13) are essential for Brucella pathogenesis in mice and suggest that the integrity of the virB operon is required for wild-type virulence.
DISCUSSION
The mechanism of virulence of Brucella spp. is not yet fully understood. It has become evident that virulence is linked to intracellular survival in brucellae. The understanding of the molecular events that allow brucellae to reach an intracellular niche where the bacteria freely multiply without causing any cellular damage will shed light on the mechanism of pathogenesis. This intracellular compartment is out of the reach of antibiotics and bactericidal plasma factors such as complement and antibodies and may be determinant of the chronic nature of this worldwide zoonosis.
In some other bacteria, pathogenesis has been associated with the integrity of a type IV secretion system. In A. tumefaciens, a type IV secretion system participates in delivering oncogenic T-DNA from the bacterium to the plant cell, whereas in B. pertussis it participates in the secretion of pertussis toxin. It is interesting to note that these type IV secretion systems display high similarity to E. coli Tra proteins, a cluster of genes that code for the conjugal pilus and mating pore. The tra region is organized in a manner similar to that of the A. tumefaciens, B. abortus, B. suis, and B. pertussis virB regions. TraL has high similarity to VirB1, a gene product that is absent in B. pertussis, L. pneumophila, Rickettsia prowazekii, and H. pylori.
Recently, O'Callaghan et al. described the presence of a virB region of B. suis 1330 (17) containing 11 genes highly similar to the 11 virB genes of A. tumefaciens and an extra ORF12 that shares homology with an adhesin of Pseudomonas fluorescens. Between the virB1 and virB2 homologues and at the end of ORF12, palindromic RS characteristic of intergenic regions of the Brucella genome were noted. This finding raises the question whether the virB1 gene is part of the same transcript as the rest of the virB genes. The authors reported that mutants with mutations in virB5, virB9, or virB10 were highly attenuated in an in vitro infection model whereas an ORF12 mutant was not and stressed that they do not know whether the mutants analyzed were affected in only one protein or whether they were polar.
Here, we demonstrate that B. abortus 2308 virB is indeed an operon, transcribed from the virB1 promoter, and show that, besides being required for intracellular survival, the operon constitutes a major determinant of virulence in mice. Moreover, the β-galactosidase fusion experiments showed that the operon is turned on at the beginning of the stationary phase of vegetative growth. Recently, Robertson and Roop described a Brucella hfq gene required for stationary-phase stress resistance and intracellular multiplication (21). They pointed out that stationary-phase gene expression might be important for virulence. This hypothesis may be applicable to the virB operon, although the pattern of virB induction in an intracellular environment remains to be characterized.
The requirement of an active virB operon for intracellular survival of brucellae may have two possible explanations: (i) the virB operon is essential to reach a competent intracellular replication niche or (ii) the virB operon is needed for replication once the intracellular replication niche has been established. Future investigation will focus on the elucidation of these questions.
Sola-Landa et al. described a B. abortus mutant in a two-component regulatory system, bvrR-bvrS, that displayed a reduced ability to enter into HeLa cells (26). This is not the case for virB mutants, since they have the same penetration rate as the wild-type strain, thus indicating that the virB operon does not participate in the mechanism of entry into the eukaryotic cell.
Homologues of A. tumefaciens virB10 and virB11 are present in type IV transport mechanisms of E. coli, B. pertussis, H. pylori, R. prowazekii, and L. pneumophila, while homologues of other components are not (7), indicating that these two proteins may play a common role in all these systems. Recently, Segal et al. reported that a type IV secretion system is required for plasmid RSF1010 conjugation in L. pneumophila (lvh genes) (24) and suggested that it may have an origin different from that of the dot-icm system. Both systems have conserved VirB10 and VirB11 homologues. VirB11 also has homologues in some type II secretion systems and pilin biosynthesis components. It has been proposed previously that there is a minimum core structure necessary for an active type IV transport system (7) that includes VirB4, VirB7, VirB9, VirB10, VirB11, and VirD4. The B. abortus virB operon contains homologues to all these proteins except for VirD4. In vitro cell infection assays showed that both polar and nonpolar B. abortus virB10 mutants failed to survive intracellularly, although some differences were observed. The results shown in Fig. 5 demonstrate that virB10 plays an essential role in intracellular survival in HeLa cells whereas virB11-ORF13-ORF12 do not. Moreover, both mutants showed different behaviors in mice depending on the nature of the mutation introduced in virB10. A polar mutation preventing the transcription of virB10-virB11-ORF13-ORF12 renders Brucella unable to establish an infection process in mice, being completely cleared from the spleen. The virB10 nonpolar mutant showed a 1,000-fold reduction in spleen counts compared with those of the wild-type strain. This phenotype is probably a consequence of defective secretion of putative effector molecules, due to the presence of a type IV secretion system that works deficiently in the absence of virB10.
The evolutionary linkage between DNA transfer and protein export systems has already been established; in some cases, they are functionally related and some components are capable of substituting for each other. One example is the A. tumefaciens T-DNA transport system, which is able to deliver IncQ plasmids to recipient cells (4). In view of the similarities that exist between the organization of the A. tumefaciens and B. abortus virB operons and considering that they are closely related species, the question whether the B. abortus type IV secretion system is competent to deliver non-self-transmissible DNA remains to be answered.
Although this report shows that the B. abortus virB operon is essential for intracellular multiplication and virulence, the effector molecule secreted by this system remains unknown. Future investigations directed toward the elucidation of the nature of the effector molecules will let us know the molecular mechanism underlying the infection process of this fascinating pathogen.
ACKNOWLEDGMENTS
We thank A. J. Parodi and J. J. Cazzulo for critical reading of the manuscript, J. E. Ugalde for helpful suggestions and for providing the gentamicin nonpolar resistance cassette, and F. Fraga for technical assistance.
This work was supported by grant PICT97-01767 from the Agencia Nacional de Promoción Científica y Tecnológica, Argentina. D.S. and R.A.U. are members of the Consejo Nacional de Investigaciones Científicas y Técnicas, CONICET, Argentina.
R.S. and D.J.C. contributed equally to the work.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Becker A, Schmidt M, Jager W, Puhler A. New gentamicin-resistance and lacZ promoter-probe cassettes suitable for insertion mutagenesis and generation of transcriptional fusions. Gene. 1995;162:37–39. doi: 10.1016/0378-1119(95)00313-u. [DOI] [PubMed] [Google Scholar]
- 3.Brand B C, Sadosky A B, Shuman H A. The Legionella pneumophila icm locus: a set of genes required for intracellular multiplication in human macrophages. Mol Microbiol. 1994;14:797–808. doi: 10.1111/j.1365-2958.1994.tb01316.x. [DOI] [PubMed] [Google Scholar]
- 4.Christie P J. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comerci D J, Pollevick G D, Vigliocco A M, Frasch A C, Ugalde R A. Vector development for the expression of foreign proteins in the vaccine strain Brucella abortus S19. Infect Immun. 1998;66:3862–3866. doi: 10.1128/iai.66.8.3862-3866.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbel J M. Brucellosis: an overview. Emerg Infect Dis. 1997;2:213–221. doi: 10.3201/eid0302.970219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Covacci A, Telford J L, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 8.Das A, Xie Y H. Construction of transposon Tn3phoA: its application in defining the membrane topology of the Agrobacterium tumefaciens DNA transfer proteins. Mol Microbiol. 1998;27:405–414. doi: 10.1046/j.1365-2958.1998.00688.x. [DOI] [PubMed] [Google Scholar]
- 9.Halling S M, Bricker B J. Characterization and occurrence of two repeated palindromic DNA elements of Brucella spp.: Bru-RS1 and Bru-RS2. Mol Microbiol. 1994;14:681–689. doi: 10.1111/j.1365-2958.1994.tb01306.x. [DOI] [PubMed] [Google Scholar]
- 10.Iñon de Iannino N, Briones G, Tolmasky M, Ugalde R A. Molecular cloning and characterization of cgs, the Brucella abortus cyclic β(1-2) glucan synthetase gene: genetic complementation of Rhizobium meliloti ndvB and Agrobacterium tumefaciens chvB mutants. J Bacteriol. 1998;180:4392–4400. doi: 10.1128/jb.180.17.4392-4400.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotob S I, Hausman S Z, Burns D L. Localization of the promoter for the ptl genes of Bordetella pertussis, which encode proteins essential for secretion of pertussis toxin. Infect Immun. 1995;63:3227–3230. doi: 10.1128/iai.63.8.3227-3230.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovach M E, Elzer P H, Hill D S, Robertson G T, Farris M A, Roop R M, 2nd, Peterson K M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 13.Kuldau G A, De Vos G, Owen J, McCaffrey G, Zambryski P. The virB operon of Agrobacterium tumefaciens pTiC58 encodes 11 open reading frames. Mol Gen Genet. 1990;221:256–266. doi: 10.1007/BF00261729. [DOI] [PubMed] [Google Scholar]
- 14.Moreno E, Stackebrandt E, Dorsch M, Wolters J, Busch M, Mayer H. Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the alpha-2 subdivision of the class Proteobacteria J. Bacteriol. 1990;172:3569–3576. doi: 10.1128/jb.172.7.3569-3576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicoletti P. Relationship between animal and human disease. In: Young E J, Corbel M J, editors. Brucellosis: clinical and laboratory aspects. Boca Raton, Fla: CRC Press, Inc.; 1989. pp. 41–51. [Google Scholar]
- 16.Nunn D. Bacterial type II protein export and pilus biogenesis: more than just homologies? Trends Cell Biol. 1999;9:402–408. doi: 10.1016/s0962-8924(99)01634-7. [DOI] [PubMed] [Google Scholar]
- 17.O'Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli M L, Bourg G, Foulongne V, Frutos P, Kulakov Y, Ramuz M. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- 18.Oka A, Sugisaki H, Takamani M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981;147:217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- 19.Pizarro-Cerda J, Meresse S, Parton R G, van der Goot G, Sola-Landa A, Lopez-Goni I, Moreno E, Gorvel J P. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect Immun. 1998;66:5711–5724. doi: 10.1128/iai.66.12.5711-5724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pohlman R F, Genetti H D, Winans S C. Common ancestry between IncN conjugal transfer genes and macromolecular export systems of plant and animal pathogens. Mol Microbiol. 1994;14:655–668. doi: 10.1111/j.1365-2958.1994.tb01304.x. [DOI] [PubMed] [Google Scholar]
- 21.Robertson G T, Roop R M., II The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol Microbiol. 1999;34:690–700. doi: 10.1046/j.1365-2958.1999.01629.x. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Segal G, Purcell M, Shuman H A. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22 kb region of the Legionella pneumophila genome. Proc Natl Acad Sci USA. 1998;95:1669–1674. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segal G, Russo J J, Shuman H A. Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. Mol Microbiol. 1999;34:799–809. doi: 10.1046/j.1365-2958.1999.01642.x. [DOI] [PubMed] [Google Scholar]
- 25.Segal G, Shuman H A. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect Immun. 1997;65:5057–5066. doi: 10.1128/iai.65.12.5057-5066.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sola-Landa A, Pizarro-Cerda J, Grillo M J, Moreno E, Moriyon I, Blasco J M, Gorvel J P, Lopez-Goni I. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol Microbiol. 1998;29:125–138. doi: 10.1046/j.1365-2958.1998.00913.x. [DOI] [PubMed] [Google Scholar]
- 27.Stachel S E, Nester E W. The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 1986;5:1445–1454. doi: 10.1002/j.1460-2075.1986.tb04381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ugalde R A. Intracellular lifestyle of Brucella spp. Common genes with other animal pathogens, plant pathogens and endosymbionts. Microbes Infect. 1999;1:1211–1219. doi: 10.1016/s1286-4579(99)00240-3. [DOI] [PubMed] [Google Scholar]
- 29.Weiss A A, Johnson F D, Burns D L. Molecular characterization of an operon required for pertussis toxin secretion. Proc Natl Acad Sci USA. 1993;90:2970–2974. doi: 10.1073/pnas.90.7.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winans S C, Walker G C. Conjugal transfer system of the IncN plasmid pKM101. J Bacteriol. 1985;161:402–410. doi: 10.1128/jb.161.1.402-410.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]