Abstract
The products of two adjacent genes in the chromosome of Methanococcus jannaschii are similar to the amino and carboxyl halves of phosphonopyruvate decarboxylase, the enzyme that catalyzes the second step of fosfomycin biosynthesis in Streptomyces wedmorensis. These two M. jannaschii genes were recombinantly expressed in Escherichia coli, and their gene products were tested for the ability to catalyze the decarboxylation of a series of α-ketoacids. Both subunits are required to form an α6β6 dodecamer that specifically catalyzes the decarboxylation of sulfopyruvic acid to sulfoacetaldehyde. This transformation is the fourth step in the biosynthesis of coenzyme M, a crucial cofactor in methanogenesis and aliphatic alkene metabolism. The M. jannaschii sulfopyruvate decarboxylase was found to be inactivated by oxygen and reactivated by reduction with dithionite. The two subunits, designated ComD and ComE, comprise the first enzyme for the biosynthesis of coenzyme M to be described.
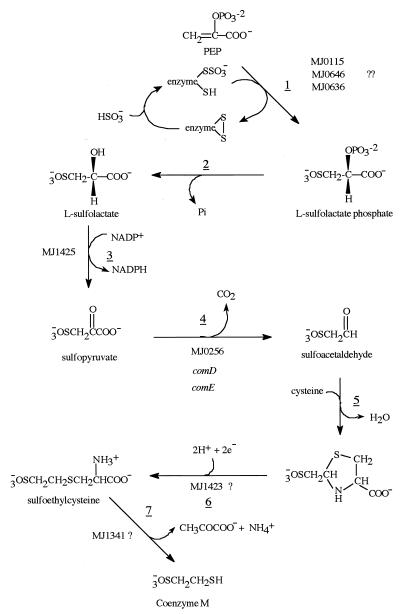
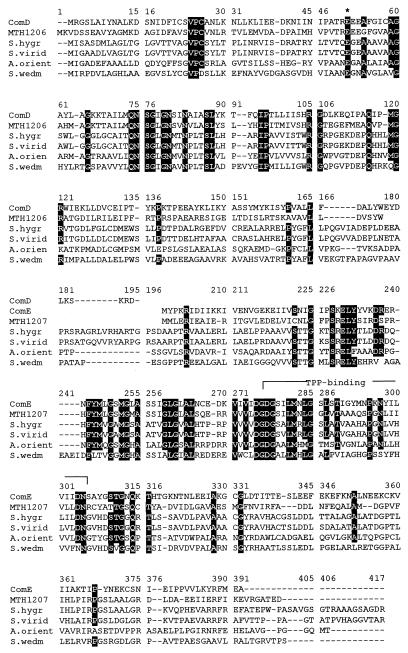
Coenzyme M (2-mercaptoethanesulfonic acid) was originally characterized as one of several coenzymes involved in the formation of methane in the methanoarchaea (14). Recently it has been shown to function as a coenzyme in the bacterial metabolism of aliphatic alkenes (1). Despite the fact that the pathway for its biosynthesis (Fig. 1) has been known for a number of years (35–37), no genes or enzymes involved in its biosynthesis have been identified. One of the steps in the biosynthesis of coenzyme M, the decarboxylation of sulfopyruvate to sulfoacetaldehyde, is chemically very analogous to the decarboxylation of phosphonopyruvate that occurs in the biosynthesis of natural products containing a C-P bond. Among these are fosfomycin (19), phosphinothricin (30), and bialaphos (24), each produced by various species of Streptomyces. The sequence homology among the Streptomyces genes for phosphonopyruvate decarboxylase (30), the genes contained in Methanococcus jannaschii MJ0256 (7), and the Methanobacterium thermoautotrophicum genes MT1206 and MT1207 (31) (Fig. 2) has prompted us to clone and overexpress the two proteins encoded by the MJ0256 open reading frame. This work has established that these two protein products catalyze the decarboxylation of sulfopyruvate to form sulfoacetaldehyde and CO2. The enzyme has been named sulfopyruvate decarboxylase, and the gene has been named comDE, to indicate that the enzymatic reaction is the fourth expected in the biosynthesis of coenzyme M from phosphoenol pyruvate and bisulfite. From sequence comparisons with other similar thiamine-PP (TPP)-dependent enzymes (Fig. 2) and from the nature of the reaction catalyzed, it is clear that this enzyme is a TPP-dependent enzyme since it contains the TPP-binding motif (DGDGSILMNLGSLSTIGYMNPKNYILVIIDN) (18). Unexpectedly, the enzyme was found to be readily inactivated by exposure to oxygen.
FIG. 1.
Pathway for the biosynthesis of coenzyme M. The underlined numbers refer to the individual reaction steps in the pathway.
FIG. 2.
Primary sequence alignments of ComD and ComE (NCBI identifier number, gi 2129192). The other five sequences shown are the putative phosphonopyruvate decarboxylase from M. thermoautotrophicum (MTH1206 [gi 2622316] and MTH1207 [gi 2622315]), the phosphonopyruvate decarboxylases of Streptomyces hygroscopicus (S. hygr) (gi 5545271), Streptomyces viridochromogenes (S. virid) (gi 3319780), Amycolatopsis orientalis (A. orien) (gi 5051794), and Streptomyces wedmorensis (S. wedm) (gi 1061008). Residues identical in all six proteins and those identical in five of the proteins are blackened. The conserved amino acids for the TPP-binding site are overlined. The protein sequences were aligned with ClustalW, version 1.7.
MATERIALS AND METHODS
Chemicals.
Sulfopyruvate and sulfoacetaldehyde were prepared as previously described (36, 37). Zhibing Lu, Department of Chemistry, University of New Mexico, Albuquerque, N.Mex., supplied the phosphonopyruvate.
Identification, cloning, and high-level expression of the gene product.
The two proteins encoded by the MJ0256 open reading frame were amplified by PCR using genomic DNA from M. jannaschii (David E. Graham, Urbana, Ill.) as the template. The synthetic oligonucleotide primers 5′-GGTGGTCATATGAGAGGTAGCTTAGCAATATAC-3′ and 5′-GATCGGATCCTTAACTCCTTTTTATCGCTTC-3′ were used in a PCR to amplify both ComD and ComE proteins simultaneously in one expression vector. The synthetic oligonucleotide primers 5′-GGTGGTCATATGAGAGGTAGCTTAGCAATTATAC-3′ and 5′-GATCTTATCCTTACTTTTCTAAATCG-3′ were used in a PCR to amplify ComD, and the synthetic oligonucleotide primers 5′-GGTGGTCATATGTATCCAAAGAGAATAG-3′ and 5′-CATCGGATCCTTAACTCCTTTTTATCGCTTC-3′ were used to amplify ComE. Each group of primers was used to insert the NdeI and BamHI restriction sites at their 5′ and 3′ ends. PCR was performed with 1 μg of genomic DNA as the template and with 20 μmol of each primer, 3.75 U of Amplitaq DNA polymerase, and 10 μl of 10× PCR buffer (Perkin-Elmer, Branchburg, N.J.) in a final volume of 100 μl. Each cycle was set for 1 min of denaturation at 95°C, 2 min of annealing at 55°C, and 3 min of extension at 72°C, and 35 reaction cycles were carried out in a DNA thermal cycler. After purification of the PCR products via absorption and desorption to a QIAquick spin column (Qiagen, Valencia, Calif.), the PCR products were digested with NdeI and BamHI and then cloned into NdeI-BamHI-digested pT7-7 plasmid vector to obtain the reconstructed plasmids. The recombinant plasmids were transformed to E. coli BL21 (DE3), which was grown in Luria-Bertani broth supplemented with 100 μg of ampicillin per ml at 37°C to an absorbance of 0.9 to 1.0 at 600 nm. Protein production was induced with 28 mM lactose. After induction for 4 h at 37°C, the cells were harvested by centrifugation (4,000 × g, 5 min) and frozen at −20°C until used. The heterologously produced protein was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% polyacrylamide) of the SDS-soluble cellular proteins.
Preparation of cell extracts and heat purification.
Cell extracts with the overproduced proteins were prepared by sonication of the cells (300 mg wet weight) suspended in 3 ml of TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid] buffer (50 mM TES, 10 mM MgCl2, 20 mM mercaptoethanol, pH 7.0) followed by centrifugation (10 min, 16,000 × g). SDS-PAGE analysis of the resulting insoluble pellets and the solubilized proteins showed that most (>90%) of the overproduced proteins were present in the insoluble pellet.
Heat purification of the solubilized proteins was performed in TES buffer or in phosphate buffer (0.15 M potassium phosphate, 25 mM TES, 5 mM MgCl2, 10 mM mercaptoethanol, pH 7.0). Heating solubilized proteins in phosphate or TES buffer at 80°C for 15 min followed by centrifugation (10 min, 16,000 × g) removed most of the Escherichia coli proteins. The resulting protein solutions were used for the activity analyses reported here. E. coli extracts not containing the overexpressed genes were treated in the same manner and were used for control experiments.
Solubilization and refolding of insoluble overexpressed proteins.
The insoluble pellets derived from the sonications of 150 mg (wet weight) of cells from the ComD and ComE overexpressions were combined and washed three times with buffers as described previously (6). Based on the SDS analyses of the samples, the desired proteins were present in a ratio of 1:1 and were essentially pure. The washed pellet was then dissolved in 30 to 40 μl of 0.1 M Tris buffer (pH 8.0) containing 6 M guanidinium HCl, 2 mM EDTA, and 0.3 M dithiothreitol (DTT). After 2 h at room temperature followed by centrifugation (16,000 × g, 45 min), the denatured proteins were refolded by rapid dilution (100-fold) into a buffer consisting of 0.1 M Tris (pH 8), 0.5 M l-arginine, 2 mM EDTA, 20 mM mercaptoethanol, 10 mM MgCl2, 10 μM TPP, 10 μM flavin adenine dinucleotide (FAD), and 50 mM potassium phosphate under an atmosphere of argon. The resulting clear solution was then kept at 10°C overnight and concentrated with an Amicon 10 Centricon concentrator. SDS-PAGE analysis of resulting refolded proteins showed only two equally heavy bands with molecular masses of 17 and 23 kDa, corresponding to ComD and ComE, respectively. Protein concentrations were determined with the Bio-Rad protein assay using bovine serum albumin as a standard.
Size exclusion chromatography and measurement of the protein flavin content.
Molecular size exclusion chromatography was performed at room temperature with a Superose 12 HR 10/30 column (Pharmacia Biotech), equilibrated in 50 mM sodium phosphate (pH 7.0)–150 mM NaCl, using a flow rate of 0.4 ml/min. Molecular weight markers consisted of apoferritin (440,000), alcohol dehydrogenase (150,000), bovine serum albumin (67,000), ovalbumin (45,000), carbonic anhydrase (29,000), and cytochrome c (12,400). The elution of the refolded sulfopyruvate decarboxylase was monitored by measurement of the absorbance at 280 nm. FAD was monitored both by absorbance (450 nm) and by fluorescence (450-nm excitation, 520-nm emission). Fractions (800 μl) were collected and analyzed for sulfopyruvate decarboxylase activity and proteins (SDS-PAGE). To increase the sensitivity for the analysis of the FAD, the individual fractions were adjusted to pH 3 and the fluorescence was remeasured at the indicated wavelengths. This procedure was expected to increase the detection limit for the FAD because of the 10-fold increase in fluorescence of FAD at this pH.
Measurement of enzymatic activities, Km, and oxygen sensitivity.
The decarboxylase activities of ComDE were generally measured in phosphate buffer (150 mM potassium phosphate, 1 mM MgCl2, and 2 mM mercaptoethanol, pH 7.0). When phosphate buffer was not used, the activity was measured in TES buffer (50 mM TES, 10 mM MgCl2, 20 mM mercaptoethanol, pH 7.0). The assay was conducted in 100 μl of the phosphate buffer containing 2.1 mM sulfopyruvate, 2 mM dithionite, and 0.2 mM methylviologen under atmospheres indicated in Tables 1 and 2. The standard assay was performed under the same conditions but under an atmosphere of argon. After the addition of the appropriately diluted enzyme solution (5 to 10 μl), the assay mixture was incubated for 15 min at 80°C. Methylviologen was added as a visual redox indicator to confirm that the solutions contained no free oxygen. The resulting incubation mixtures were derivatized with 2,4-dinitrophenylhydrazine (DNPH) for high-pressure liquid chromatography (HPLC) and/or thin-layer chromatography (TLC) analyses or purified for gas chromatography-mass spectrometry (GC-MS) analysis as described below. One unit of enzyme activity refers to 1 μmol of sulfoacetaldehyde produced per min in the standard assay.
TABLE 1.
Oxygen sensitivity of the sulfopyruvate decarboxylase (ComDE)
| Expt | Treatment of the enzyme prior to enzymatic assaya | Conditions of the enzymatic assayb | Sulfoacetaldehyde produced (nmol) | % Conversionc |
|---|---|---|---|---|
| 1 | None | Argon, SP | 208 | 99 |
| 2 | None | Air, SP | 0.0 | 0 |
| 3 | No gened | Argon, SP, dt, mv | 0.0 | 0 |
| 4 | Heated in air for 30 min at 80°C | Argon, SP | 19 | 9 |
| 5 | Heated in air for 30 min at 80°C | Argon, SP, DTT | 17 | 8 |
| 6 | Heated in air for 30 min at 80°C | Argon, SP, dt, mv | 204 | 97 |
| 7 | Heated in air for 30 min at 80°C with SP | Argon, dt, mv | 206 | 98 |
For experiments 1 and 2, an E. coli cell extract containing the ComDE enzyme was diluted in 150 mM phosphate buffer (pH 7.0) to such an extent that it was able to convert 95 to 99% of the substrate to product when incubated under the standard conditions in septated glass vials under argon (experiment 1). This same enzyme dilution was then used in experiment 2. An equivalent dilution of an E. coli cell extract without the ComDE enzyme was used for experiment 3. Each assay mixture contained 5.8 μg of protein. For experiments 4 to 7, identical dilutions of the heat-treated ComDE cell extract in 150 mM phosphate buffer (pH 7.0) were exposed to air, heated as indicated, and then assayed for activity. Each of these assay mixtures contained 1.1 μg of protein.
The compounds were added to the enzyme solutions in the order indicated as 0.1 M anaerobic solutions to produce the following final concentrations: 2.1 mM sulfopyruvate (SP), 4 mM DTT, 2 mM sodium dithionite (dt), 0.2 mM methylviologen (mv). The presence or absence of methylviologen had no effect on the assay results. After incubation for 15 min at 80°C in the indicated atmosphere, the amounts of sulfopyruvate and sulfoacetaldehyde were assayed by HPLC of the DNPH derivatives.
In each assay a measurable amount of the sulfopyruvate starting material was detected.
Cell extracts were obtained from BL21 E. coli cells that contained the plasmid without the comDE gene.
TABLE 2.
Thermostability of the sulfopyruvate decarboxylase
| Expt | Heating conditionsa | Conditions of enzymatic assayb | Sulfoacetaldehyde produced (nmol) | % of maximum conversion |
|---|---|---|---|---|
| 1 | No heating | Argon, SP, dt, mv | 208 | 99 |
| 2 | TES buffer | Argon, SP | 40 | 19 |
| 3 | TES buffer | Argon, SP, dt, mv | 38 | 18 |
| 4 | Phosphate buffer | Argon, SP, dt, mv | 206 | 98 |
| 5 | Phosphate buffer | Argon, SP | 27 | 13 |
Crude cell extracts containing the overproduced ComDE protein (58 μg of protein/ml) were heated in the indicated pH 7.0 buffer for 15 min at 80°C under an atmosphere of argon and then centrifuged; the clear liquid was transferred into a new septated glass vial under an atmosphere of argon (21 μg of protein/ml after the heat treatment).
For the determination of the enzymatic activities, the nonheated (5.8 μg of protein) and heat-purified cell extracts (1.1 μg of protein) were mixed with 2.1 mM sulfopyruvate (SP) for 15 min at 80°C under an atmosphere of argon. The addition of 2 mM dithionite (dt) was always accompanied by addition of 0.2 mM methylviologen (mv). The amounts of sulfopyruvate and sulfoacetaldehyde, as their DNPH derivatives, were analyzed by HPLC.
For the determination of the Km, the heat-purified ComDE cell extract was used for the enzyme assay. Aliquots were removed from an incubation mixture as a function of time and were immediately derivatized with the acidic DNPH solution to stop the reaction, and the DNPH derivative of the sulfoacetaldehyde generated was quantitated by HPLC. A concentration of protein was selected to give a linear time course over the 15-min duration of the assay. The sulfopyruvate concentration was varied from 0.5 to 1.8 mM, and the assay was performed in the presence of 2 mM dithionite–0.2 mM methylviologen for 5 to 15 min at 80°C. Performing the assay at higher sulfopyruvate concentrations led to substrate inhibition.
To test the oxygen sensitivity of the ComDE extract, the activity assay of the heat-purified enzyme was performed with sulfopyruvate under an atmosphere of air. For the preincubations in air, the enzyme-containing solution was exposed to air for 30 min at room temperature or at 80°C; then enzymatic activity was determined by addition of sulfopyruvate (2.1 mM) or supplementary dithionite (2 mM) and methylviologen (0.2 mM) or DTT (4 mM) under an atmosphere of argon for 15 min at 80°C. To preincubation mixtures that already contained sulfopyruvate no more substrate was added for the activity assay. Then the DNPH derivatives were analyzed by HPLC.
Testing substrate specificity of the sulfopyruvate decarboxylase.
The decarboxylation of pyruvate was measured spectrophotometrically as described previously for acetolactate synthase (25). Briefly, the ComDE extracts were incubated with 1 mM pyruvate for 2 h at 50°C in 100 μl of 0.3 M phosphate buffer (pH 7.0). The reaction was stopped by adding sulfuric acid to a final concentration of 1%. After being heated for 15 min at 60°C, the precipitated proteins were removed by centrifugation and the separated clear liquid was mixed with creatine (0.15% final concentration)–α-naphthol (1.5% final concentration)–0.3 M potassium phosphate buffer (pH 7.0) to a final volume of 900 μl. After being heated again for 15 min at 60°C, the samples were cooled to room temperature, and the absorbance was measured at 525 nm. Using acetoin as the standard, this assay method readily measured 20 nmol of product.
The decarboxylation of phosphonopyruvate to phosphonoacetaldehyde was measured under the same conditions as described for sulfopyruvate, and the phosphonoacetaldehyde formation was assayed by derivatization with DNPH as described by Nakashita et al. (24).
Product identification.
The sulfoacetaldehyde produced by the decarboxylation of sulfopyruvate was identified by three different methods: TLC and HPLC analyses of the DNPH derivative and GC-MS analysis of the methyl ester dimethyl acetal derivative. In each case a standard incubation was run with sulfopyruvate and enough enzyme to convert all of the substrate to sulfoacetaldehyde. The resulting product was then converted into the desired derivative. Known sulfoacetaldehyde samples processed in an identical manner were used to establish the identity of the generated compound.
For the formation of the DNPH derivative, 0.02 M DNPH in 2 M HCl, a 2 M excess over the amount of sulfopyruvate used in the incubation, was added to the mixture at the completion of the incubation. After 1 h at room temperature, the pH of the solution was adjusted to 6 to 7 by the addition of 2.5 M NaOH, and the unreacted DNPH and precipitated proteins were removed by centrifugation (5,000 × g, 10 min). The separated clear yellow solution was then diluted to 0.5 ml with water, and the contained DNPH derivatives were analyzed by HPLC. Samples (10 μl) were injected onto a reversed-phase HPLC column (AXXI CHROM ODS; 5 μm, 25 cm), which was protected with a guard column (RP-18 NEWGUARD; 7 μm, 1.5 cm) and controlled by the Shimadzu LC-6A HPLC system operating at a rate of 0.5 ml/min. A sodium acetate (NaOAc)-methanol gradient was used for separation. The NaOAc-methanol gradient used consisted of 25 mM NaOAc (pH 6.0; 0.02% NaN3, 5% methanol) for the first 5 min followed by the linear methanol gradient to 80% methanol over the next 40 min. The elution was monitored by measuring absorbance at 360 nm (Shimadzu SPD-6VA). The DNPH derivatives of sulfopyruvate and sulfoacetaldehyde showed retention times of 25.80 and 33.40 min, respectively, in this gradient system.
For TLC analysis, 50 μl of the DNPH-derivatized sample was placed on a preparative C18 (Waters Corporation) column (0.2 by 1 cm; 55- to 105-μm resin), and the column was then washed with 100 μl of water, which was discarded. The DNPH derivatives of sulfopyruvate and sulfoacetaldehyde were then eluted with 200 μl of water followed by 200 μl of 10% methanol. After concentration of the eluted sample by evaporation under a stream of nitrogen gas, the resulting sample was analyzed by TLC using the solvent system butanol-acetic acid-water (12:3:5 [vol/vol/vol]). The DNPH derivatives of sulfopyruvate and sulfoacetaldehyde had Rf values of 0.23 and 0.48, respectively. Unreacted DNPH had an Rf value of 0.76.
For analysis by GC-MS, an equal volume of 95% ethanol was added to an incubation mixture, which was heated for 5 min at 100°C and centrifuged (16,000 × g, 10 min) to produce a clear solution. After evaporation of the ethanol and water with a stream of nitrogen gas, the resulting residue was dissolved in 0.5 ml of water and the solution was passed through a Dowex 50-8X (H+) column (0.5 by 1.1 cm) and evaporated to dryness. The resulting sample was dissolved in methanol (100 μl), and an ether solution of diazomethane (∼300 μl) was added to produce a cloudy yellow suspension that was clarified by centrifugation. The resulting clear solution was separated, evaporated to dryness, and dissolved in methylene dichloride, and the sulfopyruvate and sulfoacetaldehyde derivatives were analyzed by GC-MS as previously described (36).
RESULTS AND DISCUSSION
The MJ0256 gene codes for two proteins.
The primers for the cloning of the MJ0256 gene were designed to clone the gene from position 241512 to position 242598 of the M. jannaschii genome (7). This region would code for an enzyme with a molecular weight of 36,000. SDS-PAGE analysis of the MJ0256-encoded overproduced protein could not confirm high overproduction of this enzyme, although enzymatic activities could be measured. Close inspection of the gene sequence for the MJ0256 gene showed the presence of a series of stop codons beginning with TAA at position 242022 about midway down the sequence. Based on this finding, we cloned the two genes and overexpressed the two individual proteins as outlined in Materials and Methods. SDS-PAGE analysis of the two individual proteins, which we call ComD and ComE, showed molecular weights of 17,000 and 23,000, respectively. Since enzymatic activity was observed only in the presence of the combined proteins, we designate the active enzyme ComDE. Thus it is clear that the MJ0256 gene encodes two different proteins. In M. thermoautotrophicum the genes homologous to the MJ0256 gene are annotated as two separate genes, MTH1206 and MTH1207, and relate to MJ0256 as shown in Fig. 2. The phosphonopyruvate decarboxylases, in contrast, are all encoded by one gene, as are the more distantly related acetolactate synthases (ALSs) and pyruvate oxidases (POXs).
Characterization of the enzyme.
Only in the presence of a mixture of both ComD and ComE could enzymatic activity be measured; the individual proteins had no measurable activity. According to the measured molecular weight of 210,000 for the active ComDE enzyme, prepared by refolding equal amounts of the two proteins, the active protein would correspond to an α6β6 dodecamer. SDS-PAGE analysis of the separated enzymatically active protein peak from the molecular size column showed only ComD and ComE protein bands of equal intensities. The fractions corresponding to the elution positions of the αβ dimer to the α4β4 octamer of the ComDE protein or the measured elution positions of the monomers of ComD and ComE showed neither enzymatic activity nor UV absorbance nor protein bands on the SDS-PAGE gel after analyses of the concentrated fractions. This observation indicated that the ComDE protein did not dissociate under the separation conditions used and existed only as the α6β6 dodecamer.
Absence of both FAD and a FAD-binding site in the enzyme.
Based on the sequence alignments of the ComDE protein with other TPP-dependent enzymes, it is clear that the ComDE protein is a member of a family of enzymes that includes acetohydroxyacid synthase (AHAS), ALS, benzoylformate decarboxylase, glyoxylate carboligase, indole pyruvate decarboxylase, pyruvate decarboxylase, the acetyl phosphate-producing POX, and the acetate-producing POX (5, 8–10, 15, 23, 33). Each of these enzymes contains a bound TPP, which serves as the coenzyme for the decarboxylation. In addition, four of these enzymes, AHAS, ALS, and both of the POXs, also require a bound FAD for activity. The one exception to this rule is the pH 6 acetolactate-forming enzyme from Aerobacter aerogenes (32). However, only for E. coli POX is the FAD directly involved in the catalysis, where it functions to transfer electrons to the quinones (13). In the remaining enzymes, the FAD appears to play only a structural role (11). Several members of this group have been shown to be oxygen sensitive and include the FAD-containing AHAS isolated from Methanococcus aeolicus (38) and the ALS isozyme II from Salmonella enterica serovar Typhimurium. (28, 29). For the M. aeolicus enzyme, the oxygen sensitivity is a function of the state of purity of the enzyme, but neither of these enzymes could be reactivated, as we have also shown for the ComDE enzyme.
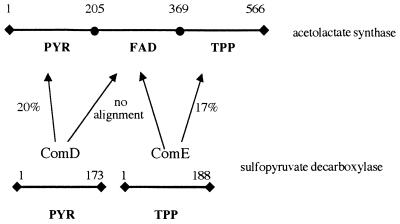
The elution profile of the molecular size column showed an A450/A280 ratio of less than 0.003 for the ComDE protein and no fluorescence for FAD, indicating the absence of FAD in the enzyme, despite the fact that the enzyme was refolded in the presence of FAD. Based on the known molecular weight of the enzyme this would correspond to less than 0.01 mol of flavin per mol of the α or β subunit. More-sensitive spectroscopic analysis of the isolated ComDE protein for FAD fluorescence also did not confirm the presence of FAD. These findings are in agreement with the sequence alignments of the ComDE protein with AHAS and POX, which show the absence of the FAD-binding domain in the enzyme (Fig. 3). Attempts to align any section of the ComD or ComE sequences to the FAD-binding region of the AHAS of E. coli failed (Fig. 3). From both these sequence alignments and the experimental results, it seems likely that the ComDE protein has no FAD-binding domain.
FIG. 3.
Schematic representation of domains of TPP-dependent enzymes. PYR represents the substrate-binding domain, FAD represents the FAD-binding domain, and TPP represents the TPP-binding domain, as was used for POX (20). Numbers above the lines, amino acids spanning the domain; percentages next to the arrows, sequence identities of the sulfopyruvate decarboxylase proteins Com D and ComE of M. jannaschii to the ALS domains of E. coli. These protein sequences were aligned with the ClustalW, version 1.7, program.
Reactions catalyzed by the ComDE protein and its oxygen sensitivity.
Extracts of the E. coli cells containing the heterologous overproduced enzyme and the enzyme regenerated from the inclusion bodies were both found to quantitatively convert sulfopyruvate to sulfoacetaldehyde when the enzyme assays were performed under an atmosphere of argon (Table 1). No conversion of the substrate to the product was detected when the activity assay was done in air under the same conditions (Table 1). This result shows that the ComDE enzyme was clearly inactivated by oxygen when heated in air at 80°C. No decarboxylation of sulfopyruvate was observed by incubation with cell extracts of E. coli strain BL-21 that contained the plasmid without the comDE genes. Exposure of the ComDE cell extract, with or without substrate, to air for 30 min at room temperature or at 80°C resulted in no loss of activity when the extract was assayed in the presence of dithionite under argon. These results show that the inactivation of the ComDE enzyme by exposure to air at 80°C is reversible after addition of dithionite. The extent of the residual activity, which could be detected in samples just by replacement of air with argon, was not very reproducible. Furthermore, the only reducing agent that was capable of reactivating the ComDE enzyme was dithionite; mercaptoethanol and DTT had no reactivating power.
The discovery that this enzyme is inactivated by exposure to oxygen and reactivated by dithionite was not a previously reported property for TPP-dependent enzymes. Many different mechanisms for how proteins can alter their activities in response to the presence or absence of oxygen have been described (3). None of these mechanisms are presently known to involve TPP-containing enzymes. We have considered and tested four possible reasons for the observed oxygen sensitivity of the ComDE enzyme: the presence of an oxidizable reduced flavin, as is known to occur in chorismate synthase (22) and possibly in the aerotaxis response system of E. coli (26); a hemin-based oxygen sensor-like protein, as occurs in rhizobia (27); an oxygen-sensitive Fe-S center, as occurs in aconitases and other Fe-S-containing enzymes (4); and, finally, an oxygen-dependent sulfenic acid or disulfide formation (2, 12). Our observations so far (White et al., unpublished results) indicate that the enzyme is inactivated by O2 by a process different from any other previously described and involves the oxidation of the hydroxyethyl-TPP intermediate as has been reported for pyruvate decarboxylase (34).
Alternate substrates.
The sequence homologies between our enzyme and the phosphonopyruvate decarboxylases (Fig. 2) and ALSs suggested that the ComDE enzyme might use phosphonopyruvate and/or pyruvate as substrates. Neither of these compounds was found to be a substrate for the ComDE enzyme. The measured activity for phosphonopyruvate decarboxylase was <0.09 μmol/min/mg of protein, and that for the ALS was <0.04 μmol/min/mg of protein. Thus the ComDE enzyme has diverged to such an extent from the phosphonopyruvate decarboxylases and ALSs that phosphonopyruvate and pyruvate can no longer serve as substrates.
Thermostability.
Since the ComDE enzyme operates in a hyperthermophilic anaerobic methanoarchaea, we expected that heating the E. coli cell extract containing the overproduced enzyme would precipitate most of the E. coli proteins and leave our desired enzyme in solution. When our extracts, present in TES buffer (pH 7.0), were heated for 15 min at 80°C either in the presence or absence of dithionite and methylviologen, most of the activity of the decarboxylation for sulfopyruvate was lost (Table 2, experiments 2 and 3). We found, however, that if the extract was made 0.15 M in potassium phosphate buffer (pH 7.0) before the heating, then all of the activity could be recovered in the soluble portion of the preparation (Table 2, experiment 4) if the activity assay was performed with dithionite and methylviologen. The phosphate did not protect the ComDE protein from oxidative destruction, since the activity was greatly reduced without the addition of dithionite (Table 2, experiment 5). Only heating in the presence of phosphate and assaying for decarboxylation in the presence of dithionite and methylviologen retained a fully active ComDE enzyme.
Kinetic constants.
The Km of sulfopyruvate for the enzyme purified by heat treatment in the presence of phosphate is 0.64 mM, and Vmax is 52 μmol/min/mg of protein.
The presence of sulfopyruvate decarboxylase in a coenzyme M gene cluster.
Having established the functional role for the MJ0256 gene products, it is important to establish if this gene could reside in a gene cluster or operon in any methanoarchaea. The discovery of such a gene cluster or operon could lead to the identification of other genes involved in the biosynthesis of coenzyme M. In M. jannaschii the only gene that is operonally connected to MJ0256 is MJ0257, a conserved protein homologous to M. thermoautotrophicum MTH1039 (31). Both the MJ0257 and MTH1039 genes could possibly encode an archaeal Fe-S oxidoreductase (21). Since a reduction must occur in step 5 of the coenzyme M biosynthetic pathway (Fig. 1), it is possible that this gene could supply an enzyme to perform this function. In M. thermoautotrophicum the genes homologous to MJ0256 are annotated as two separate genes, MTH1206 and MTH1207, which are related to MJ0256 as shown in Fig. 2. Upstream from these genes is MHT1205, which is homologous to MJ1425, a gene that has been shown to have sulfolactate dehydrogenase activity, catalyzing the NAD-dependent oxidation of sulfolactate to sulfopyruvate (reaction 3, Fig. 1) (17). Since MTH1204 is most likely the purM gene involved in purine biosynthesis (31), MTH1205, MTH1206, and MTH1207 may constitute a gene cluster for coenzyme M biosynthesis. MTH1208 encodes a DNA-dependent DNA polymerase elongation subunit and is expressed in the opposite direction.
The discovery of a clear evolutionary connection between an enzyme involved in the biosynthesis of a coenzyme in the methanoarchaea and several of the well-established bacterial enzymes indicates that the evolution of genes for the archaeal coenzymes is closely linked to the evolution of bacterial enzymes. This has now been observed for the enzymes involved in the biosynthesis of coenzyme B (20) and supports the idea that methanogenesis in the methanoarchaea is not an ancient process (16).
ACKNOWLEDGMENT
This work was supported by the National Science Foundation grant MCB963086.
REFERENCES
- 1.Allen J R, Clark D D, Krum J, Ensign S A. A role for coenzyme M (2-mercaptoethanesulfonic acid) in a bacterial pathway of aliphatic epoxide carboxylation. Proc Natl Acad Sci USA. 1999;96:8432–8437. doi: 10.1073/pnas.96.15.8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison W S. Formation and reactions of sulfenic acid in proteins. Accounts Chem Res. 1976;9:293–299. [Google Scholar]
- 3.Bauer C E, Elsen S, Bird T H. Mechanisms for redox control of gene expression. Annu Rev Microbiol. 1999;53:495–523. doi: 10.1146/annurev.micro.53.1.495. [DOI] [PubMed] [Google Scholar]
- 4.Beinert H, Kennedy M C, Stout C D. Aconitase as iron-sulfur protein, enzyme, and iron-regulatory protein. Chem Rev. 1996;96:2335–2373. doi: 10.1021/cr950040z. [DOI] [PubMed] [Google Scholar]
- 5.Bowen T L, Union J, Tumbula D L, Whitman W B. Cloning and phylogenetic analysis of the genes encoding acetohydroxyacid synthase from the archaeon Methanococcus aeolicus. Gene. 1997;188:77–84. doi: 10.1016/s0378-1119(96)00779-2. [DOI] [PubMed] [Google Scholar]
- 6.Buchner J, Pastran I, Brinkmann U. A method for increasing the yield of properly folded recombinant fusion proteins: single chain immunotoxins from renaturation of bacterial inclusion bodies. Anal Biochem. 1992;205:263–270. doi: 10.1016/0003-2697(92)90433-8. [DOI] [PubMed] [Google Scholar]
- 7.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 8.Chang Y Y. Common ancestry of Escherichia coli pyruvate oxidase and the acetohydroxy acid synthase of the branched-chain amino acid biosynthetic pathway. In: Mortlock R P, editor. The evolution of metabolic function. Boca Raton, Fla: CEC Press; 1992. pp. 81–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang Y Y, Cronan J E., Jr Common ancestry of Escherichia coli pyruvate oxidase and the acetohydroxy acid synthases of the branched-chain amino acid biosynthetic pathway. J Bacteriol. 1988;170:3937–3945. doi: 10.1128/jb.170.9.3937-3945.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang Y Y, Wang A Y, Cronan J E., Jr Molecular cloning, DNA sequencing, and biochemical analyses of Escherichia coli glyoxylate carboligase. An enzyme of the acetohydroxy acid synthase-pyruvate oxidase family. J Biol Chem. 1993;268:3911–3919. [PubMed] [Google Scholar]
- 11.Chipman D, Barak Z, Schloss J V. Biosynthesis of 2-aceto-2-hydroxy acids: acetolactate synthases and acetohydroxyacid synthases. Biochim Biophys Acta. 1998;1385:401–419. doi: 10.1016/s0167-4838(98)00083-1. [DOI] [PubMed] [Google Scholar]
- 12.Claiborne A, Yeh J I, Mallett T C, Luba J, Crane E J, Charrier V, Parsonage D. Protein-sulfenic acids: diverse roles for an unlikely player in enzyme catalysis and redox regulation. Biochemistry. 1999;38:15407–15416. doi: 10.1021/bi992025k. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham C C, Hager L P. Reactivation of the lipid-depleted pyruvate oxidase system from Escherichia coli with cell envelope neutral lipids. J Biol Chem. 1975;250:7139–7146. [PubMed] [Google Scholar]
- 14.DiMarco A A, Bobik T A, Wolfe R S. Unusual coenzymes of methanogenesis. Annu Rev Biochem. 1990;59:355–394. doi: 10.1146/annurev.bi.59.070190.002035. [DOI] [PubMed] [Google Scholar]
- 15.Dobritzsch D, Konig S, Schneider G, Lu G. High resolution crystal structure of pyruvate decarboxylase from Zymomonas mobilis. Implications for substrate activation in pyruvate decarboxylases. J Biol Chem. 1998;273:20196–20204. doi: 10.1074/jbc.273.32.20196. [DOI] [PubMed] [Google Scholar]
- 16.Fitz-Gibbon S T, House C H. Whole genome-based phylogenetic analysis of free-living microorganisms. Nucleic Acids Res. 1999;27:4218–4222. doi: 10.1093/nar/27.21.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graupner M, Xu H, White R H. Identification of an archeal 2-hydroxy acid dehydrogenase catalyzing reactions involved in coenzyme biosynthesis in methanoarchaea. J Bacteriol. 2000;182:3688–3692. doi: 10.1128/jb.182.13.3688-3692.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawkins C F, Borges A, Perham R N. A common structural motif in thiamin pyrophosphate-binding enzymes. FEBS Lett. 1989;255:77–82. doi: 10.1016/0014-5793(89)81064-6. [DOI] [PubMed] [Google Scholar]
- 19.Hidaka T, Goda M, Kuzuyama T, Takei N, Hidaka M, Seto H. Cloning and nucleotide sequence of fosfomycin biosynthetic genes of Streptomyces wedmorensis. Mol Gen Genet. 1995;249:274–280. doi: 10.1007/BF00290527. [DOI] [PubMed] [Google Scholar]
- 20.Howell D M, Harich K, Xu H, White R H. The α-keto acid chain elongation reactions involved in the biosynthesis of coenzyme B (7-mercaptoheptanoylthreonine phosphate) in methanogenic Archaea. Biochemistry. 1998;37:10108–10117. doi: 10.1021/bi980662p. [DOI] [PubMed] [Google Scholar]
- 21.Koonin E V, Mushengian A R, Galperin M Y, Walker D R. Comparison of archaeal and bacterial genomes: computer analysis of protein sequences predicts novel functions and suggests a chimeric origin for the Archaea. Mol Microbiol. 1997;25:619–637. doi: 10.1046/j.1365-2958.1997.4821861.x. [DOI] [PubMed] [Google Scholar]
- 22.Macheroux P, Schonbrunn E, Svergun D I, Volkov V V, Koch M H J, Bornemann S, Thorneley R N F. Evidence for a major structural change in Escherichia coli chorismate synthase induced by flavin and substrate binding. Biochem J. 1998;335:319–327. doi: 10.1042/bj3350319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller Y A, Schulz G E. Structure of the thiamine- and flavin-dependent enzyme pyruvate oxidase. Science. 1993;259:965–967. doi: 10.1126/science.8438155. [DOI] [PubMed] [Google Scholar]
- 24.Nakashita H, Watanabe K, Hara O, Hidaka T, Seto H. Studies on the biosynthesis of bialaphos. Biochemical mechanism of C-P bond formation: discovery of phosphonopyruvate decarboxylase which catalyzes the formation of phosphonoacetaldehyde from phosphonopyruvate. J Antibiot. 1997;50:212–219. [PubMed] [Google Scholar]
- 25.Pang S S, Duggleby R G. Expression, purification, characterization, and reconstitution of the large and small subunits of yeast acetohydroxyacid synthase. Biochemistry. 1999;38:5222–5231. doi: 10.1021/bi983013m. [DOI] [PubMed] [Google Scholar]
- 26.Rebbapragada A, Johnson M S, Harding G P, Zuccarelli A J, Fletcher H M, Zhulin I B, Taylor B L. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci USA. 1997;94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodgers K R. Heme-based sensors in biological systems. Curr Opin Chem Biol. 1999;3:158–167. doi: 10.1016/S1367-5931(99)80028-3. [DOI] [PubMed] [Google Scholar]
- 28.Schloss J V, Van Dyk D E, Vasta J F, Kutny R M. Purification and properties of Salmonella typhimurium acetolactate synthase isozyme II from Escherichia coli HB101/pDU9. Biochemistry. 1985;24:4952–4959. doi: 10.1021/bi00339a034. [DOI] [PubMed] [Google Scholar]
- 29.Schloss J V, Ciskanik L, Pai E F, Thrope C. Acetolactate synthase: a deviant flavoprotein. In: Gurti B, Ronchi S, Zanetti G, editors. Flavins and flavoproteins. Berlin, Germany: Walter de Gruyter; 1991. pp. 907–914. [Google Scholar]
- 30.Schwartz D, Recktenwald J, Pelzer S, Wohlleben W. Isolation and characterization of the PEP-phosphomutase and the phosphonopyruvate decarboxylase genes from the phosphinothricin tripeptide producer Streptomyces viridochromogenes Tu494. FEMS Microbiol Lett. 1998;163:149–157. doi: 10.1111/j.1574-6968.1998.tb13039.x. [DOI] [PubMed] [Google Scholar]
- 31.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J, Rice P, Nölling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stormer F C. The pH 6 acetolactate-forming enzyme from Aerobacter aerogenes. J Biol Chem. 1968;243:3740–3741. [PubMed] [Google Scholar]
- 33.Tsou A Y, Ransom S C, Gerlt J A, Buechter D D, Babbitt P C, Kenyon G L. Mandelate pathway of Pseudomonas putida: sequence relationships involving mandelate racemase, (S)-mandelate dehydrogenase, and benzoylformate decarboxylase and expression of benzoylformate decarboxylase in Escherichia coli. Biochemistry. 1990;29:9856–9862. doi: 10.1021/bi00494a015. [DOI] [PubMed] [Google Scholar]
- 34.Vovk A I, Khripko S S, Muraveva I V. Pyruvate decarboxylase inactivation by interaction with substrate and molecular oxygen. Ukr Biokhim Zh. 1992;64:42–47. [PubMed] [Google Scholar]
- 35.White R H. Biosynthesis of coenzyme M (2-mercaptoethanesulfonic acid) Biochemistry. 1985;24:6487–6493. [Google Scholar]
- 36.White R H. Intermediates in the biosynthesis of coenzyme M (2-mercaptoethanesulfonic acid) Biochemistry. 1986;25:5304–5308. [Google Scholar]
- 37.White R H. Characterization of the enzymatic conversion of sulfopyruvate and l-cysteine into coenzyme M (mercaptoethanesulfonic acid) Biochemistry. 1988;27:7458–7462. [Google Scholar]
- 38.Xing R, Whitman W B. Purification of the oxygen-sensitive acetohydroxy acid synthase from the archaeabacterium Methanococcus aeolicus. J Bacteriol. 1994;176:1207–1213. doi: 10.1128/jb.176.5.1207-1213.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]