Abstract
Similarities in DNA base sequence indicate that pSC101 and R1162 encode related systems for conjugal mobilization, although these plasmids are otherwise very different. The mob region of pSC101 was cloned, and two genes that are required for transfer were identified. One gene, mobA, encodes a protein similar in amino acid sequence to the DNA processing domain of the R1162 MobA protein. The other gene, mobX, is within the same transcriptional unit as the pSC101 mobA and is located just downstream, at the same position occupied by mobB in R1162. Despite this, the MobB and MobX proteins do not appear to be closely related based on a comparison of their amino acid sequences. Complementation analysis indicated that neither of the pSC101 Mob proteins could substitute for, or be replaced by, their R1162 counterparts, nor were they active together at the R1162 origin of transfer (oriT). However, the full set of R1162 Mob proteins did recognize the pSC101 oriT. A hybrid system for mobilization, active at the R1162 oriT site, was constructed. This system consists of MobX and a chimeric protein made up of the DNA cleaving-ligating domain of the R1162 MobA protein joined to a fragment of pSC101 MobA. Previous results suggested that MobB and a region of MobA distinct from the DNA processing domain together formed a functional unit in transfer. The present results support this model because the chimeric MobA, although active on R1162 oriT, requires the pSC101 protein MobX for efficient plasmid mobilization.
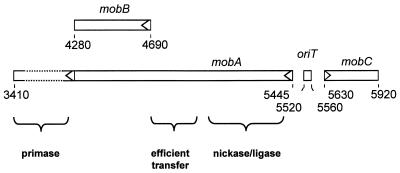
R1162, a broad-host-range plasmid essentially the same as RSF1010, is efficiently comobilized during conjugal transfer of R751, RK2 (RP4), and other members of the IncP-1 plasmid group (29). The proteins required for mobilization are encoded by a cluster of three genes, mobABC (Fig. 1), which are transcribed divergently from promoters adjacent to the origin of transfer (oriT) (27). MobA, the principal DNA processing protein, binds at oriT and cleaves one of the strands, which is then unwound and passed into a recipient cell during mating. MobA must partially disrupt the DNA duplex at oriT in order to cleave the strand (30). A second, auxiliary protein, encoded by mobC, assists in strand separation, thus increasing the efficiency of cleavage (31). MobA remains covalently linked to the 5′ end of the strand during transfer and subsequently rejoins the ends of this DNA (4, 25, 26). A third gene, mobB, embedded in a different reading frame within mobA (27), is also required for efficient mobilization (22).
FIG. 1.
Genetic organization of the mob region of R1162. Numerical coordinates are distances (in base pairs) from the unique EcoRI site, based on the sequence of the virtually identical plasmid RSF1010 (27). The arrowheads are at start sites for translation. The coding regions for the different functional domains of MobA are indicated; the section encoding the primase, which is also synthesized separately by internal initiation within the gene (27), has been truncated in the figure.
The R1162 mob genes belong to a family of transfer systems that appear to be related, with similar oriTs and MobA-like proteins. The members of this family are widely distributed in different plasmids and are found in a diverse group of bacteria, including Agrobacterium tumefaciens, Thiobacillus ferrooxidans, Staphylococcus aureus, and Streptococcus agalactiae (11, 13, 14, 28). It is likely that in each case specific characteristics of the transfer system have evolved, reflecting the properties of the plasmid host. This is clearly true for R1162, where mobA has become extended through the neighboring, downstream mobB and has become fused to the gene encoding the replicative primase of the plasmid (Fig. 1) (27). This arrangement increases the frequency of mobilization, probably because the MobA-linked primase is delivered more efficiently to the replicative origin, where it initiates synthesis of the complement to the transferred strand (17).
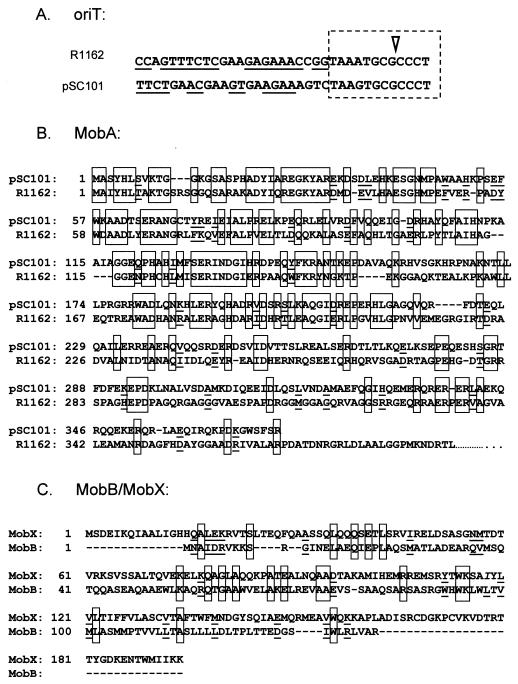
In order to investigate the kinds of variation possible among the R1162 family of transfer systems, we have begun to characterize the mob region of the mobilizable plasmid pSC101 (12). The base sequences of the R1162 and pSC101 oriTs are nearly identical in the region of the cleavage site, whereas the adjacent inverted repeats have very different sequences and potential secondary structures (Fig. 2A). In addition, pSC101 encodes a protein that is clearly homologous to MobA in the amino-terminal region containing the nickase-ligase activity (Fig. 2B). In contrast, an initial inspection of the potential open reading frames (ORFs) of pSC101 revealed no obvious homologs to the accessory proteins MobB and MobC. Moreover, the systems for replication of the two plasmids are completely different, and the pSC101 MobA-like protein is not fused to a primase or other protein.
FIG. 2.
(A) Base sequences of the R1162 oriT (8) and of the presumptive pSC101 oriT. The region with a nearly identical base sequence is boxed by the dotted broken line, and the location of the cleavage site in the R1162 oriT is shown by the inverted triangle. Bases potentially involved in intrastrand base pairing to form a hairpin loop are underlined. (B) Amino acid sequence of MobA proteins of R1162 and pSC101, aligned to show similarities. Identical amino acids are boxed; similar amino acids are underlined. Only the amino-terminal region of R1162 MobA, encoded by that portion of the gene extending up to the overlap with mobB (Fig. 1), is shown. (C) Alignment of MobB and MobX. Amino acid sequences in panels B and C were determined from plasmid DNA base sequences (3, 27).
I have identified the mob genes of pSC101 and characterized how they interact with the related mob genes of R1162. I have found that although no single protein is interchangeable, the R1162 Mob proteins can together activate the pSC101 oriT for transfer. Moreover, for both pSC101 and R1162, a domain of MobA and a second Mob protein function as a matched pair to promote efficient transfer. The R1162 pair can be replaced by the one from pSC101, resulting in a hybrid MobA active on the R1162 oriT. Therefore, this pair forms a functional unit distinct from the DNA processing domain of MobA.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The donor strains used in bacterial matings are all derivatives of MV10 (C600 ΔtrpE5) (18). The recipient strain in mating experiments was DF1019 (16), a C600 derivative resistant to nalidixic acid. The derivations of the plasmids used in this study are outlined in Table 1. All donor strains contained the IncP-1 plasmid R751 (19) as the mobilizing vector. Bacteria were mated on semisolid medium as described previously (7). All mating frequencies are the results of duplicate experiments (variations of <1 order of magnitude).
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant characteristic(s)a | Construction and/or reference |
|---|---|---|
| pUT1309 | ΔoriTR1162 | Bal31-generated 48-bp deletion in R1162 (22) |
| pUT1371 | ΔmobBR1162 | 22 |
| pUT1617 | oriTR1162 | Becker and Meyer (submitted) |
| pUT1621 | MobpSC101+ | Insertion of 2,754-bp BstY fragment from pSC101 into BamHI site of pAYC184 (10) |
| pUT1626 | mobApSC101::Km | Km cassette (21) inserted into pUT1621 at RsaI site (bp 3845 of pSC101 DNA) |
| pUT1628 | ΔMobpSC101 (ORFs E and F) | Deletion of small EcoRV fragment of pUT1621 (Δ4344–5021 pSC101 DNA) |
| pUT1642 | mobApSC101::(14 bp) | CTCGAGGCCTCGAG inserted into pUT1621 at HpaI site (bp 3097 of pSC101 DNA) |
| pUT1643 | mobApSC101::(6 bp) | CTCGAG inserted into pUT1621 at HpaI site by removing small XhoI fragment GAGGCCTC from pUT1642 |
| pUT1654 | mobXpSC101::Km | Km cassette (21) inserted into pUT1621 at XmnI site (bp 2840 of pSC101 DNA) |
| pUT1657 | mobBR1162+ | HindIII-SalI fragment containing mobB cloned from pUT221 (7) into pBR322 |
| pUT1663 | mobX mobA′pSC101 ′mobAR1162 | Insertion of 14-bp linker (above) containing ScaI site at SspI site of pUT208 (7). An NruI-DraI fragment (containing bp 2268 to 3619 pSC101 DNA) was then cloned into this ScaI site. |
| pUT1678 | Φ(mobA′pSC101+′mobAR1162) | pUT1663 digested with AatII and EcoRI and then partially digested with Bal31 exonuclease, filled in with Klenow fragment, and ligated |
| pUT1679 | Φ(mobA′pSC101+′mobAR1162) | Same as pUT1678 |
| pUT1680 | Φ(mobA′pSC101+′mobAR1162) mobX::Km | By replacement of BamHI-DraI fragment of pUT1678 (bp 2268 to 3619 pSC101 DNA) with similar fragment (containing a Km insertion at bp 2840) from pUT1654 |
| pUT1681 | Φ(mobA′pSC101+′mobAR1162) mobX::Km | Same as pUT1680, but with replacement of BamHI-DraI fragment of pUT1679 |
| pUT1693 | oriTR1162oriTpSC101 | pSC101 DNA (bp 4069 to 4199) amplified by PCR and cloned into pUT1617 |
The plasmid source of the gene or property is indicated by a subscript. The coordinates for pSC101 are from Bernardi and Bernardi (3).
Other procedures.
Plasmids encoding an active, hybrid MobA were constructed by cleaving approximately 1.3 μg of pUT1663 DNA with AatII and EcoRI (see Fig. 5) and then digesting this DNA at 30 C with 0.1 U of Bal31 exonuclease in 50-μl final volume. Samples (10 μl) were taken at 1-min intervals for 5 min, and the reaction was stopped by the addition of 36 μl of 26 mM EGTA. These samples were then pooled and further incubated with Klenow fragment in buffer containing 33 μM concentrations of each deoxynucleoside triphosphate. The DNA was finally ligated overnight at room temperature and then used to transform MV10(R751). Cells containing mobilizable plasmids were identified both by direct mating of transformed cells and by matings with pooled populations of transformants.
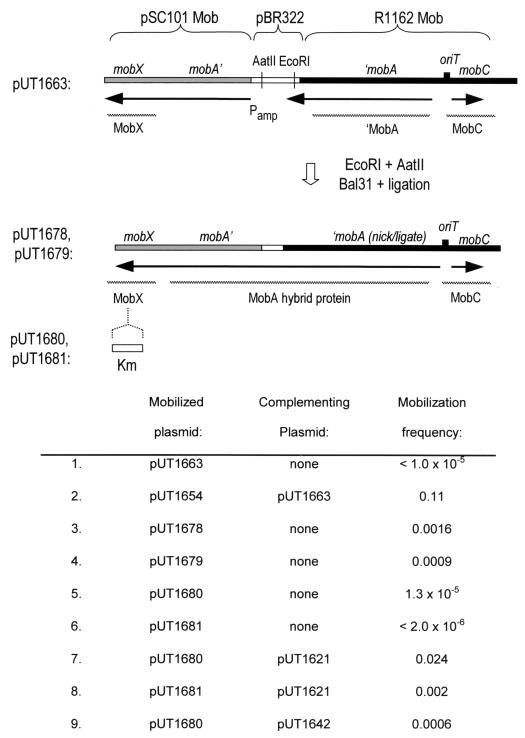
FIG. 5.
(Top) Structure of pUT1663 in a region containing mob DNA from pSC101 and R1162. The genetic manipulations used to generate the derivatives pUT1678 and pUT1679, which have chimeric mobA genes, are also indicated. The origin and direction of transcription, as well as the translation products, deduced from genetic analysis and from previously published information are also shown. (Bottom) Frequencies of mobilization for different plasmids.
RESULTS
A cloned fragment of pSC101 sufficient for conjugal mobilization.
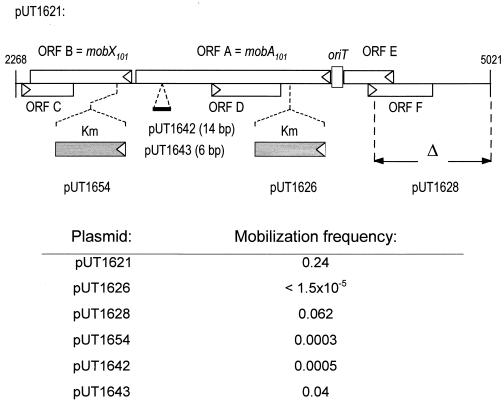
Like the IncQ plasmid R1162, pSC101 is efficiently mobilized by IncP-1 plasmids (29). The sequence of pSC101 (3) reveals a large ORF that would encode a protein with an amino acid sequence similar to that of the R1162 MobA protein, as well as an adjacent locus likely to be the origin of transfer (Fig. 2). We cloned a 2,754-bp BstYI fragment containing this DNA into the BamHI site of pACYC184 (12); the resulting plasmid, pUT1621, was efficiently mobilized by R751 (Fig. 3). In the cloned DNA there are six ORFs that would encode a protein greater than 10 kDa (Fig. 3). The 43-kDa, potentially MobA-like protein would be encoded by the largest of these, ORF A. A nonpolar cassette encoding kanamycin resistance (21) was inserted into this ORF to create the plasmid pUT1626 (Fig. 3). This plasmid was no longer mobilizable by R751 (Fig. 3), indicating that ORF A does indeed encode a protein required for transfer. ORF A was therefore designated as pSC101 mobA.
FIG. 3.
(Top) Mob region of pSC101, cloned in pACYC184 to generate pUT1621. The rectangles indicate the potential ORFs, with the arrowheads showing the direction of translation. Mutations in this DNA and the resulting derivatives of pUT1621 are also shown. (Bottom) Mobilization frequencies (number of transconjugants per donor cell) of pUT1621 and derivatives are presented.
The pSC101 mobA is adjacent to the presumptive pSC101 oriT and thus occupies the same position as the R1162 mobA (Fig. 1). ORFs E and F are on the opposite side of the pSC101 oriT. ORF F has a position and orientation similar to that of mobC of R1162 (Fig. 1). However, both ORF E and ORF F could be deleted without significantly affecting transfer (pUT1628; Fig. 3). ORF B is downstream from the pSC101 mobA, in the same position as mobB in R1162. Insertion of the kanamycin cassette into this ORF resulted in a lower frequency of mobilization (pUT1654; Fig. 3). We call this ORF mobX, since its product does not show an obvious relationship to MobB (Fig. 2C). We have not tested whether the small ORFs C and D are involved in mobilization. These ORFs overlap mobX and mobA, respectively, but would be translated in the opposite direction (Fig. 3).
In R1162, mutations in the region of mobA just upstream from mobB result in a lower level of transfer, although unlike the mutations inactivating the nickase-ligase domain of the protein, transfer is not completely abolished (Fig. 1) (23). A 14-bp insertional mutation in the region of the pSC101 mobA adjacent to mobX reduced the mobilization frequency of pUT1621 (pUT1642; Fig. 3). The decrease in transfer could be due to either a change in the structure of the MobA protein or a polar effect of the frameshift on mobX. Indeed, an in-frame, 6-bp insertion at the same position has at most a small effect on the mobilization frequency (pUT1643; Fig. 3). This question is examined in more detail below.
Complementation between the R1162 and pSC101 mob systems.
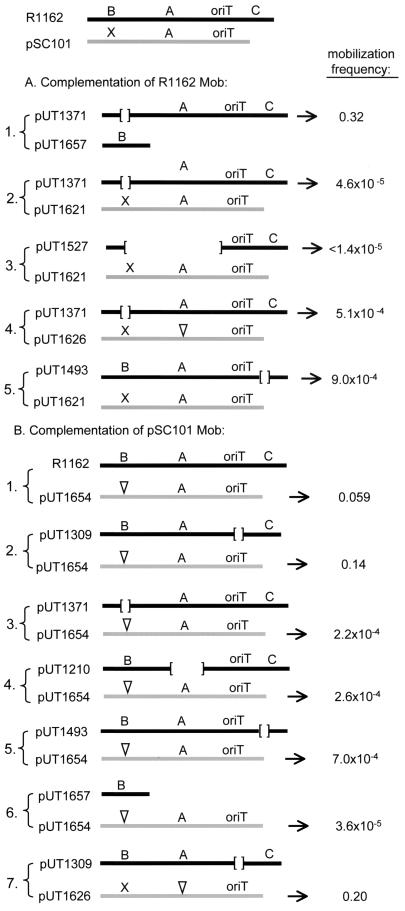
At least two genes, mobA and mobX, occupying positions similar to those occupied by mobA and mobB of R1162, are required for mobilization of pSC101. In view of the apparent relatedness of the mob systems of the two plasmids, we tested for genetic complementation. The R1162 derivative pUT1371 contains an in-frame deletion in mobB and is mobilized at a frequency of 1.8 × 10−5 (22). The mutation was complemented in trans by the R1162 mobB, cloned in the plasmid pUT1657, but not by any of the pSC101 mob genes (plasmid pairs 1 and 2; Fig. 4A). The lack of complementation by the pSC101 mob system was not due to interfering competition between the MobA proteins of the two plasmids, since there was also no complementation when either the R1162 or the pSC101 mobA genes were inactivated (plasmid pairs 3 and 4; Fig. 4A). It was also possible that the pSC101 MobA and MobX could activate the R1162 oriT, but only when MobC, which appears to be missing in the pSC101 mob system, is absent. However, the failure of complementation was also not due to interference by MobC (plasmid pair 5; Fig. 4A). Thus, the two mob systems have diverged sufficiently so that the pSC101 mob proteins cannot substitute, either singly or as a group, for those encoded by R1162.
FIG. 4.
Complementation of mutations in the mob regions of R1162 (A) and pSC101 (B). The pairs of plasmids in the donor strains (which also contained R751) are indicated by the left braces. In each case, the arrow adjacent to one of the plasmids points to its mobilization frequency from the donor. Simplified maps of the mob regions are given for each plasmid, with the parental maps at the top of the figure. The letters refer to the mob genes present in each case; the brackets designate deletions, and the inverted triangles show an inactivating insertion.
It was possible that the pSC101 Mob proteins were not complementing in trans because they were binding preferentially to the pSC101 oriT, located in cis with respect to the mob genes. To test this possibility, we first confirmed that pUT1617, a pBR322 derivative containing a cloned copy of the R1162 oriT (Table 1), was poorly mobilized (<10−5 transconjugants per donor) from cells containing pUT1621. However, when the pSC101 oriT was cloned into this molecule to generate pUT1693 (Table 1), the mobilization frequency increased to 0.6 transconjugants per donor. Thus, like the R1162 system (8), the pSC101 Mob genes can activate the pSC101 oriT in trans, even when there is a competing oriT in cis.
Different results were obtained when we tested for complementation of mutations in the pSC101 mob region. The plasmid pUT1654, poorly mobilizable because of an insertion inactivating mobX (Fig. 3), was mobilized at high frequency when R1162 was also present in the donor cells (plasmid pair 1; Fig. 4B). This complementation did not require the R1162 oriT (plasmid pair 2; Fig. 4B) and thus was not due to plasmid “piggybacking” by recombination at oriT during conjugal mobilization (9). However, complementation did require all three of the R1162 Mob proteins, since inactivation of any one of these resulted in loss of complementation (plasmid pairs 3 to 5; Fig. 4B). In addition, the mutation in mobX was not complemented by MobB alone (plasmid pair 6; Fig. 4B). These results indicate that the R1162 Mob proteins can assemble at the pSC101 oriT and activate this origin for transfer. This includes the actual nicking and ligation of the pSC101 oriT DNA, since pUT1626, which contains a large insertion completely abolishing the pSC101 MobA, was also complemented for transfer (plasmid pair 7; Fig. 4B).
A hybrid system for mobilization active on the R1162 oriT.
The complementation analysis indicated that the pSC101 Mob proteins cannot substitute for the R1162 Mob proteins to activate the R1162 oriT for transfer. We have shown elsewhere that the R1162 MobA contains, in addition to the nickase-ligase and primase regions, a third domain required for efficient mobilization (23). This domain is encoded by the region of mobA adjacent to mobB and just upstream in the direction of translation (Fig. 1), and deletions here have only a small effect on nicking and ligation at oriT (23). We therefore asked whether this domain could be substituted by the analogous region of the pSC101 MobA, even though this protein is inactive at the R1162 oriT. We previously cloned into pBR322 a fragment of R1162 mob DNA that included mobC, oriT, and the part of mobA that encodes the amino-terminal domain required to cleave and ligate oriT (4, 7). We cloned into this plasmid, at the SspI site within the vector DNA, mobX and the adjacent segment of the pSC101 mobA. This places the mobA fragments from pSC101 and R1162 near each other and oriented in the same way but separated by a small fragment of pBR322 DNA (Fig. 5). The resulting plasmid (pUT1663) complemented pUT1654 (Fig. 5), indicating that mobX was expressed. This was due to transcription initiated from the intact amp promoter of the vector (24), as well as to any of the R1162 mobA transcripts that reach mobX. However, pUT1663 itself transferred poorly (Fig. 5). We next attempted to fuse the two mobA fragments, by cleaving the DNA in the intervening pBR322 segment and then partially digesting this DNA with Bal31 nuclease (Materials and Methods). Plasmids were obtained that were now transferred at higher frequency, and two that had been isolated independently, pUT1678 and pUT1679 (Fig. 5), were characterized further. DNA base sequencing revealed that in both plasmids the two mobA fragments were unchanged, but part of the intervening pBR322 DNA had been deleted. In pUT1678, the two mobA fragments were now linked in frame, joined by 37 codons derived from pBR322 DNA and the linker DNA used in the cloning (Table 1). In pUT1679, there were 121 bp of pBR322 remaining, and synthesis of the MobA polypeptide was apparently terminated at TAA within this DNA. However, we think that mobilization of pUT1679 also depends on a hybrid, fused protein (see Discussion). Mobilizable plasmids with larger deletions extending into either the pSC101 or R1162 mobA gene fragments were not obtained, suggesting that both fragments were required for efficient transfer.
The activity of the fusion proteins indicated that MobB and a region of R1162 MobA could be replaced by protein encoded by pSC101. It was clear that MobX alone was insufficient for this replacement, since this protein was expressed in pUT1663, which nevertheless transferred poorly (Fig. 5). Alternatively, only the MobA fragment from pSC101 might be required. However, when the mobX-inactivating insertion from pUT1654 was introduced into pUT1678 and pUT1679, the resulting plasmids (pUT1680 and pUT1681) were mobilized at a much lower frequency (Fig. 5). Thus, both MobX and the fusion MobA polypeptide from pSC101 are required for active hybrid protein.
The pSC101 mobA gene fragment is probably not expressed in pUT1663, since the normal signals for initiation of translation are missing, and the fragment is not fused in frame to amp or to any other gene. Since pUT1663 can nevertheless complement pUT1654 (Fig. 5), expression of mobX does not appear to require translation of the adjacent region of the pSC101 mobA. It is therefore unlikely that the lower mobilization frequency of pUT1642, which has a 14-bp insertion in this region, is due to a polar effect of this mutation. To test this directly, we asked whether pUT1642 could complement pUT1680, which has an inactive mobX. The mobilization frequency of pUT1680 was increased in the presence of pUT1642 (Fig. 5), indicating that mobX continues to be expressed in pUT1642.
DISCUSSION
The mob region of plasmid pSC101 is located within a 2,754-bp fragment of the plasmid DNA and contains at least two genes important for transfer. One of these encodes a protein that is clearly homologous to the amino-terminal region of the R1162 protein MobA (Fig. 2). It is this region of MobA that is required for cleavage and ligation of the conjugally transferred DNA strand. The second gene, mobX, is adjacent to the pSC101 mobA and within the same transcriptional unit. Although mobX occupies a position analogous to that of mobB in the R1162 mob region, the two proteins show little similarity in amino acid sequence (Fig. 2C). However, the properties of the hybrid MobA proteins increase the likelihood that MobX is a functional analog of MobB (see below).
Although MobX and the pSC101 MobA are inactive at the R1162 oriT, the R1162 proteins will assemble at the pSC101 oriT and activate the plasmid for transfer. Efficient DNA processing at oriT requires both the inverted repeat, and the region extending from the inverted repeat and including the cleavage site (5). The outer arm of the inverted repeat is required for termination of a round of transfer but not for initiation, suggesting that its role is to restore in the transferred single strand a duplex character to the inner arm (5, 20). Single base changes in one arm of the inverted repeat and affecting DNA processing by MobA are suppressed by second mutations restoring base complementarity (2). This suggests that MobA and the inverted repeat do not interact by means of highly specific interactions between particular amino acids in the protein and corresponding bases in the DNA. Consistent with this interpretation, the inverted repeat is important in vitro for strong binding (6), but it does not determine the site of cleavage, since oriT single-stranded DNA lacking the inverted repeat is still cleaved at the correct location by the protein (26). In addition, the distance of the inverted repeat can be increased without affecting the cleavage site; instead, it is specific bases within the adjacent oriT DNA that determines the site of cleavage (E. Becker and R. Meyer, submitted for publication).
The characteristics of the interaction between MobA and the inverted repeat are consistent with the activity of the R1162 Mob proteins on the pSC101 oriT. Apparently, the inverted repeat of this oriT fits sufficiently well with the MobA molecule to be captured by the protein, despite its very different sequence from the inverted repeat of the R1162 oriT. Since the remaining part of the two oriTs are practically identical (Fig. 2A), normal DNA processing can take place. The center of symmetry of the inverted repeat in the pSC101 oriT is also located at a different distance from the normal cleavage site for the R1162 MobA protein, but this can be compatible with normal processing at the correct cleavage site (Becker and Meyer, submitted).
No ORF encoding a protein similar to MobC of R1162 has been identified in the mob region of pSC101. ORF F (Fig. 3) has the same position and orientation as mobC and would encode a protein of similar size, but this ORF can be deleted without affecting transfer (pUT1628; Fig. 3). MobC enhances the helical disruption of oriT DNA, thus promoting strand cleavage by MobA (31). It is possible that a MobC-like protein is required for transfer of pSC101, but this requirement is relieved because of fortuitously greater helical distortion of the pSC101 oriT in pUT1621. However, the R1162 MobA is active on the pSC101 oriT only in the presence of MobC (Fig. 4B). Assuming that the two MobA homologs are functionally similar, at least in requiring a partially unpaired strand prior to cleavage (30), then the pSC101 MobA appears to be more robust in strand separation at oriT.
We have shown elsewhere that a domain of MobA, distinct from that required for strand cleavage and ligation at oriT, is required for efficient mobilization and stable relaxosomes (Fig. 1) (23). One interpretation is that MobB interacts with this part of MobA. When the comparable region in the pSC101 MobA is disrupted (pUT1642; Fig. 3), transfer is affected, and thus MobX and this domain could form a similar interacting pair. The properties of the R1162/pSC101 hybrid MobA proteins encoded by pUT1678 and pUT1679 (Fig. 5) support the model that MobB or MobX and the adjacent region of the corresponding MobA protein form a matched pair. The hybrid mobilization systems were active on the R1162 oriT only when both MobX and the pSC101 MobA domain were present. This suggests that the MobB (or MobX) protein and a domain on the cognate MobA together make up a functional unit. Normally, the MobA domain is part of the whole protein, and this is also the case in pUT1678, where the DNA sequence would indicate that a single, hybrid MobA protein is produced. In pUT1679, the two mobA gene fragments are not linked in frame. However, it is unlikely that the pSC101 MobA fragment is synthesized separately and is active in mobilization. First, there are no potential upstream initiation codons in the remaining pBR322 DNA. Second, if the fragment were synthesized due to a cryptic initiation codon, either upstream or within the gene itself, then pUT1663 would be active in transfer, and this is not the case (Fig. 5). More likely, a hybrid protein is produced by internal frameshifting within the pBR322 DNA. At the site for termination of translation, the RNA has the sequence UUU UUC UAA. This is a particularly “slippery” sequence, causing −1 frameshifts (1, 15), which would then place translation in the correct reading frame for the downstream pSC101 mobA fragment. In any case, it is important to note that the functional unit represented by the pSC101 mobA fragment is distinct from the cleavage-ligation domain of MobA and, at the very least, can be separated from it by a polypeptide bridge.
If MobB and MobX function similarly within related mob systems, then why are they not more alike? One possibility is that MobB has undergone unique selective pressure because of its location just upstream from the replicative primase of R1162. Since the MobA-primase fusion in R1162 increases the efficiency of transfer (17), mutations that increased readthrough in an ancestral plasmid, where the genes were separate, would be selected. These would include deletions reducing the size of mobB, and indeed MobB is much smaller than MobX (Fig. 2C). In addition, there would be point mutations eliminating stop codons in the fusion reading frame, and these would frequently generate amino acid changes in MobB. One would also expect compensating changes in the interacting domain of MobA. In agreement with this, the similarity of MobA and its pSC101 homolog is substantially less in this region than in the nickase-ligase domain (Fig. 4B).
ACKNOWLEDGMENTS
I thank Eric Becker for helpful discussions and assistance.
This work was supported by Public Health Service grant GM37462.
REFERENCES
- 1.Atkins J, Nichols B, Thompson S. The nucleotide sequence of the first externally suppressible-1 frameshift mutant, and of some nearby leaky frameshift mutants. EMBO J. 1983;2:1345–1350. doi: 10.1002/j.1460-2075.1983.tb01590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barlett M M, Erickson M J, Meyer R J. Recombination between directly repeated origins of conjugative transfer cloned in M13 bacteriophage DNA models ligation of the transferred plasmid strand. Nucleic Acids Res. 1990;18:3579–3586. doi: 10.1093/nar/18.12.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernardi A, Bernardi F. Complete sequence of pSC101. Nucleic Acids Res. 1984;12:9415–9426. doi: 10.1093/nar/12.24.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharjee M K, Meyer R J. A segment of a plasmid gene required for conjugal transfer encodes a site-specific, single-strand DNA endonuclease and ligase. Nucleic Acids Res. 1991;19:1129–1137. doi: 10.1093/nar/19.5.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharjee M, Rao X M, Meyer R J. Role of the origin of transfer in termination of strand transfer during bacterial conjugation. J Bacteriol. 1992;174:6659–6665. doi: 10.1128/jb.174.20.6659-6665.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharjee M K, Meyer R J. Specific binding of MobA, a plasmid-encoded protein involved in the initiation and termination of conjugal DNA transfer, to single-stranded oriT DNA. Nucleic Acids Res. 1993;21:4563–4568. doi: 10.1093/nar/21.19.4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brasch M A, Meyer R J. Genetic organization of plasmid R1162 DNA involved in conjugative mobilization. J Bacteriol. 1986;167:703–710. doi: 10.1128/jb.167.2.703-710.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brasch M A, Meyer R J. A 38-base-pair segment of DNA is required in cis for conjugative mobilization of broad host-range plasmid R1162. J Mol Biol. 1987;198:361–369. doi: 10.1016/0022-2836(87)90286-5. [DOI] [PubMed] [Google Scholar]
- 9.Broome-Smith J. RecA independent, site-specific recombination between ColE1 or ColK and a miniplasmid they complement for mobilization and relaxation: implications for the mechanism of DNA transfer during mobilization. Plasmid. 1980;4:51–63. doi: 10.1016/0147-619x(80)90082-7. [DOI] [PubMed] [Google Scholar]
- 10.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Climo M, Sharma V, Archer G. Identification and characterization of the origin of conjugative transfer (oriT) and a gene (nes) encoding a single-stranded endonuclease on the staphylococcal plasmid pGO1. J Bacteriol. 1996;178:4975–4983. doi: 10.1128/jb.178.16.4975-4983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen S, Chang A. Revised interpretation of the origin of the pSC101 plasmid. J Bacteriol. 1977;132:734–737. doi: 10.1128/jb.132.2.734-737.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cook D, Farrand S. The oriT region of the Agrobacterium tumefaciens Ti plasmid pTiC58 shares DNA sequence identity with the transfer origins of RSF1010 and RK2/RP4 and with T region borders. J Bacteriol. 1992;174:6238–6246. doi: 10.1128/jb.174.19.6238-6246.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drolet M, Zanga P, Lau P C K. The mobilization and origin of transfer regions of a Thiobacillus ferrooxidans plasmid: relatedness to plasmids RSF1010 and pSC101. Mol Microbiol. 1990;4:1381–1391. doi: 10.1111/j.1365-2958.1990.tb00717.x. [DOI] [PubMed] [Google Scholar]
- 15.Dunn J, Studier F. Complete nucleotide sequence of bacteriophage T7 DNA and the location of T7 genetic elements. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 16.Figurski D, Meyer R, Miller D, Helinski D R. Generation in vitro of deletions in the broad host range plasmid RK2 using phage Mu insertions and a restriction endonuclease. Gene. 1976;1:107–119. doi: 10.1016/0378-1119(76)90010-x. [DOI] [PubMed] [Google Scholar]
- 17.Henderson D, Meyer R. The primase of broad-host-range plasmid R1162 is active in conjugal transfer. J Bacteriol. 1996;178:6888–6894. doi: 10.1128/jb.178.23.6888-6894.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hershfield V, Boyer H W, Yanofsky C, Lovett M A, Helinski D R. Plasmid ColE1 as a molecular vehicle for cloning and amplification of DNA. Proc Natl Acad Sci USA. 1974;71:3455–3459. doi: 10.1073/pnas.71.9.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jobanputra R S, Datta N. Trimethoprim R factors in enterobacteria from clinical specimens. J Med Microbiol. 1974;7:169–177. doi: 10.1099/00222615-7-2-169. [DOI] [PubMed] [Google Scholar]
- 20.Kim Y J, Lin L S, Meyer R J. Two domains at the origin are required for replication and maintenance of broad-host-range plasmid R1162. J Bacteriol. 1987;169:5870–5872. doi: 10.1128/jb.169.12.5870-5872.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menard R, Sansonetti P, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perwez T, Meyer R. MobB protein stimulates nicking at the R1162 origin of transfer by increasing the proportion of complexed plasmid DNA. J Bacteriol. 1996;178:5762–5767. doi: 10.1128/jb.178.19.5762-5767.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perwez T, Meyer R. Stabilization of the relaxosome and stimulation of conjugal transfer are genetically distinct functions of the R1162 protein MobB. J Bacteriol. 1999;181:2124–2131. doi: 10.1128/jb.181.7.2124-2131.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russell D, Bennett G. Characterization of the beta-lactamase promoter of pBR322. Nucleic Acids Res. 1981;9:2517–2533. doi: 10.1093/nar/9.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scherzinger E, Lurz R, Otto S, Dobrinski B. In vitro cleavage of double- and single-stranded DNA by plasmid RSF1010-encoded mobilization proteins. Nucleic Acids Res. 1992;20:41–48. doi: 10.1093/nar/20.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scherzinger E, Kruft V, Otto S. Purification of the large mobilization protein of plasmid RSF1010 and characterization of its site-specific DNA-cleaving/DNA-joining activity. Eur J Biochem. 1993;217:929–938. doi: 10.1111/j.1432-1033.1993.tb18323.x. [DOI] [PubMed] [Google Scholar]
- 27.Scholz P, Haring V, Wittmann-Liebold B, Ashman K, Bagdasarian M, Scherzinger E. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. 1989;75:271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- 28.Wang A, Macrina F. Streptococcal plasmid pIP501 has a functional oriT site. J Bacteriol. 1995;177:4199–4206. doi: 10.1128/jb.177.15.4199-4206.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willetts N, Crowther C. Mobilization of the nonconjugative IncQ plasmid RSF1010. Genet Res. 1981;37:311–316. doi: 10.1017/s0016672300020310. [DOI] [PubMed] [Google Scholar]
- 30.Zhang S, Meyer R J. Localized denaturation of oriT DNA within relaxosomes of the broad-host-range plasmid R1162. Mol Microbiol. 1995;17:727–735. doi: 10.1111/j.1365-2958.1995.mmi_17040727.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhang S, Meyer R. The relaxosome protein MobC promotes conjugal plasmid mobilization by extending DNA strand separation to the nick site at the origin of transfer. Mol Microbiol. 1997;25:509–516. doi: 10.1046/j.1365-2958.1997.4861849.x. [DOI] [PubMed] [Google Scholar]