Abstract
Myeloid and lymphoid neoplasms associated with FGFR1 abnormalities (MLN-FGFR1 abnormalities) are rare hematologic malignancies associated with chromosome 8p11.2 abnormalities. Translocations of 8p11.2 were detected in 10 of 17,039 (0.06%) unique patient cytogenetic studies performed at nine institutions in Japan. No inversions or insertions of 8p11.2 were detected. Among the 10 patients with 8p11.2 translocations, three patients were diagnosed with MLN-FGFR1 abnormalities, which were confirmed by FISH analysis. Peripheral blood eosinophilia was observed in all three patients, and all progressed to AML or T-lymphoblastic lymphoma/leukemia. The prevalence of 8p11.2 translocations in clinical practice and the proportion of MLN-FGFR1 abnormalities in patients with 8p11.2 translocations in Japan were consistent with those in previous reports from Western countries.
Keywords: 8p11, FGFR1, Eosinophilia
Introduction
Chromosomal 8p11 abnormalities with Fibrocyte growth factor receptor 1 (FGFR1) rearrangement is a rare hematopoietic disorder with widespread clinical and pathologic features, including myeloproliferative neoplasms (MPN), myelodysplastic syndrome (MDS), and MPN/MDS, often associated with eosinophilia, lymphadenopathy, usually T-lymphoblastic lymphoma/leukemia (T-LBL), and progression to acute myeloid leukemia (AML) [1]. The disease progresses rapidly, usually to acute leukemia within one year, and the prognosis is poor. In the 5th WHO classification of hematopoietic and lymphoid neoplasms, it was classified as “myeloid and lymphoid neoplasms associated with FGFR1 abnormalities (MLN-FGFR1 abnormalities)” in the “myeloid and lymphoid neoplasms with eosinophilia and tyrosine kinase gene fusions (MLN-TK)” [2].
FGFR1 is a member of the receptor tyrosine kinase superfamily, and the molecular pathogenesis of MLN-FGFR1 abnormalities is characterized by the generation of fusion proteins with an intact kinase domain of FGFR1 [3]. Currently, it has been reported that 17 FGFR1 gene rearrangements exist in MLN-FGFR1 abnormalities, including 15 translocations, one insertion, and one inversion [1]. The difference of partner genes with FGFR1 rearrangement is thought to be the cause of the variety of phenotypes of MLN-FGFR1 abnormalities, which makes it difficult to list MLN-FGFR1 abnormalities as a differential diagnosis. Chromosomal analysis is one of the standard examinations initially performed for the diagnosis of hematologic malignancies, and the chromosomal abnormalities involving 8p11.2 are one of the initial keys for the diagnosis of MLN-FGFR1 abnormalities. The purpose of this study is to describe the prevalence of 8p11.2 abnormalities in Japanese hematology practices.
Materials and methods
Cytogenetic analysis was performed as part of routine patient care at each institution. Twenty metaphases were analyzed per patient, and karyotypes were described according to the International System for Human Cytogenomic Nomenclature. Fluorescence in-situ hybridization (FISH) analysis of interphase nuclei was performed using dual-color split probes for the 8p11.2 loci (Leica Biosystems Inc., Nußloch, Germany).
The cytogenetic database from 2010 to 2022 in nine institutions was screened for 8p11.2 translocations, inversions, and insertions. For patients with 8p11.2 abnormalities, medical records were reviewed to collect data including clinical diagnosis, peripheral blood analysis, and their clinical course.
The study was carried out following the guidelines for the ethical guidelines for medical and health research involving human subjects and approved by the institutional review boards at each participating institution.
Statistical analysis
The required sample size was calculated as follows: n ≧ (z^2 * p * (1-p)) / (e^2), where n is the sample size, z is the z-score, p is the population proportion, and e is the margin of error. Values of 95% and 0.04% were used for z and e, respectively. If the prevalence of 8p11.2 translocations in Japan is 0.06%, which is similar to that in Western countries, the required sample size is 13,397.
Results
From 2010 to 2022, 17,039 unique patient cytogenetic studies were performed in nine institutions. Among them, chromosome 8p11.2 translocations were identified in 10 patients (0.06%). There are no cases of inversion or insertion of 8p11.2.
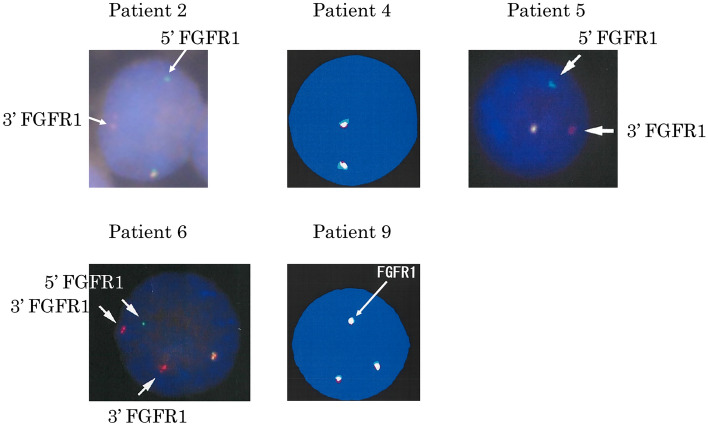
Among the 10 patients with 8p11.2 translocations, three patients (patient 2, patient 5, and patient 6) showed typical clinical manifestations of MLN-FGFR1 abnormalities (Table 1). Patient 2 was previously reported as a case report [4]. He was initially diagnosed with angioimmunoblastic T-cell lymphoma (AITL) associated with eosinophilia and achieved a first complete remission (CR) after treatment with cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP). His AITL relapsed four years later and he underwent CHASE therapy (cyclophosphamide, cytosine arabinoside, etoposide, and dexamethasone), followed by autologous peripheral blood stem cell transplantation. The second CR status lasted for approximately one year. MDS/MPN features appeared in BM samples and AML occurred. Patient 5 initially had features of atypical chronic myeloid leukemia (aCML). Two years later, she developed lymphoadenopathy and eosinophilia and was diagnosed with T-lymphoblastic lymphoma (LBL). The third patient (patient 6) was diagnosed with MLN with FGFR1 rearrangement accompanied by eosinophilia. AML developed six months later. FISH studies demonstrated disruption of FGFR1 gene regions in all three patients (Fig. 1). Eosinophilia in the peripheral blood was observed in all three cases with disruption of FGFR1 gene regions. Dysplasia in the eosinophils was not observed in patients 5 and 6; however, hypogranular eosinophils were detected in patient 2.
Table 1.
Characteristics of patients with 8p11.2 translocations
| Patient number | Age/Sex | Karyotypes | FISH | Diagnosis associated with 8p11.2 translocation | Absolute cell count (× 109/L) | Antecedent disease | Subsequent transformation | |
|---|---|---|---|---|---|---|---|---|
| Eosinophils | Monocyte | |||||||
| 1 | 70/F | 46, XX, t(8;15)(p11.2;q11.2), t(14;18)(q32;q21) [2] | n.d | Follicular lymphoma grade 3A | 0.2 | 0.26 | ||
| 2 | 61/M |
46, XY, t(8;13)(p11.2;q12) [4] 46, XY [16] |
split signal | AITL | 2.8 | 1.9 | MDS/MPN, AML | |
| 3 | 48/F |
46, XX, t(8;16)(p11.2;p13.3) [18] 46, XX [2] |
n.d | AML M5b | 0.2 | 0.28 | ||
| 4 | 69/F |
46, XX, t(1;7;8)(p13;q32;p11.2), t(6;9)(p21;p24), add(13)(q32) [4] 46, XX, idem, add(4)(p11) [7] 46, XX [9] |
no split signal | DLBCL | 0.09 | 0.62 | ||
| 5 | 70/F |
46, XX, t(8;13)(p11.2;q12) [18] 46, XX [2] |
split signal | T-LBL | 19.9 | 2.3 | aCML | |
| 6 | 55/M | 48, XY, t(8;13)(p11.2;q12), + der(13)t(8;13), + 21 [17] | split signal | MLN-FGFR1 translocation, T-lymphoblastic leukemia/lymphoma | 5.2 | 4.6 | AML | |
| 7 | 73/M |
59, XY, -X, dic (1;8)(p13;p11.2), -6, add(7)(p15), -8, -10, -12, -13, add(14)(q32), + add(15)(q22),-16, -17, -18, + 19, -20, -22 [2] 59, XY, -X, dic (1;8)(p13;p11.2), -6, add(7)(p15), -8, -10, -12, -13, + add(15)(q22), -16, -17, -18, + 19, -20, -21, -22, + mar [2] 58, XY, -X, dic (1;8)(p13;p11.2), -6, add(7)(p15), -8, -10, -12, -13, + add(15)(q22), -16, -17, -18, -18, + 19, -20, -21, -22, + mar [1] 58, XY, -X, dic (1;8)(p13;p11.2), -6, add(7)(p15), -8, -10, -12, -13, + add(15)(q22), -16, -17, -18, + 19, -20, -21, -22 [1] 46, XY [14] |
n.d | Multiple myeloma | 0.1 | 3.1 | ||
| 8 | 72/M |
46, XY, der(8)t(1;8)(q12;p11.2), t(10;14)(p11.2;q32) [2] 46, idem, -16, + 22 [1] 46, XY [17] |
n.d | NHL, marginal zone lymphoma | 0.08 | 0.16 | ||
| 9 | 53/F |
43, XX, der(3)t(1;3)(q25;p13),-7, dic(8;?)(p11.2;?), -15, -18 [8] 61, idem, + 1, + 2. + 2, + 3, + 6, + 6, + 7, + 8, + i(8)(q10), + 14, + 14, + 15, + 16, + 18, + 19, + 19, + 20, + 20, + 21 [3] |
3 signals | Acute erythroblast leukemia | 0.06 | 0.7 | MDS | |
| 10 | 59/M | 46, XY, idic(8)(p11.2), t(9;22)(q34;q11.2) [20] | n.d | Ph-positive ALL | 0 | 0.03 | ||
Abbreviations: FISH, fluorescence in situ hybridization; N.D., not done; AITL, angioimmunoblastic T-cell lymphoma; AML, acute myeloid leukemia; DLBCL, diffuse large B-cell lymphoma; T-LBL, T-cell lymphoblastic lymphoma/lymphoma; MLN-FGFR1, myeloid and lymphoid neoplasms associated with FGFR1 abnormalities; NHL, non-Hodgkin’s lymphoma; Ph, Philadelphia chromosome; ALL, acute lymphoblastic leukemia; aCML, atypical chronic myeloid leukemia; MDS, myelodysplastic syndrome
Fig. 1.
FGFR1 cleavage FISH on interphase nuclei. 5ʹ FGFR1 FISH probe was labeled green and 3ʹ FGFR1 FISH probe was labeled red. Fusion signal (red/green or yellow, white) indicates unaffected FGFR1. Splitting of the fusion signal (separated red or green signal) marks the rearrangement involving the FGFR1 locus. A split FGFR1 signal was detected in patient 2, patient 5, and patient 6. Amplification of the FGFR1 signal was detected in case 9
The other seven patients did not show typical features of MLN with FGFR1 rearrangement. Among them, patient 3 was diagnosed as AML M5b with t(8;16)(p11.2;p13.3). Patient 9 was diagnosed with MDS, and two years later, she developed AML with severe hemophagocytosis. The other five patients were clinically diagnosed with lymphoid malignancies, including follicular lymphoma, diffuse large-cell B-cell lymphoma, multiple myeloma, marginal zone lymphoma, and Philadelphia chromosome-positive ALL. FISH analysis was performed in two patients (patient 4 and patient 9). No disruption of the FGFR1 gene was observed in patient 4, and the FGFR1 gene was amplified in patient 9 (Fig. 1). In addition, there were 15 cases with eosinophilia that underwent FISH examination using dual-color split probes for the 8p11.2 loci before routine chromosomal analysis, and no case showed disruption of FGFR1 gene regions.
To examine whether there is a regional bias in the frequency of the subtypes of hematological diseases in this study, we compared the patient number of major hematological diseases between the Japanese Society of Hematology (JSH) registry data ( http://www.jshem.or.jp/modules/member/index.php?content_id=29) and seven institutions from this study. These data were unavailable for the remaining two institutions. The disease distribution is almost similar between JSH registry data and our study, however, proportion test revealed that the proportions of AML and malignant lymphoma are lower, and those of MDS, MPN, and immune thrombocytopenic purpura are higher in our study compared to JSH registry data.
Discussion
We showed that the prevalence of chromosome 8p11.2 abnormality in Japanese clinical practice was 0.06%, which is similar to that reported by Mayo Hospital of 0.06%; suggesting that there is no ethnical difference in the frequency of 8p11.2 abnormalities in hematological diseases [5]. In addition, not all patients with 8p11.2 abnormalities were diagnosed with MLN-FGFR1 abnormalities. About one third of them were categorized as MLN-FGFR1 abnormalities in our study, which was also consistent with a previous report in which four of 14 patients with 8p11.2 translocations and in another report, four of 12 patients with 8p11.2 translocations were diagnosed as MLN-FGFR1 abnormalities [5, 6].
Although the prevalence of chromosome 8p11.2 abnormalities in Japan was similar to that in Western countries, the disease types were different. The majority of Japanese patients with 8p11.2 abnormalities had lymphoid malignancies, whereas those in Western countries had myeloid malignancies. However, we cannot find a reasonable explanation for this discrepancy.
All three cases with MLN-FGFR1 abnormalities in this report harbored t(8;13)(p11.2;q12), which is the most common chromosomal abnormality in MLN-FGFR1 abnormalities [1]. In this translocation, the partner gene with FGFR1 is ZNF198 (previously reported as ZMYM2, FIM, and RAMP) located at 13q11-12, and the zinc finger domain of ZNF198 is fused to the tyrosine kinase domain of FGFR1. Recipient mice transplanted with bone marrow cells transfected with ZMYM2::FGFR1 developed myeloproliferation and intestinal T-cell lymphoma, and in immunodeficient mice, human cord blood CD34+ cells transfected with ZMYM2::FGFR1 showed expansion of multiple myeloid cell lineages and accumulation of blasts in BM [7, 8]. In patients with t(8;13)(p11.2;q12), lymphadenopathy and hepatosplenomegaly are often the first symptoms, and most patients are diagnosed with T-LBL/T-lymphoma [1]. Consistent with this, all three patients in our study developed T-cell malignancies in their clinical course.
Umino et al. reported the characteristics and prognosis of 45 cases with MLN-FGFR1 abnormalities collected from a computerized search of the medical literature using PubMed® [9]. At diagnosis, 31% and 69% of patients were in chronic phase as MPN or MDS and in advanced phase as acute leukemia or lymphoblastic lymphoma, respectively. Approximately half of patients in chronic phase transformed to advanced blast phase within one year, and the median OS from diagnosis was 9 months if allogeneic HSCT was not performed before transformation to advanced blast phase. The OS for patients in advanced phase was dismal, and the 1-year OS was 29.8%. Since FGFR1 rearrangement leads to constitutive activation of tyrosine kinase, which induces abnormal proliferation, inhibitors targeting FGFR1 kinase activity have been developed [10, 11]. Among them, pemigatinib, a reversible ATP-competitive FGFR inhibitor, has been recently approved as a novel drug for treatment of myeloid/lymphoid neoplasms with FGFR1 rearrangement in the US and Japan. To select appropriate patients for pemigatinib therapy, FISH analysis demonstrating FGFR1 gene disruption would be desirable in addition to chromosomal 8p11.2 abnormalities for patients who do not exhibit the typical clinical features of MLN-FGFR1 abnormalities, because approximately one third of patients with 8p11.2 abnormalities were diagnosed as MLN-FGFR1 abnormalities in our study and previous reports [5, 6].
Of other seven patients without typical features of MLN with FGFR1 rearrangement, one patient was diagnosed as AML M5b with t(8;16)(p11.2;p13.3). MYST3 (MYST histone acetyltransferase 3) is disrupted by the chromosomal 8p11.2 translocation, as is FGFR1, and the most common translocation partner gene for MYST3 is CEBBP (CREB-binding protein), located on chromosomal 16p13.3 [12]. As the chromosomal translocation t(8;16)(p11.2;p13.3) is reported to be associated with an aggressive form of AML M4/M5, the 8p11.2 abnormality detected in this patient is speculated to be a MYST3-CREBBP rearrangement, although we cannot show it by FISH analysis due to lack of residual BM samples [13]. Another patient whose MDS progressed to AML had FGFR1 amplification. The overexpression or amplification of FGFR1 has been reported in 10–20% of breast cancer, head and neck squamous cell carcinoma, and lung squamous cell carcinoma, and its association with poor survival has been reported in breast cancer, head and neck squamous cell carcinoma, but not in lung cancer [14–16]. In contrast, overexpression or amplification of FGFR1 was rare in hematological malignancies. For the remaining four patients (patients 1, 7, 8, and 10), FISH examination using dual-color split probes for the 8p11.2 loci was not performed because of the lack of residual samples; therefore, it is difficult to completely exclude them from having MLN with FGFR1 rearrangement. The breakpoint of partner genes rearranged with 8p11.2, in these four cases included 1p13, 1q12, 8p11.2, and 15q11.2. As these chromosomal breakpoints have not been previously reported to generate FGFR1 fusion genes and cause MLN with FGFR1 rearrangement [1], the possibility of their diagnosis as MLN with FGFR1 rearrangement might be low, along with the absence of the typical clinicopathological features of MLN with FGFR1 rearrangement.
In conclusion, the prevalence of 8p11.2 abnormalities in Japanese clinical practice and the proportion of MLN-FGFR1 abnormalities among them were almost consistent with those in previous reports from Western countries.
Funding
Open access funding provided by University of Miyazaki.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Li T, Zhang G, Zhang X, Lin H, Liu Q. The 8p11 myeloproliferative syndrome: genotypic and phenotypic classification and targeted therapy. Front Oncol. 2022;12:1015792. doi: 10.3389/fonc.2022.1015792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36(7):1703–19. [DOI] [PMC free article] [PubMed]
- 3.Katoh M, Nakagama H. FGF receptors: cancer biology and therapeutics. Med Res Rev. 2014;34(2):280–300. doi: 10.1002/med.21288. [DOI] [PubMed] [Google Scholar]
- 4.Nonaka M, Kawano N, Hisakata N, Mizutani S, Kugimiya H, Takigawa K, et al. Eight p11 myeloproliferative syndrome (EMS) that initially manifested as angioimmunoblastic T-cell lymphoma (AITL) and subsequently transformed into MDS/MPN complicating with t (8;13) (p11.2; q12). J Japanese Soc Lab Hematol. 2023;24(3):440–7.
- 5.Patnaik MM, Gangat N, Knudson RA, Keefe JG, Hanson CA, Pardanani A, et al. Chromosome 8p11.2 translocations: prevalence, FISH analysis for FGFR1 and MYST3, and clinicopathologic correlates in a consecutive cohort of 13 cases from a single institution. Am J Hematol. 2010;85(4):238–42. [DOI] [PubMed]
- 6.Baldazzi C, Luatti S, Paolini S, Papayannidis C, Marzocchi G, Ameli G, et al. FGFR1 and KAT6A rearrangements in patients with hematological malignancies and chromosome 8p11 abnormalities: biological and clinical features. Am J Hematol. 2016;91(3):E14–E16. doi: 10.1002/ajh.24276. [DOI] [PubMed] [Google Scholar]
- 7.Roumiantsev S, Krause DS, Neumann CA, Dimitri CA, Asiedu F, Cross NC, et al. Distinct stem cell myeloproliferative/T lymphoma syndromes induced by ZNF198-FGFR1 and BCR-FGFR1 fusion genes from 8p11 translocations. Cancer Cell. 2004;5(3):287–298. doi: 10.1016/S1535-6108(04)00053-4. [DOI] [PubMed] [Google Scholar]
- 8.Agerstam H, Jaras M, Andersson A, Johnels P, Hansen N, Lassen C, et al. Modeling the human 8p11-myeloproliferative syndrome in immunodeficient mice. Blood. 2010;116(12):2103–2111. doi: 10.1182/blood-2009-05-217182. [DOI] [PubMed] [Google Scholar]
- 9.Umino K, Fujiwara SI, Ikeda T, Toda Y, Ito S, Mashima K, et al. Clinical outcomes of myeloid/lymphoid neoplasms with fibroblast growth factor receptor-1 (FGFR1) rearrangement. Hematology. 2018;23(8):470–477. doi: 10.1080/10245332.2018.1446279. [DOI] [PubMed] [Google Scholar]
- 10.Subbiah V, Iannotti NO, Gutierrez M, Smith DC, Feliz L, Lihou CF, et al. FIGHT-101, a first-in-human study of potent and selective FGFR 1–3 inhibitor pemigatinib in pan-cancer patients with FGF/FGFR alterations and advanced malignancies. Ann Oncol. 2022;33(5):522–533. doi: 10.1016/j.annonc.2022.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Verstovsek S, Gotlib J, Vannucchi AM, Rambaldi A, Reiter A, Shomali W, et al. FIGHT-203, an ongoing phase 2 study of pemigatinib in patients with Myeloid/Lymphoid Neoplasms (MLNs) with Fibroblast Growth Factor Receptor 1 (FGFR1) Rearrangement (MLNFGFR1): a focus on centrally reviewed clinical and cytogenetic responses in previously treated patients. Blood. 2022;140(Suppl 1):3980–3982. doi: 10.1182/blood-2022-163099. [DOI] [Google Scholar]
- 12.Borrow J, Stanton VP, Jr, Andresen JM, Becher R, Behm FG, Chaganti RS, et al. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14(1):33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 13.Gervais C, Murati A, Helias C, Struski S, Eischen A, Lippert E, et al. Acute myeloid leukaemia with 8p11 (MYST3) rearrangement: an integrated cytologic, cytogenetic and molecular study by the groupe francophone de cytogenetique hematologique. Leukemia. 2008;22(8):1567–1575. doi: 10.1038/leu.2008.128. [DOI] [PubMed] [Google Scholar]
- 14.Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Can Res. 2010;70(5):2085–2094. doi: 10.1158/0008-5472.CAN-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Y, Ai LS, Zhou LQ. Prognostic value of FGFR1 expression and amplification in patients with HNSCC: a systematic review and meta-analysis. PLoS ONE. 2021;16(5):e0251202. doi: 10.1371/journal.pone.0251202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell PA, Yu Y, Young RJ, Conron M, Wainer Z, Alam N, et al. Prevalence, morphology, and natural history of FGFR1-amplified lung cancer, including squamous cell carcinoma, detected by FISH and SISH. Mod Pathol. 2014;27(12):1621–1631. doi: 10.1038/modpathol.2014.71. [DOI] [PubMed] [Google Scholar]