Abstract
The stringent response utilizes hyperphosphorylated guanine [(p)ppGpp] as a signaling molecule to control bacterial gene expression involved in long-term survival under starvation conditions. In gram-negative bacteria, (p)ppGpp is produced by the activity of the related RelA and SpoT proteins. Mycobacterium tuberculosis contains a single homolog of these proteins (RelMtb) and responds to nutrient starvation by producing (p)ppGpp. A relMtb knockout strain was constructed in a virulent strain of M. tuberculosis, H37Rv, by allelic replacement. The relMtb mutant displayed a significantly slower aerobic growth rate than the wild type in synthetic liquid media, whether rich or minimal. The growth rate of the wild type was equivalent to that of the mutant when citrate or phospholipid was employed as the sole carbon source. These two organisms also showed identical growth rates within a human macrophage-like cell line. These results suggest that the in vivo carbon source does not represent a stressful condition for the bacilli, since it appears to be utilized in a similar RelMtb-independent manner. In vitro growth in liquid media represents a condition that benefits from RelMtb-mediated adaptation. Long-term survival of the relMtb mutant during in vitro starvation or nutrient run out in normal media was significantly impaired compared to that in the wild type. In addition, the mutant was significantly less able to survive extended anerobic incubation than the wild-type virulent organism. Thus, the RelMtb protein is required for long-term survival of pathogenic mycobacteria under starvation conditions.
The global burden of 8 million active tuberculosis cases annually is overshadowed by the fact that fully one-third of the global population of humans is asymptomatically infected with tubercle bacilli (67). Many cases of clinically active tuberculosis arise from reactivation of an infection acquired years before the onset of overt symptoms, and these reflect the reemergence of actively growing organisms from an apparently nonreplicative state (4, 40, 65). Not all patients latently infected will develop active disease, and the fraction that ultimately develops tuberculosis through reactivation versus reinfection is a topic of current investigation (69). Conventional antibiotic therapy is sufficient to sterilize sputa and bronchoalveolar lavage specimens, whereupon bacilli cannot be detected by either acid-fast staining or culture. DNA from Mycobacterium tuberculosis can still be detected in such samples, and premature termination of therapy often results in recrudescence of disease, suggesting the continued presence of viable organisms (20, 26, 35, 51, 52, 71). During such a latent infection, the bacteria are thought to remain isolated in masses of lymphoid cells called granulomas, wherein they are encased in an impermeable caseous material by a thin layer of activated macrophages, with restricted access to nutrients or oxygen (15, 60).
The details of the dormant state are unclear, but the mechanism probably falls short of true spore formation. Instead, the processes involved in long-term survival within an asymptomatic patient probably reflect the result of bacterial responses to relatively simple signals from the environment. Adaptation of bacilli to long-term survival under these conditions may therefore be associated with coordinated alterations in patterns of gene expression in specific metabolic networks. The stringent response is a broad transcriptional program encompassing at least 80 genes in Escherichia coli that mediates prokaryotic adaptation to survival under conditions of starvation (9). Induction of the stringent response in E. coli stimulates polyphosphate synthesis (58), increases fatty acid cyclopropanation (19), inhibits fatty acid and phospholipid synthesis (27), upregulates glycogen synthesis (8, 18), upregulates the stationary-phase sigma factor RpoS (44), and inhibits stable RNA synthesis (9). The stringent response can be induced by amino acid, carbon, nitrogen, or phosphorous starvation. Additionally, UV light exposure and fatty acid starvation induce the response (39, 61). The stringent response is mediated by increased intracellular levels of hyperphosphorylated guanine nucleotides, specifically the 3′-pyrophosphate derivative of GDP (ppGpp) and the 3′-pyrophosphate derivative of GTP (pppGpp), the mixture of both being referred to as (p)ppGpp.
In E. coli and other gram-negative bacteria, two proteins have been identified that are responsible for the synthesis of (p)ppGpp, RelA, and SpoT (9). RelA, or ppGpp synthase I, is ribosome associated and is activated by binding uncharged tRNAs to the ribosome upon depletion of amino acids (52). SpoT, ppGpp synthase II, is not associated with ribosomes and has both (p)ppGpp synthetic and hydrolytic activities (21, 22, 28). E. coli strains lacking RelA or lacking both RelA and SpoT cannot grow in minimal media and survive poorly in the stationary phase (9). Among many gram-positive organisms, including the actinomycetes, there appears to be only one protein with homology to both RelA and SpoT that coordinates the metabolism of (p)ppGpp. This protein has been called Rel and has been identified in Streptomyces coelicolor and Streptomyces antibioticus as well as in Corynebacterium glutamicum and Streptococcus equisimilis (10, 11, 32, 46–49, 76). In both species of Streptomyces, the stringent response and Rel protein activity appear to be tightly coupled to stationary-phase adaptation and production of unique and specific antibiotics, such as actinomycin D and actinorhodine. Antibiotic production typically occurs coincident with the formation of aerial mycelia and entry into stationary phase (12). Disruption of the rel gene destroys the ability of both Streptomyces species to produce antibiotics and interferes with spore formation (32, 46).
M. tuberculosis also has only a single RelA or SpoT homolog (Rv2583c), designated RelMtb (13). The RelMtb protein of M. tuberculosis has the most identity to the Rel protein of three other gram-positive actinomycetes, C. glutamicum (67% identity), S. antibioticus (66% identity), and S. coelicolor (62% identity). It has significantly less homology to non-actinomycete gram-positive organisms, for example, the RelA or SpoT homolog of Bacillus subtilis (43% identity), and even less to gram-negative organisms such as Escherichia coli (39% identity to spoT and 37% identity to relA). Purified recombinant RelMtb is a protein of 738 amino acids that is an ATP:GTP/GDP/ITP 3′-pyrophosphoryltransferase as well as an Mn2+-dependent (p)ppGpp 3′-pyrophosphorylhydrolase (2).
MATERIALS AND METHODS
Nucleotide labeling and analysis.
Bacteria were grown to early log phase (optical density [OD] [650 nm] of 0.2 to 0.3) in Difco Middlebrook 7H9-ADC-Tween medium (containing [per liter] ammonium sulfate, 0.5 g; l-glutamate, 0.5 g; sodium citrate, 0.1 g; pyridoxine, 1 mg; biotin, 0.5 mg; disodium phosphate, 2.5 g; monopotassium phosphate, 1 g; ferric ammonium citrate, 40 mg; magnesium sulfate, 50 mg; calcium chloride, 0.5 mg; zinc sulfate, 1 mg; and copper sulfate, 1 mg; with 5 g of albumin [fraction V; bovine], 2 g of dextrose, 0.2% glycerol, and 0.05% Tween 80 added after sterilization). Where indicated, this medium was made with a lowered phosphate concentration to increase 32P incorporation containing 1/25 normal phosphate levels (1 mM final concentration) and with 40 mM MOPS (morpholinepropanesulfonic acid) added to maintain pH at 6.6. Bacilli were grown for 48 h in low-phosphate medium, and then 32Pi was added at 1 mCi/ml and cells were labeled for 3 to 4 h. Labeled M. tuberculosis was pelleted by centrifugation, washed once with Tris-buffered saline with Tween (TBST) (50 mM Tris-HCl [pH 7], 150 mM NaCl, 0.05% Tween 20), and then resuspended in the desired medium. Time points were generally taken at 0, 20, 40, 60, 90, and 120 min. Samples were pelleted by centrifugation and then lysed in 1 M formic acid by bead beating with 0.1-mm-diameter glass beads (BioSpec Products, Bartlesville, Okla.) with three pulses of 40 s each. After high-speed centrifugation to remove cell debris, samples were then directly spotted onto anion-exchange (polyethylenimine-cellulose) plates (EM Science, Gibbstown, N.J.). The two-dimensional thin-layer chromatography (2D-TLC) system employed was adapted from Bochner and Ames (5). First-dimension separation (by charge) was performed in 0.75 M Tris-HCl–25 mM EDTA (pH 8). The second dimension (by base content) solvent was saturated ammonium sulfate plus 5 mM EDTA, with the pH adjusted to 3.5 with sulfuric acid. Plates were dehydrated by a 5-min immersion in methanol before and between developments. Radioactivity was detected with a Storm 860 PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). Nonradioactive standards (Sigma) were run and visualized by fluorescence quenching to identify spots.
Bacterial strains, media, and growth conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. Unless otherwise indicated, M. tuberculosis was grown with constant stirring in 7H9-ADC-Tween and on Middlebrook 7H11-ADC as the solid medium (containing [per liter] pancreatic digest of casein, 1 g; l-glutamate, 0.5 g; sodium citrate, 0.4 g; pyridoxine, 1 mg; biotin, 0.5 mg; ferric ammonium citrate, 40 mg; ammonium sulfate, 0.5 g; disodium phosphate, 1.5 g; monopotassium phosphate, 1.5 g; magnesium sulfate, 50 mg; Bacto agar, 15 g; and malachite green, 1 mg; with 5 g of albumin [fraction V; bovine], 2 g of dextrose, and 0.2% glycerol added after sterilization). E. coli DH5α was grown in Luria-Bertani broth or on agar. Kanamycin and hygromycin were used at 50 and 200 μg/ml, respectively, for selection in E. coli and at 20 and 50 μg/ml, respectively, for selection in M. tuberculosis. Suicide plasmids for targeted gene knockout were electroporated into M. tuberculosis as described by Gordhan and Parish (25). Analysis of cell wall composition and mycolic acid subclass distribution was conducted as previously described (80).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | supE44 ΔlacU169 (Δ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Laboratory collection |
| M. tuberculosis | ||
| H37Rv | Virulent laboratory strain (ATCC 27294) | Laboratory collection |
| SJA14 | Single-crossover (SCO) recombinant with pSA1; relA ΔrelA::hyg lacZ Hygr | This work |
| SJA16 | Double-crossover (DCO) recombinant with pSA1 containing integrated copy of pSA1; ΔrelA::hyg lacZ Hygr | This work |
| SJA32 | SCO recombinant with pSA3; relA+ sacB Hygr | This work |
| SJA33 | DCO recombinant obtained with pSA3; ΔrelA::hyg Hygr | This work |
| CDC1551 | Virulent M. tuberculosis clinical isolate (CSU93) | 68 |
| Plasmids | ||
| pGEM3Z(+)f | E. coli cloning vector; Apr | Promega |
| Pac6 | Cosmid from pYUB328::H37Rv carrying relA | This work |
| pIJ963 | E. coli cloning vector; Apr Hygr | 29 |
| pSMT3 | E. coli-Mycobacterium shuttle vector carrying mycobacterial hsp60 promoter; Hygr | 55 |
| pSMT3lacZ | pSMT3 derivative carrying lacZ cloned in BamHI site; hsp60-lacZ | This work |
| p2NIL | E. coli cloning vector with unique PacI site; Kmr | 56 |
| pGOAL13 | E. coli cloning vector with hsp60-sacB cassette carried on PacI fragment; Apr | 56 |
| pRelΔS | pGEM3Z(+)f derivative carrying ΔrelA allele lacking internal SphI fragment | This work |
| pRelΔSH | pRelΔS derivative carrying ΔrelA marked with hyg gene from pIJ963 inserted in BglII site of ΔrelA (ΔrelA::hyg) | This work |
| pSA1 | pRelΔSH carrying hsp60-lacZ | This work |
| pSA3 | p2NIL derivative carrying ΔrelA::hyg allele from pRelΔSH and PacI cassette from pGOAL13; hsp60-sacB; Kmr Hygr | This work |
| pMV306K | E. coli-Mycobacterium shuttle vector; integrates into the L5 attB site in Mycobacteria; Kmr | 41 |
| pMV306K-rel | pMV306K derivative carrying relMtb | This work |
| pMV306K-hsp60-luc | pMV306K carrying the NotI-SalI luc fragment from pGL3 (Promega) and the KpnI-HindIII groEL promoter from pMV261 | 16 |
Construction of the ΔrelMtb::hyg allele.
DNA manipulations were carried out according to standard procedures (59). A cosmid, Pac6, containing the relMtb gene was isolated by screening the pYUB328::H37Rv cosmid library of M. tuberculosis (3) with an internal relMtb probe that was generated by PCR amplification with the degenerate primers RELA1 [5′-CATGGATCCAACGG-(GC)TACGAG(AT)(GC)(GC)(AC)T(GC)CACAC] and RELA2 [5′-CATGGATCCGTGTG(GC)A(CT)(GC)GCGTA(GC)GCGAAGTC]. PCR amplifications were carried out on a Hybaid PCRExpress thermal cycler with TaqI DNA polymerase (Roche Molecular Biochemicals) at an annealing temperature of 55°C. The plasmid pRelΔS, carrying the ΔrelMtb allele, which encodes an in-frame-deleted form of RelMtb lacking the N-terminal region between His94 and Ala413, was constructed by ligating the 1,865-bp BamHI-SphI fragment from Pac6 (containing 1,581 bp of upstream sequence and 284 bp of relMtb coding sequence) and the 1,749-bp SphI-KpnI fragment from Pac6 (containing 1,136 bp of coding sequence and 613 bp of downstream sequence) in BamHI-KpnI-pGEM3Z(+)f. A hyg marker, carried on a 1,739-bp BamHI-BglII fragment from pIJ963 (29), was cloned in the unique BglII site, located 219 bp into the relMtb coding region, to form pRelΔSH. In this construct, the hyg gene contained in the marked ΔrelMtb::hyg allele was codirectional with the ΔrelMtb gene. The ΔrelMtb::hyg allele could theoretically encode a polypeptide comprising the N-terminal 73 amino acids of RelMtb fused to a 72-amino-acid sequence derived from the 5′-untranslated region of hyg. This fusion is unlikely to possess Rel-associated synthetase or phosphohydrolase activities (22). Expression of the C-terminal part of the mutilated RelMtb as a fusion of the His94-Ala95 region to the His412-Ala413 region would require the improbable presence of a promoter downstream of the hyg gene terminator and a start codon (see Fig. 2).
FIG. 2.
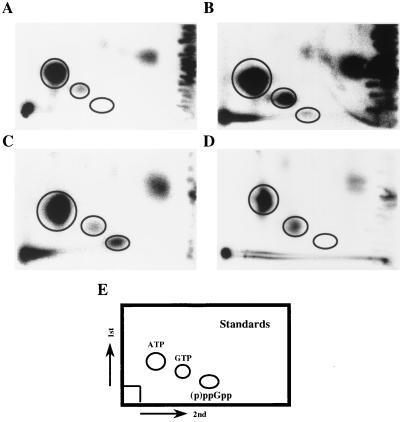
Analysis of intracellular (p)ppGpp in M. tuberculosis by 2D-TLC. (A) Control nucleotides extracted from 32Pi-labeled M. tuberculosis H37Rv aerobically grown in normal 7H9 medium during the log phase. (B) Nucleotides extracted from stationary-phase culture of H37Rv (2-week-old culture). These cultures label significantly less well than logarithmically growing cultures, and, thus, this panel has been exposed more heavily. (C) Nucleotides extracted from H37Rv cells labeled and then placed under starvation conditions (resuspension in TBST). (D) Sample prepared as in panel C, but with the SJA16 Δrel strain of H37Rv. (E) Schematic representation of nonradioactive standards run in the same 2D-TLC system. Complementation of the ΔrelMtb strain with the relMtb gene restored the ability to produce (p)ppGpp under these conditions.
Construction of ΔrelMtb::hyg mutant strains.
The hsp60-lacZ reporter from pSMT3lacZ, in which the E. coli lacZ gene was expressed under the control of the Mycobacterium bovis BCG groEL promoter of pSMT3 (55), was cloned as a 3,100-bp XbaI-HindIII fragment into the unique XbaI and HindIII sites of pRelΔSH to create pSA1. M. tuberculosis was electroporated with 1 to 5 μg of UV-irradiated pSA1 (25, 30, 56), and cells were plated on 7H11-ADC agar containing hygromycin and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal [50 μg/ml]). Chromosomal DNA was extracted from Hygr transformants for Southern blot analysis, as previously described (24). A second knockout vector containing the Bacillus subtilis sacB counterselectable marker (57) was constructed as follows. The 5,361-bp BamHI-KpnI fragment, containing the partially deleted ΔrelMtb::hyg allele, was excised from pSA1 and cloned in p2NIL (25; T. Parish, B. Gordhan, D. A. Smith, G. J. Bancroft, V. Mizrahi, R. A. McAdam, and N. G. Stoker, Fourth Int. Conf. Pathog. Mycobacterial Infect., abstr. 01, 1999). The PacI fragment from pGOAL13 (Parish et al., Fourth Int. Conf. Pathog. Mycobacterial Infect.), containing an hsp60-sacB cassette in which the sacB gene is expressed by the M. bovis BCG groEL promoter, was then introduced into this vector to produce pSA3. This vector was electroporated into M. tuberculosis following UV irradiation, as described above. Transformants were selected on 7H11-ADC agar containing hygromycin and kanamycin. Since kanamycin selects for the hyg marker contained in the p2NIL-based vector, colonies growing on kanamycin plus hygromycin are predicted to be the result of a site-specific single-crossover event. Hygr Kmr transformants were streaked on 7H11-ADC plates containing hygromycin only, to allow double-crossover events to occur. The positive selection option presented by the presence of the sacB gene was utilized to select for double-crossover recombinants by plating on 7H11-ADC containing hygromycin and 2% sucrose. Hygr Sucr (sucrose-resistant) colonies were patched onto fresh medium to test for kanamycin sensitivity, since this would be indicative of vector loss. Hygr Sucr Kms colonies obtained by this two-step process were selected as candidate allelic exchange mutants.
Construction of a relMtb-complemented strain.
We first PCR amplified the upstream region with a small amount of the N terminus of the relMtb gene. The N-terminal primer was 5′-AATATGGATCCGACGAAAATGTATGCGGTGA-3′, which included a BamHI site, and the C-terminal primer was 5′-CTCGTCGCAAGATCGACAGGTC-3′, which included a BglII site. The relMtb gene was then PCR amplified alone. The N-terminal primer for this amplification was 5′-ACGTCATATGACCGCCCAGCGCAGCACCACCAAT-3′, which included an NdeI site, and the C-terminal primer was 5′-ACGTGGTACCCTACGCGGCCGAGGTCACCCGGTA-3′, which included a KpnI site. The relMtb gene was then cut with BglII and ligated to the upstream region to reconstruct the complete relMtb gene with intact upstream sequence. This fragment was then ligated into pMV306 by using the BamHI and KpnI sites. This vector, which carries the attP site of mycobacteriophage L5, integrates site specifically in single copy at the L5 phage integration site, attB (41). The ΔrelMtb strain was electroporated with pMV306K-rel, and Kmr Hygr colonies were selected. The complemented strain thus has a single chromosomal copy of the relMtb gene under the native promoter.
Growth curves.
M. tuberculosis strains were grown in 7H9-ADC-Tween as the reference media. Log-phase cultures (OD of 0.1 to 1) were used to inoculate test media to be analyzed for growth. Bacteria were cultured in ∼50-ml volumes in 250-ml bottles rolling in a 37°C incubator. Individual ingredients were added or removed from the 7H9-ADC-Tween base medium as indicated in the figures. Growth was measured by determining the A650 of the culture. The growth rate in Table 3 is a linear fit to the OD versus time in days at the most rapid growth phase; these results were repeated at least twice in each case with similar results. Dipalmitoylphosphatidylcholine (DPPC) was obtained from Sigma.
TABLE 3.
Growth of the ΔrelMtb strain in liquid media
| Mediuma | Growth
rateb
|
Maximum
ODc
|
||
|---|---|---|---|---|
| Wild-type Rv | rel knockout mutant | Wild-type Rv | rel knockout mutant | |
| Standard 7H9 | 0.62 | 0.13 | 2.4 | 0.92 |
| +212 μM OA | 0.54 | 0.185 | 2.04 | 1.0 |
| +424 μM OA | 0.424 | 0.133 | 2.24 | 1.08 |
| 0.5% Peptone | 0.76 | 0.21 | 3.1 | 1.6 |
| +1% Glycerol | 0.56 | 0.18 | 3.6 | 1.2 |
| +1% Glycerol + 0.5% Peptone | 0.80 | 0.28 | 4.0 | 2.0 |
| No glycerol | 0.084 | 0.046 | 1.3 | 0.42 |
| No glycerol and no glucose | 0.088 | 0.060 | 0.38 | 0.26 |
| 0.2 mM DPPC | 0.160 | 0.165 | 0.60 | 0.65 |
Standard Middlebrook 7H9 has 0.2% glycerol, no peptone, no oleic acid (OA), and 5.8 mM sulfate. All media are modifications of 7H9. Low-sulfate medium contained 0.5 mM sulfate. DPPC-containing medium had no glucose or glycerol. Each condition was evaluated at least twice with similar results.
Growth rate is a linear fit to the OD (A650) versus time in days at the most rapid growth phase.
Highest OD for that culture.
Competition starvation survival assay.
The ΔrelMtb (Hygr) strain and H37Rv transformed to Kanr with the pMV306K vector were grown to early log phase in 7H9-ADC-Tween. Bacilli were then pelleted by centrifugation and washed twice with 0.1% Tween 80 to remove traces of medium. Bacteria were then resuspended in TBST, the OD of each culture was adjusted to 0.2, and then the cultures were mixed at a 1:1 ratio in a 50-ml final volume and incubated by rolling at 37°C. Bacterial clumping was minimized by vortexing a 1-ml aliquot of the culture for 2 to 3 min in a 1.5-ml tube containing ∼300 mg of glass beads. In control experiments, this procedure increased the apparent number of CFU of a clumpy M. smegmatis culture, but did not lower the number of CFU of a log-phase culture. For long-term nutrient run-out experiments with 7H9-ADC-Tween, bacteria were diluted to approximately 104 CFU/ml in fresh 7H9-ADC-Tween and grown aerobically at 37°C. Aliquots were removed at the indicated times and plated as described above. For anaerobic survival assays, bacteria were diluted to an OD of 0.05 in fresh 7H9-ADC-Tween and then mixed in a 1:1 ratio. This mixture was aliquoted into 1.5-ml screw-cap tubes with rubber septa with no headspace and incubated stationary at 37°C. Control tubes with methylene blue indicator dye demonstrated that all oxygen was consumed after 4 to 5 days. For all experiments, the number of CFU per milliliter was determined by plating in triplicate at 3 to 5 dilutions onto 7H11-ADC plates containing hygromycin or kanamycin on the indicated days. Cell viability was determined by plating 10-fold dilutions in 7H9-ADC-Tween onto 7H11-ADC plates with kanamycin or hygromycin selection and counting colonies approximately 3 weeks after inoculation at the indicated times.
Ribosome isolation.
Two liters of cells of wild-type M. tuberculosis and ΔrelMtb M. tuberculosis was grown to an OD of 1.3 in 7H9-ADC-Tween, pelleted, and resuspended in TBST for 24 h. The cells were repelleted and resuspended in buffer 1 (10 mM Tris-HCl [pH 7.6], 30 mM KCl, 15 mM MgCl2, 6 mM β-mercaptoethanol) before being lysed with a bead beater and centrifuged at 15,000 × g followed by 30,000 × g each for 20 min. The supernatant was then centrifuged at 150,000 × g for 3 h. The pellet containing ribosomes was again resuspended in buffer 1 and layered over buffer 2 (buffer 1 containing 30% [wt/vol] sucrose) and centrifuged at 150,000 × g for 15 h. This pellet was washed and resuspended in ribosome buffer (50 mM Tris-HCl [pH 7.6], 50 mM KCl, 10 mM MgCl2). Ribosome content was estimated by using the conversion factor of 1 OD260 unit = 25 pmol of 70S ribosomes in 1 ml of distilled water.
Macrophage infection.
Bacterial inocula were prepared as previously described from a 1:400 dilution into RPMI 10 medium (GIBCO/BRL) of a well-dispersed OD of 0.2 7H9-ADC-Tween culture (79). The human macrophage-like cell line THP-1 (ATCC 202-TIB) was seeded at 8 × 105 cells per well in 24-well plates. A 250-μl-per-well bacterial suspension was applied to the adherent cells (for a final multiplicity of infection of approximately 1), and these cells were incubated at 37°C for 1 h. Extracellular bacteria were removed by four washes with 0.5 ml of RPMI 10, and macrophages were subsequently fed with 1 ml of RPMI 10 per well and incubated at 37°C under 5% CO2. Luciferase-reporting strains were generated by transformation with pMV306K-hsp60-luc, in which the strong constitutive M. tuberculosis groEL promoter drives luciferase expression (16). At each time point, macrophages were lysed with 250 μl of 0.1% Triton X-100, and then 50 μl was removed and mixed with 50 μl of assay buffer (140 μg of luciferin per ml [Molecular Probes] and 0.4% Triton X-100 in distilled water). Macrophages were cultured in triplicate identical wells, and luciferase assays were read in duplicate with a TopCount 96-well reader (Hewlett Packard).
RESULTS
Starvation induces the formation of intracellular (p)ppGpp in M. tuberculosis.
To assess whether M. tuberculosis utilized a conventional stringent response involving (p)ppGpp, we labeled the total cellular nucleotides of M. tuberculosis with 32Pi during growth in vitro in low-phosphate media. Labeled bacilli were then shifted to various conditions, and the intracellular nucleotide responses were analyzed by 2D-TLC (see Fig. 2). M. tuberculosis was found to have a very low basal level of (p)ppGpp when grown aerobically and in log phase in minimal medium. (Middlebrook 7H9-ADC-Tween medium contains no complex source of nutrients [see Materials and Methods]). These 2D-TLCs do not differentiate between ppGpp and pppGpp. Several published 1D-TLC systems were tried, but incorporation of label was low in any medium, and unambiguous determination of the ppGpp/pppGpp ratio was not possible due to heavy sample loading requirements. Inhibition of the respiratory chain by azide treatment (5 mM) for 120 min and complete starvation by incubation in Tris-buffered saline with Tween for 120 min strongly increased (p)ppGpp levels (Table 2). The frontline drugs isoniazid and ethambutol at concentrations well above the MIC had no significant effect over similar time intervals. In contrast to the (p)ppGpp induction observed in E. coli, resuspension of log-phase M. tuberculosis in serine hydroxamate, a competitive inhibitor of the seryl-tRNA charging enzyme, had no effect on (p)ppGpp levels, whereas it induces the accumulation of (p)ppGpp in E. coli. While this may imply a significant divergence in amino acid metabolism, M. tuberculosis may simply be impermeable to serine hydroxamate, or the M. tuberculosis tRNA-Ser charging enzyme may be insensitive to such reagents.
TABLE 2.
(p)ppGpp formation under various conditionsa
| Condition | (p)ppGpp formation |
|---|---|
| Control | − |
| 1% Oxygen | − |
| 5 mM Sodium azide | + |
| Stationary phase | + |
| 20 mM H2O2 | − |
| Carbon starvation | +/− |
| Complete starvation | +++ |
| 1-mg/ml Serine hydroxamate | − |
| 1-μg/ml Isoniazid | − |
| 10-μg/ml Ethambutol | − |
M. tuberculosis cells were labeled as described in Materials and Methods, and nucleotides were resolved by 2D-TLC. (p)ppGpp formation was assessed by autoradiographic analysis and compared with that of authentic unlabeled (p)ppGpp.
Construction and genotypic characterization of ΔrelMtb::hyg mutants.
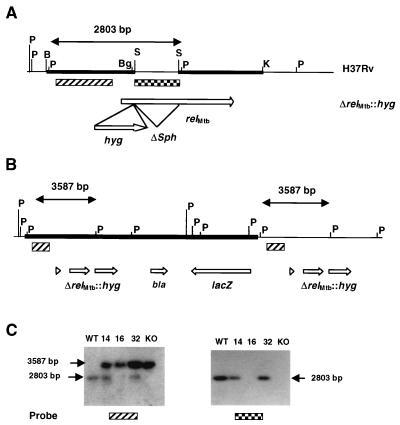
To directly demonstrate the linkage between the RelMtb protein and the production of (p)ppGpp, the gene encoding RelMtb was deleted from the H37Rv strain by allelic exchange. Construction of a deletion-replacement construct allowed the substitution of the relMtb gene for a hygromycin resistance determinant (hyg). The Southern blot analysis used to determine the genotype of transformants obtained with pSA1 is shown in Fig. 1. PvuII digestion of the wild-type strain produces a 2,803-bp relMtb-containing fragment that cross-hybridizes with the upstream 2,247-bp SmaI probe used in this analysis. Site-specific integration of pSA1 in the chromosome by a single-crossover event would result in the formation of a second, 3,587-bp PvuII fragment containing the ΔrelMtb::hyg allele from pSA1, which cross-hybridizes to the same probe. Allelic exchange by double crossover would therefore result in the disappearance of the wild-type allele and its replacement by the larger mutant allele. Southern blot analysis of the DNA extracted from 10 transformants obtained with pSA1 revealed that in one clone, SJA16, the wild-type allele was absent. Further analysis of SJA16 confirmed that the new PvuII allele contained the hyg gene. In addition, probing with the 945-bp SphI fragment that had been removed from the relMtb gene to generate the ΔrelMtb allele confirmed its absence from the genome of SJA16. However, plating of SJA16 on indicator medium containing X-Gal resulted in the unexpected formation of blue colonies, indicating the presence of lacZ in this clone and ruling out the possibility that SJA16 was the product of simple allelic exchange by double crossover. Additional Southern blot analysis suggested that a site-specific double-crossover event had occurred, but had been followed by a subsequent crossover event resulting in the integration of a second copy of pSA1 (Fig. 1). This may have been due to the relatively large amount of UV-irradiated plasmid DNA (5 μg) used in the electroporation experiment that yielded this transformant. Since the second vector copy could theoretically be lost from SJA16 by a further crossover event, the strain was grown, serially diluted, and plated on indicator media in order to identify white colonies. However, no white colonies were found among 108 colonies that were screened.
FIG. 1.
Construction and genotypic characterization of ΔrelMtb::hyg mutants. (A) Locus of the relMtb gene showing the restriction sites that were used to construct the ΔrelMtb::hyg allele contained in pSA1 and pSA3. The BamHI-SphI and SphI-KpnI chromosomal DNA fragments that were used to form the ΔrelMtb allele contained in pRelΔS are denoted by thick, horizontal lines. B, BamHI; Bg, BglII; K, KpnI; P, PvuII; S, SphI. (B) Schematic representation of the genotype of the ΔrelMtb::hyg mutant strain, SJA16, showing two copies of the ΔrelMtb::hyg allele and the absence of a wild-type relMtb allele. The additional copy of the pSA1 vector, carrying the lacZ and bla genes, is shown as a thick horizontal line. (C) Southern blot analysis of homologous recombinants obtained with pSA1 and pSA3. Chromosomal DNA was digested with PvuII and probed either with the upstream 2,247-bp SmaI fragment (▨) showing the wild-type allele of 2,803 bp and mutant allele of 3,587 bp or with the internal 945-bp SphI fragment ( ) showing the presence of this fragment in the wild type (WT) and single crossovers, but its absence from the ΔrelMtb::hyg mutants. KO, knockout. Lanes: 1, wild-type H37Rv; 2, SJA14 (single crossover carrying pSA1 integrated at the relMtb locus); 3, SJA16 (ΔrelMtb::hyg mutant carrying additional, integrated copy of pSA1); 4, SJA32 (site-specific single crossover carrying pSA3 integrated at the relMtb locus); 5, SJA33 (ΔrelMtb::hyg mutant).
A positive selection marker was therefore introduced into the knockout construct to allow double crossovers to be identified. Single-crossover recombinants obtained with the sacB-containing vector pSA3 were plated on sucrose, and of the 23 colonies thus obtained, 14 were Kms and had thus lost the vector, whereas 9 corresponded to spontaneous sacB mutants. Genotypic analysis confirmed that all 14 Kms clones were ΔrelMtb::hyg mutants. One of these, SJA33, was further characterized for loss of the SphI fragment and for site specificity of recombination downstream and upstream of the site of gene disruption. This analysis confirmed that both SJA16 and SJA33 lacked the internal segment of relMtb located between the SphI sites and that the replacement of the wild type by the mutant allele had occurred site specifically (not shown). The only difference between the two was the presence of an integrated copy of pSA1 at the ΔrelMtb::hyg locus in SJA16 (Fig. 1). We therefore concluded that both strains would be indistinguishable in terms of the relMtb-associated phenotype, and all experiments reported in this paper were carried out with strain SJA16.
Phenotypic analysis of the ΔrelMtb::hyg strain in vitro.
As expected, the ΔrelMtb strain failed to synthesize (p)ppGpp in response to starvation (Fig. 2). Complementation of the ΔrelMtb::hyg mutant with an intact relMtb gene on a single-copy-integrating plasmid restored the ability of the strain to produce (p)ppGpp (data not shown). There were no significant differences in gross surface morphology or apparent growth rate on several types of solid media. The cell wall permeability of the mutant appeared grossly similar to that of the wild type, since the MIC for the ΔrelMtb strain was the same as the wild type with a variety of compounds in the broth microdilution assay, including isoniazid, rifampin, cycloserine, hydrochloric acid, and hydrogen peroxide. One exception was that the mutant was twofold more sensitive to azide, which also appeared to induce (p)ppGpp synthesis (Table 2). There were also no significant differences in the mycolic acid subclass distribution or modification in either log- or stationary-phase bacilli, suggesting that, unlike in gram-negative bacteria, cyclopropanation of fatty acids (mycolic acids in this case) is not under relMtb control (19, 80).
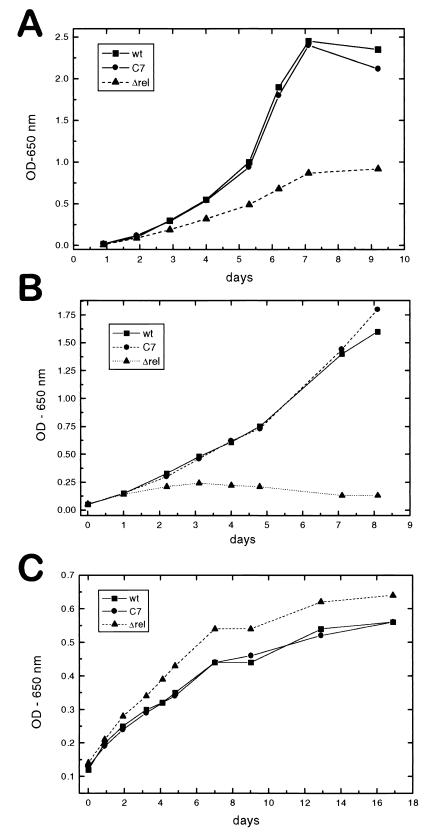
The ΔrelMtb strain had a significantly lower growth rate when grown in a variety of synthetic liquid medium compositions (Fig. 3A and Table 3). The mutant strain demonstrated a growth rate approximately one-third that of the wild type and a maximum OD of one-half independent of nutrient concentrations. Middlebrook 7H9 medium requires supplementation with an albumin-dextrose complex and glycerol to support mycobacterial growth. Nonetheless, this remains fairly a minimal medium. Supplementing this medium with additional glycerol, 0.5% peptone, or supplemental oleic acid had no significant effects on the rate of growth of either the wild type or the ΔrelMtb strain (Table 3) (supplemental material available at http://www.niaid.nih.gov/dir/labs/lhd/barry.htm). Addition of both glycerol and peptone increased the growth rate and doubled the final ODs of both strains, but they remained in a constant ratio with respect to each other. Omitting the normal 0.2% glycerol supplement decreased both the growth rate and the maximal OD, but again, the two strains displayed the same relative differences. Omitting both glycerol and dextrose attenuated growth even further, but still maintained the relative difference. This suggests that RelMtb (or RelMtb products) has a constitutive role in determining the growth of M. tuberculosis under these conditions.
FIG. 3.
In vitro growth curves. (A) Growth of wild-type H37Rv (wt) (■), the Δrel mutant (▴), and the complemented strain C7 (●) in 7H9 medium at 37°C, determined by OD650. Panel B is the same, but at 42°C. (C) Growth at 37°C in 7H9 with the carbon sources of glucose and glycerol replaced with 0.2 mM DPPC.
One condition under which the ΔrelMtb strain did not appear to suffer a decrease in growth rate (and may even have had a very slight growth advantage) was when the organisms were grown on lipid as the sole carbon source. Although the overall growth rate was slower and the final culture density was lower, the knockout strain appeared to do as well as the wild type upon growth on DPPC (Fig. 3C).
The ΔrelMtb mutant strain was thermosensitive and was incapable of growth at 42°C (Fig. 3B). The ΔrelMtb strain complemented with a single copy of relMtb was restored to wild-type growth rates under all conditions, indicating that there is no polar effect resulting from insertion of hyg (Fig. 3). Although the reason for the thermosensitivity of the knockout organism was not clear, it was also reversed by restoring RelMtb function, suggesting some role for RelMtb in facilitating adaptation to that condition.
The ΔrelMtb::hyg strain is impaired for survival during long-term starvation.
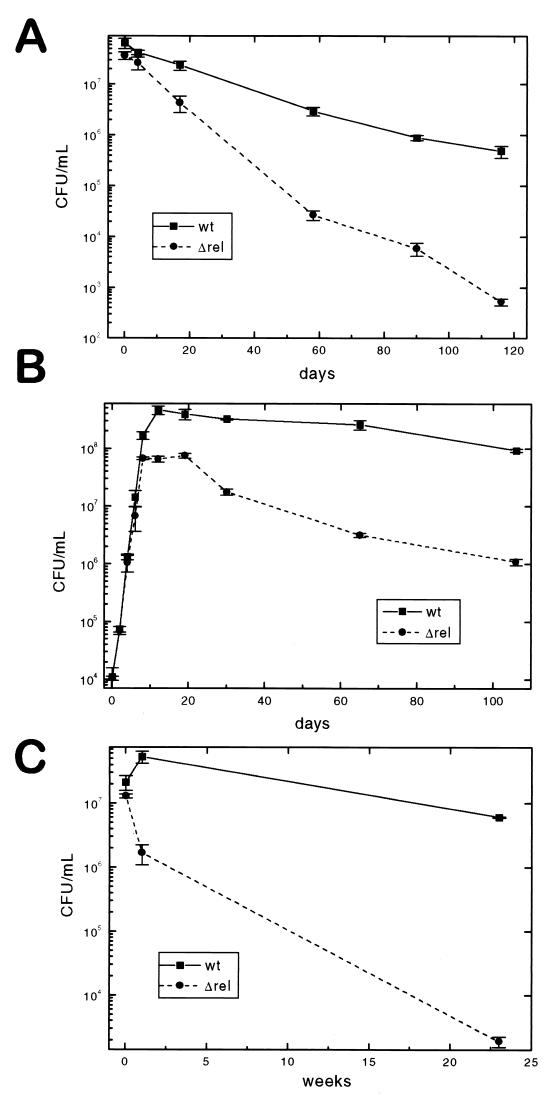
The ability of the ΔrelMtb::hyg strain to survive under conditions of long-term nutrient deprivation was also investigated. Preliminary experiments comparing CFU determined from separate cultures of antibiotic-marked wild-type H37Rv and the relMtb mutant revealed higher-than-acceptable error due to clumping of both strains. Although the degree of clumping did not appear to be different between the two strains, it did limit the accuracy of the CFU determinations. To circumvent this problem, equal amounts of a Kmr wild-type strain (created by integration of a single-copy control plasmid into H37Rv) and the Hygr ΔrelMtb mutant were mixed under starvation conditions and coincubated. Identical samples were removed at various time points, vortexed with sterile glass beads, and plated separately onto both antibiotics. When starved for several months by resuspension in TBST to induce sudden starvation, the ΔrelMtb strain lost viability much more rapidly than wild-type H37Rv (Fig. 4A). The mutant rate of loss of viability is higher than the wild-type rate across the entire experiment, suggesting that an initial adaptation event accounts for the disparity in survival.
FIG. 4.
Starvation survival. (A) Kanamycin-resistant wild-type H37Rv(pMV306K) (wt) and Δrel mutant (hygromycin resistant) were mixed in a competition starvation assay in TBST at an initial wild-type/Δrel ratio of 1.75. The number of CFU per milliliter was determined by plating in triplicate at 3 to 5 dilutions onto 7H11 plates containing hygromycin or kanamycin on the indicated days. Control plates at low dilution with both hygromycin and kanamycin gave no colonies. (B) As in panel A, both organisms were simultaneously inoculated to an initial cell density of 104 CFU/ml, and at sequential time points, the cells were removed and dilutions were plated onto selective media. (C) Anaerobic survival of wild-type H37Rv(pMV306K) and the Δrel mutant in sealed tubes. A mid-log-phase culture was divided into replicate sealed tubes at an initial cell density of 107 CFU/ml, and at the indicated time points, the tubes were unsealed and viable CFU were determined by plating the cells onto selective media.
A more gradual adaptation to the stationary phase by nutrient depletion of normal growth media occurs over the course of a prolonged agitated aerobic culture. Under these conditions, wild-type M. tuberculosis grew into the stationary phase and then retained essentially complete viability after up to 106 days of incubation (Fig. 4B). During the same interval, the ΔrelMtb mutant, which never achieves as high a culture density, lost significant viability. Thus, in this more gradual adaptation model, there is a more modest advantage of the wild type over the mutant strain of M. tuberculosis.
An abrupt transition to hypoxic conditions of a log-phase culture in nutrient-rich media was obtained by sealing culture aliquots in gas-impermeable tubes with very limited headspace (73, 75). These oxygen-deprived cultures were incubated at 37°C and then unsealed and plated to determine the numbers of viable bacilli at various time points over a 23-week period (Fig. 4C). Again, under these conditions, the mutant showed a dramatic survival disadvantage and lost almost 4 logs of viability over the course of 6 months, while the wild type declined only slightly.
ΔrelMtb::hyg has a wild-type growth rate in macrophages.
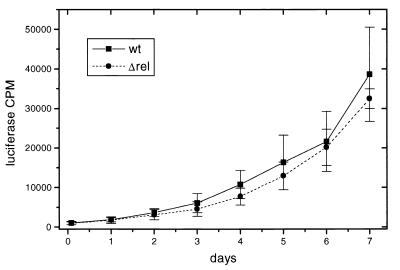
Wild-type H37Rv and ΔrelMtb strains were analyzed for growth in the THP-1 human macrophage-like cell line (66). To facilitate evaluation of the growth rates of these organisms, we first integrated a groEL promoter-driven firefly luciferase gene (conferring kanamycin resistance on the mutant) into the L5 phage integration site (79). Growth was monitored both by CFU determination on plates and by luciferase activity from a strong constitutive promoter construct. Whole-cell luciferase activities in M. tuberculosis have been shown to accurately report culture and intracellular viability (1, 6, 63). In two independent experiments, there was no significant difference in the growth rates between wild-type H37Rv(pMV306-hsp60-luc) and H37RvΔrelMtb::hyg(pMV306-hsp60-luc) in THP-1 cells (Fig. 5). The growth rate determined by plating for CFU also showed no significant difference between the two strains (data not shown).
FIG. 5.
Growth curve in macrophages. Growth of wild-type H37Rv (wt) and Δrel mutant strains in THP-1 cells was analyzed by using constitutive luciferase-expressing strains. Shown are data from one experiment, with a second experiment having similar results. In a control in vitro experiment, H37Rv grew faster than the Δrel mutant (data not shown), consistent with growth determined by absorbance (A).
DISCUSSION
Our understanding of the adaptation of mycobacteria to long-term survival within the eukaryotic host has been guided by our estimation of the likely characteristics of their environment during persistence. To date, these efforts have focused on two hypothetical characteristics of this environment: oxygen limitation and nutrient deprivation.
Oxygen limitation induces some dramatic and specific changes in mycobacteria, including enhanced resistance to some drugs (like isoniazid and rifampin) and sensitivity to nitroaromatic compounds, such as metronidazole (43, 75). Oxygen limitation also upregulates the synthesis of specific proteins, such as α-crystallin and a flavohemoglobin, and induces enzymes such as glycine dehydrogenase and alanine dehydrogenase (33, 36, 74, 78). Such organisms become thermotolerant, decrease overall protein synthesis, have lost acid fastness and the ability to grow on malachite green-containing media, and have ultramicroscopically thickened cell walls (14, 23, 35, 70). Adaptation to low oxygen tension is a coordinated activity at many levels, and rapid alteration can lead to cell death (71, 73). The coordination of these events is especially evident in that reoxygenation of microaerophilic cultures results in synchronous replication (17, 72).
Nutrient deprivation, in the form of amino acid and carbohydrate depletion, is also likely to coincide with the formation of intact granulomas in a process thought to be essential for curtailing growth of the microorganism (14, 37). Recent speculation has centered on the role of lipid metabolism in maintaining mycobacterial viability in the absence of robust growth, since the caseous intragranulomar environment is likely to be lipid rich and mycobacteria appear well endowed with suitable enzymes for utilizing lipids as a sole carbon source (the so-called “lipolytic hypothesis”) (13). There may also be shifts between the primary source of nutrients during infection, because M. tuberculosis growing within a macrophage phagolysosome may well prove to have access to different nutrients than M. tuberculosis persisting within a caseous granuloma.
To adapt to such changing situations, M. tuberculosis may utilize a variety of transcriptional control mechanisms. Some of this environment-dependent gene expression (including expression of α-crystallin) is controlled by the expression of an alternative sigma factor, SigF (45, 51). A second sigma factor, SigB, has been shown to be associated with stationary-phase adaptation and general stress (34). The interplay between these two regulons may prove important for distinguishing components important specifically for persistence. In addition to the use of such alternative sigma factors for such adaptation, many microorganisms translate the stress of amino acid or carbon source depletion into an intracellular accumulation of (p)ppGpp, leading to alterations in gene expression that suppress the synthesis of stable RNA species (rRNA and tRNA), induce degradative pathways, activate certain stationary-phase genes, and modulate genes that regulate DNA replication and growth rate (7, 9–11, 64, 77).
In this study, we have demonstrated that there is a very low basal level of (p)ppGpp in log-phase wild-type M. tuberculosis strains CDC1551 and H37Rv grown axenically on glucose with accumulation starting in early stationary-phase cultures and reaching a maximum in long-term stationary-phase cultures. When log-phase cultures were shifted into isotonic buffer with no nutrients (TBST), (p)ppGpp began to accumulate within 20 min, peaked by 40 to 60 min, and declined to a new steady state by 90 to 120 min (data not shown). This temporal pattern is similar to that determined from the kinetic studies of the metabolism of (p)ppGpp in E. coli, where the stringent response occurs on a more abbreviated time scale, being induced within seconds of starvation (9, 50). No (p)ppGpp is detectable in the M. tuberculosis H37Rv strain with a deleted relMtb gene (Fig. 2).
Adaptation to nutrient deprivation is essential for long-term survival of nongrowing cells. M. tuberculosis is capable of surviving under nongrowing starvation conditions for up to 2 years in vitro while retaining the ability to resuscitate (54). In M. smegmatis it has been reported that carbon, nitrogen, or phosphorus-starved organisms retain viability for over 650 days, with an initial 2- to 3-log drop in CFU followed by long-term maintenance of stable numbers (62). These authors speculate that sensing and responding to carbon starvation allowed an adaptation to stationary phase that facilitated long-term bacterial survival. In this study, we demonstrate that an effect of such carbon limitation is to elevate levels of (p)ppGpp in the cell through the action of the RelMtb protein. RelMtb-mediated production of (p)ppGpp presumably induces proteins involved in stationary-phase survival. Furthermore, late-log-phase cultures of the ΔrelMtb strain that were subjected to starvation stress contained five times more ribosomes per unit of protein than the wild-type, highlighting the inability of this strain to adapt to the stationary phase (data not shown). This failure to reduce the amount of stable RNA and ribosomal proteins supports the notion that the stringent response is required for growth rate control (7, 42). Thus, as we previously speculated (2), in the absence of this response, M. tuberculosis fails to adapt to the stationary phase and long-term survival is severely compromised.
The RelMtb-deficient strain consistently grows more slowly than the wild type or the complemented strain in normal growth media, suggesting that during aerobic growth under these conditions, some level of (p)ppGpp is required for optimum growth. This apparent defect is not complemented by addition of additional sources of nutrients such as peptone or glycerol. Addition of such supplements does, however, effectively double the growth rate and the maximal OD of the ΔrelMtb cultures, suggesting some of the growth defect may occur in response to uncharged tRNA concentrations, an observation consistent with biochemical data (H. Rubin and D. Avarbock, unpublished results). The wild-type strain shows more modest effects of these additives, suggesting that the effect is not strictly due to provision of a general growth factor. In light of the aforementioned theory that the major intracellular carbon source for persistent bacteria is lipid, it is particularly interesting that the RelMtb-deficient strain is not different in growth rate from the wild type under only two conditions: when grown within macrophages and when grown upon lipid. These results were surprising because of the connection of Rel with amino acid starvation in gram-negative organisms and the known defect in growth of amino acid auxotrophs of M. tuberculosis in cultured macrophages (31, 38). It is noteworthy that organisms with genetic deletion of isocitrate lyase, an enzyme essential for growth upon lipid substrates, are defective for persistence in mouse models of latent tuberculosis (J. McKinney, Rockefeller University, personal communication). Taken together, these data are consistent with the hypothesis that intracellular growth relies primarily on lipid catabolism as the major source of carbon.
The evaluation of this mutant in animal models of persistent infection will allow the direct assessment of the relevance of this and other proteins involved in stationary-phase survival to latency and long-term persistence. Validation of the importance of this (p)ppGpp-mediated response network in animal models of persistence would lead to consideration of the RelMtb protein as a target for the development of chemotherapeutic agents with unique activity in the treatment of asymptomatic tuberculosis infections.
ACKNOWLEDGMENTS
This work was supported by NIH grant R01-AI43420 (to H.R.). V.M. was also supported by grants from the National Research Foundation, the Medical Research Council of South Africa, and the South African Institute for Medical Research.
We thank Mike Cashel (NIH) for ppGpp and pppGpp standards and discussions; Beth Fischer (Rocky Mountain Laboratories, NIH) for electron microscopy studies; Bhavna Gordhan for assistance with the gene knockout work; Bill Jacobs for providing the cosmid library; Tanya Parish, Peadar O'Gaora, and Stephanie Dawes for providing plasmids; and Lynn Brown for the degenerate PCR primers.
REFERENCES
- 1.Arain T M, Resconi A E, Singh D C, Stover C K. Reporter gene technology to assess activity of antimycobacterial agents in macrophages. Antimicrob Agents Chemother. 1996;40:1542–1544. doi: 10.1128/aac.40.6.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avarbock D, Salem J, Li L S, Wang Z M, Rubin H. Cloning and characterization of a bifunctional RelA/SpoT homologue from Mycobacterium tuberculosis. Gene. 1999;233:261–269. doi: 10.1016/s0378-1119(99)00114-6. [DOI] [PubMed] [Google Scholar]
- 3.Balasubramanian V, Pavelka M S, Jr, Bardarov S S, Martin J, Weisbrod T R, McAdam R A, Bloom B R, Jacobs W R., Jr Allelic exchange in Mycobacterium tuberculosiswith long linear recombination substrates. J Bacteriol. 1996;178:273–279. doi: 10.1128/jb.178.1.273-279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes P F, el-Hajj H, Preston-Martin S, Cave M D, Jones B E, Otaya M, Pogoda J, Eisenach K D. Transmission of tuberculosis among the urban homeless. JAMA. 1996;275:305–307. [PubMed] [Google Scholar]
- 5.Bochner B R, Ames B N. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem. 1982;257:9759–9769. [PubMed] [Google Scholar]
- 6.Bonay M, Bouchonnet F, Pelicic V, Lagier B, Grandsaigne M, Lecossier D, Grodet A, Vokurka M, Gicquel B, Hance A J. Effect of stimulation of human macrophages on intracellular survival of Mycobacterium bovisBacillus Calmette-Guerin. Evaluation with a mycobacterial reporter strain. Am J Respir Crit Care Med. 1999;159:1629–1637. doi: 10.1164/ajrccm.159.5.9807021. [DOI] [PubMed] [Google Scholar]
- 7.Bremer H, Ehrenberg M. Guanosine tetraphosphate as a global regulator of bacterial RNA synthesis: a model involving RNA polymerase pausing and queuing. Biochim Biophys Acta. 1995;1262:15–36. doi: 10.1016/0167-4781(95)00042-f. [DOI] [PubMed] [Google Scholar]
- 8.Bridger W A, Paranchych W. relA gene control of bacterial glycogen synthesis. Can J Biochem. 1978;56:403–406. doi: 10.1139/o78-063. [DOI] [PubMed] [Google Scholar]
- 9.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtis R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1488–1496. [Google Scholar]
- 10.Chakraburtty R, Bibb M. The ppGpp synthetase gene (relA) of Streptomyces coelicolorA3(2) plays a conditional role in antibiotic production and morphological differentiation. J Bacteriol. 1997;179:5854–5861. doi: 10.1128/jb.179.18.5854-5861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakraburtty R, White J, Takano E, Bibb M. Cloning, characterization and disruption of a (p)ppGpp synthetase gene (relA) of Streptomyces coelicolorA3(2) Mol Microbiol. 1996;19:357–368. doi: 10.1046/j.1365-2958.1996.390919.x. [DOI] [PubMed] [Google Scholar]
- 12.Chater K F. Multilevel regulation of Streptomyces differentiation. Trends Genet. 1989;5:372–377. doi: 10.1016/0168-9525(89)90172-8. [DOI] [PubMed] [Google Scholar]
- 13.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G, et al. Deciphering the biology of Mycobacterium tuberculosisfrom the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. . (Erratum, 396:190.) [DOI] [PubMed] [Google Scholar]
- 14.Cunningham A F, Spreadbury C L. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton α-crystallin homolog. J Bacteriol. 1998;180:801–808. doi: 10.1128/jb.180.4.801-808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dannenberg A M, Jr, Rook G A W. Pathogenesis of pulmonary tuberculosis: an interplay of tissue-damaging and macrophage-activating immune responses—dual mechanisms that control bacillary multiplication. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C.: American Society for Microbiology; 1994. pp. 459–483. [Google Scholar]
- 16.De Voss J J, Rutter K, Schroeder B G, Su H, Zhu Y, Barry C E., III The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosisare essential for growth in macrophages. Proc Natl Acad Sci USA. 2000;97:1252–1257. doi: 10.1073/pnas.97.3.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dick T, Lee B H, Murugasu-Oei B. Oxygen depletion induced dormancy in Mycobacterium smegmatis. FEMS Microbiol Lett. 1998;163:159–164. doi: 10.1111/j.1574-6968.1998.tb13040.x. [DOI] [PubMed] [Google Scholar]
- 18.Dietzler D N, Leckie M P. Regulation of ADP-glucose synthetase, the rate-limiting enzyme of bacterial glycogen synthesis, by the pleiotropic nucleotides ppGpp and pppGpp. Biochem Biophys Res Commun. 1977;77:1459–1467. doi: 10.1016/s0006-291x(77)80143-5. [DOI] [PubMed] [Google Scholar]
- 19.Eichel J, Chang Y-Y, Riesenberg D, Cronan J E., Jr Effect of ppGpp on Escherichia coli cyclopropane fatty acid synthesis is mediated through the RpoS sigma factor (ςs) J Bacteriol. 1999;181:572–576. doi: 10.1128/jb.181.2.572-576.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gangadharam P R. Mycobacterial dormancy. Tuber Lung Dis. 1995;76:477–479. doi: 10.1016/0962-8479(95)90521-9. [DOI] [PubMed] [Google Scholar]
- 21.Gentry D R, Cashel M. Cellular localization of the Escherichia coliSpoT protein. J Bacteriol. 1995;177:3890–3893. doi: 10.1128/jb.177.13.3890-3893.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gentry D R, Cashel M. Mutational analysis of the Escherichia colispoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol Microbiol. 1996;19:1373–1384. doi: 10.1111/j.1365-2958.1996.tb02480.x. [DOI] [PubMed] [Google Scholar]
- 23.Gillespie J, Barton L L, Rypka E W. Phenotypic changes in mycobacteria grown in oxygen-limited conditions. J Med Microbiol. 1986;21:251–255. doi: 10.1099/00222615-21-3-251. [DOI] [PubMed] [Google Scholar]
- 24.Gordhan B G, Andersen S J, De Meyer A R, Mizrahi V. Construction by homologous recombination and phenotypic characterization of a DNA polymerase domain polA mutant of Mycobacterium smegmatis. Gene. 1996;178:125–130. doi: 10.1016/0378-1119(96)00350-2. [DOI] [PubMed] [Google Scholar]
- 25.Gordhan, B. G., and T. Parish.Mycobacterium tuberculosis protocols. Methods Mol. Biol., in press.
- 26.Gupta U D, Katoch V M. Understanding the phenomenon of persistence in mycobacterial infections. Indian J Lepr. 1997;69:385–393. [PubMed] [Google Scholar]
- 27.Heath R J, Jackowski S, Rock C O. Guanosine tetraphosphate inhibition of fatty acid and phospholipid synthesis in Escherichia coli is relieved by overexpression of glycerol-3-phosphate acyltransferase (plsB) J Biol Chem. 1994;269:26584–26590. [PubMed] [Google Scholar]
- 28.Heinemeyer E A, Geis M, Richter D. Degradation of guanosine 3′-diphosphate 5′-diphosphate in vitro by the spoT gene product of Escherichia coli. Eur J Biochem. 1978;89:125–131. doi: 10.1111/j.1432-1033.1978.tb20904.x. [DOI] [PubMed] [Google Scholar]
- 29.Henderson D J, Lydiate D J, Hopwood D A. Structural and functional analysis of the mini-circle, a transposable element of Streptomyces coelicolorA3(2) Mol Microbiol. 1989;3:1307–1318. doi: 10.1111/j.1365-2958.1989.tb00112.x. [DOI] [PubMed] [Google Scholar]
- 30.Hinds J, Mahenthiralingam E, Kempsell K E, Duncan K, Stokes R W, Parish T, Stoker N G. Enhanced gene replacement in mycobacteria. Microbiology. 1999;145:519–527. doi: 10.1099/13500872-145-3-519. [DOI] [PubMed] [Google Scholar]
- 31.Hondalus M K, Bardarov S, Russell R, Chan J, Jacobs W R, Jr, Bloom B R. Attenuation of and protection induced by a leucine auxotroph of Mycobacterium tuberculosis. Infect Immun. 2000;68:2888–2898. doi: 10.1128/iai.68.5.2888-2898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoyt S, Jones G H. relA is required for actinomycin production in Streptomyces antibioticus. J Bacteriol. 1999;181:3824–3829. doi: 10.1128/jb.181.12.3824-3829.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu Y, Butcher P D, Mangan J A, Rajandream M-A, Coates A R M. Regulation of hmp gene transcription in Mycobacterium tuberculosis: effects of oxygen limitation and nitrosative and oxidative stress. J Bacteriol. 1999;181:3486–3493. doi: 10.1128/jb.181.11.3486-3493.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Y, Coates A R M. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J Bacteriol. 1999;181:469–476. doi: 10.1128/jb.181.2.469-476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Y M, Butcher P D, Sole K, Mitchison D A, Coates A R. Protein synthesis is shut down in dormant Mycobacterium tuberculosisand is reversed by oxygen or heat shock. FEMS Microbiol Lett. 1998;158:139–145. doi: 10.1111/j.1574-6968.1998.tb12813.x. [DOI] [PubMed] [Google Scholar]
- 36.Hutter B, Dick T. Increased alanine dehydrogenase activity during dormancy in Mycobacterium smegmatis. FEMS Microbiol Lett. 1998;167:7–11. doi: 10.1111/j.1574-6968.1998.tb13200.x. [DOI] [PubMed] [Google Scholar]
- 37.Imboden P, Schoolnik G K. Construction and characterization of a partial Mycobacterium tuberculosiscDNA library of genes expressed at reduced oxygen tension. Gene. 1998;213:107–117. doi: 10.1016/s0378-1119(98)00192-9. [DOI] [PubMed] [Google Scholar]
- 38.Jackson M, Phalen S W, Lagranderie M, Ensergueix D, Chavarot P, Marchal G, McMurray D N, Gicquel B, Guilhot C. Persistence and protective efficacy of a Mycobacterium tuberculosisauxotroph vaccine. Infect Immun. 1999;67:2867–2873. doi: 10.1128/iai.67.6.2867-2873.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kramer G F, Baker J C, Ames B N. Near-UV stress in Salmonella typhimurium: 4-thiouridine in tRNA, ppGpp, and ApppGpp as components of an adaptive response. J Bacteriol. 1988;170:2344–2351. doi: 10.1128/jb.170.5.2344-2351.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le H Q, Davidson P T. Reactivation and exogenous reinfection: their relative roles in the pathogenesis of tuberculosis. Curr Clin Top Infect Dis. 1996;16:260–276. [PubMed] [Google Scholar]
- 41.Lee M H, Pascopella L, Jacobs R, Jr, Hatfull G F. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc Natl Acad Sci USA. 1991;88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levine A, Vannier F, Dehbi M, Henckes G, Seror S J. The stringent response blocks DNA replication outside the ori region in Bacillus subtilis and at the origin in Escherichia coli. J Mol Biol. 1991;219:605–613. doi: 10.1016/0022-2836(91)90657-r. [DOI] [PubMed] [Google Scholar]
- 43.Lim A, Eleuterio M, Hutter B, Murugasu-Oei B, Dick T. Oxygen depletion-induced dormancy in Mycobacterium bovisBCG. J Bacteriol. 1999;181:2252–2256. doi: 10.1128/jb.181.7.2252-2256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loewen P C, Hu B, Strutinsky J, Sparling R. Regulation in the rpoS regulon of Escherichia coli. Can J Microbiol. 1998;44:707–717. doi: 10.1139/cjm-44-8-707. [DOI] [PubMed] [Google Scholar]
- 45.Manabe Y C, Chen J M, Ko C G, Chen P, Bishai W R. Conditional sigma factor expression, using the inducible acetamidase promoter, reveals that the Mycobacterium tuberculosis sigFgene modulates expression of the 16-kilodalton alpha-crystallin homologue. J Bacteriol. 1999;181:7629–7633. doi: 10.1128/jb.181.24.7629-7633.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez-Costa O H, Arias P, Romero N M, Parro V, Mellado R P, Malpartida F. A relA/spoT homologous gene from Streptomyces coelicolorA3(2) controls antibiotic biosynthetic genes. J Biol Chem. 1996;271:10627–10634. doi: 10.1074/jbc.271.18.10627. [DOI] [PubMed] [Google Scholar]
- 47.Martínez-Costa O H, Fernández-Moreno M A, Malpartida F. The relA/spoT-homologous gene in Streptomyces coelicolorencodes both ribosome-dependent (p)ppGpp-synthesizing and -degrading activities. J Bacteriol. 1998;180:4123–4132. doi: 10.1128/jb.180.16.4123-4132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mechold U, Cashel M, Steiner K, Gentry D, Malke H. Functional analysis of a relA/spoT gene homolog from Streptococcus equisimilis. J Bacteriol. 1996;178:1401–1411. doi: 10.1128/jb.178.5.1401-1411.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mechold U, Malke H. Characterization of the stringent and relaxed responses of Streptococcus equisimilis. J Bacteriol. 1997;179:2658–2667. doi: 10.1128/jb.179.8.2658-2667.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Metzger S, Schreiber G, Aizenman E, Cashel M, Glaser G. Characterization of the relA1 mutation and a comparison of relA1 with new relA null alleles in Escherichia coli. J Biol Chem. 1989;264:21146–21152. [PubMed] [Google Scholar]
- 51.Michele T M, Ko C, Bishai W R. Exposure to antibiotics induces expression of the Mycobacterium tuberculosis sigFgene: implications for chemotherapy against mycobacterial persistors. Antimicrob Agents Chemother. 1999;43:218–225. doi: 10.1128/aac.43.2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitchison D A. The Garrod Lecture. Understanding the chemotherapy of tuberculosis—current problems. J Antimicrob Chemother. 1992;29:477–493. doi: 10.1093/jac/29.5.477. [DOI] [PubMed] [Google Scholar]
- 53.Neidhardt F C. Roles of amino acid activating enzymes in cellular physiology. Bacteriol Rev. 1966;30:701–719. doi: 10.1128/br.30.4.701-719.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nyka W. Studies on the effect of starvation on mycobacteria. Infect Immun. 1974;9:843–850. doi: 10.1128/iai.9.5.843-850.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Gaora P, Barnini S, Hayward C, Filley E, Rook G, Young D, Thole J. Mycobacteria as immunogens: development of expression vectors for use in multiple mycobacterial species. Med Princ Pract. 1997;6:91–96. [Google Scholar]
- 56.Parish T, Gordhan B G, McAdam R A, Duncan K, Mizrahi V, Stoker N G. Production of mutants in amino acid biosynthesis genes of Mycobacterium tuberculosisby homologous recombination. Microbiology. 1999;145:3497–3503. doi: 10.1099/00221287-145-12-3497. [DOI] [PubMed] [Google Scholar]
- 57.Pelicic V, Reyrat J M, Gicquel B. Generation of unmarked directed mutations in mycobacteria, using sucrose counter-selectable suicide vectors. Mol Microbiol. 1996;20:919–925. doi: 10.1111/j.1365-2958.1996.tb02533.x. [DOI] [PubMed] [Google Scholar]
- 58.Rao N N, Kornberg A. Inorganic polyphosphate regulates responses of Escherichia colito nutritional stringencies, environmental stresses and survival in the stationary phase. Prog Mol Subcell Biol. 1999;23:183–195. doi: 10.1007/978-3-642-58444-2_9. [DOI] [PubMed] [Google Scholar]
- 59.Sambrook J, Fritsch E F, Maniatas T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 60.Saunders B M, Frank A A, Orme I M. Granuloma formation is required to contain bacillus growth and delay mortality in mice chronically infected with Mycobacterium tuberculosis. Immunology. 1999;98:324–328. doi: 10.1046/j.1365-2567.1999.00877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seyfzadeh M, Keener J, Nomura M. spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc Natl Acad Sci USA. 1993;90:11004–11008. doi: 10.1073/pnas.90.23.11004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smeulders M J, Keer J, Speight R A, Williams H D. Adaptation of Mycobacterium smegmatisto stationary phase. J Bacteriol. 1999;181:270–283. doi: 10.1128/jb.181.1.270-283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Snewin V A, Gares M P, Gaora P, Hasan Z, Brown I N, Young D B. Assessment of immunity to mycobacterial infection with luciferase reporter constructs. Infect Immun. 1999;67:4586–4593. doi: 10.1128/iai.67.9.4586-4593.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spira B, Silberstein N, Yagil E. Guanosine 3′,5′-bispyrophosphate (ppGpp) synthesis in cells of Escherichia coli starved for Pi. J Bacteriol. 1995;177:4053–4058. doi: 10.1128/jb.177.14.4053-4058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Starke J R. Tuberculosis in children. Curr Opin Pediatr. 1995;7:268–277. doi: 10.1097/00008480-199506000-00005. [DOI] [PubMed] [Google Scholar]
- 66.Stokes R W, Doxsee D. The receptor-mediated uptake, survival, replication, and drug sensitivity of Mycobacterium tuberculosiswithin the macrophage-like cell line THP-1: a comparison with human monocyte-derived macrophages. Cell Immunol. 1999;197:1–9. doi: 10.1006/cimm.1999.1554. [DOI] [PubMed] [Google Scholar]
- 67.Sudre P, ten Dam G, Kochi A. Tuberculosis: a global overview of the situation today. Bull W H O. 1992;70:149–159. [PMC free article] [PubMed] [Google Scholar]
- 68.Valway S E, Sanchez M P, Shinnick T F, Orme I, Agerton T, Hoy D, Jones J S, Westmoreland H, Onorato I M. An outbreak involving extensive transmission of a virulent strain of Mycobacterium tuberculosis. N Engl J Med. 1998;338:633–639. doi: 10.1056/NEJM199803053381001. . (Erratum, 338:1783.) [DOI] [PubMed] [Google Scholar]
- 69.van Rie A, Warren R, Richardson M, Victor T C, Gie R P, Enarson D A, Beyers N, van Helden P D. Exogenous reinfection as a cause of recurrent tuberculosis after curative treatment. N Engl J Med. 1999;341:1174–1179. doi: 10.1056/NEJM199910143411602. [DOI] [PubMed] [Google Scholar]
- 70.Wallace J G. The heat resistance of tubercle bacilli in the lungs of infected mice. Am Rev Respir Dis. 1961;83:866–870. doi: 10.1164/arrd.1961.83.6.866. [DOI] [PubMed] [Google Scholar]
- 71.Wayne L G. Dormancy of Mycobacterium tuberculosisand latency of disease. Eur J Clin Microbiol Infect Dis. 1994;13:908–914. doi: 10.1007/BF02111491. [DOI] [PubMed] [Google Scholar]
- 72.Wayne L G. Synchronized replication of Mycobacterium tuberculosis. Infect Immun. 1977;17:528–530. doi: 10.1128/iai.17.3.528-530.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wayne L G, Hayes L G. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosisthrough two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wayne L G, Lin K-Y. Glyoxylate metabolism and adaptation of Mycobacterium tuberculosisto survival under anaerobic conditions. Infect Immun. 1982;37:1042–1049. doi: 10.1128/iai.37.3.1042-1049.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wayne L G, Sramek H A. Metronidazole is bactericidal to dormant cells of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1994;38:2054–2058. doi: 10.1128/aac.38.9.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wehmeier L, Schafer A, Burkovski A, Kramer R, Mechold U, Malke H, Puhler A, Kalinowski J. The role of the Corynebacterium glutamicumrel gene in (p)ppGpp metabolism. Microbiology. 1998;144:1853–1862. doi: 10.1099/00221287-144-7-1853. [DOI] [PubMed] [Google Scholar]
- 77.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- 78.Yuan Y, Crane D D, Barry C E., III Stationary phase-associated protein expression in Mycobacterium tuberculosis: function of the mycobacterial α-crystallin homolog. J Bacteriol. 1996;178:4484–4492. doi: 10.1128/jb.178.15.4484-4492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yuan Y, Crane D D, Simpson R M, Zhu Y Q, Hickey M J, Sherman D R, Barry C E., III The 16-kDa alpha-crystallin (Acr) protein of Mycobacterium tuberculosisis required for growth in macrophages. Proc Natl Acad Sci USA. 1998;95:9578–9583. doi: 10.1073/pnas.95.16.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yuan Y, Zhu Y, Crane D D, Barry C E., III The effect of oxygenated mycolic acid composition on cell wall function and macrophage growth in Mycobacterium tuberculosis. Mol Microbiol. 1998;29:1449–1458. doi: 10.1046/j.1365-2958.1998.01026.x. [DOI] [PubMed] [Google Scholar]