Abstract
A novel coronavirus, known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has surfaced and caused global concern owing to its ferocity. SARS-CoV-2 is the causative agent of coronavirus disease 2019; however, it was only discovered at the end of the year and was considered a pandemic by the World Health Organization. Therefore, the development of novel potent inhibitors against SARS-CoV-2 and future outbreaks is urgently required. Numerous naturally occurring bioactive substances have been studied in the clinical setting for diverse disorders. The intricate infection and replication mechanism of SARS-CoV-2 offers diverse therapeutic drug targets for developing antiviral medicines by employing natural products that are safer than synthetic compounds. Marine natural products (MNPs) have received increased attention in the development of novel drugs owing to their high diversity and availability. Therefore, this review article investigates the infection and replication mechanisms, including the function of the SARS-CoV-2 genome and structure. Furthermore, we highlighted anti-SARS-CoV-2 therapeutic intervention efforts utilizing MNPs and predicted SARS-CoV-2 inhibitor design.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42995-023-00215-9.
Keywords: SARS-CoV-2, Marine natural products, Drug targets, Inhibitors
Introduction
Mankind has been attacked by three epidemics in the twenty-first century including coronaviruses that belong to the family Coronaviridae, which comprises a positive-sense single-stranded RNA (+ssRNA) genome. This family has a high recombination rate and genetic variability, leading to easy distribution among humans and other animals, resulting in diverse coronavirus types in human and animal populations. Coronaviruses primarily target the respiratory system and cause diseases, ranging from mild respiratory diseases to acute pneumonia and respiratory failure. Recently, there have been three severe pandemics involving respiratory diseases caused by the coronavirus. The most recent global pandemic was coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome (SARS-CoV-2), and the other two were regional epidemics, SARS-CoV and Middle East respiratory syndrome (MERS-CoV) in 2003 and 2012, respectively. The world has suffered from this pandemic and is continuing to do so. Thus, the immediate development of antiviral drugs against SARS-CoV-2 is essential. The conventional sources for treating human ailments have long been natural substances derived from plants, animals, microorganisms, and minerals. Natural product drug research has been dramatically reinvigorated by recent advancements in analytical technology, spectroscopy, and high-throughput screening with contributions from marine-based pharmaceuticals. The maritime environment is a unique resource with a vast array of biological diversity and, if properly investigated, has the potential to produce ground-breaking treatments. As more substances derived from marine sources enter clinical trials, the influence of this discipline on the pharmaceutical business grows (Shinde et al. 2019). Several compounds are produced by marine species for antiviral activity. More than 40 substances are commercially available in the pharmaceutical market, including prospective antiviral treatments or alternative antiviral medications. Many more are undergoing preclinical and clinical testing for potential antiviral medications (Yasuhara-Bell and Lu 2010). The current exploration of the marine environment for compounds with significant pharmacological applications will be significantly accelerated by the growing interest in marine-derived antiviral compounds, and it will continue to be a promising strategy and new trend in contemporary medicine. Thus, this study attempted to offer insights into the implications of a novel therapeutic agents against SARS-CoV-2 using marine natural products (MNPs), a treasure trove of antiviral agents. Furthermore, the structural and functional correlation in the identification of therapeutic drug targets was thoroughly discussed and can be used as a resource for developing antiviral agents against future coronavirus infections.
Coronavirus structure
Coronaviruses belong to the kingdom Othornavirae, family Coronaviridae, and phylum Pisuviricota and are a monophyletic cluster in the order Nidovirales. Coronavirus is an enveloped virus strain that comprises approximately 30 kb of +ssRNA genetic material (Speake et al. 2020). The subfamily Orthocoronavirinae comprises four coronavirus genera: α, β, γ, and δ. SARS-CoV and SARS-CoV-2 belong to the genus Betacoronavirus. The coronavirus virion comprises four main structural proteins, as depicted in Fig. 1. Furthermore, a coronavirus does not require these proteins to prepare a functional infectious virus. It utilizes additional proteins to prepare infectious virions (Perlman and Netland 2009; Schoeman and Fielding 2019).
Fig. 1.
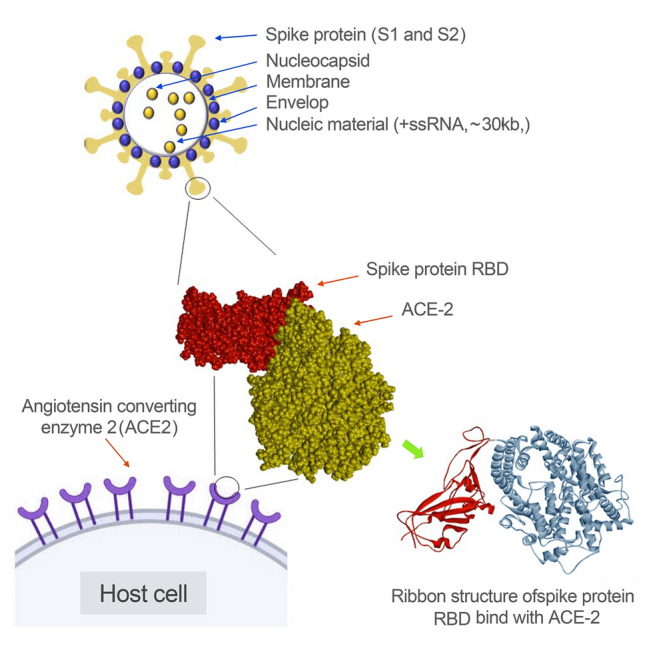
Structural proteins of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and the interaction of SARS-CoV-2 spike protein with the angiotensin-converting enzyme 2 (ACE-2)
Spike proteins have an extensive contribution to the determination of a variety of coronaviruses. The spike protein comprises two major protein subunits: S1 (amino-terminal) and S2 (carboxyl-terminal). The S1 subunit is the outermost part of the cell and plays a pivotal role in receptor binding. Meanwhile, the S2 subunit provides fusion between the coronavirus and the cellular membrane (Chen et al. 2020; Hasoksuz et al.2002). Collectively, these two protein subunits enable the virus to attach to the host cell receptor. The receptor-binding domain (RBD) in the spike protein S1 region is responsible for the initial attachment and varies among diverse types of coronaviruses. Mouse hepatitis virus (MHV) contains the RBD at the S1 N-terminus, and SARS-CoV and SARS-CoV-2 contain the RBD at the S1 C-terminus. The RBD of human coronavirus NL63 (HCoV-NL63), SARS-CoV, and SARS-CoV-2 forms an attachment with angiotensin-converting enzyme 2 (ACE-2) during host cell infection, and aminopeptidase is used as a receptor by several α-coronaviruses, such as transmissible gastroenteritis (TGEV) and diarrhea virus (PEDV). Dipeptidyl peptidase 4 (DPP4) is employed by MERS-CoV as a receptor for infection, and MHV is transmitted through the carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) receptor (Satija et al. 2007). Receptor binding provides an avenue into the host cell and employs proteolytic cleavage (acid-dependent) of spike proteins by transmembrane protease serine 2 (TMPRSS2), cathepsin (CTSs), or other protease enzymes (Bosch et al.2003).
Membrane proteins play key roles in viral assembly. They provide a surface for factors from the virus and host to join and create new virus particles. Moreover, the Golgi apparatus is targeted by this membrane protein in MHV, feline coronavirus (FCoV), SARS-CoV-2, infectious bronchitis virus (IBV), MERS-CoV, bovine coronavirus (BCoV), and SARS-CoV. Previous studies revealed that membrane proteins combine with spike glycoproteins and viral ribonucleoproteins at the budding site for virus assembly (Neuman et al. 2011).
The enveloped protein is considered a major structural protein. Elevated expression of this protein was detected during the viral replication cycle in host cells. However, the full protein is not involved in the viral envelope (Schoeman and Fielding 2019). Previous studies have revealed that the coronavirus envelope proteins play three major roles. These are the interactions between the membrane protein and envelope protein cytoplasmic tail, which emphasizes the participation of envelope proteins in viral assembly, the essentiality of hydrophobic transmembrane of envelope protein for releasing the assembled virion, and implications for virus pathogenicity (Satija and Lal 2007; Schoeman and Fielding 2019).
The coronavirus nucleocapsid is created by nucleocapsid proteins and its primary function is to bind to the virus’s genetic material. Furthermore, it plays a major role in viral genome-related processes, such as viral RNA replication and host cellular responses to the virus. Localization of nucleocapsid proteins in the endoplasmic reticulum (ER) provides functions via assembly and budding. Moreover, a significant increase in the production of virus-like particles (VLPs) in some coronaviruses has been found to be due to nucleocapsid protein expression (Chen et al. 2020).
SARS-CoV-2 genome
The SARS-CoV-2 genome comprises +ssRNAs (Wu et al. 2020). The NCBI database contains the complete genome sequence under accession No. NC_045512.2. The genome (~approximately 29.9 kb) encodes numerous open reading frames (ORFs) (13–15 ORFs), with 12 functional ORFs comprising approximately 30,000 nucleotides, including 11 protein-coding genes. Furthermore, high similarity in genetic arrangement has been reported among SARS-CoV-2, MERS-CoV, and SARS-CoV, with 89% sequence identity (Lu et al. 2015; Rota et al. 2003). The encoded proteins are predominantly divided into two groups: non-structural (NSPs) and structural proteins, which play critical roles in the entry, fusion, replication, and survival of host cells (Tong 2009). The entire SARS-CoV-2 genome encodes a polyprotein containing 7096 residues, which comprises several structural proteins and NSPs. There are two major polyproteins that can be found called pp1a and pp1ab, which are encoded by ORF1a and ORF1ab, respectively. The ribosomal frameshift mechanism of 1b was used to encode polyprotein pp1ab. The proteinases encoded by the viral genome further process these polyproteins and generate 16 proteins that are important for the viral life cycle and are conserved in coronaviruses of the same family (Fig. 2A). SARS-CoV-2 exhibits an elevated level of infection compared to SARS-CoV and MERS-CoV due to its different epidemiological dynamics and the successful utilization of other mammalian species as amplifying or intermediate hosts and acquiring mutations for efficient human transmission (Graham and Baric 2020).
Fig. 2.
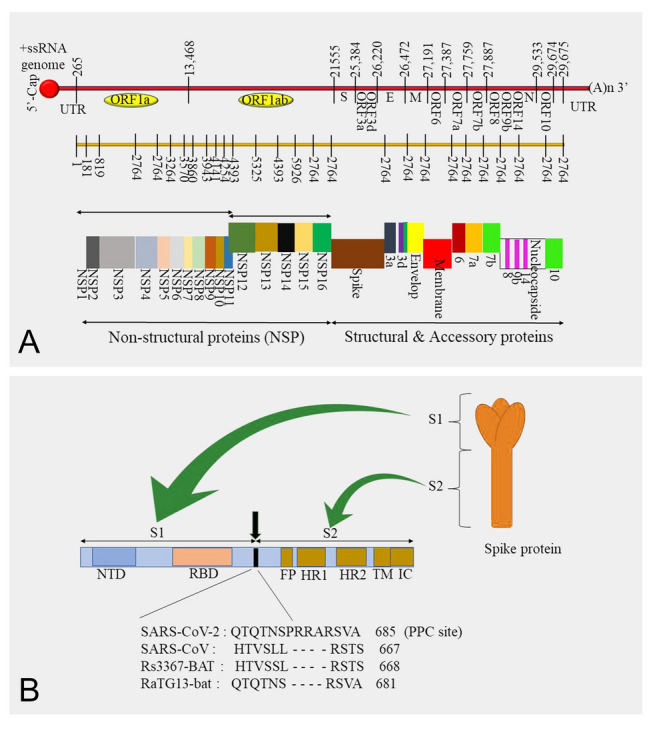
The structure of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome and SARS-CoV-2 spike protein. A Order of the open reading frames (ORFs) and their expression to produce structural and non-structural proteins and B SARS-CoV-2 spike protein structure and the comparison of proprotein convertase sites in diverse coronaviruses
NSPs play specific roles in viral replication and assembly in host cells by engaging in viral pathogenesis via gene transactivation, modulating helicase activity, countering antiviral responses, early transcription regulation, and immunomodulation (Xue et al. 2014) (EA and Jones 2019; Muller et al. 2018; Tang et al. 2020). The major functions of these NSPs are summarized in Table 1. In addition to the conserved regions, accessory genes can also be found in the SARS-CoV-2 genome, which expresses notable variability among the coronavirus groups. Nine accessory proteins were found to be encoded by at least five accessory genes (Fig. 2A).
Table 1.
The functions of non-structural proteins of SARS-CoV-2
| No | Range | Protein name and ID | Role | Proposed function |
|---|---|---|---|---|
| 1 | 1–180 |
Nsp1 |
Nsp1 is the N-terminal product of the viral replicase | Leader protein host translation inhibitor. Mediates RNA replication and processing. Involved in mRNA degradation |
| 2 | 181–818 |
Nsp2 |
Nsp2 is a replicase product essential for proofreading viral replication | Modulation of host cell survival signaling pathway by interacting with host PHB and PHB2 |
| 3 | 819–2763 |
Nsp3 |
Nsp3 is a papain-like proteinase, contains several domains | Functions as a protease to separate the translated polyprotein into its distinct proteins |
| 4 | 2764–3263 |
Nsp4 |
A membrane-spanning protein, contains transmembrane domain 2 (TM2) | Believed to anchor the viral replication–transcription complex to modified ER membranes |
| 5 | 2364–3569 |
Nsp5 |
3C-like proteinase and main proteinase | Involved in viral polyprotein processing during replication |
| 6 | 3570–3859 |
Nsp6 |
Putative transmembrane domain | Plays a role in the initial induction of autophagosomes from host endoplasmic reticulum |
| 7 | 3860–3942 |
Nsp7 |
Nsp7 is an RNA-dependent RNA polymerase | It forms a hexadecameric super-complex with nsp8 that adopts a hollow cylinder-like structure implicated in replication |
| 8 | 3943–4140 |
Nsp8 |
Multimeric RNA polymerase; replicase | It forms a hexadecameric super-complex with nsp7 that adopts a hollow cylinder-like structure implicated in replication |
| 9 | 4141–4253 |
Nsp9 |
A single-stranded RNA-binding viral protein | Participates in viral replication by acting as an ssRNA-binding protein |
| 10 | 4254–4392 |
Nsp10 |
Growth-factor-like protein contains two zinc-binding motifs | In viral transcription by stimulating both nsp14 3′–5′ exoribonuclease and nsp16 2′-O-methyltransferase activities. Therefore, plays an essential role in viral mRNAs cap methylation |
| 11 | 4393–5324 |
Nsp12 |
RNA-dependent RNA polymerase (Pol/RdRp) |
Responsible for replication and transcription of the viral RNA genome |
| 12 | 5325–5925 |
Nsp13 |
Zinc-binding domain, NTPase/helicase domain, RNA 5′-triphosphatase | A helicase core domain that binds ATP. Zinc-binding domain is involved in replication and transcription |
| 13 | 5926–6452 |
Nsp14 |
Proofreading Exoribonuclease domain (ExoN/nsp14) | Exoribonuclease activity acting in a 3′ to 5′ direction and N7-guanine methyltransferase activity |
| 14 | 6453–6798 |
Nsp15 |
EndoRNAse; nsp15-A1 and nsp15B-NendoU | Mn(2+)-dependent endoribonuclease activity |
| 15 | 6799–7096 |
Nsp16 |
2′-O-ribose methyltransferase | Methyltransferase that mediates mRNA cap 2′-O-ribose methylation to the 5′-cap structure of viral mRNAs |
| 16 | 4393–4405 | Nsp11 YP_009725312.1 | Made of 13 amino acids (sadaqsflngfav) and identical to the first segment of nsp12 | Unknown |
Cell entry mechanism
Determination of coronaviruses’ cell entry mechanism is important for evaluating SARS-CoV-2 pathogenicity and infectivity (Li 2016; Perlman and Netland 2009). Moreover, it is a key target for intervention strategies and host immune surveillance (Du et al. 2009; Perlman and Netland 2009). The cell entry mechanism can be divided into three major phases. Initially, viral attachment is conducted by a coronavirus via binding to the cell surface receptor of the host cells, which then enters the endosome and fuses lysosomal and viral membranes (Du et al. 2009; Perlman and Netland 2009). The mature coronavirus spike protein is present as a trimer that comprises three receptor-binding S1 heads placed on top of the S2 stalk. According to previous SARS-CoV studies, ACE-2 is recognized as its receptor by SARS-CoV S1 RBD (Li 2015; Li et al. 2005, 2003; Walls et al. 2019). RBD follows a specific mechanism that switches regularly between standing up and lying down positions to evade the host cell’s immune response (Gui et al. 2017; Yuan et al. 2017). The SARS-CoV spike protein requires proteolytic activation at the S1/S2 boundary to fuse the membranes. Here, the S1 disunites and S2 undergoes structural changes. The lysosomal proteases cathepsin and cell surface protease TMPRSS2 play major roles in activating the spike protein and cell entry mechanism (Belouzard et al. 2012; Heald-Sargent and Gallagher 2012). These specific factors of the cell entry mechanism led to severe symptoms, rapid spread, and high fatality rates in infected patients (Bolles et al.2011; Frieman and Baric 2008; Li 2013).
Recent studies have revealed that SARS-CoV-2 also recognizes ACE-2 as a receptor for cell entry, and these studies have guided the identification of critical SARS-CoV and SARS-CoV-2 functional characteristics in receptor recognition (Letko et al. 2020; Xu et al. 2020; Zhou et al. 2020). The RBD of SARS-CoV-2 expresses a significantly higher binding affinity for ACE-2 than for SARS-CoV (Shang et al. 2020a, b). However, there are conflicting reports from diverse studies on the SARS-CoV-2 RBD binding affinity to ACE-2. As mentioned above, RBD follows standing up and laying down states to evade the immune response, and these states are associated with receptor-binding affinity. An interesting study regarding the cryo-electron microscopy structure of SARS-CoV-2 revealed that RBD was mostly in the lying down state, which demonstrates ineffective binding with ACE-2 (Walls et al. 2020; Wrapp et al. 2020). Moreover, studies have verified the role of TMPRSS-2 and lysosomal proteases as protease activators in the SARS-CoV-2 cell entry mechanism (Hoffmann et al. 2020; Ou et al. 2020).
Proprotein convertase (PPC) is a protein family responsible for activating other proteins. The PPC motif of surface glycoproteins plays a key role in the pathogenesis of viruses, such as avian influenza. However, previous studies have reported that there is no specific function of the PPC motif in enhancing SARS-CoV-2 cell entry (Letko et al. 2020). However, novel studies have emphasized the PPC motif function by investigating protease activation and receptor binding of SARS-CoV and SARS-CoV-2 as a comparison study. The results of this study revealed the important factors of the cell entry mechanism that play pivotal roles in cell infectivity, immune evasion, and virus spread in the host. The identified cleavage site is located at the boundary between S1 and S2 (Fig. 2B). Moreover, SARS-CoV does not have this cleavage site. Cell entry assays for SARS-CoV, SARS-CoV-2, and SARS-CoV-2 with a mutated cleavage site were conducted. This assay revealed that SARS-CoV-2 exhibited significantly higher cell entry than SARS-CoV and SARS-CoV-2-mutated strains. In addition, PPC inhibitors were used to evaluate the effects of PPC on SARS-CoV and SARS-CoV-2. SARS-CoV-2 cell entry was significantly downregulated by the PPC inhibitor, and there was no significant difference in SARS-CoV cell entry. These results verify that SARS-CoV-2 requires prior PPC cleavage for cell entry (Shang et al. 2020a, b).
SARS-CoV-2 replication in the host cell
Fusion of the SARS-CoV-2 spike protein with the ACE-2 receptor causes subtle conformational modifications, and the viral nucleocapsid is released into the host cell cytosol. Several host factors can be found to support these processes, including TMPRSS-2 and cathepsin-L. Immediately after releasing the nucleocapsid into the cytosol, +ssRNA acts as a functional messenger RNA (mRNA) for ORF1a and ORF1b, which encode the polyprotein pp1a (440–500 kDa) and pp1ab (740–810 kDa), respectively. Furthermore, compared to pp1ab, pp1a is expressed 1.2- to 2.2-fold more in host cells due to the high efficiency of frameshift between the ORF1 and ORF1b genes (Finkel et al. 2021). The autoproteolytic process leads to the processing of these 2 polyproteins and produces 16 NSPs, which collectively form a replication-transcription complex (RTC) for the synthesis of viral RNA. The set of sgRNAs results from this functional RTC via discontinuous transcription (V’kovski et al. 2021). RTC formation causes molecular processes in the passage guide for synthesizing numerous viral RNA copies. This negative-sense single-stranded RNA (−ssRNA) acts as an intermediate template. Simultaneously, during −ssRNA synthesis, the polymerase switches templates at short motifs called transcription-regulated sequences (TRS) to generate many 5-nested negative-sense sgRNA sets, which, in turn, are utilized as templates for the formation of 3′-nested positive-sense sgRNAs. Consequently, they interact with the host ribosomes and synthesize numerous structural and accessory proteins that build multiple viral structures (Sola et al.2015).
Free cytosolic ribosomes are responsible for the translation of nucleocapsid proteins in the host cells. Furthermore, proteins associated with spike, membrane, and envelope proteins are synthesized by ribosomes bound to the ER. Subsequently, these proteins undergo post-translational modifications (PTMs). The endoplasmic reticulum–Golgi intermediate compartment (ERGIC) is the virion assembly site. Here, scaffolding and orchestrate virion morphogenesis are provided by membrane proteins via heterotrophic interactions with other structural proteins, such as membrane spike and membrane envelope proteins provide molecular incorporation. Moreover, membrane–nucleocapsid interactions facilitate nucleocapsid condensation with the envelope along with the envelope protein (V’kovski et al. 2021). After molecular assembly, virion progenies are carried in smooth-wall vesicles and transported by secretory pathways to the plasma membrane, eventually exiting through exocytosis, upon which they spread to other parts of the body (Astuti and Ysrafil 2020; Naqvi et al. 2020; V’kovski et al. 2021).
The structure, genome, cell entry, and replication mechanisms provide many potential drug targets for inhibiting SARS-CoV-2. The traditional sources for treating human ailments have long been natural substances derived from plants, animals, microorganisms, and minerals. Natural product drug research has been dramatically reinvigorated by recent advancements in analytical technology, spectroscopy, and high-throughput screening with contributions from marine-based pharmaceuticals. The maritime environment is a unique resource with a vast array of biological diversity and, if properly investigated, has the potential to produce ground-breaking treatments. As more substances derived from marine sources enter clinical trials, the influence of this discipline on the pharmaceutical industry has grown. Herein, we discuss the potential drug targets of SARS-CoV-2 and their inhibition by MNPs.
MNPs against SARS-CoV-2
Although the development of vaccines to eliminate or limit its effects never ceases, SARS-CoV-2 continues to spread rapidly across the globe. Insufficient production to meet the global demand, specificity with the desired viral strain, continuous SARS-CoV-2 genome mutation, and subsequent novel strains necessitate the investigation of drugs that can potentially prevent COVID-19. Furthermore, many efforts have been made to repurpose US Food and Drug Administration (FDA)-approved drugs against SARS-CoV-2. The potential drugs, antibodies, and compounds that demonstrate antiviral effects against SARS-CoV-2 are summarized in Table 2.
Table 2.
Repurposed drugs against SARS-CoV-2 and their activities
| No | Drug/compound/antibody | Targets of drug on SARS-CoV-2 |
|---|---|---|
| 1 | Chloroquine and formoterol |
Target Papain-like protease (PLpro) Interferes with viral replication Chloroquine targets the terminal glycosylation of ACE-2 Interferes with the spike protein and ACE-2 |
| 2 | Remdesivir (nucleotide analog) |
Target RNA-dependent RNA polymerase (RdRp) Interferes with the nascent viral RNA |
| 3 | Bananin (adamantane derivative) |
Targets helicase (NSP13) Interferes with viral replication |
| 4 | Pyridone-containing α-ketoamides |
Targets chymotrypsin-like protease (3CLpro) Interferes with viral replication |
| 5 | β-D-N4-Hydroxycytidine (ribonucleoside analog) | Inhibits viral replication |
| 6 | Ebselen | Reduces COVID-19 by 20.3 fold |
| 7 | Ivermectin |
Targets nuclear transporter and Impα/β1 heterodimer, binds and destabilizes it. This prevents its binding with viral cargo protein and its translocation to nucleus Interferes with the suppression of antiviral responses and viral load (reduce by 5000 fold) |
| 8 | Zidovudine |
Targets nucleocapsid phosphoprotein Binds with nucleocapsid phosphoprotein and provides antiviral effect |
| 9 | Camostat mesylate and bromhexine hydrochloride |
Targets TMPRRS-2. Acts as a TMPRSS-2 inhibitor Interferes with viral entry |
| 10 | CR3022 (monoclonal antibody) |
Targets RBD of spike protein Interferes with the cellular interaction of virus |
Natural compounds are becoming increasingly enticing in pharmaceuticals, cosmeceuticals, nutraceuticals, and functional foods because people admit that naturally occurring compounds are more secure than artificially synthesized compounds. Currently, COVID-19 control has become a global health emergency owing to the unavailability of antiviral drugs against SARS-CoV-2. Therefore, the repurposing of WHO-approved drugs, including remdesivir (Ebola), chloroquine and hydroxychloroquine (malaria), and lopinavir and ritonavir (HIV), against SARS-CoV-2 has been investigated (Kupferschmidt and Cohen 2020).
Hydroxychloroquine was approved by the FDA for treating COVID-19. The results demonstrated a significant reduction in viral load by hydroxychloroquine in infected patients as a combination treatment with azithromycin. In a further study, hydroxychloroquine was proven to be more effective in COVID-19 treatment than chloroquine (Gautret et al. 2020). However, statistical data from hospitalized COVID-19 patients revealed that there was no beneficial effect of hydroxychloroquine on the recovery rate compared to standard COVID-19 care (Therapy 2021, March 5). A study of treatments with lopinavir–ritonavir also demonstrated that there was no significant difference in standard care for COVID-19 (Cao et al. 2020). However, lopinavir–ritonavir combined with interferon β-1b and ribavirin demonstrated effective antiviral activity by alleviating and shedding symptoms in patients with mild-moderate COVID-19. Notably, interferon β-1b was not included in the control group in this study, and a placebo control group was not included in this study to compare the treatment’s efficacies (Hung et al. 2020). Further studies are required to verify this.
The ocean contains many resources that yield a variety of natural products. It encompasses more than 70% of the surface of the Earth and is home to more than 300,000 identified plant and animal species (Pomponi 1999). The ocean is described as a vast treasure awaiting the discovery of numerous valuable compounds that can be used against diseases. Unique and extreme conditions, such as ecological pressure, are responsible for the evolution of these secondary metabolites with numerous biological activities (Ireland et al. 2000). The significance and use of these secondary metabolites have been extensively evaluated for many years. Experts worldwide have discovered more than 12,000 novel compounds from marine animals and plants within the last 30–40 years (König et al. 1994) and have evaluated them in diverse fields to discover useful applications, including anti-cancer, anti-inflammatory, antiviral, antibacterial, and antifungal properties (Donia and Hamann 2003; Sanjeewa et al. 2016).
An antiviral agent that can prevent SARS-CoV-2 infection remains under research and development, and repurposing of FDA-approved drugs can cause several failures and side effects, including many major handling failures, side effects, resistance, long-term treatment, and cell toxicity (Khan et al. 2021). The available vaccines against SARS-CoV-2 predominantly target structural proteins of the virus; however, regulating NSPs of SARS-CoV-2 using marine natural products may cause significant antiviral activity against SARS-COV-2. Furthermore, the complex infection and replication mechanisms of SARS-CoV-2 have provided diverse therapeutic drug targets (Table 3).
Table 3.
Potential therapeutic drug targets against SARS-CoV-2
| SARS-CoV-2 | Host |
|---|---|
| Spike protein | Proteases of host |
| Envelope protein | Proprotein convertase (PPC) |
| Membrane protein | Toll-like receptors (TLRs) |
| Nucleoprotein | High-density lipoprotein (HDL) scavenger receptor B type 1 (SR-B1) |
| Replicase protein | Glycosaminoglycans (GAGs) on the cell surface |
| Main protease (Mpro or 3CLpro) | |
| Papine-like protease | |
| RNA-dependent RNA polymerase | |
| Non-structural proteins |
MNPSs as potent SARS-CoV-2 spike protein inhibitors
The SARS-CoV-2 spike protein plays a pivotal role in the infectivity and pathogenesis of coronaviruses (Du et al. 2009; Hofmann et al. 2004). It comprises 1273 amino acids created by 2 primary subunits: S1 and S2. These subunits undergo structural changes during viral–host membrane fusion. The RBD of S1 binds with ACE-2 via Glu394 of RBD and Lys31 of ACE-2, and these residues play an important role in viral–host interactions (Zhang et al. 2020). The RBD receptor-binding motif demonstrates high variation, which causes variations in coronavirus pathogenesis. The S2 subunit causes membrane fusion between the host and viral membranes. Therefore, S2 had three conformations. The pre-fusion native state was the initial phase. It then extends to the hairpin intermediate state and becomes a post-fusion hairpin state. Discovering this changing conformation of the spike protein has led to the establishment of therapeutic agents for alleviating SARS-CoV-2. Novel studies have revealed that the binding site of RBD on SARS-COV-2 comprises six residues (Phe486, Leu455, Gln493, Tyr505, Asn501, and Ser494), which play an important role in ACE-2 binding (Naqvi et al. 2020). The SARS-CoV-2 spike protein also comprises a homologous trimeric spike protein structure similar to SARS-CoV and MERS-CoV. This conformation comprises three chains (A, B, and C), but the N-terminus of chain B has a unique conformation compared to the other two chains (Alexandra et al. 2019; Walls et al. 2020). These similarities and alterations could be exploited as key targets in vaccine production for antiviral drug development. Vaccines are considered a major solution to control COVID-19 spread; the risk of morbidity and mortality has encouraged the development of vaccines against SARS-CoV-2. The SARS-CoV-2 spike protein is targeted by several vaccines owing to its immunogenicity. However, mutations in the spike protein may affect the efficacy of the vaccine; for example, a vaccine produced against SARS-CoV-2 isolated from Wuhan, China may not be effective against the United Kingdom variant (VUI202012/01) due to its D614G mutation in the spike protein. Therefore, the development of vaccines or drug candidates that do not depend on these mutations is vital. Thus, the interaction between the spike protein and ACE-2 RBD can be identified as an important drug target (Aatif et al. 2021).
The computational approach to evaluate MNPSs against the spike protein of the SARS-COV-2 VUI202012/01 strain provided effective drug candidates against SARS-COV-2. In this study, 1110 unique compounds with known biological activities, including anti-microbial, antiviral, anti-cancer, and anti-inflammatory activities, from the Seaweed Metabolite Database were investigated. According to these results, dieckol was identified as a successful inhibitor of the interaction between SARS-CoV-2 RBD and ACE-2. However, structural analysis of dieckol has revealed that it does not possess drug-like properties; therefore, it cannot be used as a lead inhibitor. Dieckol derivatives have been proposed as alternative 8–3-hydroxy-4-(7-hydroxynaphthalen-2-yl) oxy-phenoxy-1,4-benzodioxin-5-ol inhibitors. It successfully binds to the RBD and interferes with RBD-ACE-2 binding. However, in vitro and in vivo studies are required for further evaluation (Aatif et al. 2021).
Previous studies have demonstrated that the SARS-CoV-2 spike protein initially interacts with GAGs, including HS (Kim et al. 2020; Lindahl and Li 2020). Moreover, the results of these studies revealed that the SARS-CoV-2 RBD spike protein binds tightly with immobilized heparin compared to SARS-CoV and MERS-CoV. Brown seaweeds comprise sulfated polysaccharides, which have a structure similar to GAGs. These polysaccharides demonstrated strong binding abilities with SARS-COV-2 and inhibit its interaction with immobilized heparin (Kwon et al. 2020). The structure–activity relationship of the binding ability of polysaccharides with the SARS-CoV-2 spike protein was evaluated in a previous study using surface plasmon resonance. The results demonstrated that two polysaccharides, sulfated galactan and glucuronomannan, strongly inhibited the interaction between the spike protein and ACE-2 via the interaction between the SARS-CoV-2 spike protein and heparin (Jin et al. 2020).
Main protease (Mpro) inhibition by MNPSs
Mpro is one of the most attractive drug targets for SARS-CoV-2 owing to its specific role in polyprotein processing. This is the most vastly studied and well-validated drug target for SARS-CoV-2. Mpro is a key enzyme in the viral replication cycle. Some essential enzymes, such as RdRp for replication, require a prior proteolytic release for complete functioning [86]. Mpro inhibition can downregulate infectious viral particle production, which leads to the reduction of disease symptoms (Anand et al. 2003). Mpro studies have revealed a close structural relationship with the Mpro of other coronaviruses. Amino acid sequence alignment results demonstrated 99% sequence identity with BatCoV RaTG13 Mpro, 50% with MERS-CoV Mpro, and 96% with SARS-CoV Mpro. Sequence alignment data of Mpro revealed a 96% similarity between SARS-CoV and SARS-CoV-2 (Xu et al. 2020). Structural analysis of Mpro also demonstrated great similarity, except for the 12 amino acids in the surface proteins. Furthermore, the Mpro structures of SARS-CoV and SARS-CoV-2 are similar to those of cysteine proteases, which comprise a catalytic dyad (His41 and Cys145) in the active site and a stable water molecule that forms at least three hydrogen bonds with the surrounding residues instead of a third residue (Anand et al. 2003). Previous research has demonstrated that the high structural similarity of Mpro among SARS-CoV, SARS-CoV-2, and MERS-CoV guides drug development against SARS-CoV-2 Mpro, based on previously developed compounds against Mpro of SARS-CoV and MERS-CoV (Ullrich and Nitsche 2020).
Mpro belongs to the cysteine protease family of enzymes. Normally, serine and cysteine proteases contain a catalytic triad; whereas Mpro contains a catalytic dyad (histidine and cysteine) in its active site. The proteolytic mechanism of Mpro is considered a multistep mechanism. Histidine imidazole abstracts the cysteine side chain protons, and the amide bonds of the substrate are attacked by the resulting thiolate nucleophile. The proton abstraction from histidine releases N-terminal peptide products, after which the release of C-terminal products and restoration of the catalytic dyad occurs by thioester hydrolyzation (Pillaiyar et al. 2016).
The initial auto-cleavage of Mpro between NSP6 and NSP7 is required for polyprotein processing of pp1a and pp1ab at 11 cleavage sites (Du et al. 2004). The Mpro monomer is the inactive form and the primary active species is the homodimer. Two orthogonally aligned protomers can be found, and each protomer contains three domains. The domains Ι (residues 8–101) and ΙΙ (residues 102–184) of SARS-CoV and SARS-CoV-2 contain an antiparallel β-barrel, which is similar to the trypsin-like serine protease. The domain ΙΙΙ (residues 201–306) comprises a cluster of α-helices and is connected to domain ΙΙ using a longer loop region (residues 185–200). The N-terminal fingers are used to bind protomers with each other, which involves creating a substrate-binding site located in a cleft between the domains Ι and ΙΙ (Ullrich et al. 2020). The Ile286Ala, Thr285Ala, and Ser284Ala mutations in SARS-CoV-2 Mpro cause a 3.6-fold increase in catalytic activity than SARS-CoV Mpro (Lim et al. 2014). SARS-CoV and SARS-CoV-2 active sites were highly similar, except for the minor mutation S64A. Mutation influences the size, shape, plasticity, and flexibility of the active site, and further studies are required to use this for inhibitor design (Bzowka et al. 2020). In addition, as previously mentioned, the inactive Mpro monomer requires dimerization for activation. This emphasizes the need for a drug target against Mpro and SARS-CoV-2.
The in silico approach for evaluating inhibitors of MNPs against SARS-CoV-2 Mpro revealed great potential to inhibit SARS-CoV-2 (Gentile et al. 2020; Khan et al. 2021). Structural data demonstrated that the average volume of SARS-CoV-2 Mpro is half that of SARS-CoV Mpro. Furthermore, the SARS-CoV Mpro binding cavity is highly flexible and demonstrates significant changes in volume and shape after binding to the ligand (Anand et al. 2003). These features can also be used to design inhibitors or to convert suitable substrates into strong inhibitors. For example, the N3 inhibitor, a computer-aided designed peptide inhibitor is a successful Mpro inhibitor designed by mimicking natural substrates (Pillaiyar et al. 2016).
In a previous study, evaluation of phlorotannins isolated from Ecklonia cava as SARS-CoV Mpro inhibitors revealed the great potential of MNPs. Nine phlorotannins (phloroglucinol, triphloretol A, dioxinodehydroeckol, eckol, 7-phloroeckol, 2-phloroeckol, dieckol, fucodiphloroethol G, and phlorofucofuroeckol A) were evaluated. All phlorotannins, except phloroglucinol, demonstrated significant and dose-dependent inhibition against SARS-CoV Mpro; whereas dieckol demonstrated the highest activity (Park et al. 2013). The structural similarity of Mpro between SARS-CoV and SARS-CoV-2, including its structural and functional relationship, should be exploited for further development of these compounds against SARS-CoV-2 Mpro.
The crystal structure of the SARS-CoV-2 Mpro (PDB ID. 6LU7) is available in PBD as a complex with an N3 inhibitor bound to the Cys145 residue. In a SARS-CoV-2 Mpro study, 14,064 compounds from the MNPs library were evaluated against SARS-COV-2 Mpro. All the molecules were screened in molecular docking assay, and 17 molecules (hydroxypentafuhalol A, heptafuhalol A, phlorethopentafuhalol A, pentaphlorethol A, phlorethopentafuhalol B, pentaphlorethol B, resinoside B, pseudopentafuhalol C, aeruginosin 98B, pseudotheonamide C, pseudotheonamide D, Dieckol, 6,6′-bieckol, apigenin-7-O-neohesperidoside, 8,8′-bieckol, luteolin-7-rutinoside, and tunichrome An2) were identified as potential SARS-CoV-2 Mpro inhibitors. Among these compounds, 8,8′-bieckol, 6,6′-bieckol, and dieckol were found to be the most active inhibitors; however, future in vitro and in vivo studies are required for further evaluation. On a positive note, these results emphasize another important factor that these phlorotannins, which have been used as therapeutic agents in anti-oxidant, anti-cancer, anti-inflammatory, anti-diabetic, and anti-hypertensive treatments, could potentially be used for treating COVID-19 (Gentile et al. 2020). Another study assessed five MNPs (fistularin-3/11-epi-fistularin-3 and 15-methyl-9(Z)-hexadecenoic acid isolated from sponges of the family Aplysinidae; (hexadecyloxy) propane,1,2-diol isolated from the soft coral Pterogorgia citrine; 15-α-methoxypuupehenol and puupehedione isolated from Petrosia Strongylophora) against SARS-CoV-2 Mpro. According to the results, fistularin-3/11-epi-fistularin-3 demonstrated the highest inhibitory activity. Furthermore, 15-methyl-9(Z)-hexadecenoic acid demonstrated considerable binding to the SARS-CoV-2 Mpro catalytic dyad. These in silico studies suggest the great potential of MNPs as SARS-COV-2 Mpro inhibitors. Further studies are required to evaluate these inhibitors and develop therapeutic drugs against COVID-19.
Papain-like protease (PLpro) inhibition by MNPs
The SARS-CoV-2 genome encodes another protease, PLpro, which is responsible for polyprotein processing. NSP1, NSP2, and NSP3 are cleavage sites used by PLpro and the other 13 NSPs are processed by Mpro. This leads to the viral replicase complex assembly on the host cell membrane, initiating replication and viral genome transcription (Baez-Santos et al. 2015). In addition to polyprotein processing, PLpro inhibits host innate immune responses, such as interferon responses, which create an antiviral state in the host cell using interferon-stimulated genes (ISGs). These responses lead to the detection of viral threats and subsequent responses (Berlin et al. 2020).
Antagonizing ubiquitin and ubiquitin-like modifications is a common mechanism for regulating innate immune responses by viral proteases (Heaton et al. 2016). Many ubiquitin chain formations encode both degradative and non-degradative functions. This results in complex protein ubiquitination (Yau and Rape 2016). Inflammatory signaling pathways use specific ubiquitin signals in human cells, including interferon-stimulated gene 15 (ISG15), a secreted protein (17kDa) encoded by ISG15. The viral infection leads to this ubiquitin-like (Ubl) ISG15 modification (Dzimianski et al. 2019). Several cellular enzymes participate in this process, enabling danger signals caused by viral infections. Viruses often use their proteases as deubiquitinases (DUBs) and deISGylases to avoid this. PLpro acts as a DUB and disturbs the antiviral response of host cells by inhibiting Ubl-ISG15 modification. (Klemm et al. 2020) (Békés et al. 2015). Thus, three specific PLpro substrates can be identified: antiviral ISG15 signals, degradative Lys48-polyubiquitin, and viral polyprotein. These activities are important for viral maturation, replication, and survival of the host which make PLpro an excellent candidate for antiviral drug development. Thus, researchers are conducting studies to inhibit PLpro and develop antiviral drugs against SARS-CoV-2. This was verified by evaluating the inhibitory activity of GRL0617 against SARS-CoV-2 PLpro. According to the results, GRL0617 blocked the binding of ISG15 or ubiquitin with PLpro and significantly inhibited polyprotein processing (Fu et al. 2021).
A study was conducted to evaluate PLpro inhibition using ilimaquinone, a marine sponge metabolite isolated from a marine sponge called Hippospongia metachromia, in comparison with hydroxychloroquine, ivermectin, remdesivir, azithromycin, and favipiravir. The results demonstrated that ivermectin had the highest binding ability, followed by ilimaquinone. According to these in silico results, ilimaquinone is a promising drug candidate against SARS-CoV-2 PLpro (Surti et al. 2020). Another computational simulation was conducted to identify MNPs as SARS-CoV-2 PLpro inhibitors to discover potential MNPs that can significantly bind to the PLpro active site and inhibit its activity. Compounds from the MNPs library were filtered using their drug-like properties, including the number of hydrogen bonds, donors, and acceptors. The results demonstrated that 14 MNPs expressed higher binding affinity than the positive controls, lopinavir and ritonavir (Kumar et al.2021).
RNA-dependent RNA polymerase (RdRp) inhibition by MNPs
NSP12, also called RdRp, contributes to the viral genome and protein synthesis, together with helicase. Furthermore, they play an essential role in the viral lifecycle. The production of the viral genome progeny depends on RdRp, which must be synthesized to continue the process. RdRp is a common function among diverse virus genera and is the most conserved enzyme across several viral species, including influenza, hepatitis C, Zika, and coronaviruses (Venkataraman et al. 2018). The protein sequence similarity of RdRp between SARS-CoV and SARS-CoV-2 is 96% and existing structural differences can be found in catalytically inactive areas (Morse et al. 2020).
RdRp is a central enzyme used for viral replication. During viral genome synthesis, a complementary negative RNA strand is synthesized by RdRp based on +ssRNA. Based on the primer dependence, two conceivable methods are identified in genomic RNA synthesis as primer-dependent or independent (Ferrer-Orta et al.2006). Furthermore, cellular ribonucleotide triphosphates (rNTPs), CTP, ATP, UTP, and GTP afford template substrates, as observed by RdRp. The divalent metal ions manganese (Mn) and magnesium (Mg), which act as essential co-factors, promote reactions with rNTPs and catalytic aspartates are coordinated (Ogden et al. 2012).
Results from previous studies indicate that similar structures and catalytic mechanisms are shared by all RNA polymerases. In addition, this provides insights into the relationship between RdRp function and structure (Steitz 1998; Venkataraman et al. 2018). When studying the RdRp structure, a large, grooved domain resembling a cupped right hand is evident in the core RdRp structure, which is connected by “fingers,” “thumb,” and “palm” subdomains surrounding the active site cavity. (Fig. 3A). The catalytic process is affected by the structural motifs of RdRp, which are pitched in these domains. (Venkataraman et al. 2018). The subdomains play a role in the entry of nucleotide triphosphates, binding templates, and polymerization. The finger subdomain contains an active site and plays an important role in RNA binding and polymerization (Gao et al. 2020). Inhibition of this enzyme strongly affects the viral life cycle and activity. Therefore, the inhibition of RdRp has a promising effect on SARS-CoV-2 inhibition. Sargassum cristaefolium, S. echinocarpum, and Padina australis are three different brown seaweeds that are utilized to isolate MNPs. From these results, 99 compounds were evaluated and among them, 20 demonstrated a strong binding affinity with SARS-CoV-2 RdRp. Rhamnetin, a compound available in Sargassum spp., has the highest affinity with the RdRp finger subdomain, which is responsible for the active site (Firdaus et al. 2020). Moreover, another in silico approach revealed that compounds identified from the MNP library bind to the RdRp catalytic pocket using the same or slightly different residues as remdesivir. Among these compounds, moniloside A, isolated from Formia monilis, demonstrated a strong binding affinity with the RdRp active site. These studies demonstrate the potential of MNPs as promising drug candidates against SARS-CoV-2 through RdRp.
Fig. 3.
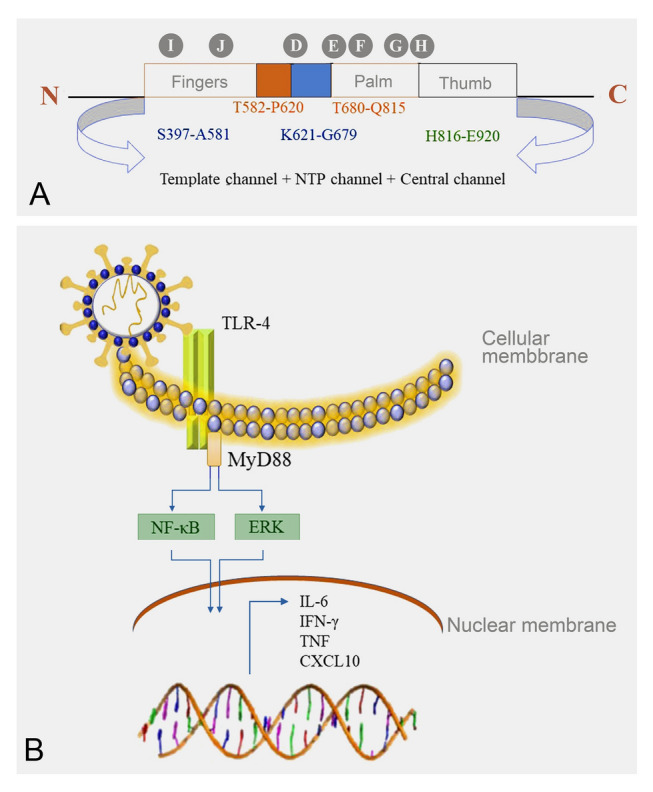
RNA-dependent RNA polymerase (RdRp) structure and mechanism of SARS-CoV-2 induce cytokine storm. A Structure of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA-dependent RNA polymerase (RdRp) enzyme and B SARS-CoV-2-induced Toll-like receptor (TLR)-mediated inflammatory signaling pathways
Nucleoside analogs are another approach to inhibit RdRp activity. Nucleosides are elements that make up nucleic acids and are involved in important biological activities, such as nucleotide formation (Seley-Radtke and Yates 2018). Previous studies have revealed significant antiviral activity of nucleoside analogs (Anjum et al. 2016). Spongouridine, spongothymidine, mycalisine A, and mycalisine B, isolated from marine sponges Cryptotethya and Mycale spp., respectively (Bergmann and Feeney 2002), are some examples of nucleoside analogs isolated from marine creatures that can be used against SARS-CoV-2.
Transmembrane protease serine 2 (TMPRSS-2) inhibition by MNPs
The SARS-CoV-2 cell entry mechanism strongly influences its pathogenicity and infectivity with support of host proteases. According to recent studies, TMPRSS-2 (Iwata-Yoshikawa et al. 2019) and cathepsin B and L (Simmons et al. 2005) were identified as the major host proteases for viral cell entry and membrane fusion. The role of TPMRSS-2 in SARS-CoV-2 infection remains obscure. In humans, TMPRSS-2 is vastly expressed in epithelial tissues, including the bronchi, epithelial lining of the upper airways, and lungs (Bugge et al.2009). The similarity in the TMPRSS-2 protein sequence between humans and mice was 78%. Moreover, a previous study on mouse embryos and adults revealed that TMPRSS-2 was expressed in the respiratory tract (bronchi and bronchioles), epithelial lining of the gastrointestinal tract, and urogenital system, but not in the alveolar epithelium (Vaarala et al. 2001). Furthermore, mice with depleted TMPRSS-2 demonstrated no abnormalities in organ histology, function, development, or survival (Kim et al.2006). The in vivo results of a previous study using TMPRSS-2 knockout mice and wild-type mice revealed that TMPRSS-2-depleted mice had less pronounced coronavirus replication in the lungs, especially in bronchioles (Iwata-Yoshikawa et al. 2019). The physiological function of TMPRSS-2 was not 100% revealed. However, the involvement of TMPRSS-2 in sodium current regulation by proteolytic cleavage of the epithelial sodium channel in lung epithelial cells has been described in earlier studies (Donaldson et al. 2002). The coronavirus cell entry mechanism predominantly involves two distinct pathways: entering the cell via either the cell surface (TMPRSS-2) or the endosome (Cathepsin-L). According to previous studies, TMPRSS-2 (serine proteases) facilitates SARS-CoV spread more than cathepsin-L (cysteine protease) (Zhou et al. 2015). Overall, the role of host proteases on the spike proteins is crucial for cell entry. Thus, inhibition of this priming process could be an effective way to regulate infectivity, emphasizing the potential of host proteases as drug targets against SARS-CoV-2. This was further verified by studies conducted with a serine protease inhibitor, camostat mesylate, which partially inhibited TMPRSS-2 and SARS-CoV-2 cell entry. The cell entry mechanism was fully inhibited by the addition of camostat mesylate together with E-64d, a cathepsin B/L inhibitor.
A previous study investigated the inhibitory activities of pseudotheonamide C and D and aeruginosin 98B isolated from the marine sponges Theonella swinhoei and Microcystis aeruginosa, respectively, on TPMRSS-2 (Nakao et al. 1999) (Ersmark et al. 2008). In addition, both compounds contain a guanidine group that mimics the arginine substrate of the enzyme (Buchanan et al. 2008). This result reveals an interesting factor in serine protease inhibition. Compounds that contain a guanidine group that can mimic the arginine substrate may be potential SARS-CoV-2 cell entry inhibitors. Another study revealed that gallinamide A, a selective inhibitor of human cathepsin-L isolated from marine cyanobacteria (Schizothrix spp.), can bind to the active site of this cysteine protease and block its activity (Miller et al. 2014). This would be useful for inhibiting SARS-CoV-2 cell entry.
Glycosaminoglycans (GAGs) on the cell surface inhibition by MNPs
GAGs are a linear polysaccharide family found on the cell surface that comprises repeating disaccharide units containing hexosamine, sulfated galactose residues, or uronic acid. GAGs are involved in several important biological processes, including pathogenesis, immunity, and cellular signaling (Lindahl et al.2015). According to a novel study, the binding kinetic results between both monomeric and trimetric spike proteins of SARS-CoV-2 and GAGs revealed that the GAG binding motif of spike proteins significantly binds with heparin sulfate (HS). According to the results, virions initially land on the surface of airway epithelial cells by binding to HS using a spike protein. The proteoglycans on the cell surface wrap the trimeric spike protein using their long HS chains. This leads to an interaction between the SARS-CoV-2 spike protein and ACE-2 (Kim et al. 2020). Therefore, inhibition of these processes strongly disturbs the entry of SARS-CoV-2 into the cell. The positive charge of the SARS-CoV-2 spike protein preferentially binds with long structures that are heavily sulfated (Kim et al. 2020). Fucoidan, a polysaccharide primarily obtained from brown seaweed, contains significant quantities of l-fucose and sulfate ester groups. These polysaccharides exhibit a vast range of biological activities, including anti-inflammation, immunomodulatory, and anti-oxidant activities (Jayawardena et al. 2022, 2020; Wang et al. 2020). Furthermore, these polysaccharides demonstrated remarkable antiviral potential against various envelope proteins, including herpes simplex, dengue, and respiratory syncytial viruses. The production cost, availability, and low toxicity of fucoidan are additional advantages. Fucose serves as the primary monomeric module in polymers, known as fucoidans. Monomeric monomers are linked together by either alpha-(1–2) or alpha-(1–3). Other potential sugar residues include galactose, mannose, xylose, and glucuronic acid. The polymer also contained acetyl groups. l-Fucopyranosyl residues often have the sulfate component substituted at the C2 or C4 and occasionally at the C3 position (Damonte et al. 2004). Thus, they have the potential to interfere with the interaction between spike protein and host cell receptors by mimicking GAGs. Several studies have verified the antiviral potential of sulfated polysaccharides against SARS-CoV-2 (Jin et al. 2020; Kwon et al. 2020; Song et al. 2020). This was further verified in a study conducted using a polysaccharide series isolated from Saccharina japonica. According to the results, sulfated galactofucan and glucuronomannan demonstrated significantly high inhibition, which directly interfered with the interaction between the spike protein and SARS-CoV-2 RBD (Jin et al. 2020). The specific role and significance of GAGs in SARS-CoV-2 cell entry make GAGs a potential drug target for SARS-CoV-2.2
Host cell translational mechanism inhibition by MNPs
Numerous host proteins play central roles in the SARS-CoV-2 life cycle. Some host proteins are essential for viral translation and replication. Viruses are entirely dependent on host translational mechanisms and cell pathways, which are essential for viral replication. Thus, these pathways are considered alternative antiviral approaches (Wong and Damania 2021). The binding of the 43S pre-initiation complex with the 5′ cap on mRNA initiates the translational process, and this complex begins the mRNA scanning from the start codon and recruits the 60S ribosomal subunit. This leads to ribosome assembly and initiates elongation. Viruses exploit host translational mechanisms by mimicking host mRNAs (Jaafar and Kieft 2019). Ribosomal entry of mRNA is blocked by NSP1 of SARS-CoV-2, which prevents its translation. Furthermore, a reporter gene that encodes the 5′ untranslated region (UTR) of SARS-CoV-2 mRNA exhibits a fivefold upregulation of gene expression compared to the host cell 5′ untranslated region (UTR) (Schubert et al. 2020). This revealed that SARS-CoV-2 hijacks the host translational process and depends on it for producing viral proteins. Plitidepsin, a chemical compound isolated from the sea squirt Aplidium albicans, which targets eukaryotic translation elongation factor 1a (eEF1A) of the host, has antiviral activity against SARS-CoV-2. Plitidepsin treatment in mice resulted in a significant reduction in viral titer in the lungs compared to remdesivir. Furthermore, the viral nucleocapsid protein expression was also significantly lower in plitidepsin-treated cells than in remdesivir-treated cells, despite the same amount of nucleocapsid sgRNA in both cells. This verified the inhibitory effect of plitidepsin on viral nucleocapsid protein translation (White et al. 2021). The safety profile of plitidepsin has also been verified in multiple cancer clinical trials (Wong and Damania. 2021). These findings suggest that translational inhibitors are promising antiviral drugs against SARS-COV-2.
Future perspectives to inhibit SARS-CoV-2 using MNPs
As mentioned under the “Cell entry mechanism” section, the SARS-CoV-2 cell entry mechanism depends on serine and cysteine proteases and requires spike protein cleavage by PPC. Furin activity and type of PPC are essential for SARS-CoV-2 cell entry, and the furin cleavage site does not exist in other β-coronavirus subtypes. Furin is present in the trans-Golgi network and is activated under acidic conditions. Precursor proteins with a specific PPC motif are cleaved and activated by furin. Sequence alignment of spike protein S1 and S2 sites among diverse coronaviruses demonstrates that the SARS-CoV-2 spike cleavage site contains a specific redundant amino acid sequence “PRRA” that is not expressed in other coronaviruses. This is a furin cleavage site. The higher infectivity of SARS-COV-2 compared to that of SARS-CoV and BAT-CoVRaTG13 is mediated by furin cleavage, and furin inhibition may significantly decrease infectivity (Devi et al. 2022). This hypothesis suggests that furin activity is predominant in the virus infection cycle and could be an effective drug target against SARS-CoV-2. According to previous studies, several furin inhibitors were discovered, such as α1-antitrypsin Portland inhibiting the HIV, D-Arg-based peptides (Anderson et al. 1993), and decanoyl-Arg-Val-Lys-Arg-chloromethylketone. Presently, pure peptides, peptide mimics, and non-peptide compounds are used as furin inhibitors. However, these peptides have numerous obstacles, including degradation by proteases, low stability without additional modifications, opsonization, and agglutination (Bruno et al. 2013). The chloromethyl ketone-derived molecule, dec-RVKR-cmk, is a vastly used furin inhibitor that inhibits catalytic site binding. Chemical modification elevates the inhibitory activity of these compounds, such as the C-terminal modification of dek-RVKS-cmk with dicarboxylated arginine mimetics (Imran et al. 2019). Recently published data revealed that a synthetic peptide mimetic named MI-1851 inhibits the use of furin to regulate SARS-CoV-2 infection. Furthermore, MI-1851 combined with the TMPRSS-2 inhibitor T-ex5 PPMO demonstrated remarkable SARS-CoV-2 inhibition (Bestle et al. 2020). Based on these results, several important factors regarding furin behavior were identified, such as the amino acid side chains in the active site that play a pivotal role in cleavage. An Arg residue in the P1 and P2 positions, at least two residues, either Lys or Arg in the P4 and P6 positions, and the P1 position should be free from hydrophobic or aliphatic amino acids. These facts provide insights into the design of inhibitors against furin, for example, α1-PDX/hf (Dufour et al. 2001; Jean et al. 1998). Marine organisms, including vertebrates and invertebrates, exhibit a significant diversity. The peptides or proteins isolated from them also demonstrated highly diverse biological activities depending on the organism and body part. Furthermore, peptides derived from marine organisms are more stable against gastrointestinal proteases than peptides from other sources (Pavlicevic et al. 2020). Thus, peptides isolated from marine organisms have elevated potential for use against SARS-CoV-2.
SARS-CoV-2 is associated with immune-mediated pathology via inappropriately regulated cytokine responses in the lungs and other tissues (Merad and Martin 2020). The activation mechanism and upstream signaling pathways of this hyperactive cytokine response are yet to be completely elucidated. This hyperinflammatory response is known as a cytokine storm and is responsible for severe complications and death. The MYD88 adaptor protein plays a crucial role in TLR-mediated downstream signaling pathways for inflammatory cytokines production (Fig. 3B) (Sariol and Perlman 2021). A novel study revealed the involvement of TLR-4 in this hyperinflammatory condition. Therefore, TLR-4 modulators can successfully control COVID-19 complications, revealing their potential as drug targets against SARS-CoV-2 (Kaushik et al. 2021). The anti-inflammatory activity of secondary metabolites isolated from marine organisms has been vastly studied (Mayer et al. 2011; Nagahawatta et al. 2022a, b, c). Many compounds and peptides isolated from marine organisms attenuate TLR-mediated NF-κB and MAPK signaling pathways, which inhibit the production of inflammatory mediators, such as iNOS and COX-2, and inflammatory cytokines, such as TNF-α, IL-6, and IL-1β (Gonzalez et al. 2013; Ko et al. 2016; Sanjeewa et al. 2020). As reported by recent studies, Eastern Asian countries that have commonly consumed seaweed-rich diets demonstrated fewer disasters caused by SARS-CoV-2 than Western countries (Pereira and Critchley 2020; Tamama 2020). Thus, compounds isolated from Phaeophyta, such as polyphenolic compounds, can be a great source for regulating the cytokine storm caused by SARS-CoV-2 infection.
SR-B1 is a high-density lipoprotein (HDL) receptor located on the cell surface that is responsible for the selective uptake of cholesterol esterase and other lipid components (Shen et al. 2018a, b). This transportation system can be found in isolated hepatocytes, adipocytes, fibroblasts, macrophages, ovarian cells, testicular Leydig cells, and adrenal cells (Shen et al. 2018a, b). Furthermore, SR-B1 is expressed in alveolar ΙΙ cells and is responsible for the uptake of vitamin E preferentially from HDL (Kaushik et al. 2021). According to a novel study, the SARS-CoV-2 spike protein comprises six putative amino acid consensus motifs responsible for cholesterol recognition. Furthermore, the results of this study revealed that SARS-CoV-2 S1 binds to cholesterol and interacts with HDL or its components, but SARS-CoV-2 S2 does not. One motif was present in the spike protein RBD. SR-B1 cannot directly bind to the SARS-CoV-2 spike protein, but HDL attachment to the spike protein and SARS-CoV-2 entry are both significantly enhanced by SR-B1 expression. Moreover, SR-B1 co-expression with ACE-2 significantly increases the susceptibility of cells to SARS-CoV-2. These findings revealed that SR-B1 facilitates cellular attachment of SARS-CoV-2. This observation further verified that SR-B1 silencing inhibits SARS-CoV-2 entry (Wei et al. 2020). SR-B1 acts as a cofactor for SARS-CoV-2 via a cell entry mechanism. This emphasizes the potential of SR-B1 as a therapeutic target for SARS-CoV-2 infection. As reported by Wen-Jun et al. (2018), some structural characteristics are crucial for its functionality. For typical receptor oligomerization and lipid transportation of SR-B1, the N-terminal transmembrane glycine (Gly) dimerization motif (Gly15, Gly18, and GLY25l) is essential (Shen et al. 2018a, b). Dante et al. provided a structural framework for this protein family through the crystal structure of LIMP-2 which is homologous to SR-B1 (Neculai et al. 2013). Nearly seven cysteine residues are conserved in mammals, including mice, hamsters, pigs, cows, and humans. Furthermore, four SR-B1 cysteines (Cys280, Cys321, Cys323, and Cys324) participate in binding to HDL (Shen et al. 2018a, b). These results provide insights into the development of inhibitors using MNPs through in silico evaluations.
Many researchers have discovered or are attempting to discover drug agents against diverse therapeutic targets for SARS-CoV-2. Herein, we considered the potential of a drug agent with a multi-target approach against different SARS-CoV-2 drug targets. If any drug agent has this multi-target potential, it will be able to inhibit SARS-CoV-2 by multiple approaches. Based on this hypothesis, we recently conducted a study that developed inhibitors against two drug targets, including 3CLpro and PLpro of SARS-CoV-2. In this study, authors evaluated 16 compounds isolated from marine seaweeds using molecular docking and conducted further evaluations. According to the study results, four polyphenolic compounds isolated from brown seaweeds were identified: ishophloroglucin A, diphlorethohydroxycarmalol, dieckol, and eckmaxol, which significantly inhibit 3CLpro and PLpro proteolytic activity and have the potential to develop into inhibitors with a multi-target approach against SARS-CoV-2 (Nagahawatta et al. 2022a, b, c, d). We are currently observing the inhibitory potential of these compounds against the interaction between SARS-CoV-2 ACE-2 and spike protein.
The results of these studies have revealed the potential of these compounds as therapeutic SARS-CoV-2 inhibitors. The natural marine compounds, structures, source of origin, and method of inhibition are summarized in Supplementary Table 1.
Conclusion
The coronavirus disease (COVID-19) pandemic has caused major global health concerns. Communities and scientists have a social and ethical responsibility to work collectively to defeat SARS-CoV-2. Vaccination is the major tool used to combat SARS-CoV-2 infection. Furthermore, several studies have been conducted to repurpose FDA-approved drugs. The discussion on the limitations of these attempts highlights the need for alternative therapeutic drugs against SARS-CoV-2. The authors described the structural and functional relationships in complex infections and replication of SARS-CoV-2 and revealed the potential of MNPs as inhibitors of diverse drug targets. Moreover, this analysis identified the potential of MNPs in developing promising therapeutic drugs against SARS-CoV-2 and offered extensive information to researchers for future COVID-19-related studies.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This research was part of the project “Development of functional food products with natural materials derived from marine resources (no. 20170285)”, funded by the Ministry of Oceans and Fisheries, Korea.
Author contributions
Conceptualization, DPN and JYJ; methodology, DPN; software, DPN; validation, DPN; formal analysis, DPN; investigation, DPN; resources, DPN AND JYJ; data collection, DPN, NML, TUJ, HHACKJ, and SHJ; writing—original draft preparation, DPN; writing—review and editing, DPN, JYJ, and HJK; supervision, JYJ; project administration, JYJ; funding acquisition, JYJ. All authors have read and agreed to the published version of the manuscript.
Data availability
The data that support the findings of this study are included in this published article (and its supplementary information files).
Declarations
Conflict of interest
The authors declare no conflict of interest. Author You-Jin Jeon is one of the Editorial Board Members, but he was not involved in the journal’s review of, or decision related to, this manuscript.
Animal and human rights statement
This article does not contain any studies with human participants or animals performed by the authors.
Contributor Information
Hyung-Jun Kwon, Email: hjkwon@kribb.re.kr.
You-Jin Jeon, Email: youjin2014@gmail.com.
References
- Aatif M, Muteeb G, Alsultan A, Alshoaibi A, Khelif BY. Dieckol and its derivatives as potential inhibitors of SARS-CoV-2 spike protein (UK strain: VUI 202012/01): a computational study. Mar Drugs. 2021;19:242. doi: 10.3390/md19050242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Anderson ED, Thomas L, Hayflick JS, Thomas G. Inhibition of HIV-1 gp160-dependent membrane fusion by a furin-directed alpha 1-antitrypsin variant. J Biol Chem. 1993;268:24887–24891. doi: 10.1016/S0021-9258(19)74548-7. [DOI] [PubMed] [Google Scholar]
- Anjum K, Abbas SQ, Shah SA, Akhter N, Batool S, Hassan SS. Marine sponges as a drug treasure. Biomol Ther (seoul) 2016;24:347–362. doi: 10.4062/biomolther.2016.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astuti I, Ysrafil Y. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diabetes Metab Syndr. 2020;14:407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baez-Santos YM, St John SE, Mesecar AD. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Békés M, Rut W, Kasperkiewicz P, Mulder MPC, Ovaa H, Drag M, Lima CD, Huang TT. SARS hCoV papain-like protease is a unique Lys48 linkage-specific di-distributive deubiquitinating enzyme. Biochem J. 2015;468:215–226. doi: 10.1042/BJ20141170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann W, Feeney RJ. Contributions to the study of marine products. Xxxii. The Nucleosides of Sponges. I.1. J Org Chem. 2002;16:981–987. doi: 10.1021/jo01146a023. [DOI] [Google Scholar]
- Berlin DA, Gulick RM, Martinez FJ. Severe covid-19. N Engl J Med. 2020;383:2451–2460. doi: 10.1056/NEJMcp2009575. [DOI] [PubMed] [Google Scholar]
- Bestle D, Heindl MR, Limburg H, Van Lam T, Pilgram O, Moulton H, Stein DA, Hardes K, Eickmann M, Dolnik O, Rohde C, Klenk H, Garten W, Steinmetzer T, Bottcher-Friebertshauser E. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance. 2020;3:e202000786. doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolles M, Donaldson E, Baric R. SARS-CoV and emergent coronaviruses: viral determinants of interspecies transmission. Curr Opin Virol. 2011;1:624–634. doi: 10.1016/j.coviro.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno BJ, Miller GD, Lim CS. Basics and recent advances in peptide and protein drug delivery. Ther Deliv. 2013;4:1443–1467. doi: 10.4155/tde.13.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan MS, Carroll AR, Wessling D, Jobling M, Avery VM, Davis RA, Feng Y, Xue Y, Öster L, Fex T, Deinum J, Hooper JNA, Quinn RJ. Clavatadine A, a natural product with selective recognition and irreversible inhibition of factor XIa. J Med Chem. 2008;51:3583–3587. doi: 10.1021/jm800314b. [DOI] [PubMed] [Google Scholar]
- Bugge TH, Antalis TM, Wu Q. Type II transmembrane serine proteases. J Biol Chem. 2009;284:23177–23181. doi: 10.1074/jbc.R109.021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzowka M, Mitusinska K, Raczynska A, Samol A, Tuszynski JA, Gora A. Structural and evolutionary analysis indicate that the SARS-CoV-2 Mpro is a challenging target for small-molecule inhibitor design. Int J Mol Sci. 2020;21:3099. doi: 10.3390/ijms21093099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, Ruan L, Song B, Cai Y, Wei M, Li X, Xia J, Chen N, Xiang J, Yu T, Bai T, Xie X, Zhang L, Li C, Yun Y, et al. A trial of lopinavir-ritonavir in adults hospitalized with severe covid-19. N Engl J Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damonte EB, Matulewicz MC, Cerezo AS. Sulfated seaweed polysaccharides as antiviral agents. Curr Med Chem. 2004;11:2399–2419. doi: 10.2174/0929867043364504. [DOI] [PubMed] [Google Scholar]
- Devi KP, Pourkarim MR, Thijssen M, Sureda A, Khayatkashani M, Cismaru CA, Neagoe IB, Habtemariam S, Razmjouei S, Khayat Kashani HR. A perspective on the applications of furin inhibitors for the treatment of SARS-CoV-2. Pharmacol Rep. 2022;74:425–430. doi: 10.1007/s43440-021-00344-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson SH, Hirsh A, Li DC, Holloway G, Chao J, Boucher RC, Gabriel SE. Regulation of the epithelial sodium channel by serine proteases in human airways. J Biol Chem. 2002;277:8338–8345. doi: 10.1074/jbc.M105044200. [DOI] [PubMed] [Google Scholar]
- Donia M, Hamann MT. Marine natural products and their potential applications as anti-infective agents. Lancet Infect Dis. 2003;3:338–348. doi: 10.1016/S1473-3099(03)00655-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du QS, Wang SQ, Zhu Y, Wei DQ, Guo H, Sirois S, Chou KC. Polyprotein cleavage mechanism of SARS CoV Mpro and chemical modification of the octapeptide. Peptides. 2004;25:1857–1864. doi: 10.1016/j.peptides.2004.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour EK, Denault JB, Bissonnette L, Hopkins PC, Lavigne P, Leduc R. The contribution of arginine residues within the P6–P1 region of alpha 1-antitrypsin to its reaction with furin. J Biol Chem. 2001;276:38971–38979. doi: 10.1074/jbc.M102959200. [DOI] [PubMed] [Google Scholar]
- Dzimianski JV, Scholte FEM, Bergeron E, Pegan SD. ISG15: It's complicated. J Mol Biol. 2019;431:4203–4216. doi: 10.1016/j.jmb.2019.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EA JA, Jones IM. Membrane binding proteins of coronaviruses. Future Virol. 2019;14:275–286. doi: 10.2217/fvl-2018-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersmark K, Del Valle JR, Hanessian S. Chemistry and biology of the aeruginosin family of serine protease inhibitors. Angew Chem Int Ed Engl. 2008;47:1202–1223. doi: 10.1002/anie.200605219. [DOI] [PubMed] [Google Scholar]
- Ferrer-Orta C, Arias A, Escarmis C, Verdaguer N. A comparison of viral RNA-dependent RNA polymerases. Curr Opin Struct Biol. 2006;16:27–34. doi: 10.1016/j.sbi.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Finkel Y, Mizrahi O, Nachshon A, Weingarten-Gabbay S, Morgenstern D, Yahalom-Ronen Y, Tamir H, Achdout H, Stein D, Israeli O, Beth-Din A, Melamed S, Weiss S, Israely T, Paran N, Schwartz M, Stern-Ginossar N. The coding capacity of SARS-CoV-2. Nature. 2021;589:125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- Firdaus M, Nurdiani R, Artasasta IN, Mutoharoh S, Pratiwi O. Potency of three brown seaweeds species as the inhibitor of RNA-dependent RNA polymerase of SARS-CoV-2. Rev Chim. 2020;71:80–86. doi: 10.37358/RC.20.11.8376. [DOI] [Google Scholar]
- Frieman M, Baric R. Mechanisms of severe acute respiratory syndrome pathogenesis and innate immunomodulation. Microbiol Mol Biol Rev. 2008;72:672–685. doi: 10.1128/MMBR.00015-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Huang B, Tang J, Liu S, Liu M, Ye Y, Liu Z, Xiong Y, Zhu W, Cao D, Li J, Niu X, Zhou H, Zhao YJ, Zhang G, Huang H. The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery. Nat Commun. 2021;12:488. doi: 10.1038/s41467-020-20718-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, Wang T, Sun Q, Ming Z, Zang L, Ge J, Zheng L, Zhang Y, Wang H, Zhu Y, Zhu C, Hu T, Hua T, Zhang B, Yang X, et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautret P, Lagier JC, Parola P, Hoang VT, Meddeb L, Mailhe M, Doudier B, Courjon J, Giordanengo V, Viera VE, Dupont HT, Honoré S, Colson P, Chabrière E, Scola B, Rolanin J, Brouqui P, Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020;56:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gentile D, Patamia V, Scala A, Sciortino MT, Piperno A, Rescifina A. Putative inhibitors of SARS-CoV-2 main protease from a library of marine natural products: a virtual screening and molecular modeling study. Mar Drugs. 2020;18:225. doi: 10.3390/md18040225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez Y, Doens D, Santamaria R, Ramos M, Restrepo CM, Barros de Arruda L, Lleonart R, Gutiérrez M, Fernandez PL. A pseudopterane diterpene isolated from the octocoral Pseudopterogorgia acerosa inhibits the inflammatory response mediated by TLR-ligands and TNF-alpha in macrophages. PLoS ONE. 2013;8:e84107. doi: 10.1371/journal.pone.0084107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RL, Baric RS. SARS-CoV-2: combating coronavirus emergence. Immunity. 2020;52:734–736. doi: 10.1016/j.immuni.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M, Song W, Zhou H, Xu J, Chen S, Xiang Y, Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasoksuz M, Sreevatsan S, Cho KO, Hoet AE, Saif LJ. Molecular analysis of the S1 subunit of the spike glycoprotein of respiratory and enteric bovine coronavirus isolates. Virus Res. 2002;84:101–109. doi: 10.1016/S0168-1702(02)00004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald-Sargent T, Gallagher T. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses. 2012;4:557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton SM, Borg NA, Dixit VM. Ubiquitin in the activation and attenuation of innate antiviral immunity. J Exp Med. 2016;213:1–13. doi: 10.1084/jem.20151531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrier G, Wu NH, Nitsche A, Muller MA, Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H, Hattermann K, Marzi A, Gramberg T, Geier M, Krumbiegel M, Kuate S, Uberia K, Niedrig M, Pohlmann S. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J Virol. 2004;78:6134–6142. doi: 10.1128/JVI.78.12.6134-6142.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung IF, Lung KC, Tso EY, Liu R, Chung TW, Chu MY, Ng YY, Lo J, Chan J, Tan AR, Shum HP, Chan V, Ku AK, Sin KM, Leung WS, Law WL, Lung DC, Sin S, Yeung P, Yip CC, et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imran M, Saleemi MK, Chen Z, Wang XG, Zhou DY, Li YC, Zhao Z, Zheng B, Li Q, Cao S, Ye J. Decanoyl-Arg-Val-Lys-Arg-Chloromethylketone: an antiviral compound that acts against flaviviruses through the inhibition of furin-mediated prM cleavage. Viruses-Basel. 2019;11:1011. doi: 10.3390/v11111011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland CM, Copp BR, Foster MP, McDonald LA, Radisky DC, Swersey JC. Bioactive compounds from the sea Marine and freshwater products handbook. Boca Raton: CRC Press; 2000. pp. 641–661. [Google Scholar]
- Iwata-Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol. 2019;93:e01815–01818. doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaafar ZA, Kieft JS. Viral RNA structure-based strategies to manipulate translation. Nat Rev Microbiol. 2019;17:110–123. doi: 10.1038/s41579-018-0117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardena TU, Sanjeewa KKA, Nagahawatta DP, Lee HG, Lu YA, Vaas APJP, Abeytunga DTU, Nanayakkara CM, Lee DS, Jeon YJ. Anti-inflammatory effects of sulfated polysaccharide from Sargassum swartzii in macrophages via blocking TLR/NF-Κb signal transduction. Mar Drugs. 2020;18:601. doi: 10.3390/md18120601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardena TU, Nagahawatta DP, Fernando IPS, Kim YT, Kim JS, Kim WS, Lee JS, Jeon YJ. A review on fucoidan structure, extraction techniques, and its role as an immunomodulatory agent. Mar Drugs. 2022;20:755. doi: 10.3390/md20120755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean F, Stella K, Thomas L, Liu G, Xiang Y, Reason AJ, Thomas G. alpha1-Antitrypsin Portland, a bioengineered serpin highly selective for furin: application as an antipathogenic agent. Proc Natl Acad Sci USA. 1998;95:7293–7298. doi: 10.1073/pnas.95.13.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Zhang W, Mitra D, McCandless MG, Sharma P, Tandon R, Zhang F, Linhardt RJ. The structure-activity relationship of the interactions of SARS-CoV-2 spike glycoproteins with glucuronomannan and sulfated galactofucan from Saccharina japonica. Int J Biol Macromol. 2020;163:1649–1658. doi: 10.1016/j.ijbiomac.2020.09.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik D, Bhandari R, Kuhad A. TLR4 as a therapeutic target for respiratory and neurological complications of SARS-CoV-2. Expert Opin Ther Targets. 2021;25:491–508. doi: 10.1080/14728222.2021.1918103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MT, Ali A, Wang Q, Irfan M, Khan A, Zeb MT, Zhang YJ, Chinnasamy S, Wei DQ. Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2-a molecular dynamic study. J Biomol Struct Dyn. 2021;39:3627–3637. doi: 10.1080/07391102.2020.1769733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TS, Heinlein C, Hackman RC, Nelson PS. Phenotypic analysis of mice lacking the Tmprss2-encoded protease. Mol Cell Biol. 2006;26:965–975. doi: 10.1128/MCB.26.3.965-975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Jin W, Sood A, Montgomery DW, Grant OC, Fuster MM, Fu L, Dordick JS, Woods R, Zhang F, Linhardt RJ. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antivir Res. 2020;181:104873. doi: 10.1016/j.antiviral.2020.104873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm T, Ebert G, Calleja DJ, Allison CC, Richardson LW, Bernardini JP, Lu BG, Kuchei NW, Grohmann C, Shibata Y, Gan ZY, Cooney JP, Doerflinger M, Au AE, Blackmore TR, Noort GJ, Geurink PP, Ovaa H, Newman J, Tunnicliffe AR, et al. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020;39:e106275. doi: 10.15252/embj.2020106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko W, Sohn JH, Jang JH, Ahn JS, Kang DG, Lee HS, Kim JS, Kim YC, Oh H. Inhibitory effects of alternaramide on inflammatory mediator expression through TLR4-MyD88-mediated inhibition of NF-small ka, CyrillicB and MAPK pathway signaling in lipopolysaccharide-stimulated RAW264.7 and BV2 cells. Chem Biol Interact. 2016;244:16–26. doi: 10.1016/j.cbi.2015.11.024. [DOI] [PubMed] [Google Scholar]
- König GM, Wright AD, Sticher O, Angerhofer CK, Pezzuto JMJPM. Biological activities of selected marine natural products. Planta Med. 1994;60:532–537. doi: 10.1055/s-2006-959565. [DOI] [PubMed] [Google Scholar]
- Kumar V, Parate S, Yoon S, Lee G, Lee KW. Computational simulations identified marine-derived natural bioactive compounds as replication inhibitors of SARS-CoV-2. Front Microbiol. 2021;12:647295–647295. doi: 10.3389/fmicb.2021.647295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupferschmidt K, Cohen J. Race to find COVID-19 treatments accelerates. Science. 2020;367:1412–1413. doi: 10.1126/science.367.6485.1412. [DOI] [PubMed] [Google Scholar]
- Kwon PS, Oh H, Kwon SJ, Jin W, Zhang F, Fraser K, Hong JJ, Linhard RJ, Dordick JS. Sulfated polysaccharides effectively inhibit SARS-CoV-2 in vitro. Cell Discov. 2020;6:50. doi: 10.1038/s41421-020-00192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Receptor recognition and cross-species infections of SARS coronavirus. Antivir Res. 2013;100:246–254. doi: 10.1016/j.antiviral.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol. 2015;89:1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Lim L, Shi J, Mu Y, Song JJPO. Dynamically-driven enhancement of the catalytic machinery of the SARS 3C-like protease by the S284–T285-I286/A mutations on the extra domain. PLoS ONE. 2014;9:e101941. doi: 10.1371/journal.pone.0101941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl U, Li JP. Heparin - an old drug with multiple potential targets in Covid-19 therapy. J Thromb Haemost. 2020;18:2422–2424. doi: 10.1111/jth.14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl U, Couchman J, Kimata K, Esko JD. Proteoglycans and sulfated glycosaminoglycans. 3. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2015. [PubMed] [Google Scholar]
- Lu R, Wang Y, Wang W, Nie K, Deng Y, Zhou W, Li Y, Wang H, Wang W, Ke C, Ma X, Wu G, Tan W. Complete genome sequence of Middle East Respiratory Syndrome Coronavirus (MERS-CoV) from the first imported MERS-CoV Case in China. Genome Announc. 2015;3:e00818–e1815. doi: 10.1128/genomeA.00818-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer AMS, Rodríguez AD, Berlinck RGS, Fusetani N. Marine pharmacology in 2007–8: marine compounds with antibacterial, anticoagulant, antifungal, anti-inflammatory, antimalarial, antiprotozoal, antituberculosis, and antiviral activities; affecting the immune and nervous system, and other miscellaneous mechanisms of action. Comp Biochem Physiol Part C Toxicol Appl Pharmacol. 2011;153:191–222. doi: 10.1016/j.cbpc.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller B, Friedman AJ, Choi H, Hogan J, McCammon JA, HookV GWH. The marine cyanobacterial metabolite gallinamide A is a potent and selective inhibitor of human cathepsin L. J Nat Prod. 2014;77:92–99. doi: 10.1021/np400727r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse JS, Lalonde T, Xu S, Liu WR. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiocehm. 2020;21:730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller C, Schulte FW, Lange-Grunweller K, Obermann W, Madhugiri R, Pleschka S, Ziebuhr J, Hartmann RK, Grunweller A. Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona- and picornaviruses. Antivir Res. 2018;150:123–129. doi: 10.1016/j.antiviral.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahawatta DP, Kim HS, Jee Y-H, Jayawardena TU, Ahn G, Namgung J, Yeo I, Sanjeewa KKA, Jeon YJ. Sargachromenol isolated from Sargassum horneri inhibits particulate matter-induced inflammation in macrophages through toll-like receptor-mediated cell signaling pathways. Mar Drugs. 2022;20:28. doi: 10.3390/md20010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahawatta DP, Liyanage NM, Jayawardhana HHACK, Jayawardena TU, Lee HG, Heo MS, Jeon YJ. Eckmaxol isolated from Ecklonia maxima attenuates particulate-matter-induced inflammation in MH-S lung macrophage. Mar Drugs. 2022;20:766. doi: 10.3390/md20120766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahawatta DP, Liyanage NM, Jayawardhana HHACK, Lee HG, Jayawardena TU, Jeon YJ. Anti-fine dust effect of fucoidan extracted from Ecklonia maxima laves in macrophages via inhibiting inflammatory signaling pathways. Mar Drugs. 2022;20:413. doi: 10.3390/md20070413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahawatta DP, Liyanage NM, Je JG, Jayawardhana HHACK, Jayawardena TU, Jeong SH, Kwon HJ, Choi CS, Jeon YJ. Polyphenolic compounds isolated from marine algae attenuate the replication of SARS-CoV-2 in the host cell through a multi-target approach of 3CLpro and PLpro. Mar Drugs. 2022;20:786. doi: 10.3390/md20120786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao Y, Masuda A, Matsunaga S, Fusetani N. Pseudotheonamides, serine protease inhibitors from the marine sponge Theonella swinhoei 1. J Am Chem Soc. 1999;121:2425–2431. doi: 10.1021/ja9831195. [DOI] [Google Scholar]
- Naqvi AAT, Fatima K, Mohammad T, Fatima U, Singh IK, Singh A, Atif SM, Hariprasad G, Hasan GM, Hassan MI. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165878. doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neculai D, Schwake M, Ravichandran M, Zunke F, Collins RF, Peters J, Neculai M, Plumb J, Loppnau P, Pizarro JC, Seitova A, Trimble WS, Saftig P, Grinstein S, Dhe-Paganon S. Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36. Nature. 2013;504:172–176. doi: 10.1038/nature12684. [DOI] [PubMed] [Google Scholar]
- Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, Connelly S, Droese B, Klaus JP, Makino S, Sawicki SG, Siddell SG, Stamou DG, Wilson IA, Kuhun P, Buchmeier MJ. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden KM, Ramanathan HN, Patton JT. Mutational analysis of residues involved in nucleotide and divalent cation stabilization in the rotavirus RNA-dependent RNA polymerase catalytic pocket. Virology. 2012;431:12–20. doi: 10.1016/j.virol.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, Xiang Z, Mu Z, Chen X, Chen J, Hu K, Jin Q, Wang J, Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Kim JH, Kwon JM, Kwon HJ, Jeong HJ, Kim YM, Kim D, Lee WS, Ryu YB. Dieckol, a SARS-CoV 3CL(pro) inhibitor, isolated from the edible brown algae Ecklonia cava. Bioorg Med Chem. 2013;21:3730–3737. doi: 10.1016/j.bmc.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlicevic M, Maestri E, Marmiroli M. Marine bioactive peptides-an overview of generation, structure and application with a focus on food sources. Mar Drugs. 2020;18:424. doi: 10.3390/md18080424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Critchley AT. The COVID 19 novel coronavirus pandemic 2020: seaweeds to the rescue? Why does substantial, supporting research about the antiviral properties of seaweed polysaccharides seem to go unrecognized by the pharmaceutical community in these desperate times? J Appl Phycol. 2020;32:1875–1877. doi: 10.1007/s10811-020-02143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar T, Manickam M, Namasivayam V, Hayashi Y, Jung SH. An overview of Severe Acute Respiratory Syndrome-Coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J Med Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomponi SA. The bioprocess-technological potential of the sea. In: Osinga R, Tramper J, Burgess JG, Wijffels RH, editors. Progress in industrial microbiology. Amsterdam: Elsevier; 1999. pp. 5–13. [Google Scholar]
- Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Penaranda S, Bankamp B, Maher K, Chen MH, Tong S, Tamin A, Lowe L, Frace M, Derisi JL, Chen Q, Wang D, Erdman DD, Peret TC, Burns C, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Sanjeewa KKA, Kim EA, Son KT, Jeon YJ. Bioactive properties and potentials cosmeceutical applications of phlorotannins isolated from brown seaweeds: a review. J Photochem Photobiol B Biol. 2016;162:100–105. doi: 10.1016/j.jphotobiol.2016.06.027. [DOI] [PubMed] [Google Scholar]
- Sanjeewa KKA, Nagahawatta DP, Yang HW, Oh JY, Jayawardena TU, Jeon YJ, Zoysa M, Whang I, Ryu B. Octominin inhibits LPS-induced chemokine and pro-inflammatory cytokine secretion from RAW 264.7 macrophages via blocking TLRs/NF-κB signal transduction. Biomolecules. 2020;10:511. doi: 10.3390/biom10040511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariol A, Perlman S. SARS-CoV-2 takes its toll. Nat Immunol. 2021;22:801–802. doi: 10.1038/s41590-021-00962-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija N, Lal SK. The molecular biology of SARS coronavirus. Ann N Y Acad Sci. 2007;1102:26–38. doi: 10.1196/annals.1408.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler LA, Leibundgut M, Thiel V, Mühlemann O, Ban N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol. 2020;27:959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- Seley-Radtke KL, Yates MK. The evolution of nucleoside analogue antivirals: a review for chemists and non-chemists. Part 1: Early structural modifications to the nucleoside scaffold. Antivir Res. 2018;154:66–86. doi: 10.1016/j.antiviral.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerabach A, Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WJ, Asthana S, Kraemer FB, Azhar SJJOLR. Scavenger receptor B type 1: expression, molecular regulation, and cholesterol transport function. JLR. 2018;59:1114–1131. doi: 10.1194/jlr.R083121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WJ, Azhar S, Kraemer FB. SR-B1: a unique multifunctional receptor for cholesterol influx and efflux. Annu Rev Physiol. 2018;80:95–116. doi: 10.1146/annurev-physiol-021317-121550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde P, Banerjee P, Mandhare A. Marine natural products as source of new drugs: a patent review (2015–2018) Expert Opin Ther Pat. 2019;29:283–309. doi: 10.1080/13543776.2019.1598972. [DOI] [PubMed] [Google Scholar]
- Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I, Almazán F, Zúñiga S, Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Peng H, Wang Q, Liu Z, Dong X, Wen C, Ai C, Zhang Y, Wang Z, Zhu B. Inhibitory activities of marine sulfated polysaccharides against SARS-CoV-2. Food Funct. 2020;11:7415–7420. doi: 10.1039/D0FO02017F. [DOI] [PubMed] [Google Scholar]
- Speake H, Phillips A, Chong T, Sikazwe C, Levy A, Lang J, Scalley B, Speers DJ, Smith DW, Effler P, McEvoy SP. Flight-associated transmission of Severe Acute Respiratory Syndrome Coronavirus 2 corroborated by whole-genome sequencing. Emerg Infect Dis. 2020;26:2872–2880. doi: 10.3201/eid2612.203910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- Surti M, Patel M, Adnan M, Moin A, Ashraf SA, Siddiqui AJ, Snoussi M, Deshpande S, Reddy MNJRA. Ilimaquinone (marine sponge metabolite) as a novel inhibitor of SARS-CoV-2 key target proteins in comparison with suggested COVID-19 drugs: designing, docking and molecular dynamics simulation study. RSC Adv. 2020;10:37707–37720. doi: 10.1039/D0RA06379G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamama K. Potential benefits of dietary seaweeds as protection against COVID-19. Nutr Rev. 2020;79:814–823. doi: 10.1093/nutrit/nuaa126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Deng Z, Li X, Yang M, Tian Z, Chen Z, Wang G, Wu W, Feng WH, Zhang G, Chen Z. Helicase of type 2 porcine reproductive and respiratory syndrome virus strain HV reveals a unique structure. Viruses. 2020;12:215. doi: 10.3390/v12020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therapy REoC (2021) Recovery trial closes recruitment to colchicine treatment for patients hospitalised with COVID-19. https://www.recoverytrial.net/news/recovery-trial-closes-recruitment-to-colchicine-treatment-for-patients-hospitalised-with-covid-19
- Tong TR. Drug targets in severe acute respiratory syndrome (SARS) virus and other coronavirus infections. Infect Disord Drug Targets. 2009;9:223–245. doi: 10.2174/187152609787847659. [DOI] [PubMed] [Google Scholar]
- Ullrich S, Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorg Med Chem Lett. 2020;30:127377. doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaarala MH, Porvari KS, Kellokumpu S, Kyllönen AP, Vihko PT. Expression of transmembrane serine protease TMPRSS2 in mouse and human tissues. J Pathol. 2001;193:134–140. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH743>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Venkataraman S, Prasad BVLS, Selvarajan R. RNA dependent RNA polymerases: Insights from structure, function and evolution. Viruses. 2018;10:76. doi: 10.3390/v10020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Xiong X, Park YJ, Tortorici MA, Snijder J, Quispe J, Cameroni E, Gopal R, Dai M, Lazavecchia A, Zambon M, Rey FA, Veesler D. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–1039. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SH, Huang CY, Chen CY, Chang CC, Huang CY, Dong CD, Chang JS. Structure and biological activity analysis of fucoidan isolated from Sargassum siliquosum. ACS Omega. 2020;5:32447–32455. doi: 10.1021/acsomega.0c04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Wan L, Yan Q, Wang X, Zhang J, Yang X, Zhang Y, Fan C, Li D, Deng Y, Sun J, Gong J, Yang X, Wang Y, Wang X, Li J, Yang H, Li H, Zhang Z, Wang R, et al. HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat Metab. 2020;2:1391–1400. doi: 10.1038/s42255-020-00324-0. [DOI] [PubMed] [Google Scholar]
- White KM, Rosales R, Yildiz S, Kehrer T, Miorin L, Moreno E, Jangra S, Uccellini MB, Rathnesinghe R, Coughlan L, Romareo C, Batra J, Rojc A, Bouhaddou M, Fabius JM, Dejosez MD, Guillen MJ, Losada A, Aviles P, Schotsaert M, et al. Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A. Science. 2021;371:926–931. doi: 10.1126/science.abf4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JP, Damania B. SARS-CoV-2 dependence on host pathways. Science. 2021;371:884–885. doi: 10.1126/science.abg6837. [DOI] [PubMed] [Google Scholar]
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Peng C, Shi Y, Zhu Z, Mu K, Wang X, Zhu W (2020) Nelfinavir was predicted to be a potential inhibitor of 2019-nCov main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation. BioRxiv 2020.2001.2027.921627
- Xue B, Blocquel D, Habchi J, Uversky AV, Kurgan L, Uversky VN, Longhi S. Structural disorder in viral proteins. Chem Rev. 2014;114:6880–6911. doi: 10.1021/cr4005692. [DOI] [PubMed] [Google Scholar]
- Yasuhara-Bell J, Lu Y. Marine compounds and their antiviral activities. Antivir Res. 2010;86:231–240. doi: 10.1016/j.antiviral.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol. 2016;18:579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Cao D, Zhang Y, Ma J, Qi J, Wang Q, Lu G, Wu Y, Yan J, Shi Y, Zhang X, Gao GF. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Vedantham P, Lu K, Agudelo J, Carrion R, Jr, Nunneley JW, Barnard D, Pöhlmann S, McKerrow JH, Reslo AR, Simmons G. Protease inhibitors targeting coronavirus and filovirus entry. Antivir Res. 2015;116:76–84. doi: 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Yan Z, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jianh RD, Liu MQ, Chen Y, Shen XR, Wang X, Shuang X, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are included in this published article (and its supplementary information files).


