Summary
Background
Gastrointestinal stromal tumors (GISTs) represent the most prevalent type of subepithelial lesions (SELs) with malignant potential. Current imaging tools struggle to differentiate GISTs from leiomyomas. This study aimed to create and assess a real-time artificial intelligence (AI) system using endoscopic ultrasonography (EUS) images to differentiate between GISTs and leiomyomas.
Methods
The AI system underwent development and evaluation using EUS images from 5 endoscopic centers in China between January 2020 and August 2023. EUS images of 1101 participants with SELs were retrospectively collected for AI system development. A cohort of 241 participants with SELs was recruited for external AI system evaluation. Another cohort of 59 participants with SELs was prospectively enrolled to assess the real-time clinical application of the AI system. The AI system's performance was compared to that of endoscopists. This study is registered with Chictr.org.cn, Number ChiCT2000035787.
Findings
The AI system displayed an area under the curve (AUC) of 0.948 (95% CI: 0.921–0.969) for discriminating GISTs and leiomyomas. The AI system's accuracy (ACC), sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) reached 91.7% (95% CI 87.5%–94.6%), 90.3% (95% CI 83.4%–94.5%), 93.0% (95% CI 87.2%–96.3%), 91.9% (95% CI 85.3%–95.7%), and 91.5% (95% CI 85.5%–95.2%), respectively. Moreover, the AI system exhibited excellent performance in diagnosing ≤20 mm SELs (ACC 93.5%, 95% CI 0.900–0.969). In a prospective real-time clinical application trial, the AI system achieved an AUC of 0.865 (95% CI 0.764–0.966) and 0.864 (95% CI 0.762–0.966) for GISTs and leiomyomas diagnosis, respectively, markedly surpassing endoscopists [AUC 0.698 (95% CI 0.562–0.834) for GISTs and AUC 0.695 (95% CI 0.546–0.825) for leiomyomas].
Interpretation
We successfully developed a real-time AI-assisted EUS diagnostic system. The incorporation of the real-time AI system during EUS examinations can assist endoscopists in rapidly and accurately differentiating various types of SELs in clinical practice, facilitating improved diagnostic and therapeutic decision-making.
Funding
Science and Technology Commission Foundation of Shanghai Municipality, Science and Technology Commission Foundation of the Xuhui District, the Interdisciplinary Program of Shanghai Jiao Tong University and the Research Funds of Shanghai Sixth people’s Hospital.
Keywords: Gastrointestinal subepithelial lesions, Gastrointestinal stromal tumors, Leiomyomas, Endoscopic ultrasound, Artificial intelligence
Research in context.
Evidence before this study
We conducted a PubMed search up to August 31, 2023, for research articles containing the terms of “(artificial intelligence OR deep learning OR machine learning OR convolutional neural network) AND “(endoscopic OR endoscopy)” AND “(ultrasound OR endosonography OR echo-endosonography)” AND “(gastrointestinal subepithelial tumor OR subepithelial tumor OR subepithelial lesion OR stromal tumor OR GIST)”, without any date or language restrictions. We found previous studies focused on developing screening systems for gastrointestinal SELs using EUS images, but these primarily restricted to classifying of lesions or determining the malignant potential of GIST. There was no real-time AI-aid EUS diagnostic system that could be directly applied in clinical setting.
Added value of this study
Here, we successfully developed a real-time AI-aid EUS diagnostic system, which can automate the identification, localization and diagnosis of SELs in clinical setting. To the best of our knowledge, this is the first study reporting the application of a real-time AI-aid EUS diagnostic system with excellent performance in clinical setting. Inclusion of AI system during EUS procedures significantly enhanced the diagnostic efficiency, especially for small SELs.
Implications of all the available evidence
The incorporation of the real-time AI system during EUS examinations can assist endoscopists in rapidly and accurately differentiating various types of SELs in clinical practice, facilitating improved diagnostic and therapeutic decision-making.
Introduction
Subepithelial lesions (SELs) are commonly encountered in routine gastrointestinal tract screening endoscopy, occurring in approximately 0.8–3.1% of patients.1, 2, 3 Most SELs, typically measuring <20 mm in diameter, are incidentally discovered.3,4 Gastrointestinal stromal tumors (GISTs) are the predominant type of gastrointestinal SELs and are deemed potentially malignant in biological behavior.4,5 Small GISTs, with a diameter <2 cm, constitute a subset, and their reported incidence ranges from 35% to 40.6%.6,7 While small GISTs are often asymptomatic and clinically benign, those displaying high-risk features may manifest malignant behaviors such as rapid progression or metastasis.8,9 Consequently, surgical or endoscopic resection, or long-term endoscopic monitoring, is recommended.10,11
Endoscopic ultrasonography (EUS) presently stands as the most effective imaging modality for SELs evaluation, relying on echogenic features, shape, origin layer, and size.4,12 However, distinguishing GISTs from other SELs, particularly leiomyomas, remains challenging for endoscopists due to their similar EUS image features. Furthermore, the subjective interpretation of EUS image features, reliant on endoscopists' experience, results in insufficient accuracy for human diagnosis, ranging from 45.5% to 66.7%.13,14 Consequently, histopathology remains the “gold standard” for accurate diagnosis. Despite the use of EUS-guided fine-needle biopsy (EUS-FNB) or mucosal cutting biopsy for confirmation, diagnostic accuracy remains low, especially for small SELs.15,16 Moreover, the invasive nature of these procedures, associated risks of bleeding and peritoneal seeding, along with varying global availability and increased medical expenses, hinder their clinical application.15,17 Hence, there is an urgent need for a more reliable and non-invasive diagnostic method to distinguish GISTs from leiomyomas.
Recently, AI-based image recognition has shown promise in endoscopic diagnosis of gastrointestinal mucosal lesions and SELs.18,19 AI-assisted EUS diagnostic models have demonstrated promising results.20, 21, 22, 23 However, these studies were limited by small case numbers, single-institution involvement, retrospective designs, or the absence of real-time application in clinical scenarios.
Therefore, this study aims to develop a real-time AI-assisted EUS diagnostic system for distinguishing GISTs from leiomyomas and to assess the utility and accuracy of the AI system in a clinical setting.
Methods
Ethics statement
The study follows the Standards for Reporting of Diagnostic Accuracy (STARD) reporting guideline and was approved by the ethics committees of all participating hospitals in China (Shanghai JiaoTong University Affiliated Sixth People's Hospital, Shanghai General People's Hospital, Ruijin Hospital, Huadong Hospital, and Shanghai East Hospital) (No. 2020-146, dated August 25, 2020). The research adhered to the principles outlined in the Declaration of Helsinki. Informed consent was obtained from all participants for prospective clinical trial. Registration for this study was completed at the Chinese Clinical Trial Registry (Registration number: ChiCTR2000035787).
Participants and study design
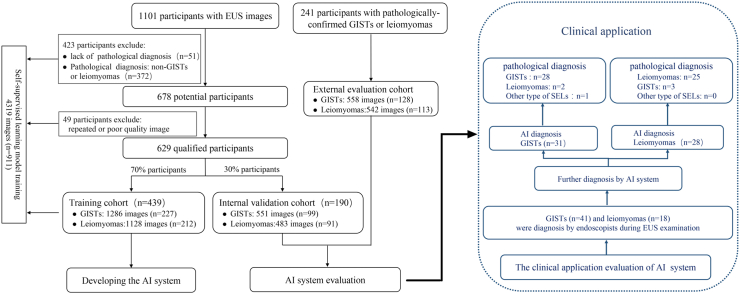
3 cohorts of participants with SELs were collected for development and testing (including the AI system development cohort, the external evaluation cohort and the prospective clinical evaluation cohort) of the AI system. The study flowchart is presented in Fig. 1.
Fig. 1.
Flow chart depicting the study process. The illustration outlines participant enrollment in the AI model development cohort and clinical application.
In the AI system development cohort, participants with gastrointestinal SELs were retrospectively identified from the aforementioned five tertiary hospitals in China, spanning January 2013 to December 2020. The inclusion criteria for SELs patients were: (1) age between 18 and 75 years; (2) prior completion of an EUS examination with comprehensive EUS image data; (3) availability of complete clinical data. Ultimately, 1101 participants with SELs were enrolled, and all pertinent medical information was digitally recorded. Among these participants, 629 individuals met the inclusion criteria, with pathologically confirmed diagnoses of GISTs or leiomyomas. They were randomly assigned to the training cohort and the internal validation cohort in a 7:3 ratio. The training cohort comprised 439 patients (1128 EUS images from 212 patients with leiomyomas and 1286 EUS images from 227 patients with GISTs). The internal validation cohort included 190 patients (483 EUS images from 91 leiomyoma cases and 551 EUS images from 99 GISTs). Image data from 472 participants lacking confirmed pathological diagnoses, having pathological diagnoses as other types of SELs, or possessing poor EUS images (poor echogenic quality, blurred, or artifacts) were used for self-supervised learning model training, along with data from 439 participants in the training cohort. In total, 5419 EUS images from 1101 subjects were utilized for the development of the AI model.
In the external evaluation cohort, an additional cohort of 241 participants with SELs was recruited from five centers between January 2021 and December 2022. Inclusion criteria were as follows: (1) age between 18 and 75; (2) complete EUS image data; (3) confirmed pathological diagnosis of GISTs or leiomyomas; (4) complete clinical data. The exclusion criteria were as follows: (1) the clinical data and histopathological data are incomplete; (2) poor echogenic quality couldn't meet the data processing requirement. The external evaluation cohort comprised 1100 EUS images (542 EUS images from 113 leiomyoma cases and 558 EUS images from 128 GIST cases). This cohort was also utilized to compare the performance of the AI model with that of endoscopists. Six endoscopists, comprising three senior endoscopists (with 5–10 years of experience in EUS) and three junior endoscopists (with less than 5 years of experience in EUS), independently evaluated EUS images from each case within this cohort. They were informed that each case was pathologically confirmed to have either GIST or leiomyoma, however, they were blinded to the information of the pathological results. The endoscopists were required to classify each case as either GIST or leiomyoma after assessment of all anonymized EUS images. Regardless of their expertise, all endoscopists was trained using EUS images obtained from 10 patients with GIST or leiomyoma not included in the study sample. An average method was used to combine assessment results within sub-groups with different experienced endoscopists in the study. The diagnostic results of the AI system and the endoscopists were then compared with the final pathological results. Furthermore, subgroup analyses by size (≤20 mm and >20 mm) or pathological type (GISTs and leiomyomas) were conducted to investigate the performance of the AI system in the external evaluation cohort.
Data preprocessing
The EUS images collected for AI model development underwent meticulous selection and review by two expert endoscopists. Representative images for each patient were chosen, and all EUS images were retrieved in JPEG format. Identical information, including patient identity, number, name, and acquisition time, was cropped out from the EUS images. The region of interest (ROI) containing a GIST or leiomyoma in the training dataset was manually delineated using Labelme software (version 4.2.10, https://github.com/wkentaro/labelme) and saved as a JSON file. Annotation was performed independently by each expert, and consensus was reached. All labeled lesion images were resized to 256 × 256 pixels before AI model training. To enhance the diversity of training data and improve the model's generalization ability, while minimizing the risk of overfitting, image augmentation techniques were applied. These techniques included flipping, rotation (random rotation between 0 and 60° for each image), and random cropping (cropping size of 224 × 224 pixels). To mitigate the impact of image data from different ultrasound endoscopes and probes at various centers on AI model development, the image data from different centers were randomly divided into the training cohort and internal validation cohort in a 7:3 ratio for AI model development.
AI system development
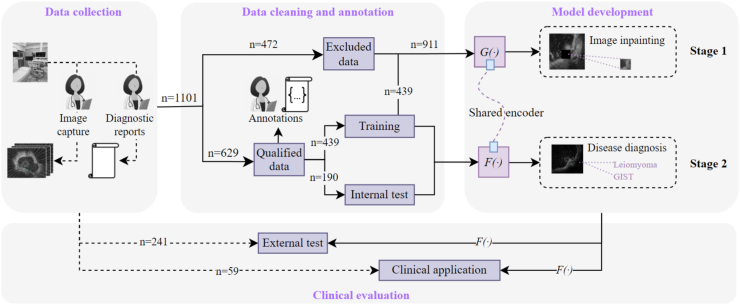
The comprehensive development process of the AI system is illustrated in Fig. 2. Briefly, a two-stage training approach was employed to fully leverage the potential semantic information in the EUS images (see Supplementary Figure S1). In the first stage, a self-supervised learning approach was utilized. In the second stage, input images underwent a region of interest (ROI) detection model, FROI (·), to identify the tumor ROI. The feature encoder Fe (·) replicated the parameters of Ge (·) from the first stage and, combined with a fully connected layer classifier (Classifier), performed a binary classification task. Through this two-stage training approach, a deep learning model capable of accurately extracting and predicting tumor information based on EUS images was successfully established. During the development of the AI system, given the potential diagnostic correlations among multiple images from the same patient, we employed a voting mechanism to consolidate the AI analysis results of these images. Specifically, multiple consecutive endoscopic images from the same patient (patient-level assessment) were integrated to obtain the AI analysis results. If the majority of images from a patient are categorized as a specific lesion, then the patient is diagnosed with that lesion. Specific details regarding the development of the AI system are outlined in Supplementary Materials.
Fig. 2.
Comprehensive overview of the AI system model development. Data collection (Left) involves the selection of representative EUS images by endoscopists from 1101 SELs patients. Data cleaning and annotation (Middle) manually segments and delineates the Region of Interest (ROI) of lesions for model training and validation. Model development and clinical evaluation (Right).
Prospective clinical application evaluation
To further assess the diagnostic performance of the AI system in clinical application, a prospective clinical evaluation was conducted at Shanghai JiaoTong University School of Medicine Affiliated Sixth People's Hospital from January 2023 to August 2023. Inclusion criteria were as follows: (1) age between 18 and 75; (2) endoscopists identified subepithelial lesions as potential GISTs or leiomyomas during EUS examination; (3) patients consented to endoscopic resection and were followed up until a definitive pathological diagnosis was obtained. The exclusion criteria were as follows: (1) patients who underwent upper gastrointestinal emergency examination or treatment for symptoms such as hematemesis or abdominal pain; (2) pregnant or lactating individuals; (3) potential participants with gastrointestinal submucosal bulges that were determined to be normal vessels, organ oppression, or extra-gastrointestinal lesions. During the application evaluation, a separate monitor with the AI system was installed adjacent to the original EUS monitor. The AI system was linked to the endoscopy generator, and the video stream was captured synchronously. If the SEL was suspected to be a GIST or leiomyoma during EUS examination, the endoscopists were required to classify it as one of the two types. Herein, two endoscopists with over 5 years of experience in EUS participated in the prospective evaluation. In cases of disagreement, a consensus was reached through discussion with a third reviewer who had over 10 years of experience in EUS. Subsequently, the AI system was turned on and applied, the system processed each frame and displayed the location of the detected SELs with a rectangular tracing box on monitor and provided a tumor type with diagnostic confidence. Concurrently, an independent data collector recorded the diagnostic data from the endoscopists and AI system. The AI system conducted real-time video analysis at a minimum of 30 frames per second (30fps), with the detection delay being barely noticeable for the endoscopists.
Sample size estimation
The sample size for the prospective clinical application evaluation was determined using Tests for Two ROC Curves by the NCSS PASS software (version 15.0). Based on previous research indicating that the AUC for the diagnostic yield for SELs of EUS-AI and EUS experts was 0.861–0.965 and 0.684–0.739, respectively.24 The AUC of the AI system and endoscopists in this study was estimated to be 0.900 and 0.650, respectively. The sample size of 29 patients in each trial group would be needed for the primary outcome based on an alpha level of 0.05, power of 0.9, and 15% dropout rate. In the end, a total of 59 patients with potential GISTs or leiomyomas, as identified by endoscopists, were enrolled.
Statistical analysis
Statistical analyses were performed using SPSS 22.0 (IBM, CA, USA) and R software (version 4.1.3). Normally distributed continuous variables were presented as mean ± standard deviation (SD), while non-normally distributed variables were expressed as median and range. Kolmogorov–Smirnov test and Q–Q plots were used to evaluate and check normality. One-way ANOVA was used for comparing continuous variables that adhere to a normal distribution, while the Kruskal–Wallis test was employed for comparing continuous variables that do not conform to a normal distribution, and categorical variables were compared using the Chi-square test or Fisher's exact test. The Wilson method was employed to calculate 95% confidence intervals (95% CI). The performance of the AI system was assessed using accuracy, sensitivity, specificity, PPV, and NPV. The overall performance was evaluated through the ROC curves and AUC. DeLong's test was utilized to compare differences in ROC curves. Calibration curves were generated to evaluate diagnostic performance in both the model development and internal validation cohorts. Decision curve analysis (DCA) was conducted to assess the clinical utility of the model. Interobserver agreement of the endoscopists was assessed using intraclass correlation coefficient (ICC). ICC values range from 0 to 1, with higher values indicating higher consistency; generally, ICC ≥0.75 indicates good consistency and ICC <0.4 indicates poor consistency. All statistical tests were two-sided, and a two-tailed significance level of 0.05 was considered.
Role of the funding source
The funder of the study had no role in study design, data collection and analysis, data interpretation, preparation of the manuscript, or decision to publish.
Results
Clinical characteristics of participants
A total of 870 participants [mean (Standard Deviation, SD) age, 55.5 (11.3) years; 505 females (58.0%)] with pathologically confirmed diagnosis of GISTs or leiomyomas were included in the AI system training and evaluation cohort. GISTs (n = 454) and leiomyomas (n = 416) accounted for 52.2% and 47.8%, respectively. 84.7% of GISTs and leiomyomas had a size ≤20 mm. The majority of SELs were located in the stomach. The training cohort consisted of 439 participants, with 261 (59.5%) being female. The mean (SD) age for this cohort was 55.7 (11.1) years. The internal validation cohort included 190 participants (53.2% female) with a mean (SD) age of 54.9 (11.0) years. The external evaluation cohort comprised 241 participants (59.3% female) with a mean (SD) age of 55.9 (11.9) years. Additionally, the prospective clinical application cohort included 59 participants (61% female) with a mean (SD) age of 59 (10.5) years. Detailed information of sample distribution within each cohort is shown in Table 1. No significant differences were noted in age, sex, tumor location, tumor size on EUS images, and pathological type among the training cohort, internal validation cohort and the external evaluation cohort.
Table 1.
Baseline characteristics of patients in the AI model development and evaluation cohort.
| Characteristics | AI development |
External evaluation |
P valuea | Prospective clinical application evaluation cohort (n = 59) | ||
|---|---|---|---|---|---|---|
| Overall (n = 870) | Training cohort (n = 439) | Internal validation cohort (n = 190) | External evaluation cohort (n = 241) | |||
| Gender, n (%) | ||||||
| Male | 365 (42.0%) | 178 (40.5%) | 89 (46.8%) | 98 (40.7%) | 0.303 | 23 (39%) |
| Female | 505 (58.0%) | 261 (59.5%) | 101 (53.2%) | 143 (59.3%) | 36 (61%) | |
| Age, years, mean (SD) | 55.5 (11.3) | 55.7 (11.1) | 54.9 (11.0) | 55.9 (11.9) | 0.617 | 59.1 (10.5) |
| Size, mm, mean (range) | 13.7 (5.0–55.0) | 13.4 (6.0–50.0) | 14.3 (6.0–55.0) | 13.5 (5.0–50.0) | 0.292 | 1.4 (7.0–50.0) |
| Size, n (%) | ||||||
| ≤20 mm | 737 (84.7%) | 372 (84.7%) | 166 (87.4%) | 199 (82.6%) | 0.635 | 46 (78.0%) |
| >20 mm | 133 (15.3%) | 67 (15.3%) | 24 (12.4%) | 42 (17.4%) | 13 (22.0%) | |
| Tumor location, n (%) | ||||||
| Esophagus | 235 (27.0%) | 111 (25.3%) | 49 (25.8%) | 75 (31.1%) | 0.582 | 11 (18.6%) |
| Stomach | 607 (69.8%) | 316 (72.0%) | 134 (70.5%) | 157 (65.1%) | 48 (81.4%) | |
| Duodenum | 9 (1.0%) | 4 (0.9%) | 3 (1.6%) | 2 (0.8%) | / | |
| Colorectum | 19 (2.2%) | 8 (1.8%) | 4 (2.1%) | 7 (2.9%) | / | |
| Pathological type, n (%) | ||||||
| GISTs | 454 (52.2%) | 227 (51.7%) | 99 (52.1%) | 128 (53.1%) | 0.940 | 29 (49.1%) |
| Leiomyomas | 416 (47.8%) | 212 (48.3%) | 91 (47.9%) | 113 (46.9%) | 28 (47.5%) | |
| Other | / | / | / | / | 2 (3.4%) | |
Comparison among the training cohort, the internal validation cohort and the external evaluation cohort.
The performance of AI system in the internal validation and external evaluation cohort
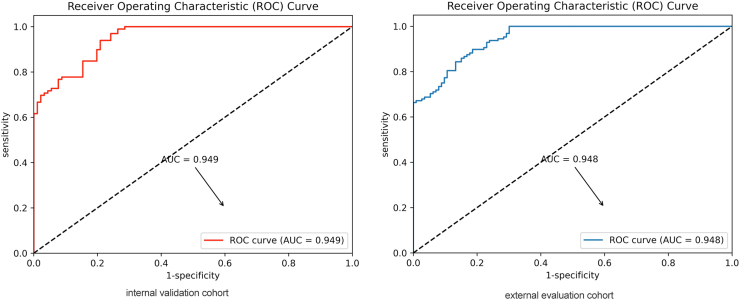
The overall diagnostic performance of the AI system was assessed in both the internal validation and external evaluation cohorts. In the internal validation cohort, the AI system accurately diagnosed 177 out of 190 tumors, achieving an accuracy of 93.1% (95% CI, 88.6%–96.0%). The AUC of the AI system was 0.949 (95% CI: 0.918–0.973). Calibration curves demonstrated that the model was well-calibrated (see Supplementary Figure S3). Additionally, Decision Curve Analysis (DCA) indicated the added benefits of using the AI system (Supplementary Figure S4). In the external evaluation cohort, the AI system correctly diagnosed 221 out of 241 tumors, resulting in an accuracy of 91.7% (95% CI, 87.5%–94.6%). The AI system showed sensitivity, specificity, PPV, and NPV of 90.3% (83.4%–94.5%), 93.0% (87.2%–96.3%), 91.9% (85.3%–95.7%), and 91.5% (85.5%–95.2%), respectively. The AUC of the AI system in the external evaluation cohort was 0.948 (95% CI: 0.921–0.969) (see Table 2). The ROC curves for the internal validation and external evaluation cohorts are depicted in Fig. 3.
Table 2.
Diagnostic performance of AI system in internal validation and external evaluation cohort.
| Formulas | Internal validation cohort % (95% CI) | Formulas | External evaluation cohort % (95% CI) | |
|---|---|---|---|---|
| Accuracy | 177/190 | 93.1% (88.6%–96.0%) | 221/241 | 91.7% (87.5%–94.6%) |
| Sensitivity | 85/91 | 94.4% (86.4%–96.9%) | 102/113 | 90.3% (83.4%–94.5%) |
| Specificity | 92/99 | 93.0% (86.1%–96.5%) | 119/128 | 93.0% (87.2%–96.3%) |
| PPV | 85/92 | 92.4% (85.1%–96.3%) | 102/111 | 91.9% (85.3%–95.7%) |
| NPV | 92/98 | 93.8% (87.3%–97.2%) | 119/130 | 91.5% (85.5%–95.2%) |
| AUC | 0.949 (0.918–0.973) | 0.948 (0.921–0.969) | ||
Fig. 3.
Diagnostic efficiency of the AI system in the internal and external test cohort. ROC curves for GISTs and leiomyomas diagnosis in the internal test set (Left) and the external test set (Right).
Comparison on the performance between AI systems and endoscopists
To assess diagnostic performance, three senior and three junior endoscopists, who were not involved in the AI model development, analyzed 1100 EUS images from 241 cases in the external evaluation cohort. The accuracy, sensitivity, specificity, PPV, NPV and AUC of each endoscopist for the diagnosis of SELs are shown in Supplementary Table S1 and Supplementary Figure S5. In addition, the confusion matrix for the per-category diagnostic performance of the AI system and endoscopists is also shown in Supplementary Table S1. All endoscopists showed a diagnostic ability of less than 0.75 for distinguishing GISTs from leiomyomas (accuracy range, 67.2%–74.3%). The sensitivity for the diagnosis of SELs ranged from 58.4% to 69.0%, and the specificity ranged from 72.7% to 80.5%. In comparison with the AI system, the endoscopists' performance was unsatisfactory. The highest diagnostic accuracy among endoscopists was only 74.3%, significantly lower than that of the AI system (91.7%, 95% CI 87.5%–94.6%, P < 0.001). As shown in Supplementary Table S2, the interobserver agreement among endoscopist for distinguishing GISTs from leiomyoma was high agreement (ICC = 0.931 for junior endoscopists and ICC = 0.885 for senior endoscopists).
The performance of AI system for different size- and type- SELs in external evaluation cohort
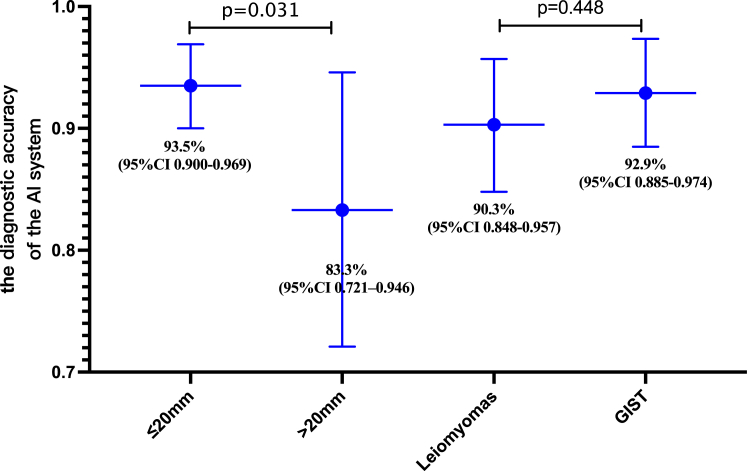
Given that over 80% of SELs in this study had a size of ≤20 mm, we considered the potential impact of SEL size on the diagnostic accuracy of the AI system. The diagnostic performance of the AI system for SELs with size ≤20 mm and >20 mm was further analyzed. As depicted in Fig. 4, the diagnostic accuracy of the AI system for SELs with size ≤20 mm reached 93.5% (95% CI 0.900–0.969), which was significantly higher than that for SELs with size >20 mm [ACC 83.3%, 95% CI (0.721–0.946), (P = 0.031)]. Furthermore, the diagnostic performance of the AI system for different types of tumors was also examined. The results demonstrated that the diagnostic accuracy of the AI system for leiomyomas and GISTs reached 90.3% (95% CI 0.848–0.957) and 92.9% (95% CI 0.885–0.974), respectively. No significant difference was observed between the diagnostic accuracies for leiomyomas and GISTs (P = 0.448).
Fig. 4.
Diagnostic performance of the AI system for different size- and type- SELs in the external evaluation cohort. The accuracy of AI system for diagnosing ≤20 mm SELs is significantly higher (P = 0.031). There is no significant difference in accuracy for different types of SELs (P = 0.448).
The application assessment of AI system in a prospective cohort
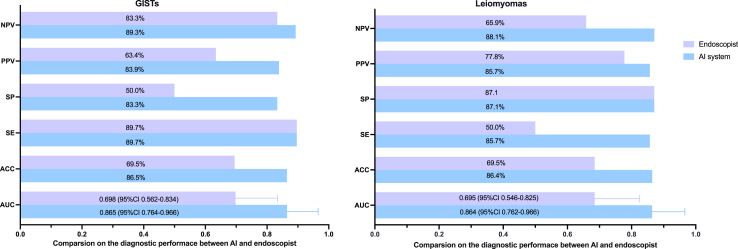
Following evaluation in an external cohort, the AI model was transformed into a real-time application system for use during EUS procedures, named the AI-aid EUS diagnostic system (Supplementary Figure S6). The diagnostic performance of the AI-aid EUS diagnostic system was assessed in a prospective cohort of 59 patients. SELs identified as potential GISTs or leiomyomas by endoscopists during EUS procedures were further diagnosed by the AI-aid EUS diagnostic system. The diagnostic results of the AI-aid EUS diagnostic system and endoscopists were then compared with the final histopathological results. Results indicated that the AI-aid EUS diagnostic system exhibited excellent discrimination between GISTs (AUC: 0.865, 95% CI: 0.782–0.977) and leiomyomas (AUC: 0.864, 95% CI 0.762–0.966) (Fig. 5). The corresponding accuracy, sensitivity, specificity, PPV, and NPV of the AI system for GISTs were 86.5%, 89.7%, 83.3%, 83.9%, and 89.3%, respectively. In contrast, the AUC of the endoscopist was only 0.698 (95% CI 0.562–0.834). The DeLong test revealed that the diagnostic efficiency of the AI system was significantly superior to that of the endoscopist (P = 0.010). Similar results were found in the diagnosis of the AI system for leiomyomas. The diagnostic confidence of the AI system (Supplementary Figure S7) for GISTs and leiomyomas, along with the real-time application video in a clinical setting (video 1), can be found in the supplementary information.
Fig. 5.
Real-time application assessment of the AI system in a prospective clinical cohort. Performance metrics (including AUC, ACC, SE, SP, PPV, NPV) in diagnosing GISTs and leiomyomas evaluated in a prospective clinical cohort. The AI system demonstrates superior discrimination in clinical settings.
Discussion
GISTs and leiomyomas are the most prevalent SELs in the gastrointestinal tract, with an increasing incidence. However, no definitively specific imaging features exist to reliably differentiate GISTs from leiomyomas. Despite improvements in diagnostic yield through tissue acquisition methods like EUS-FNB or mucosal cutting biopsy, variations in endoscopists' experience in EUS image interpretation and procedural techniques limit diagnostic accuracy.10,11,25 Recently, AI has been widely applied to the field of digestive endoscopy and demonstrated promising performance.26,27 Previous studies have highlighted the effectiveness of AI in diagnosing SELs: Hirai et al.18 developed an AI system for multi-category classifications by retrospective analysis of 631 SELs, the accuracy of the AI system for differentiating SELs achieved 86.1%. Yoon et al.28 established a convolutional neural network computer-aided diagnosis system based on 212 EUS images, the diagnostic accuracy for distinguishing GISTs was 79.2%. However, the limited case numbers, retrospective designs and the absence of prospective external evaluation were the common drawbacks in these studies. Yang et al.20 have published a prospective study on an AI-assisted EUS system for distinguishing GIST from leiomyomas, while the AUC of their AI system was only 0.642 in the external testing set. Cai MY et al.29 established an automatically optimized radiomics modeling system for small SELs based on 383 SELs, while the accuracy for identifying GISTs was only 74.2%. The poor performance may hinder the practical implementation of these AI models in clinical settings.
Compared with these previous studies, the current study had several notable novelties. First, a multicenter dataset comprising of 1401 patients in the current study were employed to develop the AI-assisted EUS diagnostic system, to our understanding, which is the largest dataset reported in this field. Second, the AI system was successfully implemented in real-time during clinical practice and evaluated in a prospective cohort, to the best of our knowledge, this is the first study reporting the application of a real-time AI- aid EUS diagnostic system in a prospective clinical trial. Third, the AI system demonstrated superior performance in both internal and external evaluation cohorts as well as in clinical practice.
In the current study, the AI system demonstrated an accuracy of 93.1%, sensitivity of 94.4%, and specificity of 93.0% in distinguishing GISTs from leiomyomas in the internal validation cohort. When the AI system was evaluated in an external evaluation cohort, it achieved an accuracy of 91.7%, sensitivity of 90.3%, and specificity of 93.0%, surpassing the performance of previous studies.18,20,23,29 Consistent with these studies,18,20,23,29 the diagnostic efficiency of the AI system (ACC 91.7%) was significantly superior to that of endoscopists (67.2%–74.3%) in distinguishing GISTs from leiomyomas.
It is noteworthy that the majority of SELs are small in size, with this study revealing that over 80% of SELs were ≤20 mm. The overall diagnostic accuracy of EUS in discriminating small SELs remains suboptimal. Even with the addition of tissue diagnosis, the reported diagnostic yield for small SELs ranges only from 62% to 82%.30,31 Moreover, there is a significant discrepancy in guidelines regarding the optimal management strategy for small SELs. The American College of Gastroenterology (ACG) recommends endoscopic follow-up for small GISTs,32 while Asian guidelines and the European Society of Medical Oncology (ESMO) prefer endoscopic resection.11,33 In contrast, the European Society of Gastrointestinal Endoscopy (ESGE) suggests limiting endoscopic resection to patients with pathologically confirmed GIST.34 The lack of consensus on management and the absence of effective diagnostic methods posed a challenge for endoscopists when encountering <20 mm SELs. Previous studies have reported that EUS-based AI models have higher diagnostic accuracy for larger SELs (93.3–96.3% for SELs ≥20 mm).18,20 In this study, it is encouraging that the AI system demonstrated better diagnostic performance for small SELs (93.5% for SELs ≤20 mm, 83.3% for SELs >20 mm). Cai MY et al. established an automatically optimized radiomics modeling system for small SELs based on 383 SELs with <20 mm in size; however, the accuracy for identifying GISTs was only 74.2%,29 significantly lower than the performance of the AI system in this study. Possible reasons include: 1) the larger sample size of small SELs (≤20 mm) in the present study; 2) the EUS images of small SELs exhibit relatively well-defined boundaries and higher contrast with normal tissues than those of large SELs.
In the prospective evaluation cohort, the AI system exhibited favorable performance in discriminating between GISTs and leiomyomas. The real-time application of the AI system suggests its potential in mitigating disparities in diagnostic proficiency across various regions, particularly benefiting patients in medically underserved areas and contributing to the scientific diagnosis and treatment of gastrointestinal SELs. Moreover, the AI system is intended to serve as an assisting decision-making system. Drawing from results in the prospective evaluation cohort, the real-time AI-assisted system have the potential to improve the diagnostic accuracy even for experienced endoscopists and would therefore, change the treatment decisions. In the future, we anticipate that the utilization of AI system can significantly enhance the capabilities of endoscopists in EUS procedures. This will be especially valuable for endoscopists with lower procedural volumes to help reduce their variability and maintain high standards of EUS evaluation for patients. In terms of training, experienced endoscopists may have more confident in allowing trainee endoscopists to perform EUS examination on their behalf under direct or oversight supervision. For trainees, having the opportunity to perform more endoscopic examinations could also potentially hasten their path to proficiency and expedite their advancement towards competence.
Despite the promising performance demonstrated by the AI system, certain limitations should be acknowledged. Firstly, the AI system was exclusively employed for the binary classification of GISTs and leiomyomas, requiring the involvement of experienced endoscopists during its utilization. Attempts were made to incorporate other types of SELs into the development of the AI system. However, due to their low prevalence and the risk of overfitting without sufficient data, a dependable model for the multi-classification of SELs could not be trained. Addressing these challenges necessitates larger-scale multicenter or international studies. Secondly, the real-time evaluation of the AI system was conducted solely at one center with a limited sample size. Therefore, a prospective study involving multiple centers with a more extensive sample size is required to further validate the real-time diagnostic efficiency of the AI system. Thirdly, although we have endeavored to ensure the representativeness of the cohorts and the universality of the research findings in this study. It is possible that they may not fully encompass all possible clinical scenarios and patient characteristics, Future studies will further seek to broaden the diversity of samples and enhance the representativeness of cohorts to strengthen the universality of research conclusions. Furthermore, the present study focused on the real-time diagnostic efficiency of the AI system. However, it is also important to assess its impact on clinical outcomes, such as patient management decisions, treatment strategies, and long-term prognosis. Future research could involve prospective studies to assess the clinical usefulness and cost-effectiveness of integrating the AI system into routine practice. Lastly, the interpretability of the AI system's decisions remains a challenge. Despite its excellent performance, the ability to provide explanations for its diagnoses would greatly enhance trust and acceptance among healthcare providers. Advancements in explainable AI techniques should be pursued to address this limitation.
In conclusion, we successfully developed a real-time AI-aid EUS diagnostic system that demonstrated superior diagnostic performance in discriminating between GISTs and leiomyomas compared to experienced endoscopists. This novel tool could provide a relatively accurate, convenient, and noninvasive method, serving to assist endoscopists in real-time differentiation of various types of SELs in clinical practice and facilitating improved diagnostic and therapeutic decision-making.
Contributors
ZXD, HZ, JSB and XJW designed the research goals and aims. HYZ, HBZ, BJQ and JSB designed the model. XYZ, ZXD and DFC designed the evaluation methodology. HYZ, HBZ, BJQ and JSB developed the software for the deep learning model. ZXD, XYZ, HBZ, HYZ, DFC, JC, ZLX, YWS, QZ, SW, JX, MN, HZ curated the datasets. ZXD, HBZ and HYZ performed the analysis. ZXD, HBZ, HYZ and XYZ wrote the manuscript with the assistance and feedback of all other co-authors. XJW, XYZ, ZXD conceived and directed the project. XJW, HZ and JSB have accessed and verified data. All authors have read and agreed to publish the paper.
Data sharing statement
In order to safeguard patient privacy, EUS image datasets and other patient-related data are not publicly accessible. But all data and algorithm source codes utilized in this study are available upon a reasonable request email to the corresponding author. To obtain access, data requestors will be required to sign a data-access agreement.
Declaration of interests
All the authors disclose no competing interest conflicts.
Acknowledgements
This work was supported by grants from the Science and Technology Commission Foundation of Shanghai Municipality (Grant No. 19411951500), Science and Technology Commission Foundation of the Xuhui District (Grant No. 2021-013), the Interdisciplinary Program of Shanghai Jiao Tong University (project number YG2021QN99) and the Research Funds of Shanghai Sixth people’s Hospital (yngh202018 and ynhg202323). We would like to express our gratitude to Dr. XiangTian Yu, PhD, and Mei Kang, PhD, for their valuable contributions in editing a draft of this manuscript. Additionally, we extend our thanks to MedSci (https://www.medsci.cn) for providing expert linguistic services.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2024.102656.
Contributor Information
Hui Zhou, Email: mdzhouhui@sjtu.edu.cn.
Jinsong Bao, Email: bao@dhu.edu.cn.
Xinjian Wan, Email: slwanxj2019@sjtu.edu.cn.
Appendix A. Supplementary data
Real-time application video of the AI system in a clinical setting. The red rectangular box shows the identified tumor location by the AI system during EUS procedure, with state, tumor type, and diagnostic confidence displayed in the top left corner.
References
- 1.Kim M.Y., Jung H.Y., Choi K.D., et al. Natural history of asymptomatic small gastric subepithelial tumors. J Clin Gastroenterol. 2011;45(4):330–336. doi: 10.1097/MCG.0b013e318206474e. [DOI] [PubMed] [Google Scholar]
- 2.Lee J.H., Lee H.L., Ahn Y.W., et al. Prevalence of gastric subepithelial tumors in Korea: a single center experience. Korean J Gastroenterol. 2015;66(5):274–276. doi: 10.4166/kjg.2015.66.5.274. [DOI] [PubMed] [Google Scholar]
- 3.Abe K., Tominaga K., Yamamiya A., et al. Natural history of small gastric subepithelial lesions less than 20 mm: a multicenter retrospective Observational study (NUTSHELL20 study) Digestion. 2023;104(3):174–186. doi: 10.1159/000527421. [DOI] [PubMed] [Google Scholar]
- 4.Standards of Practice C., Faulx A.L., Kothari S., et al. The role of endoscopy in subepithelial lesions of the GI tract. Gastrointest Endosc. 2017;85(6):1117–1132. doi: 10.1016/j.gie.2017.02.022. [DOI] [PubMed] [Google Scholar]
- 5.Li J., Ye Y., Wang J., et al. Chinese consensus guidelines for diagnosis and management of gastrointestinal stromal tumor. Chin J Cancer Res. 2017;29(4):281–293. doi: 10.21147/j.issn.1000-9604.2017.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miettinen M., Sobin L.H., Lasota J. Gastrointestinal stromal tumors of the stomach: a clinicopathologic, immunohistochemical, and molecular genetic study of 1765 cases with long-term follow-up. Am J Surg Pathol. 2005;29(1):52–68. doi: 10.1097/01.pas.0000146010.92933.de. [DOI] [PubMed] [Google Scholar]
- 7.Kawanowa K., Sakuma Y., Sakurai S., et al. High incidence of microscopic gastrointestinal stromal tumors in the stomach. Hum Pathol. 2006;37(12):1527–1535. doi: 10.1016/j.humpath.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Pang T., Zhao Y., Fan T., et al. Comparison of safety and outcomes between endoscopic and surgical resections of small (</= 5 cm) primary gastric gastrointestinal stromal tumors. J Cancer. 2019;10(17):4132–4141. doi: 10.7150/jca.29443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coe T.M., Fero K.E., Fanta P.T., et al. Population-based epidemiology and mortality of small malignant gastrointestinal stromal tumors in the USA. J Gastrointest Surg. 2016;20(6):1132–1140. doi: 10.1007/s11605-016-3134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishida T., Blay J.Y., Hirota S., Kitagawa Y., Kang Y.K. The standard diagnosis, treatment, and follow-up of gastrointestinal stromal tumors based on guidelines. Gastric Cancer. 2016;19(1):3–14. doi: 10.1007/s10120-015-0526-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casali P.G., Abecassis N., Aro H.T., et al. Gastrointestinal stromal tumours: ESMO-EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29(Suppl 4):iv68–iv78. doi: 10.1093/annonc/mdy095. [DOI] [PubMed] [Google Scholar]
- 12.Baysal B., Masri O.A., Eloubeidi M.A., Senturk H. The role of EUS and EUS-guided FNA in the management of subepithelial lesions of the esophagus: a large, single-center experience. Endosc Ultrasound. 2017;6(5):308–316. doi: 10.4103/2303-9027.155772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karaca C., Turner B.G., Cizginer S., Forcione D., Brugge W. Accuracy of EUS in the evaluation of small gastric subepithelial lesions. Gastrointest Endosc. 2010;71(4):722–727. doi: 10.1016/j.gie.2009.10.019. [DOI] [PubMed] [Google Scholar]
- 14.Hwang J.H., Saunders M.D., Rulyak S.J., Shaw S., Nietsch H., Kimmey M.B. A prospective study comparing endoscopy and EUS in the evaluation of GI subepithelial masses. Gastrointest Endosc. 2005;62(2):202–208. doi: 10.1016/s0016-5107(05)01567-1. [DOI] [PubMed] [Google Scholar]
- 15.Minoda Y., Chinen T., Osoegawa T., et al. Superiority of mucosal incision-assisted biopsy over ultrasound-guided fine needle aspiration biopsy in diagnosing small gastric subepithelial lesions: a propensity score matching analysis. BMC Gastroenterol. 2020;20(1):19. doi: 10.1186/s12876-020-1170-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Moura D.T.H., McCarty T.R., Jirapinyo P., et al. EUS-guided fine-needle biopsy sampling versus FNA in the diagnosis of subepithelial lesions: a large multicenter study. Gastrointest Endosc. 2020;92(1):108–119.e3. doi: 10.1016/j.gie.2020.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osoegawa T., Minoda Y., Ihara E., et al. Mucosal incision-assisted biopsy versus endoscopic ultrasound-guided fine-needle aspiration with a rapid on-site evaluation for gastric subepithelial lesions: a randomized cross-over study. Dig Endosc. 2019;31(4):413–421. doi: 10.1111/den.13367. [DOI] [PubMed] [Google Scholar]
- 18.Hirai K., Kuwahara T., Furukawa K., et al. Artificial intelligence-based diagnosis of upper gastrointestinal subepithelial lesions on endoscopic ultrasonography images. Gastric Cancer. 2022;25(2):382–391. doi: 10.1007/s10120-021-01261-x. [DOI] [PubMed] [Google Scholar]
- 19.Byrne M.F., Chapados N., Soudan F., et al. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. 2019;68(1):94–100. doi: 10.1136/gutjnl-2017-314547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X., Wang H., Dong Q., et al. An artificial intelligence system for distinguishing between gastrointestinal stromal tumors and leiomyomas using endoscopic ultrasonography. Endoscopy. 2022;54(3):251–261. doi: 10.1055/a-1476-8931. [DOI] [PubMed] [Google Scholar]
- 21.Oh C.K., Kim T., Cho Y.K., et al. Convolutional neural network-based object detection model to identify gastrointestinal stromal tumors in endoscopic ultrasound images. J Gastroenterol Hepatol. 2021;36(12):3387–3394. doi: 10.1111/jgh.15653. [DOI] [PubMed] [Google Scholar]
- 22.Niikura R., Aoki T., Shichijo S., et al. Artificial intelligence versus expert endoscopists for diagnosis of gastric cancer in patients who have undergone upper gastrointestinal endoscopy. Endoscopy. 2022;54(8):780–784. doi: 10.1055/a-1660-6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J., Huang J., Song Y., et al. Differentiating gastrointestinal stromal tumors from leiomyomas of upper digestive tract using convolutional neural network model by endoscopic ultrasonography. J Clin Gastroenterol. 2023 doi: 10.1097/MCG.0000000000001907. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 24.Minoda Y., Ihara E., Komori K., et al. Efficacy of endoscopic ultrasound with artificial intelligence for the diagnosis of gastrointestinal stromal tumors. J Gastroenterol. 2020;55(12):1119–1126. doi: 10.1007/s00535-020-01725-4. [DOI] [PubMed] [Google Scholar]
- 25.Yoshinaga S., Hilmi I.N., Kwek B.E., Hara K., Goda K. Current status of endoscopic ultrasound for the upper gastrointestinal tract in Asia. Dig Endosc. 2015;27(Suppl 1):2–10. doi: 10.1111/den.12422. [DOI] [PubMed] [Google Scholar]
- 26.Messmann H., Bisschops R., Antonelli G., et al. Expected value of artificial intelligence in gastrointestinal endoscopy: European society of gastrointestinal endoscopy (ESGE) position statement. Endoscopy. 2022;54(12):1211–1231. doi: 10.1055/a-1950-5694. [DOI] [PubMed] [Google Scholar]
- 27.Xie X., Xiao Y.F., Zhao X.Y., et al. Development and validation of an artificial intelligence model for small bowel capsule endoscopy video review. JAMA Netw Open. 2022;5(7) doi: 10.1001/jamanetworkopen.2022.21992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim Y.H., Kim G.H., Kim K.B., et al. Application of A Convolutional neural network in the diagnosis of gastric mesenchymal tumors on endoscopic ultrasonography images. J Clin Med. 2020;9(10) doi: 10.3390/jcm9103162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai M., Song B., Deng Y., et al. Automatically optimized radiomics modeling system for small gastric submucosal tumors (<2 cm) discrimination based on endoscopic ultrasound images. Gastrointest Endosc. 2023;99:537. doi: 10.1016/j.gie.2023.11.006. [DOI] [PubMed] [Google Scholar]
- 30.Larghi A., Fuccio L., Chiarello G., et al. Fine-needle tissue acquisition from subepithelial lesions using a forward-viewing linear echoendoscope. Endoscopy. 2014;46(1):39–45. doi: 10.1055/s-0033-1344895. [DOI] [PubMed] [Google Scholar]
- 31.Akahoshi K., Oya M., Koga T., et al. Clinical usefulness of endoscopic ultrasound-guided fine needle aspiration for gastric subepithelial lesions smaller than 2 cm. J Gastrointestin Liver Dis. 2014;23(4):405–412. doi: 10.15403/jgld.2014.1121.234.eug. [DOI] [PubMed] [Google Scholar]
- 32.Jacobson B.C., Bhatt A., Greer K.B., et al. ACG clinical guideline: diagnosis and management of gastrointestinal subepithelial lesions. Am J Gastroenterol. 2023;118(1):46–58. doi: 10.14309/ajg.0000000000002100. [DOI] [PubMed] [Google Scholar]
- 33.Koo D.H., Ryu M.H., Kim K.M., et al. Asian consensus guidelines for the diagnosis and management of gastrointestinal stromal tumor. Cancer Res Treat. 2016;48(4):1155–1166. doi: 10.4143/crt.2016.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deprez P.H., Moons L.M.G., O'Toole D., et al. Endoscopic management of subepithelial lesions including neuroendocrine neoplasms: European Society of Gastrointestinal Endoscopy (ESGE) Guideline. Endoscopy. 2022;54(4):412–429. doi: 10.1055/a-1751-5742. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Real-time application video of the AI system in a clinical setting. The red rectangular box shows the identified tumor location by the AI system during EUS procedure, with state, tumor type, and diagnostic confidence displayed in the top left corner.