Abstract
Transcription factor (TF)-based biosensors that connect small-molecule sensing with readouts such as fluorescence have proven to be useful synthetic biology tools for applications in biotechnology. However, the development of specific TF-based biosensors is hindered by the limited repertoire of TFs specific for molecules of interest since current construction methods rely on a limited set of characterized TFs. In this study, we present an approach for engineering the specificity of TFs through a computation-based workflow using molecular docking that enables targeted alteration of TF ligand specificity. Using this method, we engineer the LysR family BenM TF to alter its specificity from its cognate ligand cis,cis-muconic acid to adipic acid through a single amino acid substitution identified by our computational workflow. When implemented in a cell-free system, the engineered biosensor shows higher ligand sensitivity, expanding the potential applications of this circuit. We further investigate ligand binding through molecular dynamics to analyze the substitution, elucidating the impact of modulating a single amino acid position on the mechanism of BenM ligand binding. This study represents the first application of biomolecular modeling methods for altering BenM specificity and for gaining insights into how mutations influence the structural dynamics of BenM. Such methods can potentially be applied to other TFs to alter specificity and analyze the dynamics responsible for these changes, highlighting the applicability of computational tools for informing experiments. In addition, our developed adipic acid biosensor can be applied for the identification and engineering of enzymes to produce adipic acid.
Keywords: Biosensors, Muconic acid, Adipic acid, Docking, Protein engineering, Molecular dynamics
Graphical Abstract
Highlights
-
•
Established a computational workflow for ligand specificity engineering.
-
•
Computation-assisted protein engineering of the binding pocket of BenM.
-
•
Ligand docking can identify hotspot residues for ligand specificity.
-
•
Cell-free systems can improve sensitivity.
-
•
Molecular dynamics can be used to analyze the impact of mutations on TFs.
1. Introduction
Microbe-based bioproduction of value-added chemicals has been pursued as an attractive alternative to traditional chemical synthesis, owing to the use of renewable carbon sources and milder reaction conditions [1]. 1,6-hexanedioic acid or adipic acid (AA), is one such example of a value-added chemical with industrial interest [2]. AA is used in the manufacturing of nylon 66, a polymer involved in a variety of products such as textiles, with a global production of over 3.3 million tons/year [2]. However, conventional processes require the use of petroleum-based feedstocks and result in the release of greenhouse gases [3]. Instead, microbial production of AA is a promising alternative to address the sustainability issues of current processes. A variety of pathways for the bioproduction of adipic acid have been investigated and implemented [4], [5]. We have demonstrated the bioproduction of adipic acid via a synthetic pathway to cis,cis-muconic acid (CCM) that culminates in the reduction of CCM to AA using an enoate reductase [6], [7]. However, significant improvements to the involved organisms and enzymes are needed to reach commercially viable production levels.
Generally, the engineering of microbial pathways and enzymes toward efficient bioproduction requires extensive optimization in the design-build-test-learn cycle. Although large libraries can be designed and built with current advancements in DNA design and synthesis methods, the testing phase causes a major bottleneck in the cycle [8], [9]. To address the bottleneck, transcription factor (TF)-based protein biosensors have been developed to couple sensing of small molecules or effectors to gene regulation through DNA binding [8]. In particular, microbial biosensors utilizing single-component TFs are attractive due to their simplistic and modular design [10]. These TF-based biosensors consist of a TF that has a ligand binding domain (LBD) that binds to specific ligands, and a DNA-binding domain (DBD) that allows for control of downstream gene expression depending on the ligand binding state of the LBD. In response to the detection of the ligand by the TF, the DNA binding of the TF is modulated and results in changes in the expression of a downstream gene in a dose-dependent manner [11]. When linked to a readout such as a fluorescent protein, large libraries of variant strains and proteins can be screened for the production of biotechnologically relevant compounds [12]. Combined with fluorescence-activated cell sorting (FACS), individual strains can be isolated, allowing for higher-order screening [9]. Screening large libraries using biosensors with an easily detectable output allows for rapid characterization, providing a rapid alternative to approaches like liquid chromatography-mass spectrometry (LC-MS). Biosensors can also be incorporated into cell-free systems, allowing for rapid and efficient in vitro screening of enzymes, pathways, and components, especially when combined with automation or microfluidics for droplet-based FACS [13].
While TF-based biosensors have demonstrated their potential in a variety of applications, their widespread development and deployment is still limited by the availability of fully characterized TFs that can detect target chemicals of interest. According to our survey of the literature, the set of publicly characterized biosensors numbers in the 100’s, while there are on the order of hundreds of thousands of putative microbial TF proteins that can be utilized as biosensors with additional testing and characterization [14]. While many putative TFs may be redundant, the large repertoire available suggests that there may exist numerous TFs with unique specificities from existing biosensors. Identifying novel biosensors requires extensive mining and characterization [15]. The lack of accessible TFs with characterized specificities of interest hinders the development of biosensors and limits the scope of potential applications in biology and engineering. An alternative to characterizing new TFs and biosensors is to engineer the TF itself to alter the ligand specificity of the TF.
The modular nature of TFs makes them promising starting points for protein engineering. A range of methods, including rational engineering and directed evolution through mutagenesis and sorting, have been successfully applied to altering TF specificity [16]. However, these methods require extensive laboratory resources and multiple rounds of mutation, selection, and characterization. The use of algorithms such as AutoDock for protein-ligand docking and Rosetta for protein design have provided valuable tools for model-guided protein engineering for altering ligand-binding specificity [17], [18], [19]. Such existing computational tools provide a repertoire of methods to generate protein variants. For example, docking can be used to guide the selection of residues for semi-rational engineering strategies, as was done for the LysR family TF LysG to alter ligand specificity [20]. Combining computational methods into high-throughput computational workflows targeted to TFs can potentially generate promising variants while searching a large sequence space. The combination of computational methods such as homology modeling and docking can be used to guide experimental tests through computational results. Focusing on computational workflows also exchanges expensive lab time and resources for computational resources.
BenM, the LysR family TF from Acinetobacter sp. ADP1, regulates the benABCDE operon for benzoate degradation through CCM binding [21]. Upon ligand binding, the BenM activator undergoes small conformational changes through movements in the helices of the ligand binding domain, and changes in the oligomeric interfaces, resulting in increased activation of transcription [22]. Recently, BenM has been engineered to alter its response curve characteristics and its specificity from the cognate ligand CCM to AA using error-prone PCR-based directed evolution and continuous hypermutation in S. cerevisiae [23], [24]. An engineered BenM variant was shown to be responsive to AA in addition to CCM, and was also shown to be functional in the model bacterial chassis E. coli [24]. The TF PcaR has also been engineered in P. putida to be responsive to AA [25]. In these cases, recognition of the ligand CCM can still be observed due to the lack of negative selection to eliminate CCM specificity. Further reducing such ligand recognition crosstalk is needed for applications requiring high specificity, such as screening for improved enzyme variants. There also remains a need to develop highly specific BenM biosensors in other contexts, like in bacterial systems.
Since the BenM TF has been well-characterized, including through a variety of published crystal structures, such as the structure in complex with the cognate ligand CCM, we were interested in the development of a new computational workflow to engineer the ligand specificity of this TF since previous BenM engineering have been based on random mutations and sorting [21], [22], [26], [27]. In this study, the specificity of BenM is modified through guidance from a computational workflow utilizing high-throughput molecular modeling and docking simulations, which enabled the discovery of BenM variants that possess altered specificity experimentally in whole-cell biosensors. When implemented in a cell-free reaction system, our best-performing BenM variant exhibits increased sensitivity towards AA. Our results demonstrate that amino acid substitutions at a single residue of the binding pocket could completely alter the ligand specificity of BenM, and reduction of the recognition of the natural ligand CCM, thus highlighting the application of this computational approach for identifying hotspot residues for specificity. BenM is then further studied using molecular dynamics to gain insights into the impact of mutations on structural dynamics and ligand specificity. This study provides a framework for using computational and structural insights for TF engineering, guiding the alteration of TF specificity towards molecules of interest.
2. Results
2.1. Development of a TF specificity engineering pipeline
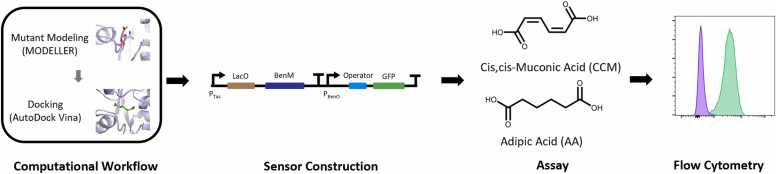
We developed a high-throughput workflow for homology model predictions of protein variants and docking predictions of ligand binding poses followed by experimental construction and testing (Fig. 1). The computational methodology involves high-throughput generation of 3D models of a targeted site-saturation variant library of the target protein using the MODELLER program [28], geometry restraints preparation using AutoDock Tools, energy minimization with Chimera [29], and flexible docking with AutoDock Vina using supercomputing resources. While many computational tools can be utilized in the pipeline, open-source tools that could be readily scripted for high-throughput execution were selected. The workflow only requires a protein structure and a selection of residue targets, making it possible to execute this workflow for any protein of interest. Following these steps, the experimental testing phase involves flow cytometry evaluation of the TF variant-driven expression of a downstream reporter gene, in our case, GFP [30].
Fig. 1.
Overview of computational and experimental workflow. The computational workflow utilized MODELLER and AutoDock Vina [25], [27]. Variants of interest were created and tested in an in vivo sensor with cis,cis-muconic acid and adipic acid using flow cytometry.
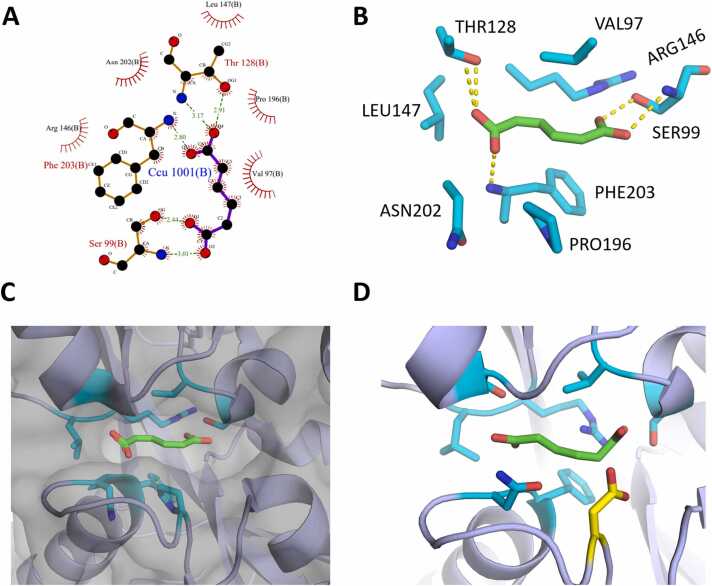
To advance our goal of altering the specificity of the BenM TF from CCM to AA via our developed pipeline, we initiated the in silico work with the crystal structure of BenM bound to CCM (PDB: 2F7A) [22]. Using this model, residues in the binding pocket known to contribute interactions according to PDBsum were targeted to create a site-saturation variant library in MODELLER (Fig. 2A) [31]. The residues selected were positions 97, 99, 128, 146, 147, 196, 202, and 203, which are all critical residues for interactions in the binding pocket, with positions 99, 128, and 203 participating in hydrogen bonds with CCM, and 97, 146, 147, 196, and 202 participating in van der Waals/hydrophobic interactions with the ligand (Figs. 2B, 2 C). The BenM structure was incorporated into the computational workflow, producing a site-saturation variant library of the selected residues, resulting in 150 variants, accounting for single canonical amino acid substituted variants minus two variants that could not be generated due to clashes at the selected parameters. Each 3D model in the in silico saturation variant library was then docked with both CCM and AA to generate docking scores.
Fig. 2.
(A) PDBsum result of PDB 2F7A. (B) View of critical residue interactions between the selected residues (cyan) and CCM (green). (C) View of the binding pocket of BenM bound to CCM. (D) Variant BenMP196D (yellow) docked with AA (green).
2.2. Molecular docking results reveals hotspot residues for specificity
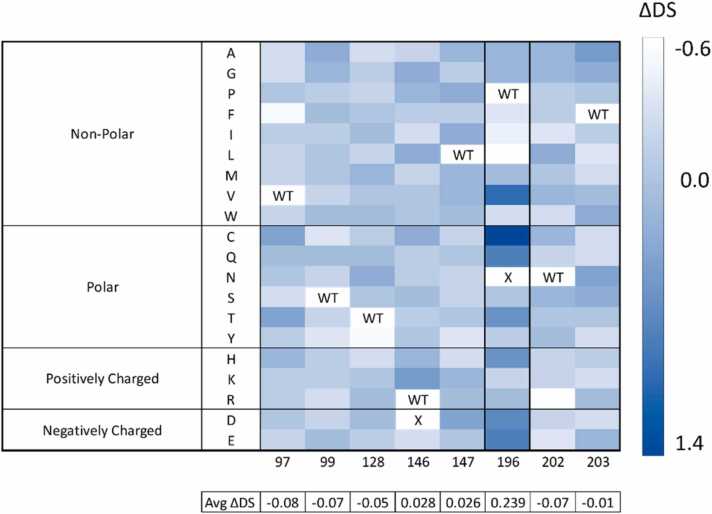
To compare in silico results, the docking score of AA was subtracted from the docking score of the cognate molecule CCM (resulting in a metric referred to as ΔDS here) for each of the mutants in the site-saturation library (Fig. 3). With this comparative scoring method, more positive differences are desirable, indicating that the pose produces a more favorable docking score with AA. When the ΔDS were averaged across the substitutions per each residue, our results indicated that there were specific hotspot residues that, when substituted, resulted in the largest changes in the computed docking scores. In particular, residue 196 showed the highest average of ΔDS of 0.239, while the average ΔDS across all variants at all 8 residues was only 0.002.
Fig. 3.
Heatmap of the difference of docking scores between the saturation variant library with AA and CCM. The ΔDS scale at the right is the AA score subtracted from the respective CCM docking score (kcal/mol). “X” indicates that the computational workflow was not able to generate a feasible variant or pose. The average of the ΔDS calculated for all the available mutants is listed below each residue.
The top seven variants of residue 196 resulted in ΔDS scores above 0.5 (Table 1 and Table S1). Not only are these the top substitutions at that residue, but they were also the variants with the highest difference scores among the entire in silico library. This suggested that residue 196 is a hotspot for conferring ligand specificity according to our computational results. Variant BenMP196C resulted in the highest ΔDS of 1.4 kcal/mol followed by BenMP196V, BenMP196Q and BenMP196E, BenMP196D, BenMP196H, and BenMP196T. Based on the docking results, those top variants in terms of highest ΔDS were chosen for experimental verification.
Table 1.
Docking Scores Ordered by ΔDS.
| Mutation | CCM Docking Score (kcal/mol) | AA Docking Score (kcal/mol) | ΔDS (CCM-AA) |
|---|---|---|---|
| P196C | -4.6 | -6 | 1.4 |
| P196V | -4.8 | -5.8 | 1 |
| P196Q | -4.9 | -5.7 | 0.8 |
| P196E | -5.2 | -6 | 0.8 |
| P196D | -5.4 | -6.1 | 0.7 |
| P196H | -5 | -5.6 | 0.6 |
| P196T | -4.7 | -5.3 | 0.6 |
| WT | -5.9 | -5.5 | -0.4 |
2.3. Experimental validation of BenM variants of interest
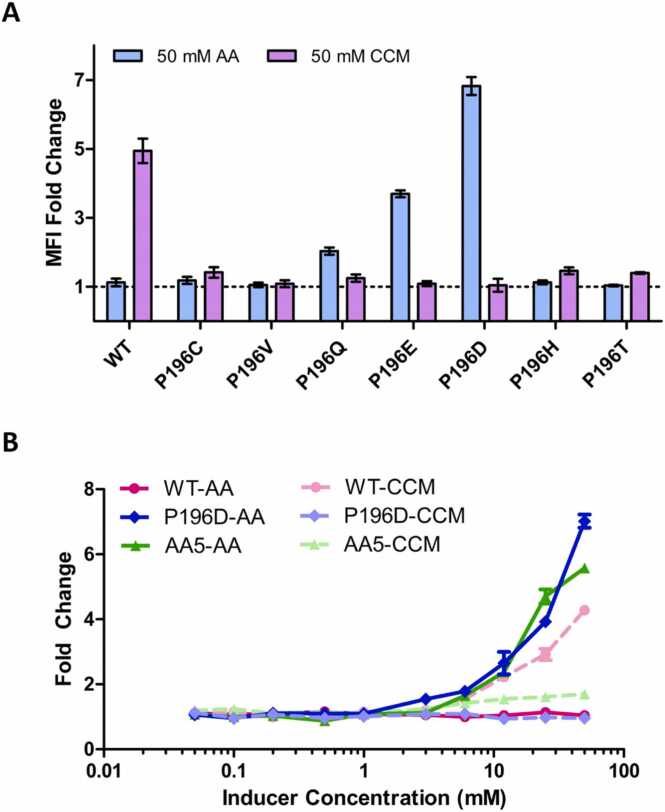
Biosensors including WT BenM and selected BenM variants were constructed and tested by flow cytometry for fluorescence protein (GFP) signal to verify the docking predictions (Fig. 4A). Within a previously established low-copy biosensor plasmid pRolR, we placed the BenM open reading frame upstream of the operator sequence BenO controlling GFP expression in E. coli, resulting in pBenM [24]. Out of the selected variants representing the in silico top ranked variants, variant BenMP196D showed the desired altered specificity profile and with the highest fold change. While the WT BenM sensor shows 4.4-fold more response (measured by fold change of mean fluorescence intensity, MFI) to CCM relative to AA, the BenMP196D sensor shows 6.5 more response to AA relative to CCM. Importantly, BenMP196D showed essentially no response to CCM at this tested concentration, as indicated by the MFI value of 1. BenMP196D is followed by the BenMP196E sensor, containing the other polar negatively charged acidic amino acid variant, with a 3.4-fold more response to AA than CCM. BenMP196Q, harboring a substitution to another polar amino acid with a flexible sidechain of similar size, also showed some specificity to AA, but with a lower fold change (1.6-fold more response to AA than CCM). Notably, the substitutions are single substitutions altering the specificity of BenM from CCM to AA compared to the WT. The BenMP196C, BenMP196H, and BenMP196T sensors demonstrated compromised response to CCM, and only minor gains in response to AA. The BenMP196V sensor demonstrated no response to either CCM or AA.
Fig. 4.
In vivo experimental characterization of mutants. (A) Specificity of top docking-predicted variants against AA and CCM. (B) Dose-response curve of WT, P196D, and AA5 literature mutant towards AA and CCM. Fold changes are relative to uninduced cultures and represent the mean of n = 3 replicates.
To further evaluate the performance of BenMP196D, the top-performing biosensor, a dose-response curve was generated across a range of ligand concentrations (Fig. 4B, S1A). For comparison, dose-response curves were also generated for the WT and AA5, a mutant previously discovered in S. cerevisiae [23]. AA5 was selected for having the highest fold change towards AA and reduced specificity towards CCM [23]. The WT BenM sensor behaves as expected with induction by CCM and not AA. The dose-response curves showed a clear change in specificity where BenMP196D is responsive to AA but not CCM or other tested dicarboxylic acids (Figure S2). Based on this data, the BenMP196D sensor can detect AA at millimolar concentrations, with distinguishable responses starting between 1 mM and 3 mM of AA. In our hands, the AA5 sensor, when constructed in E. coli, showed the expected response towards AA, but with some residual activity to higher concentrations of CCM. Overall, our experimental results showed that the in silico workflow can be used to guide our selection of variants to test that can achieve the desired complete ligand specificity change of BenM from CCM to AA in vivo.
2.4. Cell-free biosensors demonstrate orthogonal application with increased sensitivity
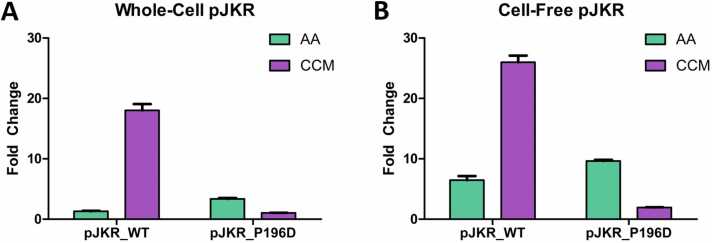
To characterize the performance of the engineered BenMP196D biosensor in other contexts and to enable future engineering, we implemented a cell-free reaction system to express the BenM biosensors. Cell-free systems provide a variety of advantages to the whole-cell sensors such as overcoming constraints like ligand uptake limitations and differences between cellular contexts [32], [33]. We implemented a cell-free system utilizing a previously developed high-copy vector expressing WT BenM, or BenMP196D, named here as pJKR_WT and pJKR_P196D, respectively [34], [35]. To validate this vector system in the context of the whole E. coli cell, the BenM sensors encoded on pJKR_WT and pJKR_P196D behaved consistently with the results of pBenMWT and pBenMP196D as expressed earlier (Fig. 4A), where pJKR_WT exhibited increased fluorescence in the presence of CCM, while pJKR_P196D exhibited higher fluorescence in the presence of AA, with inducers at 15 mM (Fig. 5A). 15 mM was selected as a concentration that produced a strong response in the whole-cell tests. In the cell-free system, when we evaluated the same vectors with 15 mM inducers, both variants exhibited behavior similar to the respective whole-cell tests, with pJKR_WT showing higher GFP signal with CCM, and pJKR_P196D showing higher GFP signal with AA (Fig. 5B, S1B). Notably, the cell-free tests showed increased sensitivity to the ligands compared to the whole-cell tests, with the mean fluorescence fold change response of pJKR_WT towards CCM at 26-fold in cell-free vs 18-fold in whole-cell. Similarly, pJKR_P196D’s fold change towards AA increased from an average of 3-fold in whole-cell to nearly 10-fold in cell-free. For pJKR_P196D, this is an approximately 3-fold improvement in sensitivity in the cell-free system compared to the whole-cell system at equimolar concentrations.
Fig. 5.
Whole-cell and cell-free tests of WT BenM and P196D BenM in the pJKR vector. (A) Whole-cell BenM tests were recorded by flow cytometry using AA and CCM at 15 mM at 20 h. (B) Cell-free BenM tests using AA and CCM at 15 mM with fold change recorded at 20 h. n = 3 for all experiments.
2.5. Molecular dynamic simulations of BenM variants uncover mechanisms of specificity
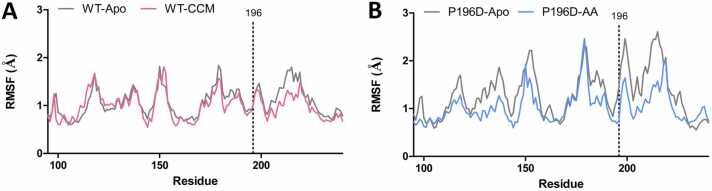
To elucidate the molecular mechanisms of its altered specificity, we utilized molecular dynamics (MD) to analyze the model of the structure of BenMP196D. The root-mean-square fluctuation (RMSF) indicator, reflective of flexibility on a per-residue basis, showed that the apo BenMP196D structure undergoes a higher level of residue fluctuation compared to the apo WT (Fig. 6 A). In particular, residue 196 displayed more flexibility in the simulation with the introduction of the P196D substitution, increasing its RMSF of 0.996 Å in the WT apo structure to 1.48 Å in the BenMP196D apo structure, consistent with the loss of the rigid proline sidechain (Fig. 6B). We also observed increases in RMSF’s in the residues surrounding the P196D variant, likely attributable to the loss of the stability that would have been provided by a proline at position 196. In both cases, the simulations with the ligand showed a decrease in RMSF, indicating the stabilizing role played by the respective ligands. Notably, the decrease in RMSF upon ligand binding is higher at position 196 in the P196D variant, reflecting that the flexibility of the aspartate variant is decreased upon ligand binding, compared to the typically rigid WT proline. In the P196D variant simulation, the RMSF at residue 196 decreases from 1.48 Å in the apo structure to 0.745 Å in the complex with AA, indicating that the aspartate at residue 196 creates strong interactions with AA and therefore plays an important role in AA binding. In the second-best performing variant experimentally, P196E, AA binding also stabilizes the RMSF, but to a lesser extent compared to P196D (Figure S3A).
Fig. 6.
(A) RMSF comparison of WT BenM with CCM and without (apo). (B) RMSF comparison of mutant P196D with and without AA.
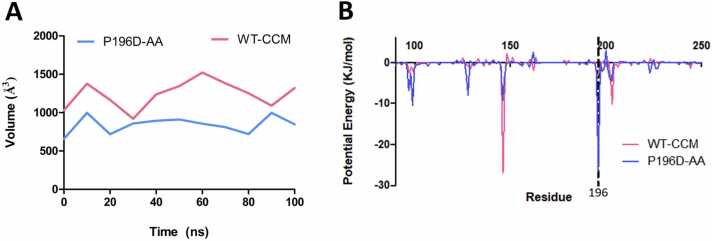
To look at the structural changes to the binding pocket as a result of the substitution, we analyzed the volume of the ligand binding pocket throughout the simulation with fpocket (Fig. 7 A) [36]. Overall, the WT structure consistently maintains a lower pocket volume when bound to CCM than P196D when bound to AA. The WT structure has an average pocket volume of 1241 Å3 over the course of the simulation compared to P196D’s average pocket volume of 843 Å3. The decrease in pocket size is likely the result of the presence of the flexible sidechain with the aspartate substitution from the WT proline. P196D also has a smaller average pocket volume compared to the second-best performing variant experimentally, P196E (Figure S3B).
Fig. 7.
(A) Binding pocket volume calculation over the simulation time shows that the WT BenM has a larger pocket volume compared to the P196D variant across the whole simulation. (B) Per-residue decomposition of potential energy from simulations of WT BenM with CCM and the P196D variant with AA. The per-residue decomposition implicates different residues as contributors to ligand binding in each case. Position 196 is marked.
In addition to the previous analysis, the Molecular Mechanics Poisson-Boltzmann Surface Area (MM/PBSA) approach was conducted to assess binding energies of the simulations [37], [38]. The potential energy (which includes van der Waal and electrostatic energy) per-residue decomposition indicates different contributions from residues of interest in each complexed simulation (Fig. 7B). The results confirm that residue 196 plays a significant role in AA binding by BenMP196D, while residue 146 plays a major role in WT binding of CCM as indicated by the significant potential energy at these positions, respectively. Overall, these findings show that the P196D substitution alters the substrate specificity through the introduction of different interactions and a decrease in volume of the ligand binding pocket, altering the binding mode of BenM.
3. Discussion
In this study, we successfully altered the specificity of the BenM TF towards AA, removing its natural recognition of CCM, with a single residue substitution identified by a computational pipeline. Instead of existing approaches using computational methods to guide the selection of residues for semi-rational engineering, we utilized our computational method for in silico screening of a semi-rational library, resulting in a focused selection for experimental testing. The high-throughput in silico amino acid substitution and docking pipeline first identified a hotspot with a significant role in ligand specificity determination and then allowed for the evaluation of amino acid substitutions focused on the identified hotspot, thereby delineating a short list of BenM variants for experimental assessment. Variant BenMP196D demonstrated completely altered specificity with a preference for AA and negligible specificity towards the cognate molecule CCM in our E. coli system. In addition to BenMP196D, the variants BenMP196Q and BenMP196E, containing substitutions with similar amino acids, also achieved notable specificity changes in the computational workflow and in experimental testing. Our experimental verification resulted in a highly specific whole-cell biosensor which appears to outperform the specificity of previously engineered BenM variants in our context and tested concentrations. The negatively charged amino acids are overrepresented in the top docking variants since both glutamate and aspartate are in the top seven variants according to the docking scores. Once we experimentally characterized a series of substitutions at position 196, it became clear that the computational pipeline effectively identified a key hotspot in BenM for the determination of ligand specificity, thereby achieving our goal of developing a method capable of reducing the set of variants requiring experimental validation. In addition, the experimental characterization validated that negatively charged amino acids at position 196 confer a significant ligand specificity swap.
While aspects of our approach have been utilized by others, the synthesis of the different components of this work is innovative, especially with respect to TF-based biosensor ligand specificity engineering [39], [40]. For example, Rosetta or MD can be used to virtually screen variants, looking at changes in predicted binding energy [41], [42]. In silico saturation mutagenesis utilizing MODELLER and docking similar to our workflow here has been demonstrated previously but without experimental verification [43]. While examples exist of virtual saturation mutagenesis libraries used to guide experimental testing [44], the comparative method presented here is the only one to our knowledge that specifically utilized the output to switch the specificity of a transcription factor. Other methods may predict hotspot residues for impactful protein variants but do not consider our objective of reducing the specificity of one molecule in favor of another. When compared to existing hotspot predictors HotSpot Wizard and SNAP2, which utilize different algorithms for determining functional consequences of protein substitutions, our method is the only one to highlight residue 196 (Figure S4) [45], [46]. This positions our workflow as a unique addition to the computational protein engineering toolkit, providing guidance for semi-rational design particularly when specificity needs to be altered.
While some substitutions identified in the computational workflow with favorable ΔDS for AA recognition were validated experimentally for this altered ligand specificity, the experimentally derived ligand specificities for some of the other computationally predicted substitutions did not correlate well, illustrating the limitations of the computational method. Based on the computational pipeline alone, the top substitutions comprised amino acids with a variety of characteristics, suggesting that the docking simulations could not provide sufficient precision in the best amino acid type to optimize AA binding. This could be due to a variety of possible reasons. These substitutions in BenM could have been deleterious due to impacts in expression, folding, or solubility of the sensor in the tested E. coli whole cell system. Although the top ranked substitutions identified by the computational workflow did not all correlate with experimental results, we did obtain experimental results that showed a successful delineation of one hotspot position out of the full protein sequence. The other residues in the binding pocket that had positive average ΔDS are positions 146 and 147, which makes them interesting candidates for further study. Residue 146 was found to be a major contributor to CCM binding in the WT molecular dynamic simulations, highlighting the importance of this position. While the residues targeted in our workflow focus on a small number in the ligand binding pocket, a full saturation library would have resulted in over 100 variants to evaluate experimentally. Our workflow brought required testing down to a handful of variants by focusing on not only a single residue, but a subset of possible amino acids for that position. Such a determination of specific amino acids to substitute in each position would be difficult rationally. Our results show the applicability of molecular docking studies to guide selection of hotspots to target for site-saturation mutagenesis, which could then be experimentally validated, thus reducing the experimental resources necessary. This is an alternative to constructing and testing a larger library targeting multiple positions throughout the protein sequence or introducing and screening through random variant libraries. However, the limitations of this method in terms of the possible TFs and ligands that could targeted are unknown. There are likely constraints in the binding pocket of TFs that would prevent a TF from being engineered towards any ligand of interest, such as size and charge. In this work, we focused on the modification of specificity towards a similar molecule in terms of class and size. Future work would need to explore such limitations.
The success rate for our computational approach could be improved as docking algorithms continue to advance, or by using machine learning-based algorithms that can learn from positive and negative data and incorporate aspects such as solubility and stability based on sequence or structure-based predictions. Using more simulations and clustering combined with rational decisions could also help facilitate the use of computational methods for guiding experiments [47]. Determining an alternative metric to rank optimal substitutions could also be used to rank the substitutions for experimental evaluation. It is worth noting that ΔDS does not capture the variation between docking scores of the different variants towards each ligand. For example, variant BenMP196T had a high difference score, but the individual scores for CCM and AA are below that of BenMWT.
In addition to establishing a variant BenM specific for AA through just a single substitution, we also demonstrate the portability of the biosensor to an orthogonal application as a cell-free biosensor. It is notable that our operational range in our in vivo test is lower than previous demonstrations in S. cerevisiae, likely due to differences in acid transport. While previous biosensors have been able to achieve the engineering of BenM towards AA to some success, the operating range of detection may be limited due to the uptake of the molecule, which can be ameliorated by the implementation of a dicarboxylic acid transporter as was done for PcaR in P. putida [23], [24], [25]. Targeting biosensor circuit components such as the ribosomal binding site for optimization could also be used to improve biosensor performance [16], [48]. Alternatively, cell-free systems allow implementation of the biosensor outside of cellular contexts that may impact biosensor response. Previous studies have observed differences in response profiles of biosensors implemented in prokaryotes versus eukaryotes [49]. Our results show that the cell-free system has an increased sensitivity compared to the whole-cell biosensor at equimolar concentrations and confirm that the observed differences in specificity upon the P196D substitution are not due to artifacts from the context of the biosensor. Our cell-free system opens new opportunities for utilizing the BenM biosensor, such as in cell-free screening of variants of enzymes that bind CCM or AA [50].
While the success of docking pipelines vary, MD simulations have been proven to give insight on the stability of binding over time and have been demonstrated to incorporate the flexibility of the ligand and receptors more effectively than docking [51], [52]. Our molecular dynamics results showed the impact of substitutions at position 196 on binding stability and highlighted that the mechanisms of specificity can be altered through a single residue substitution. In addition, the analysis suggested that changes in the volume of the pocket could also impact ligand specificity, limiting the accessibility of the binding pocket to CCM, which is less flexible compared to AA. CCM contains two double bonds, making the molecule more rigid overall compared to AA.
While our workflow relied on an existing ligand-bound protein structure, developments in protein structure prediction such as AlphaFold2 and its application for ligand-bound structures, would make it possible to generate reasonable ligand-bound structures as starting points for further simulation and analysis [53], [54]. With this in mind, the developed workflow can potentially be applied to other proteins to alter specificity towards molecules of interest.
4. Methods
4.1. Homology modeling and refinement
Variant models of the BenM ligand binding domain using the structure PDB: 2F7A were generated with a custom script based on MODELLER 9.21 model_mutate.py. The modified script allowed for the high-throughput generation of models for site-saturation variants for residues of interest. Energy minimization was performed using UCSF Chimera with default settings via a high-throughput script [29].
4.2. Molecular docking
AutoDock Vina was used to dock ligands into the previously characterized BenM active site of the energy-minimized mutant models to identify binding conformations and docking scores [30]. AutoDock Vina was run with a grid box of 20x28x30 Å3 and 1 Å spacing with an exhaustiveness of 25. Ligands and models were prepared using AutoDock Tools. Docking was run on the flexible setting, allowing for ligands and receptors to be able to rotate. Interacting residues based on PDBsum were selected to be flexible [31]. Docking and molecular dynamics simulations were run using supercomputing resources provided by the Digital Research Alliance of Canada.
4.3. Plasmid construction
E. coli strain DH5α was used for cloning and testing. Primers used in this study were purchased from IDT or Eurofins. Gene fragments were purchased from Twist Biosciences. Plasmid pRolR, which is modified from pQacR-Q2 (Addgene plasmid # 74690) [55], was obtained as a gift from Mohamed Nasr and used as our biosensor vector [56]. Variant BenM genes were inserted into the biosensor vector pBenM containing the BenM promoter and GFP using ligation-independent cloning or generated with site-directed mutagenesis using the pfu polymerase. Plasmids used in this work are listed in Table S2.
4.4. Biosensor assay and flow cytometry
E. coli cells harboring biosensor plasmids were grown overnight at 37 °C in LB media containing 50 ug/mL kanamycin. Overnight cultures were used to seed fresh 5 mL tubes of LB with kanamycin 1:100. Cultures were grown to exponential phase at OD= 0.6. Cultures were cooled to room temperature and used to inoculate media to OD= 0.2 in deep well plates containing media with 1 mM IPTG and various concentrations of ligands of interest at pH 7.0. Cultures were grown for 20 h and diluted into PBS for flow cytometry analysis. Cells were measured by the LSR II Flow Cytometer. Fluorescence of GFP was measured for 10,000 events and the mean fluorescence intensity (MFI) was determined for each population using FlowJo. Fold-induction was calculated by dividing the MFI of the induced conditions by the MFI of the uninduced condition. Data represents 3 replicates.
4.5. Cell-Free extract preparation and reaction
The cell-free extracts and reactions were prepared and conducted based on previously published methods by Silverman et. al and Kwon & Jewett [31], [57] E. coli BL21(DE3) ΔiscR was obtained from Dr. Patrick Hallenbeck and inoculated into 30 mL LB and grown in a 250 mL baffled flask at 37 °C overnight [58], [59]. After overnight growth, 1 L of 2X YT+P (16 g/L tryptone, 10 g/L yeast extract, 5 g/L sodium chloride, 7 g/L potassium phosphate dibasic, 3 g/L potassium phosphate monobasic) was inoculated with 20 mL of the overnight culture in a 2.5 L flask at 37 °C. Cultures were grown to an optical density (OD600) of 3.0 ± 0.2. Cultures were then divided and centrifuged for 10 min at 5000 g at 4 °C to obtain the cell pellet. Cell pellets were washed two times in 25 mL of wash buffer (50 mM Tris, 14 mM magnesium glutamate, 60 mM potassium glutamate, 2 mM DTT, brought to pH 7.7 with acetic acid). Pellets were flash-frozen in liquid nitrogen and thawed the next day. When thawed, the cell pellets were lysed by sonication using an amplitude of 50 with 10-second on/off cycles for a total of 10 min of sonication time, delivering a total of 20,000 J. Tubes were centrifuged at 12,000 g for 10 min at 4 °C and the supernatant was removed. For the runoff reaction, the tubes of crude lysate were covered with aluminum and incubated shaking at 200 rpm for 80 min at 37 °C. After another round of centrifugation, the supernatant was pipetted off and loaded into a 10 K MWCO Slide-A-Lyzer (Thermo) dialysis cassette. The supernatant was dialyzed against 600 mL of dialysis buffer (5 mM Tris, 14 mM magnesium glutamate, 60 mM potassium glutamate, 1 mM DTT, pH 8.2) at 4 °C for 3 h. After dialysis, the extract was removed from the cassette and centrifuged at 12,000 g for 10 min at 4 °C. The final supernatant was removed, aliquoted, and flash-frozen with liquid nitrogen for storage at − 80 °C.
Our cell-free reactions were conducted using mini-prepped plasmid concentrated using a SpeeVac to the appropriate concentrations that allow for final plasmid concentrations of 35 nM. pOPINN-GFP was a gift from Ray Owens (Addgene plasmid # 53541) and was used as a fluorescence positive control. The final cell-free reaction mix is as follows: 16 mM magnesium glutamate; 10 mM ammonium glutamate; 130 mM potassium glutamate; 1.2 mM ATP; 0.850 mM each of GTP, UTP, and CTP; 0.034 mg/mL folinic acid; 0.171 mg/mL E.coli tRNA; 2 mM of the 20 canonical amino acids; 30 mM PEP; 0.33 mM NAD; 0.27 mM CoA; 4 mM oxalic acid; 1 mM putrescine; 1.5 mM spermidine; 57 mM HEPES; 30% CFE extract by volume; 35 nM of plasmid DNA); and water. Cell-free reactions were prepared on ice in triplicate at 10 µl volumes. Solutions were pipetted into a 384-well plate, avoiding bubbles. Plates were sealed and briefly spun at 3000 RPM. GFP fluorescence was monitored at 487 nm excitation and 520 emission every 10 min on a BioRad plate reader for up to 20 h at 30 °C.
4.6. Molecular dynamics simulations
MD simulations were executed using the GROMACS package version 2020.2 [60]. The system was set inside a dodecahedron box. The CHARMM forcefield was selected along with the TIP3P water model. Na+ and Cl- ions were proportionally added to neutralize the system. An energy minimization step was conducted employing the Steepest Descent algorithm with up to 50000 steps. For the isothermal-isometric (NVT) ensemble, the leap-frog integrator algorithm was with a time step of 2 fs. Bonds were constrained using the LINCS algorithm and the long-range electrostatic interaction was calculated using the Particle Mesh Ewald (PME) method. The Maxwell distribution was used to generate the initial velocity from a random seed. For the isothermal-isobaric (NPT) ensemble, the V-rescale method was used for temperature coupling and Berendsen method for pressure coupling. The MD simulation was performed for 100 ns for apo and complex forms of the variants from the docking results. The MD simulation trajectories were analyzed using functions including the gmx rmsf commands to calculate the root-mean-square fluctuation (RMSF). The Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA) method was employed to calculate the binding free energies for ligand-bound structures using the g_mmpbsa program [38]. Pocket volume was calculated using fpocket [36]. Structure frames were extracted every 10 ns and volumes were calculated in Fpocket using default settings with the ligand removed, focusing on the known binding pocket.
CRediT authorship contribution statement
Chester Pham: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. Peter J. Stogios: Conceptualization, Methodology, Resources, Supervision, Writing – original draft, Writing – review & editing. Alexei Savchenko: Conceptualization, Supervision, Writing – original draft, Writing – review & editing. Radhakrishnan Mahadevan: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
We would like to thank Mohamed Nasr from Concordia University for providing the vector pRolR and Patrick Hallenbeck from the University of Montreal for providing the E. coli ΔiscR strain. We thank Brenda Wang and Michael Jewett from Northwestern University for providing guidance on the implementation of the cell-free system, and Katherine Picott from the University of Toronto for assistance in preparing cell-free extracts and reaction components. We also thank the staff at the Temerty Faculty of Medicine Flow Cytometry Facility at the University of Toronto, for access, training, and support for the flow cytometry experiments. This research was enabled in part by support provided by the Digital Research Alliance of Canada. This study was financially supported by Natural Sciences and Engineering Research Council of Canada (NSERC) through the Discovery Grant, the Industrial Biocatalysis Network, the Biochemicals from Cellulosic Biomass (BioCeB) grant through the Ministry of Ontario, and Genome Canada through a Genomics Applied Partnership Program (GAPP) grants to A.S. and R.M.; C.P. was supported by the NSERC Collaborative Research and Training Experience (CREATE) program.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2024.05.002.
Contributor Information
Alexei Savchenko, Email: alexei.savchenko@ucalgary.ca.
Radhakrishnan Mahadevan, Email: krishna.mahadevan@utoronto.ca.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Chen X., Zhou L., Tian K., Kumar A., Singh S., Prior B.A., et al. Metabolic engineering of Escherichia coli: a sustainable industrial platform for bio-based chemical production. Biotechnol Adv. 2013;31:1200–1223. doi: 10.1016/J.BIOTECHADV.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Polen T., Spelberg M., Bott M. Toward biotechnological production of adipic acid and precursors from biorenewables. J Biotechnol. 2013;167:75–84. doi: 10.1016/J.JBIOTEC.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Bart J.C.J., Cavallaro S. Transiting from Adipic Acid to Bioadipic Acid. 1, Petroleum-Based Processes 2014. 10.1021/ie5020734. [DOI]
- 4.Averesch N.J.H., Martínez V.S., Nielsen L.K., Krömer J.O. Toward synthetic biology strategies for adipic acid production: an in silico tool for combined thermodynamics and stoichiometric analysis of metabolic networks. ACS Synth Biol. 2018;7:490–509. doi: 10.1021/acssynbio.7b00304. https://doi.org/10.1021/ACSSYNBIO.7B00304/SUPPL_FILE/SB7B00304_SI_011.PDF. [DOI] [PubMed] [Google Scholar]
- 5.Kruyer N.S., Peralta-Yahya P. Metabolic engineering strategies to bio-adipic acid production. Curr Opin Biotechnol. 2017;45:136–143. doi: 10.1016/J.COPBIO.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Joo J.C., Khusnutdinova A.N., Flick R., Kim T., Bornscheuer U.T., Yakunin A.F., et al. Alkene hydrogenation activity of enoate reductases for an environmentally benign biosynthesis of adipic acid. Chem Sci. 2017;8:1406–1413. doi: 10.1039/c6sc02842j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raj K., Partow S., Correia K., Khusnutdinova A.N., Yakunin A.F., Mahadevan R. Biocatalytic production of adipic acid from glucose using engineered Saccharomyces cerevisiae. Metab Eng Commun. 2018;6:28–32. doi: 10.1016/J.METENO.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogers J.K., Taylor N.D., Church G.M. Biosensor-based engineering of biosynthetic pathways. Curr Opin Biotechnol. 2016;42:84–91. doi: 10.1016/J.COPBIO.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Dietrich J.A., Mckee A.E., Keasling J.D. High-throughput metabolic engineering: advances in small-molecule screening and selection. Annu Rev Biochem. 2010;79:563–590. doi: 10.1146/annurev-biochem-062608-095938. [DOI] [PubMed] [Google Scholar]
- 10.Carpenter A.C., Paulsen I.T., Williams T.C. Blueprints for biosensors: design, limitations, and applications. Genes. 2018;9 doi: 10.3390/GENES9080375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Paepe B., Peters G., Coussement P., Maertens J., De Mey M. Tailor-made transcriptional biosensors for optimizing microbial cell factories. J Ind Microbiol Biotechnol. 2017;44:623–645. doi: 10.1007/S10295-016-1862-3. [DOI] [PubMed] [Google Scholar]
- 12.Mustafi N., Grünberger A., Kohlheyer D., Bott M., Frunzke J. The development and application of a single-cell biosensor for the detection of l-methionine and branched-chain amino acids. Metab Eng. 2012;14:449–457. doi: 10.1016/J.YMBEN.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Kim W., Lee S., Sung B.H., Na J.-G., Lee J.W. Cell-free systems and genetic biosensors for accelerating enzyme and pathway prototyping. Curr Opin Syst Biol. 2024;37 doi: 10.1016/j.coisb.2023.100501. [DOI] [Google Scholar]
- 14.Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021;49:D412–D419. doi: 10.1093/NAR/GKAA913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tellechea-Luzardo J., Stiebritz M.T., Carbonell P. Transcription factor-based biosensors for screening and dynamic regulation. Front Bioeng Biotechnol. 2023;11 doi: 10.3389/fbioe.2023.1118702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pham C., Stogios P.J., Savchenko A., Mahadevan R. Advances in engineering and optimization of transcription factor-based biosensors for plug-and-play small molecule detection. Curr Opin Biotechnol. 2022;76 doi: 10.1016/J.COPBIO.2022.102753. [DOI] [PubMed] [Google Scholar]
- 17.Ray S., Panjikar S., Anand R. Structure guided design of protein biosensors for phenolic pollutants. ACS Sens. 2017;2:411–418. doi: 10.1021/acssensors.6b00843. [DOI] [PubMed] [Google Scholar]
- 18.Ray S., Panjikar S., Anand R. Design of protein-based biosensors for selective detection of benzene groups of pollutants. ACS Sens. 2018 doi: 10.1021/acssensors.8b00190. [DOI] [PubMed] [Google Scholar]
- 19.Tinberg C.E., Khare S.D., Dou J., Doyle L., Nelson J.W., Schena A., et al. Computational design of ligand-binding proteins with high affinity and selectivity. Nature. 2013;501:212–216. doi: 10.1038/nature12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Della Corte D., van Beek H.L., Syberg F., Schallmey M., Tobola F., Cormann K.U., et al. Engineering and application of a biosensor with focused ligand specificity. Nat Commun. 2020;11:1–11. doi: 10.1038/s41467-020-18400-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collier L.S., Gaines Iii G.L., Neidle E.L. Regulation of benzoate degradation in acinetobacter sp. Strain ADP1 by BenM, a LysR-type transcriptional activator. 1998;vol. 180 doi: 10.1128/jb.180.9.2493-2501.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ezezika O.C., Haddad S., Clark T.J., Neidle E.L., Momany C. Distinct effector-binding sites enable synergistic transcriptional activation by BenM, a LysR-type Regulator. J Mol Biol. 2007;367:616–629. doi: 10.1016/j.jmb.2006.09.090. [DOI] [PubMed] [Google Scholar]
- 23.Javanpour A.A., Liu C.C. Evolving small-molecule biosensors with improved performance and reprogrammed ligand preference using OrthoRep. ACS Synth Biol. 2021;10:2705–2714. doi: 10.1021/ACSSYNBIO.1C00316/SUPPL_FILE/SB1C00316_SI_003.XLSX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snoek T., Chaberski E.K., Ambri F., Kol S., Bjørn S.P., Pang B., et al. Evolution-guided engineering of small-molecule biosensors. Nucleic Acids Res. 2019;48 doi: 10.1093/nar/gkz954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin S., Jha R.K., Dale T. Tackling the Catch-22 situation of optimizing a sensor and a transporter system in a whole-cell microbial biosensor design for an anthropogenic small molecule. ACS Synth Biol. 2022 doi: 10.1021/acssynbio.2c00364. [DOI] [PubMed] [Google Scholar]
- 26.Craven S.H., Ezezika O.C., Haddad S., Hall R.A., Momany C., Neidle E.L. Inducer responses of BenM, a LysR-type transcriptional regulator from Acinetobacter baylyi ADP1. Mol Microbiol. 2009;72:881–894. doi: 10.1111/j.1365-2958.2009.06686.x. [DOI] [PubMed] [Google Scholar]
- 27.Ruangprasert A., Craven S.H., Neidle E.L., Momany C. Full-length structures of benm and two variants reveal different oligomerization schemes for LysR-Type Transcriptional Regulators. J Mol Biol. 2010;404:568–586. doi: 10.1016/j.jmb.2010.09.053. [DOI] [PubMed] [Google Scholar]
- 28.Eswar N., Webb B., Marti-Renom M.A., Madhusudhan M.S., Eramian D., Shen M., et al. Comparative protein structure modeling using modeller. Curr Protoc Bioinforma. 2006;15 doi: 10.1002/0471250953.bi0506s15. 5.6.1-5.6.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., et al. UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/JCC.20084. [DOI] [PubMed] [Google Scholar]
- 30.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function. Effic Optim multithreading. 2010 doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laskowski R.A., Hutchinson E.G., Michie A.D., Wallace A.C., Jones M.L., Thornton J.M. PDBsum: a web-based database of summaries and analyses of all PDB structures. Trends Biochem Sci. 1997;22:488–490. doi: 10.1016/S0968-0004(97)01140-7. [DOI] [PubMed] [Google Scholar]
- 32.Tinafar A., Jaenes K., Pardee K. Synthetic biology goes cell-free. BMC Biol. 2019;17:64. doi: 10.1186/s12915-019-0685-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silverman A.D., Karim A.S., Jewett M.C. Cell-free gene expression: an expanded repertoire of applications. Nat Rev Genet. 2020;21:151–170. doi: 10.1038/s41576-019-0186-3. [DOI] [PubMed] [Google Scholar]
- 34.Silverman A.D., Kelley-Loughnane N., Lucks J.B., Jewett M.C. Deconstructing cell-free extract preparation for in vitro activation of transcriptional genetic circuitry. ACS Synth Biol. 2019;8:403–414. doi: 10.1021/ACSSYNBIO.8B00430/SUPPL_FILE/SB8B00430_SI_003.XLSX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogers J.K., Church G.M. Genetically encoded sensors enable real-time observation of metabolite production. Proc Natl Acad Sci USA. 2016;113:2388–2393. doi: 10.1073/pnas.1600375113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le Guilloux V., Schmidtke P., Tuffery P. Fpocket: An open source platform for ligand pocket detection 2009. 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed]
- 37.Genheden S., Ryde U. Expert Opinion on Drug Discovery The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities 2015. 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed]
- 38.Kumari R., Kumar R., Consortium O.S.D.D., Lynn A. g_mmpbsa—a GROMACS Tool for High-Throughput MM-PBSA Calculations. J Chem Inf Model. 2014;54:1951–1962. doi: 10.1021/CI500020M. [DOI] [PubMed] [Google Scholar]
- 39.Marques S.M., Planas-Iglesias J., Damborsky J. Web-based tools for computational enzyme design. Curr Opin Struct Biol. 2021;69:19–34. doi: 10.1016/j.sbi.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 40.Sequeiros-Borja C.E., Surpeta B., Brezovsky J. Recent advances in user-friendly computational tools to engineer protein function. Brief Bioinforma. 2020;2020:1–15. doi: 10.1093/bib/bbaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang J., Dai S., Chen X., Xu L., Yan J., Yang M., et al. Alteration of chain-length selectivity and thermostability of rhizopus oryzae lipase via virtual saturation mutagenesis coupled with disulfide bond design. Appl Environ Microbiol. 2023 doi: 10.1128/AEM.01878-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Öhlknecht C., Katz S., Kröß C., Sprenger B., Engele P., Schneider R., et al. Efficient in silico saturation mutagenesis of a member of the caspase protease family. J Chem Inf Model. 2021;61:1193–1203. doi: 10.1021/acs.jcim.0c01216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiappori F., D’Ursi P., Merelli I., Milanesi L., Rovida E. In silico saturation mutagenesis and docking screening for the analysis of protein-ligand interaction: The endothelial protein C receptor case study. BMC Bioinforma. 2009;10:1–8. doi: 10.1186/1471-2105-10-S12-S3/FIGURES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo C., Lv Y., Li H., Zhou J., Xu S. De novo biosynthesis of 8-prenylnaringenin in Saccharomyces cerevisiae improved by screening and engineering of prenyltransferases and precursor pathway. Syst Microbiol Biomanuf. 2022 doi: 10.1007/S43393-022-00133-4. 2022;1:1–12. [DOI] [Google Scholar]
- 45.Hecht M., Bromberg Y., Rost B. Better prediction of functional effects for sequence variants. BMC Genom. 2015;16 doi: 10.1186/1471-2164-16-S8-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sumbalova L., Stourac J., Martinek T., Bednar D., Damborsky J. HotSpot Wizard 3.0: web server for automated design of mutations and smart libraries based on sequence input information. Nucleic Acids Res. 2018;46:W356–W362. doi: 10.1093/nar/gky417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiadò A., Bosco F., Bardelli M., Simonelli L., Pedotti M., Marmo L., et al. Rational engineering of the lccβ T. versicolor laccase for the mediator-less oxidation of large polycyclic aromatic hydrocarbons. Comput Struct Biotechnol J. 2021;19:2213–2222. doi: 10.1016/j.csbj.2021.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ding N., Zhang G., Zhang L., Shen Z., Yin L., Zhou S., et al. Engineering an AI-based forward-reverse platform for the design of cross-ribosome binding sites of a transcription factor biosensor. Comput Struct Biotechnol J. 2023;21:2929–2939. doi: 10.1016/j.csbj.2023.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nasr M.A., Timmins L.R., Martin V.J.J., Kwan D.H. A versatile transcription factor biosensor system responsive to multiple aromatic and indole inducers. ACS Synth Biol. 2022;11:1692–1698. doi: 10.1021/ACSSYNBIO.2C00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Catherine C., Lee K.-H., Oh S.-J., Kim D.-M. Cell-free platforms for flexible expression and screening of enzymes. Biotechnol Adv. 2013;31:797–803. doi: 10.1016/j.biotechadv.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 51.Guterres H., Im W. Improving protein-ligand docking results with high-throughput molecular dynamics simulations. J Chem Inf Model. 2020 doi: 10.1021/acs.jcim.0c00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Śledź P., Caflisch A. Protein structure-based drug design: from docking to molecular dynamics. Curr Opin Struct Biol. 2018;48:93–102. doi: 10.1016/j.sbi.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 53.Guo H.-B., Varaljay V.A., Kedziora G., Taylor K., Farajollahi S., Lombardo N., et al. Accurate prediction by AlphaFold2 for ligand binding in a reductive dehalogenase and implications for PFAS (per- and polyfluoroalkyl substance) biodegradation. Sci Rep. 2023;13 doi: 10.1038/s41598-023-30310-x. 1 2023;13:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nielsen A.A.K., Der B.S., Shin J., Vaidyanathan P., Paralanov V., Strychalski E.A., et al. Genetic circuit design automation. Sci (N Y, NY) 2016;352 doi: 10.1126/science.aac7341. [DOI] [PubMed] [Google Scholar]
- 56.Nasr M.A., Martin V.J.J., Kwan D.H. Divergent directed evolution of a TetR-type repressor towards aromatic molecules. Nucleic Acids Res. 2023:1–16. doi: 10.1093/nar/gkad503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kwon Y.C., Jewett M.C. High-throughput preparation methods of crude extract for robust cell-free protein synthesis. Sci Rep. 2015 doi: 10.1038/srep08663. 5:1 2015;5:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Akhtar M.K., Jones P.R. Deletion of iscR stimulates recombinant clostridial Fe–Fe hydrogenase activity and H2-accumulation in Escherichia coli BL21(DE3) Appl Microbiol Biotechnol. 2008;78:853–862. doi: 10.1007/s00253-008-1377-6. [DOI] [PubMed] [Google Scholar]
- 59.Kuchenreuther J.M., Grady-Smith C.S., Bingham A.S., George S.J., Cramer S.P., Swartz J.R. High-yield expression of heterologous [FeFe] hydrogenases in escherichia coli. PLOS ONE. 2010;5 doi: 10.1371/journal.pone.0015491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J.C. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material