Abstract
The roles of antibodies secreted by subsets of B cells in dengue virus (DENV) infection have been extensively studied, yet, the contribution of tissue-homing B cells to antiviral immunity remains unclear. In this study, we performed a comprehensive analysis of B cell subpopulations in peripheral blood samples from DENV-infected patients using single-cell RNA-sequencing (scRNA-seq) datasets and flow cytometry. We showed that plasma cells (PCs) and plasmablasts (PBs) were the predominant B cell populations during the acute phase of secondary natural DENV infection, but not in convalescent phase nor in healthy controls. Interestingly, these cells expressed proliferation, adhesion, and tissue-homing genes, including SELPLG, a homing marker of the skin, the initial infected site of DENV. Flow cytometry analysis confirmed a significant upregulation of cell surface expression of a cutaneous lymphocyte-associated antigen (CLA) encoded by SELPLG in PCs and PBs, compared to naive and memory B cells from the same patients. The analysis of an independent single-cell B-cell receptor sequencing (scBCR-seq) dataset of DENV-infected patients revealed that the peripheral blood PCs and PBs exhibited the highest clonal expansion in secondary DENV infection compared to other B cell subsets. These clonally expanded cells also expressed the highest levels of tissue-homing genes, including SELPLG. In addition, by utilizing a public scRNA-seq dataset of SARS-CoV2 infection, we demonstrated the upregulation of several tissue-homing genes in PCs and PBs. Our study provides evidence for the potential roles of tissue-homing B cell subsets in the context of immune responses against viral infections in humans.
Keywords: Single-cell RNA-Seq, Dengue virus, Viral infection, Tissue-homing, B cells
Highlights
-
•
Expansion of skin-homing CLA + antibody-secreting cells (ASCs) during acute dengue infection.
-
•
Clonally expanded ASCs expresses skin and other tissue-homing markers.
-
•
Upregulation of tissue-homing markers in ASCs responses to dengue virus and SARS-CoV2 infection.
1. Introduction
Dengue virus (DENV) is a mosquito-borne viral disease that affects an estimated 390 million people annually in over 100 countries [1]. While the majority of DENV infections are asymptomatic, up to 96 million infected individuals may experience a range of symptoms, from mild dengue fever (DF) to more severe conditions such as severe dengue hemorrhagic fever (DHF) and life-threatening dengue shock syndrome (DSS) [1,2]. Despite efforts to develop safe and effective vaccines, challenges remain due to the immunopathological nature of DENV, where pre-existing immunity against previous DENV serotypes can exacerbate subsequent heterotypic infections with different serotypes [3]. Therefore, a better understanding of protective and pathogenic immune responses in DENV infection is needed.
Antibodies produced by B cells play critical roles in both protection and pathogenesis during DENV infection [4]. While circulating neutralizing antibodies against the currently infected DENV serotype can prevent virus entry into host cells, cross-reactive non-neutralizing antibodies generated from a previous infection with a different DENV serotype can recognize and bind to the virus. However, due to their low binding affinity, these antibodies can promote the entry of immature DENV particles into host cells via Fc receptor-mediated endocytosis, leading to enhanced viral load and potentially more severe symptoms. This mechanism is known as antibody-dependent enhancement (ADE) [[5], [6], [7], [8]].
Despite their roles as antibody secreting cells, B cells could serve as a major target for DENV infection [9,10]. Srikiatkhachorn and colleagues have shown that B cells have the highest levels of positive-stranded RNA of DENV as compared to other immune cell types in peripheral blood of DENV-infected patients [11]. Moreover, during the acute phase of secondary DENV infection, there is typically a massive expansion of plasma cells (PCs) and plasmablasts (PBs), differentiated forms of B cells capable of secreting antibodies (also known as antibody secreting cells or ASCs) [12,13]. These peripheral PCs and PBs have been shown to be cross-reactive DENV-specific ASCs that represent only a minor portion of memory B cell pool [12,14]. Nevertheless, their origin, function, and tissue homeostasis remain largely unknown.
Previously, we and others have reported the possible tissue-homing of B cell subpopulations during acute DENV infection [[14], [15], [16]], including the upregulation of inflamed tissue-homing molecules, CXCR3 and CCR2 [15], and the skin-homing molecule Cutaneous lymphocyte-associated antigen (CLA) on PBs [14]. In our recent study [16], we utilized single-cell transcriptomics to study the dynamics of immune responses in the peripheral blood of DENV-infected patients over the course of infection. We showed evidence supporting the possible migration functions of B cell subsets to tissues during acute DENV infection [16]. Specifically, we observed that certain circulating PCs and PBs subsets of highly expanded lymphocytes expressed a set of tissue-homing signatures, including adhesion molecules, a cell proliferation marker, and cell-cycling genes [16]. In particular, we also observed that these B cell subsets expressed skin-homing markers such as GLG and SELPLG, which encode the CLA protein [16]. We speculated that these skin-homing expressing PCs and PBs might possess the ability to migrate to the skin, the initial site of DENV infection during the acute phase. However, this finding relied on the transcriptional expression of only two DENV-infected donors, which may not always correspond to translation or functional phenotypes. Therefore, these findings may not fully represent the broader population of DENV-infected individuals [16].
To demonstrate the possible homing functions among the major B cell subpopulations, in this study, we investigated the expression of tissue-homing molecules and surface protein expression of the skin-homing molecule, CLA, in a larger sample cohort, comparing between the acute and convalescent phases from the same DENV–infected patients, as well as healthy controls (HCs), which is not the case in previous studies [[14], [15], [16]]. Specifically, we focused on investigating the cell surface expression of the skin-homing molecule, CLA, on B cell subsets by performing flow cytometry on 16 blood samples collected from DENV-infected patients. Moreover, using independent single-cell RNA sequencing (scRNA-seq) and single-cell BCR sequencing (scBCR-seq) datasets from primary and secondary natural DENV-infected patients [17], we demonstrated a positive relationship between tissue-homing gene expression and ASC clonal expansion, with the highest clonally expanded B cell subsets observed in PCs and PBs expressing tissue-homing molecules, including a skin-homing gene, SELPLG. Additionally, we observed the increase in the number of PCs and PBs in the peripheral blood and expression of tissue-homing molecules in PCs and PBs responding to SARS-CoV2 infection using an independent scRNA-seq dataset [18]. Our findings highlight the potential roles of circulating tissue-homing PCs and PBs in DENV infection, which could also apply to other viral infections in humans.
2. Materials and methods
2.1. Blood sample collection and peripheral blood mononuclear cell (PBMC) isolation
The cohort sample collection was conducted as part of the EU/FP7/DENFREE consortium (https://cordis.europa.eu/project/id/282378/results). All the participants included in the current study were adult participants with written informed consent. Ethical approval was obtained from the Institutional Review Boards of Faculty of Medicine Vajira Hospital (No.015/12), Faculty of Tropical Medicine Mahidol University (TMEC 13041), and Faculty of Medicine, Ramathibodi Hospital, Mahidol University (MURA2016/219 and MURA2019/603). Individuals presenting with dengue-related symptoms such as fever, headache, muscle ache, or rashes at Vajira Hospital, Tropical Medicine Hospital Mahidol University, or Thasongyang Hospital underwent testing for DENV by NS1 Antigen rapid test. Viral serotype was determined by nested-RT-PCR for dengue viral RNA (serotype 1, 2, 3, or 4) and the viral load was measured by qRT-PCR [19]. Those testing positive were enrolled in the study as “index cases”. Severity of DENV infection was classified as DF or DHF according to WHO criteria 1997 [20]. Healthy controls were recruited from uninfected household members of index cases who exhibited no dengue-related symptoms and tested negative for DENV via nested RT-PCR.
Heparinized blood samples were collected once every day from the day of hospital admission until defervescence (Def), and at two week convalescence (Wk2). Samples collected before defervescence were labeled according to the number of days prior to reaching the defervescence phase. For example, samples that were labeled “Day −1” (minus one) were collected one day before defervescence. PBMCs were separated from whole blood by Ficoll-Hypaque density gradient centrifugation and kept in liquid nitrogen for subsequent uses.
2.2. Flow cytometry staining
PBMC samples were stained with the following antibodies; Anti-CD3-BV510, Anti-CD19-APC, Anti-CD27-APC-Cy7, Anti-CD38-PerCP, Anti-CD138-FITC, and Anti-CLA-PE (all from Biolegend, USA, catalog numbers are listed in the key resources table). According to the manufacturer's recommendation, cells were fixed with 1 % paraformaldehyde (Sigma-Aldrich, USA) and acquired using CytoFlex (Beckman Coulter, USA). The data analyses were performed using Flowjo v10.8.1 (TreeStar Inc, USA). The percentages of positive cells were normalized by the isotype controls from one representative sample from each condition.
2.3. 3’ scRNA-seq data processing and analysis
We utilized the previously published dataset [16] of scRNA-seq of DENV patients, which has been made publicly available at ArrayExpress under the accession number E-MTAB-9467 (https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-9467). Pre-processing and downstream analyses of scRNA-seq data was performed following the pipelines and codes (https://github.com/vclabsysbio/scRNAseq_DVtimecourse) provided in the original paper [16]. Briefly, the raw and filtered matrix files were generated using CellRanger version 3.0.2 (10x Genomics, USA) using the reference human genome GRCh38 1.2.0. Potential contamination of ambient RNAs was predicted and corrected using SoupX [21]. Low quality cells, including cells expressing mitochondrial genes higher than 10 % and doublets/multiplets, were excluded using Seurat version 3 [22], and doubletFinder [23], respectively. The data of individual samples were then integrated using the SCTransform method [24] with 3000 gene features. Principal component analysis (PCA) and clustering were performed with the Louvain algorithm with multi-level refinement algorithm [22]. Gene expression level of each cell was normalized using the LogNormalize method using Seurat version 3 [22]. Cell types were annotated using the canonical marker genes described in Ref. [16]. The processed scRNA-seq data has been deposited in Mendeley, URL: https://data.mendeley.com/datasets/6ry3x7r8hf/3.
After integration and cell type annotation, we re-clustered naive B cells, memory B cells, PCs, and PBs using the harmony package [25] with default parameters. The B cell subsets were then reannotated using established canonical marker genes described in Table S1. The processed scRNA-seq data of B cell subsets has been deposited in Mendeley, URL: https://data.mendeley.com/datasets/xmnp8c5c65/1.
For the integration of COVID-19 and DENV data, processed scRNA-seq dataset from COVID-19 patients were retrieved from FASTGenomics (https://www.fastgenomics.org/) [18]. The harmony package [25] with default parameters was used to correct batch effects in the principal component space. The first 30th corrected principal components were used for computing the dimensional reduction and shared nearest-neighbor graphs. Gene expression level of each cell was normalized by the LogNormalize function using Seurat V3 [22]. Cell types were then annotated using canonical marker genes as shown in Table S1.
Stacked bar plots were constructed using ggplot2 [26] to visualize changes of abundances in B cell subpopulations. Dot plots displaying average gene expression of tissue-homing genes and the percentages of cells expressing more than one transcript were generated using the DotPlot function in Seurat V3 [22].
2.4. 5’ scRNA-seq and scBCR sequencing data processing and analysis
Quantification matrices, “barcodes.tsv.gz”, “features.tsv.gz”, and “matrix.mtx.gz”, of 5’ gene expression data of another DENV scRNA-seq dataset [17] were retrieved from Gene Expression Omnibus (GEO) database under the accession number GSE145307. Raw files were processed as described by the authors [17]. Briefly, raw sequencing reads were demuliplexed using CellRanger mkfastq version 2.1.1 (10x Genomics, USA) and converted to fastq files using bcl2fastq2 version 2.20.0 (Illumina, USA). Fastq files were then analyzed using CellRanger count pipeline version 2.1.1 (10x Genomics, USA) against a human reference genome generated from the CellRanger mkref and the Ensembl GRCh38 v87 top-level genome FASTA and the corresponding Ensembl v87 gene GTF [17].
For BCR sequencing data, we obtained the “filtered_contig_annotations.csv” files through the courtesy of Dr. Adam Waickman [17]. Raw sequencing files were also re-analyzed as described by the authors [17]. In brief, immunoglobulin clonotype alignments and CDR3 sequences were analyzed using the CellRanger vdj pipeline (10x Genomics, USA) against a filtered human V(D)J references generated by CellRanger mkvdjref (10x Genomics, USA).
Downstream analyses were performed using Scanpy [27]. Genes that are detected in less than three cells were excluded. Cells with fewer than 200 unique genes, and/or more than 10 % of mitochondrial transcripts were discarded. The data of individual samples and BCR contig annotation files were combined into a single dataset using the anndata package version 0.8.0. A batch-corrected neighborhood graph was performed in integrated data with the BBKNN package [28] with default settings. The dimensionality reduction was performed using the scanpy. tl.umap function. Cells were clustered using the neighborhood graph using the Leiden algorithm with default parameters. Counts per cell was normalized by the total library size multiplied by 10,000, and transformed to log scales.
B cell receptors were analyzed using the Scirpy package version 1.8.0 [29]. Cells with the “no IR (immune receptor)”, “multichain”, “orphan VJ”, “orphan VDJ”, and “ambiguous” labels were excluded. Data was then re-integrated following the same pipeline as mentioned above. Cell types were assigned with the following expression of established canonical marker genes (Table S1). Clonotypes were defined based on complementarity-determining regions 3 (CDR3) nucleotide sequence similarity. Clonal expansions were then assigned based on the numbers of multiple cells expressing identical BCR.
For visualization of the expression levels and clonal expansion, Uniform Manifold Approximation and Projection (UMAP) and stacked violin plots were constructed using the scanpy. pl.umap and scanpy. pl.stacked_violin functions, respectively. Stacked bar plots of expanded clones were generated using the scirpy. pl.clonal_expansion function in Scirpy [29].
2.5. Statistical analysis of flow cytometry data
All the statistical analyses were performed in R (https://www.rstudio.com/). Statistical significance between acute and late convalescent phases from the same DENV-infected patients were calculated using the Wilcoxon signed-rank test. For the differences between CLA+ in each B cell subset during acute phase, Kruskal-Wallis test followed by Dunn's test with a Bonferroni correction method for multiple comparisons was applied. The percentages of each B cell subset comparing between the COVID-19 patients and healthy controls were calculated using Wilcoxon rank sum test (Mann-Whitney U test). ns = p > 0.05, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.0001.
3. Results
3.1. B cell subpopulation dynamics and tissue-homing gene expression during acute DENV infection
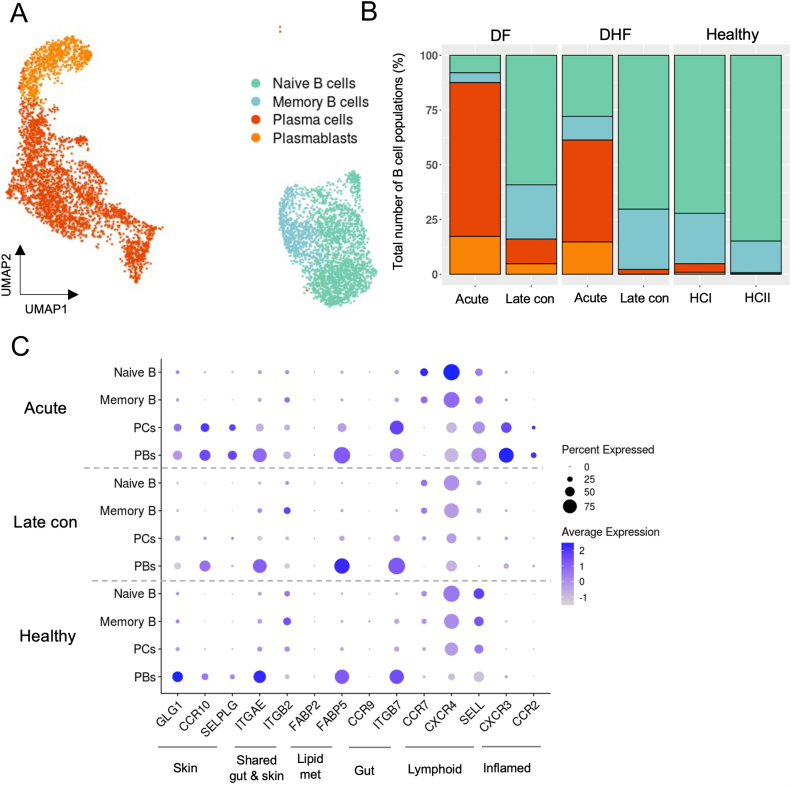
To characterize the dynamic changes among B cell subpopulations, we utilized the scRNA-seq dataset from natural secondary DENV-infected patients generated in our earlier study [16]. After quality control processes to exclude low quality cells and ambient RNAs, we subset the cells within the B cell lineage into naive B cells, memory B cells, plasma cells (PCs) and plasmablasts (PBs), as annotated according to the canonical markers (Fig. 1A and Table S1). The total number of B cells obtained for further analysis was 6,590 across 10 samples (Fig. 1A). Consistent with previous studies [12,13], large proportions of PCs and PBs in all the B cell populations expanded during the acute phase of DENV in both patients, as compared to the two-week convalescent phase (Wk2, late convalescent) and two independent healthy controls (HCs), whereas the opposite trend was observed for naive and memory B cells (Fig. 1B and Fig. S1).
Fig. 1.
Upregulation of several tissue-homing genes on plasma cells and plasmablasts during the acute phase in DENV-infected patients.
(A). Uniform Manifold Approximation and Projection (UMAP) plot showing integrated B cell subsets from the DENV-infected patients and healthy controls as colored by cell types.
(B). Relative abundances of B cell subsets during acute, late convalescent phases (two weeks after defervescence), and healthy controls.
(C). Dot plot representing the average expression values of tissue-homing genes in each B cell subset during the acute and late convalescent phases. Color intensity represents the relative expression levels and dot size represents the fraction of cells within cell type expressing a given gene.
PCs = plasma cells, PBs = plasmablasts, DF = dengue fever, DHF = dengue hemorrhagic fever, HC = healthy control, and Late con = late convalescent phase. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Importantly, these peripheral blood infiltrating PCs and PBs expressed elevated levels of gene modules associated with tissue-homing functions [16]. We observed the upregulation of genes those suggested the homing to shared gut and skin (ITGAE), lipid metabolism associated with skin-homing (FABP5) [30], gut (ITGB7), and inflamed tissues (CXCR3 and CCR2) during the acute phase of DENV infection (Fig. 1C). In contrast, naive and memory B cells exhibited the upregulation of lymphoid tissue homing genes (CCR7 and CXCR4) (Fig. 1C). Remarkably, PCs and PBs expressed homing genes associated with migration to the skin (SELPLG, GLG, and CCR10), which is the primary site of DENV infection, while this upregulation was not observed in naive and memory B cells (Fig. 1C).
3.2. Flow cytometry analysis validates B cell subset changes and a skin-homing molecule CLA expression at the protein level
To further investigate the possible homing function of B cell subsets, we performed flow cytometry analysis to assess the expression levels of a skin-specific homing molecule, cutaneous lymphocyte-associated antigen or CLA (encoded by SELPLG) on the cell surface of B cell populations obtained from the PBMC samples of additional 16 secondary DENV-infected patients and 10 HCs (Table 1 and S2). The DENV samples during the acute phase were collected one day before defervescence (Day −1), and late convalescent samples were obtained two weeks after defervescence (Wk2). The characteristics of DENV-infected donors were summarized in Table 1 and S2.
Table 1.
Summary of DENV-infected patients used in the flow cytometry experiments.
| DF | DHF | Healthy Donors | |
|---|---|---|---|
| Number of samples | 8 | 8 | 10 |
| Number of males/females | 7/1 | 3/5 | 2/8 |
| Mean age (range) | 23.2 (16–37) | 23.1 (19–49) | 41.7 (25–55) |
| Timepoints | One day before defervescence (Day −1) and late convalescent phase (Wk2) | ||
| Mean Log 10 viral load of Day −1 samples | 7.58 | 6.81 | |
| Serotype | |||
| DENV-2 | 2 | 4 | |
| DENV-3 | 4 | 3 | |
| DENV-4 | 2 | 1 | |
DF = dengue fever; DHF = dengue hemorrhagic fever; DENV = dengue virus.
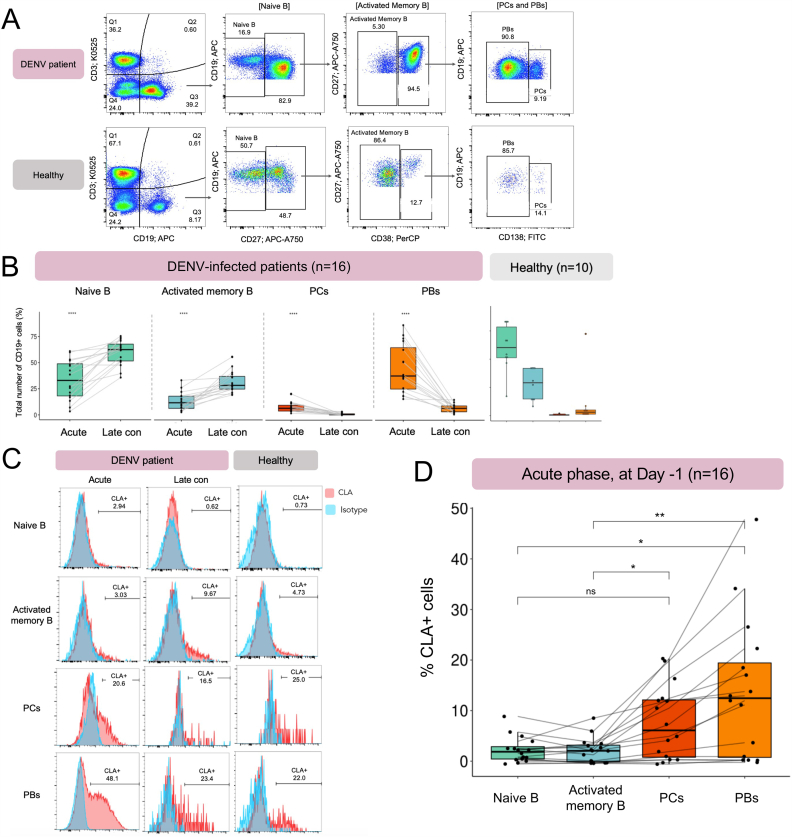
We first investigate the dynamics among the major B cell subpopulations of paired samples between acute and late convalescent phases. The B cell subsets were identified based on the expression of CD19, CD27, CD38 and CD138 as shown in Fig. 2A. In line with our scRNA-seq data and previous studies [12,15], the frequencies of PCs and PBs from acute DENV-infected patients were significantly higher than the same group of patients at late convalescence (n = 16, p-values ≤0.0001, Wilcoxon signed-rank test) and healthy donors (n = 10, p-values ≤0.0001) (Fig. 2B and Fig. S2). Conversely, the frequencies of the naive and activated memory B cells were significantly lower in DENV-infected patients during the acute phase compared to the late convalescence (n = 16, p-values ≤0.0001) (Fig. 2B and Fig. S2). These together show that during the acute DENV infection, PCs and PBs were expanded and returned to the baseline level at two-week convalescence (Fig. 2B and Fig, S2).
Fig. 2.
Significant upregulation of the skin-homing molecule CLA one day before defervescence in secondary DENV-infected patients.
(A). A representative gating strategy of B cell subsets in a DENV-infected patient (upper panel) and healthy control (lower panel). Total B cells (CD3−CD19+) were classified into naive B cells (CD3−CD19+CD27−), activated memory B cells (CD3−CD19+CD27+CD38−), plasma cells (CD3−CD19+CD27+CD38+CD138+), and plasmablasts (CD3−CD19+CD27+CD38+CD138-). The numbers represent the percentages of cells in relation to the parent gate.
(B). Comparison of percentages in each B cell subset between acute (one day before defervescence) and late convalescent (two weeks after defervescence) phases. The statistical significance from the same patients were calculated using the Wilcoxon signed-rank test.
(C). Histograms illustrate the CLA+ cells in each B cell subpopulation during acute (one day before defervescence) and late convalescent (two weeks after defervescence) phases in the DENV-infected patient and healthy control. The numbers represent the percentages of the CLA+ cells. CLA stained condition is shown in red and isotype condition is in blue.
(D). Percentages of the CLA+ cells in each B cell subset during acute DENV infection (one day before defervescence). Kruskal-Wallis test followed by Dunn's test with a Bonferroni correction method was applied for statistical significance of the differences between CLA+ cells in each B cell subset among multiple samples. Each line represents each DENV-infected patient. ns = p > 0.05, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.0001. Number of patients, n = 16, healthy controls, n = 10. PCs = plasma cells, PBs = plasmablasts, Late con = late convalescent phase. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
As we observed the upregulation of tissue-homing genes, including a skin-homing marker SELPLG on PCs and PBs from scRNA-seq data analysis [16] (Fig. 1C), we validated the expression of a CLA molecule (encoded by SELPLG) among B cell subpopulations during the acute phase (Day −1) of DENV infection (Fig. 2C and D). Remarkably, CLA was significantly upregulated in PCs and PBs, when compared to those in the naive and activated memory B cells within the same group of patients (n = 16, p-values ≤0.05 and ≤0.01, respectively) (Fig. 2C and D). The percentage of CLA+ PBs demonstrated a positive correlation with the titer of anti-DENV neutralizing antibodies against serotype 3 (DENV-3), indicating their potential involvement in the generation of neutralizing antibodies (R = 0.50, p-values = 0.05). However, no significant correlation was found between CLA+ PCs and neutralizing antibody titers (R = 0.31, p-value = 0.24) (Fig. S3).
3.3. Skin-homing plasma cells and plasmablasts exhibited clonal expansion during the acute secondary DENV infection
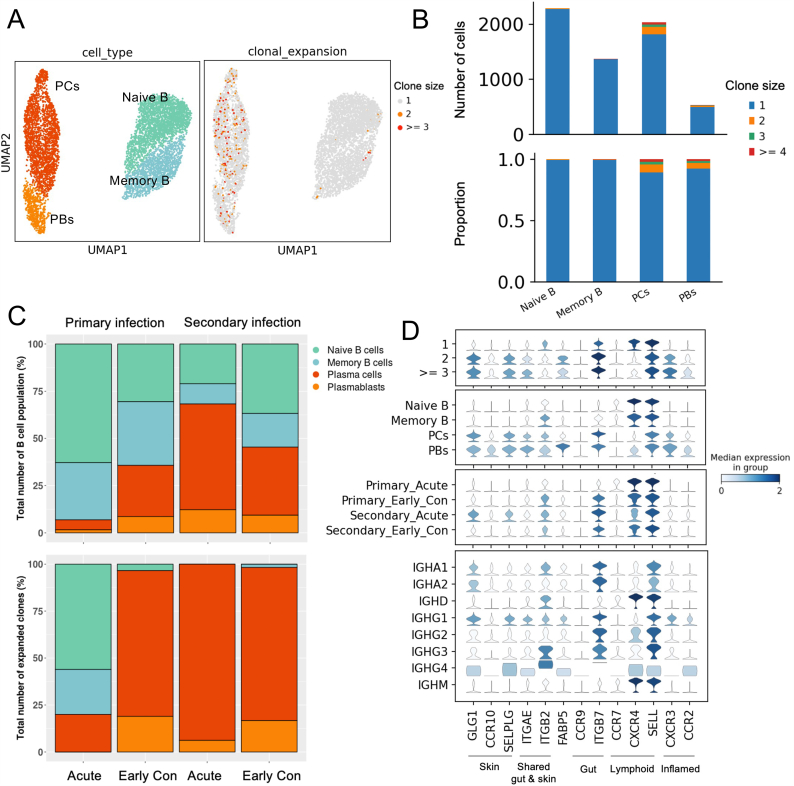
We further investigated the relationship between tissue-homing gene expression and the clonal expansion on B cell subsets using an independent publicly available scRNA-seq dataset that contains B cell receptor sequences from primary and secondary DENV-infected patients [17]. We re-annotated the cell types using the same canonical marker genes as explained earlier (Fig. 3A, right panel and Table S1). Among the B cell subpopulations, the most highly expanded clones were identified in PCs and PBs, with the clone sizes ranging from 2 to 4 cells (Fig. 3A, left panel and 3B). Based on this dataset, PCs were highly expanded in the secondary DENV-infection during acute and early convalescence phases (Day 1 - Day 7 after defervescence), whereas in the primary DENV-infection during acute phase, naive B cells were more expanded (Fig. 3C, lower panel).
Fig. 3.
Upregulation of several tissue-homing genes in natural DENV-infected patients observed in clonally expanded plasma cells and plasmablasts.
(A). UMAP plot of the integrated 5′ scRNA-seq and scBCR-seq datasets from six DENV-infected patients, colored by B cell subsets in the left panel and colored by expanded clones in the right panel. At least three B cells that expressed identical BCR clonotypes are indicated by “ ≥ 3” and at least two B cells are indicated by “2”. Only one B cell that expresses a specific BCR clonotype is indicated by “1”.
(B). Numbers (upper panel) and proportions (lower panel) of clonally expanded groups in each B cell subset in DENV-infected patients.
(C). Relative abundances of B cell subsets (upper panel) and relative abundances of expanded clones (lower panel) in primary acute (one day before defervescence), primary early convalescent (Days 1–3 after defervescence and Day 5 after defervescence), secondary acute (one day before defervescence and defervescence), and secondary early convalescent (Days 1–3 after defervescence, Day 5 after defervescence and Day 7 after defervescence).
(D). Stacked violin plot of tissue-homing genes in clonal expansion groups (top panel), B cell subsets (second panel), primary or secondary infection and clinical phases (third panel), and antibody isotypes (bottom panel). Color intensity represents the median expression level in the group. PCs = plasma cells, PBs = plasmablasts, Early con = early convalescent phase. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Interestingly, among the PCs and PBs expressing high level of tissue-homing genes, those exhibited the highest clonal expansion showed the highest up-regulations of genes associated with skin-homing (GLG1, CCR10 and SELPLG), shared gut and skin-homing (ITGAE), gut-homing (ITGB7), and inflamed tissue-homing (CXCR3 and CCR2), when compared to their non-expanded counterparts (Fig. 3D). These together suggest that the tissue-homing genes were upregulated in clonally expanded PCs and PBs. While B cell clonal expansion could be triggered by an encounter with their cognate antigens, we cannot conclude that these expanded PCs and PBs were DENV-specific from this result alone, as antigen specificity was not explored.
3.4. Upregulation of tissue-homing genes in plasma cells and plasmablasts in COVID-19 patients
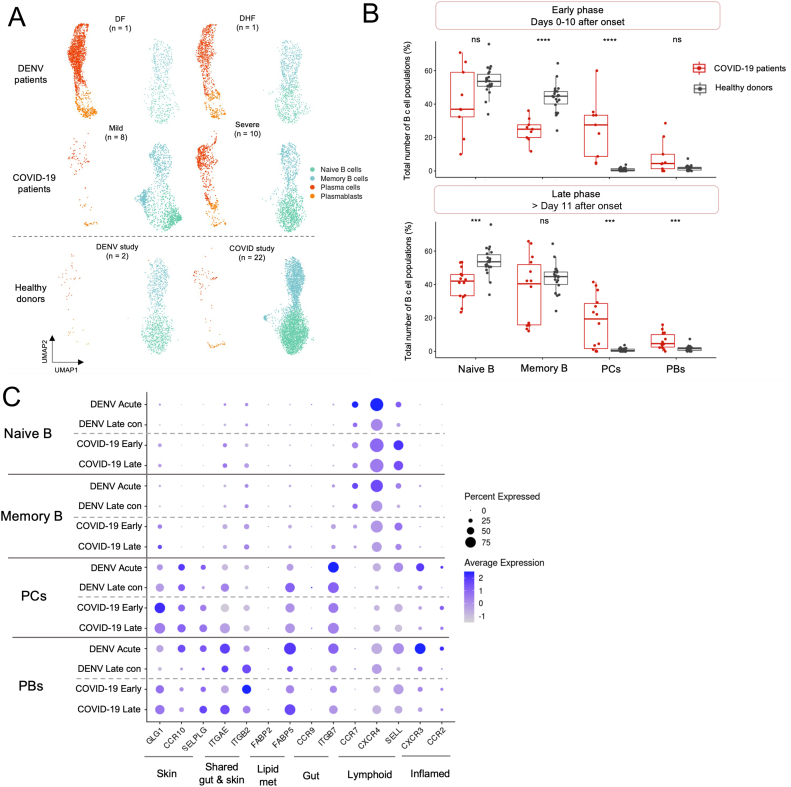
We further investigated if the expansion of circulating PCs and PBs observed in DENV patients could also be seen in other viral infections. To address this question, we compared the transcriptomic profiles of B cell populations from DENV-infected patients during acute infection described earlier, with those from a publicly available scRNA-seq dataset from COVID-19 patients (https://www.fastgenomics.org/) (Fig. 4A) [18].
Fig. 4.
Upregulation of tissue-homing genes in plasma cells and plasmablasts in DENV and COVID-19 patients.
(A). UMAP plot of integrated DENV and COVID-19 datasets (Total 14,998 cells), colored by B cell subsets. The top panel represents abundances of B cell subsets in DENV patients, whereas the middle panel shows abundances of B cells in COVID-19 patients. The bottom panel represents healthy controls from two different studies. The number represents the sample sizes.
(B). Comparison of percentages between COVID-19 patients and healthy controls in each B cell subset during the early (days 0–11 after symptom onset; COVID-19 patients, n = 9; HCs, n = 22) and late phases (>11 after symptom onset; COVID-19 patients, n = 14; HCs, n = 22). The statistical significance was calculated using the Wilcoxon rank sum test (Mann-Whitney U test). ns = p > 0.05, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.0001.
(C). Dotplot representing the average expression levels of tissue-homing genes in each B cell subset in DENV and COVID-19 datasets. Color intensity represents the relative expression values and dot size represents the fraction of cells within cell type expressing a given gene. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The enrichment of PCs and PBs were observed in both the COVID-19 and DENV patients during acute infections, but was slightly more prominent in the severe COVID-19 and DENV patients, when compared to the HCs (Fig. 4A). During the early phase of COVID-19 (Days 0–10 after symptom onset as described in the original publication [18]), the frequency of PCs was significantly higher than in HCs, whereas the proportion of memory B cell population was significantly reduced (COVID-19 patients, n = 9; HCs, n = 22; p-value ≤0.0001, Wilcoxon rank sum test) (Fig. 4B, upper panel). Moreover, we also observed that during the late convalescent phase (more than 11 days after symptom onset), the abundances of PCs and PBs remained significantly higher, while the proportion of naive B cells was significantly reduced when compared to HCs (COVID-19 patients, n = 14; HCs, n = 22; p-value ≤0.001) (Fig. 4B, lower panel).
We further looked at the expression of tissue-homing genes and found that the PCs and PBs identified from these COVID-19 and DENV datasets demonstrated the upregulations of several tissue-homing genes, including skin-homing markers (GLG1 and SELPLG), shared gut and skin-homing marker (ITGAE and ITGB2), a lipid binding molecule associated with skin homing [30] (FABP5), and gut-homing marker (ITGB7) (Fig. 4C). Notably, we observed the up-regulation of the inflamed tissue-homing gene, CXCR3, in PCs and PBs identified in DENV, but not in the COVID-19 patients (Fig. 4C).
To ensure that these tissue-homing gene modules were upregulated only in peripheral blood-infiltrating PCs and PBs during viral infection, but not general markers of PCs and PBs, we further integrated and compared the expression levels of these tissue-homing genes in PCs and PBs using another publicly available scRNA-seq dataset of healthy human bone marrow, a known niche for PCs and PBs [31]. Remarkedly, several tissue-homing markers were only upregulated in the circulating PCs and PBs during the acute viral infections, but not those from the bone marrow (Fig. S4).
4. Discussion
In this study, we have extended the current understanding of the antigen secreting cells (ASCs, including PCs and PBs) in acute dengue viral infections by investigating their potential homing activities to the tissue niches. We have confirmed at protein level that circulating PCs and PBs expressing a skin-homing molecule, CLA, significantly expanded during secondary acute DENV infection. Additionally, using an independent scRNA-seq and scBCR-seq dataset [17], we have shown that the infiltrating PCs and PBs were prominent in the acute phase of secondary DENV infection and that they had the highest clonal expansion compared to other B cell subpopulations. Finally, we have demonstrated that circulating PCs and PBs expressing tissue-homing molecules such as GLG1, CCR10, SELPLG, ITGB7, and ITGAE were also observed during acute SARS-CoV2 infection.
Our study, along with others, has provided evidence documenting the potential migratory functions of B cell subsets to tissues during acute DENV infection [[14], [15], [16]]. A previous report has demonstrated that PCs and PBs showed the up-regulation of inflamed tissue-homing molecules, CXCR3 and CCR2, on ASCs, but not CCR10, a chemokine receptor suggested for its role in skin-homing [15]. In our current study, having the advantage of whole-transcriptome sequencing, we were able to spontaneously explore the expression levels of several other skin-homing genes in addition to CCR10, such as GLG, and SELPLG, as well as lipid metabolism genes that are associated with skin homing, including FABP5 and FABP2 [30]. We found that these sets of genes concordantly upregulated in PCs and PBs during the acute DENV infection. We expanded such findings by suggesting the possibility of these PCs and PBs to migrate to other tissues such as the gut and inflamed tissue as well as exploring the relationship between tissue-homing gene expression and clonal expansion.
The roles of tissue-resident ASCs in viral infections have been highlighted in several previous studies [[32], [33], [34]]. In murine models of influenza virus immunization, Oh and colleagues have shown that secretion of IgA-expressing cells from tissue-resident ASCs in the lung after local immunization contribute to the protection of subsequent viral challenge, and that the localization of these cells required the expression of CXCR3 [34]. More recently, the localization of IgA-expressing PCs in human airway submucosal glands was suggested to be mediated by the interaction between CCL28 and CCR10 [35]. These previous studies not only provided evidence for the existence of ASCs, but also demonstrated their localization functions and locally antibody production activities in human mucosal tissues.
For the skin, there is growing evidence that certain subsets of B cells reside in and exhibit localization activities, including defense against invading antigens, homeostasis maintenance, and regulate cutaneous inflammation during chronic and acute infection [36,37]. Innate-like B cells were suggested to be found in the normal skin of humans and mice [38]. During the chronic skin inflammation, these innate-like B cells migrated to the inflamed site through α4β1-integrin interaction and secreted the anti-inflammatory cytokine IL-10 [38]. The presence of skin-resident IgG-expressing B cells in normal skin has also been previously reported [39]. These studies suggest the roles of skin-resident B cells in healthy skin and in chronic skin inflammation, yet whether DENV-reactive B cells reside in the skin, and their roles in the infection, remain unknown. Further in-situ experiments are needed to precisely identify the presence of DENV-specific B cells in the skin.
It remains unclear whether or not the circulating PCs and PBs play a protective or pathogenic role in DENV infection. In line with a previous study [12], we did not observe significant differences in the frequency of PCs and PBs between mild and severe patients. Antibodies produced by circulating PCs and PBs have been shown to be mostly cross-reactive, but can still bind to and neutralize DENV envelope (E) protein [40]. Similar to a previous report by Appana and colleagues [40], we found minimal clonal overlap between PCs/PBs and memory B cells (data not shown), suggesting that antibodies produced by PCs/PBs were directed towards different viral proteins, rather than those of memory B cells [40]. Our finding left the field with the question about the origin of these PCs and PBs, whether or not they are from that same pool of memory B cells. That is, if the progenitor of these PCs and PBs reside in tissues such as the skin; and if so, whether or not they play a significant role in dengue protection, all remain to be further addressed.
5. Limitation of the study
Our study has a few major limitations that could be addressed in the future study. The first is the limited number of donors in the DENV scRNA-seq analysis of two, one DF and one DHF, which might hinder the exploration of the factors that were associated with disease severity. A future study with more samples for each severity could potentially improve the understanding of the roles of B cells in the severity of dengue fever. Additionally, we did not investigate the antigen specificity of the skin-homing PCs and PBs identified in our current study. To address this question, we suggest the use of antigen-labeled technologies for flow cytometry analysis or scRNA-seq in order to determine the antigen specificity of tissue-homing PCs and PBs [41]. To confirm our hypothesis that the infiltrating PCs and PBs during acute DENV infection home to the skin, cell collection from the skin or induced skin blisters from DENV infected patients is required. Rivino and colleagues [42] have demonstrated the presence of dengue-specific T cells from fluid collected from skin blisters of DENV infected patients [42]. Similar methods could potentially be used to examine the presence of DENV-specific B cell subpopulations in the skin. Finally, in this work we did not identify the DENV-infected B cells. Whether or not the upregulation of tissue homing genes observed in B cell subsets was the result of direct infection with DENV needs to be further elucidated.
6. Conclusion
Our study sheds light on the behavior of B cell subsets during viral infections. We have demonstrated that highly expanded PCs and PBs in the peripheral blood express molecules associated with skin homing and undergo clonal expansion. Furthermore, our findings suggest that tissue-homing PCs and PBs are also present in SARS-CoV2 infection. Further investigation into the roles of tissue-homing lymphocytes in various viral infections holds promise for informing the development of vaccines or therapeutics strategies in the future.
Ethics statement
Sample collection and experimental processes were revealed and approved by the Institutional Review Boards of Faculty of Medicine Vajira Hospital (No.015/12), Faculty of Tropical Medicine Mahidol University (TMEC 13041), and Faculty of Medicine, Ramathibodi Hospital, Mahidol University (MURA2016/219 and MURA2019/603), as part of the DENFREE initiative (https://cordis.europa.eu/project/id/282378/results) [19]. Informed consent was acquired from the patients and the patients consented to the publishing of the results obtained from the study.
Data availability statement
-
●
Processed scRNA-seq data of B cell subsets have been deposited in Mendeley, URL: https://data.mendeley.com/datasets/xmnp8c5c65/1
Funding
JA is supported by the Royal Golden Jubilee (RGJ) Ph.D. Programme (PHD/0091/2559), through the National Research Council of Thailand (NRCT), Thailand Science Research and Innovation (TSRI) and Mahidol University (MU). VC is supported by the mid-career researcher grant from National Research Council of Thailand (NRCT) and Mahidol University (NRCT5-RSA63015-24); Mahidol University’s Basic Research Fund: fiscal year 2021 (BRF1-017/2564); National Research Council of Thailand (NRCT) (N35A640274). PM and DENFREE Thailand are supported by European Union Seventh Framework Program (EU/FP7 under Grant Agreement #282378 (DENFREE). PM lab is supported by Mahidol University (Fundamental Fund: fiscal year 2024 by National Science Research and Innovation Fund (NSRF)). VC and PM are also supported by the Program Management Unit for National Competitiveness Enhancement (PMU-C) (C10F650132) and the Ancestry Networks Grant from the Chan Zuckerberg Initiative. AO (Grant No. RGNS 65–217) is supported by Office of the Permanent Secretory, Ministry of Higher Education, Science Research and Innovation (OPS-MHESI), Thailand Science Research and Innovation (TSRI) and Mahidol University.
Consortia DENFREE Thailand
Anavaj Sakuntabhai (Functional Genetics of Infectious Diseases Unit, Institut Pasteur, Paris, France, Centre National de la Recherche Scientifique (CNRS), URM2000, Paris, France), Pratap Singhasivanon (Department of Tropical Hygiene, Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand), Swangjit Suraamornkul (Endocrinology Division, Department of Medicine, Faculty of Medicine Vajira Hospital, Navamindradhiraj University, Bangkok, Thailand), Tawatchai Yingtaweesak (Thasongyang Hospital, Tak, Thailand), Khajohnpong Manopwisedjaroen (Department of Microbiology, Faculty of Science, and Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand), Nada Pitabut (Faculty of Tropical Medicine, Mahidol University, and Faculty of Medicine, King Mongkut’s Institute of Technology Ladkrabang, Bangkok, Thailand).
CRediT authorship contribution statement
Jantarika Kumar Arora: Writing – review & editing, Writing – original draft, Visualization, Validation, Methodology, Investigation, Formal analysis. Ponpan Matangkasombut: Writing – review & editing, Writing – original draft, Supervision, Resources, Methodology, Investigation, Funding acquisition, Conceptualization. Varodom Charoensawan: Writing – review & editing, Writing – original draft, Supervision, Resources, Methodology, Investigation, Funding acquisition, Conceptualization. Anunya Opasawatchai: Writing – review & editing, Writing – original draft, Visualization, Supervision, Methodology, Investigation, Funding acquisition, Formal analysis, Conceptualization. DENFREE Thailand: Resources.
Declaration of generative AI and AI-assisted technologies in the writing process
During the preparation of this work the authors used ChatGPT developed by OpenAI to improve language and readability. After using this tool, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to express our sincere gratitude to Adam Waickman for generously sharing additional single-cell B cell receptor data to the ones already described in their study [17]. Data processing was supported by Mahidol University and the Office of the Ministry of Higher Education, Science, Research and Innovation under the Reinventing University project: the Center of Excellence in AI-Based Medical Diagnosis (AI-MD) sub-project. We thank Ataco (Chindasook Group, Thailand) and the central instrument facility (CIF), Faculty of Science, Mahidol University for instrumental support. The authors thank Waradon Sungnak, Benjamaporn Sriwilai and Natnicha Jiravejchakul for their invaluable support and insightful comments.
Footnotes
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Brilliant Violet 510TM anti-human CD3 Antibody | Biolegend | Cat#; 317,332 RRID: AB_2561943 |
| APC anti-human CD19 | Immunotools | Cat#; 302,816 RRID: AB_571977 |
| APC/Cyanine 7 anti-human CD27 Antibody | Biolegend | Cat#; 302,816 RRID: AB_571977 |
| PerCP anti-human CD38 Antibody | Biolegend | Cat#; 303,520 RRID: AB_893313 |
| FITC anti-human CD138 (Syndecan-1) Antibody | Biolegend | Cat#; 352,304 RRID: AB_10900441 |
| PE anti-human/mouse Cutaneous Lymphocyte Antigen (CLA) Antibody | Biolegend | Cat#321312; RRID: AB_2565589 |
| Biological Samples | ||
| Human PBMC samples | The Institutional Review Boards of Faculty of Medicine Vajira Hospital (No.015/12), Faculty of Tropical Medicine Mahidol University (TMEC 13041), and Faculty of Medicine, Ramathibodi Hospital, Mahidol University (MURA2016/219 and MURA2019/603), as part of the DENFREE initiative (https://cordis.europa.eu/project/id/282378/results) | N/A |
| Deposited Data | ||
| Dataset: Processed 3′ scRNA-seq datasets of dengue PBMC samples | Arora et al. (2023) | https://data.mendeley.com/datasets/6ry3x7r8hf/3 |
| Dataset: Quantification matrices of 5′ scRNA-seq and scBCR sequencing of primary and secondary dengue PBMC samples | Waickman et al. | Gene Expression Omnibus (GEO): GSE145307 |
| Dataset: Processed 3′ scRNA-seq datasets of COVID-19 PBMC samples | Schulte-Schrepping et al. | http://fastgenomics.org |
| Processed scRNA-seq datasets of B cell populations used in this study | Mendeley Data | -https://data.mendeley.com/datasets/xmnp8c5c65/1 |
| Software and Algorithms | ||
| RStudio v3.1.4 | RStudio | https://www.rstudio.com/ |
| Seurat v3.1.2 | (Stuart et al., 2019) | https://satijalab.org/seurat/ |
| ggplot2 v3.3.2 | (Wickham, 2016) | https://ggplot2.tidyverse.org/ |
| Scanpy v1.9.1 | (Wolf et al., 2018) | https://scanpy.readthedocs.io/en/stable/index.html# |
| BBKNN | (Polański et al., 2019) | https://github.com/Teichlab/bbknn |
| Scirpy v1.8.0 | (Sturm et al., 2020) | https://scirpy.scverse.org/en/latest/ |
| Flowjo v10.8.1 | TreeStar Inc | https://www.flowjo.com/ |
Contributor Information
Ponpan Matangkasombut, Email: ponpan.mat@mahidol.edu.
Varodom Charoensawan, Email: varodom.cha@mahidol.ac.th.
Anunya Opasawatchai, Email: anunya.opa@mahidol.edu.
DENFREE Thailand:
Anavaj Sakuntabhai, Pratap Singhasivanon, Swangjit Suraamornkul, Tawatchai Yingtaweesak, Khajohnpong Manopwisedjaroen, and Nada Pitabut
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bhatt S., et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Regional Office for South-East, A . WHO Regional Office for South-East Asia; New Delhi: 2011. Comprehensive Guideline for Prevention and Control of Dengue and Dengue Haemorrhagic Fever. Revised and Expanded Edition. [Google Scholar]
- 3.Torres-Flores J.M., Reyes-Sandoval A., Salazar M.I. Dengue vaccines: an update. BioDrugs. 2022;36(3):325–336. doi: 10.1007/s40259-022-00531-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rey F.A., et al. The bright and the dark side of human antibody responses to flaviviruses: lessons for vaccine design. EMBO Rep. 2018;19(2):206–224. doi: 10.15252/embr.201745302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dejnirattisai W., et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science. 2010;328(5979):745–748. doi: 10.1126/science.1185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flipse J., Wilschut J., Smit J.M. Molecular mechanisms involved in antibody-dependent enhancement of dengue virus infection in humans. Traffic. 2013;14(1):25–35. doi: 10.1111/tra.12012. [DOI] [PubMed] [Google Scholar]
- 7.Halstead S.B. Dengue antibody-dependent enhancement: knowns and unknowns. Microbiol. Spectr. 2014;2(6) doi: 10.1128/microbiolspec.AID-0022-2014. [DOI] [PubMed] [Google Scholar]
- 8.Katzelnick L.C., et al. Antibody-dependent enhancement of severe dengue disease in humans. Science. 2017;358(6365):929–932. doi: 10.1126/science.aan6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zanini F., et al. Virus-inclusive single-cell RNA sequencing reveals the molecular signature of progression to severe dengue. Proc. Natl. Acad. Sci. U. S. A. 2018;115(52):E12363–E12369. doi: 10.1073/pnas.1813819115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Upasani V., et al. Direct infection of B cells by dengue virus modulates B cell responses in a Cambodian pediatric cohort. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.594813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srikiatkhachorn A., et al. Dengue viral RNA levels in peripheral blood mononuclear cells are associated with disease severity and preexisting dengue immune status. PLoS One. 2012;7(12) doi: 10.1371/journal.pone.0051335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wrammert J., et al. Rapid and massive virus-specific plasmablast responses during acute dengue virus infection in humans. J. Virol. 2012;86(6):2911–2918. doi: 10.1128/JVI.06075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Bates T.M., et al. Association between magnitude of the virus-specific plasmablast response and disease severity in dengue patients. J. Immunol. 2013;190(1):80–87. doi: 10.4049/jimmunol.1103350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aggarwal C., et al. Immunophenotyping and transcriptional profiling of human plasmablasts in dengue. J. Virol. 2021;95(23) doi: 10.1128/JVI.00610-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pattanapanyasat K., et al. B cell subset alteration and the expression of tissue homing molecules in dengue infected patients. J. Biomed. Sci. 2018;25(1):64. doi: 10.1186/s12929-018-0467-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arora J.K., et al. Single-cell temporal analysis of natural dengue infection reveals skin-homing lymphocyte expansion one day before defervescence. iScience. 2022;25(4) doi: 10.1016/j.isci.2022.104034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waickman A.T., et al. Transcriptional and clonal characterization of B cell plasmablast diversity following primary and secondary natural DENV infection. EBioMedicine. 2020;54 doi: 10.1016/j.ebiom.2020.102733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulte-Schrepping J., et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182(6):1419–1440. e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matangkasombut P., et al. Dengue viremia kinetics in asymptomatic and symptomatic infection. Int. J. Infect. Dis. 2020;101:90–97. doi: 10.1016/j.ijid.2020.09.1446. [DOI] [PubMed] [Google Scholar]
- 20.Organization W.H. World Health Organization; 1997. Dengue Haemorrhagic Fever: Diagnosis, Treatment, Prevention and Control. [Google Scholar]
- 21.Young M.D., Behjati S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. GigaScience. 2020;9(12) doi: 10.1093/gigascience/giaa151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stuart T., et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902 e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8(4):329–337 e4. doi: 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20(1):296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korsunsky I., et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16(12):1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wickham H., Wickham H. 2016. Data Analysis. Ggplot2: Elegant Graphics for Data Analysis; pp. 189–201. [Google Scholar]
- 27.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19(1):15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polański K., et al. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics. 2019;36(3):964–965. doi: 10.1093/bioinformatics/btz625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sturm G., et al. Scirpy: a Scanpy extension for analyzing single-cell T-cell receptor-sequencing data. Bioinformatics. 2020;36(18):4817–4818. doi: 10.1093/bioinformatics/btaa611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frizzell H., et al. Organ-specific isoform selection of fatty acid-binding proteins in tissue-resident lymphocytes. Sci Immunol. 2020;5(46) doi: 10.1126/sciimmunol.aay9283. [DOI] [PubMed] [Google Scholar]
- 31.Oetjen K.A., et al. Human bone marrow assessment by single-cell RNA sequencing, mass cytometry, and flow cytometry. JCI Insight. 2018;3(23) doi: 10.1172/jci.insight.124928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allie S.R., et al. The establishment of resident memory B cells in the lung requires local antigen encounter. Nat. Immunol. 2019;20(1):97–108. doi: 10.1038/s41590-018-0260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y., et al. Clonal expansion and activation of tissue-resident memory-like TH17 cells expressing GM-CSF in the lungs of patients with severe COVID-19. Science Immunology. 2021;6(56):eabf6692. doi: 10.1126/sciimmunol.abf6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh J.E., et al. Intranasal priming induces local lung-resident B cell populations that secrete protective mucosal antiviral IgA. Science immunology. 2021;6(66):eabj5129. doi: 10.1126/sciimmunol.abj5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madissoon E., et al. A spatially resolved atlas of the human lung characterizes a gland-associated immune niche. Nat. Genet. 2023;55(1):66–77. doi: 10.1038/s41588-022-01243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geherin S.A., et al. The skin, a novel niche for recirculating B cells. J. Immunol. 2012;188(12):6027–6035. doi: 10.4049/jimmunol.1102639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Debes G.F., McGettigan S.E. Skin-associated B cells in health and inflammation. J. Immunol. 2019;202(6):1659–1666. doi: 10.4049/jimmunol.1801211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geherin S.A., et al. IL-10+ innate-like B cells are part of the skin immune system and require α4β1 integrin to migrate between the peritoneum and inflamed skin. J. Immunol. 2016;196(6):2514–2525. doi: 10.4049/jimmunol.1403246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saul L., et al. IgG subclass switching and clonal expansion in cutaneous melanoma and normal skin. Sci. Rep. 2016;6 doi: 10.1038/srep29736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Appanna R., et al. Plasmablasts during acute dengue infection represent a small subset of a broader virus-specific memory B cell pool. EBioMedicine. 2016;12:178–188. doi: 10.1016/j.ebiom.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shiakolas A.R., et al. Efficient discovery of SARS-CoV-2-neutralizing antibodies via B cell receptor sequencing and ligand blocking. Nat. Biotechnol. 2022;40(8):1270–1275. doi: 10.1038/s41587-022-01232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rivino L., et al. Virus-specific T lymphocytes home to the skin during natural dengue infection. Sci. Transl. Med. 2015;7(278):278ra35. doi: 10.1126/scitranslmed.aaa0526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
●
Processed scRNA-seq data of B cell subsets have been deposited in Mendeley, URL: https://data.mendeley.com/datasets/xmnp8c5c65/1