Summary
Background
International distribution of contaminated foods can be a source of Salmonella infections in people and can contribute to the spread of antimicrobial-resistant bacteria across countries. We report an investigation led by the United States Centers for Disease Control and Prevention, the Food and Drug Administration (FDA), and state governmental officials into a multistate outbreak of salmonellosis linked to pig ear pet treats.
Methods
Pig ear treats and companion dogs were tested for Salmonella by state officials and the FDA. Products were traced back to the country of origin when possible. Cases were defined as outbreak illnesses in people associated with one of seven Salmonella serotypes genetically related to samples from pig ear pet treats, with isolation dates from June 2015 to September 2019. Whole genome sequencing (WGS) of isolates was used to predict antimicrobial resistance.
Findings
The outbreak included 154 human cases in 34 states. Of these, 107 of 122 (88%) patients reported dog contact, and 65 of 97 (67%) reported contact with pig ear pet treats. Salmonella was isolated from 137 pig ear treats, including some imported from Argentina, Brazil, and Colombia, and from four dogs. WGS predicted 77% (105/137) of human and 43% (58/135) of pig ear treat isolates were resistant to ≥3 antimicrobial classes.
Interpretation
This was the first documented United States multistate outbreak of Salmonella infections linked to pig ear pet treats. This multidrug-resistant outbreak highlights the interconnectedness of human health and companion animal ownership and the need for zoonotic pathogen surveillance to prevent human illness resulting from internationally transported pet food products.
Funding
Animal Feed Regulatory Program Standards award. Animal and product testing conducted by FDA Vet-LIRN was funded by Vet-LIRN infrastructure grants (PAR-22-063).
Keywords: Salmonellosis, Antimicrobial resistance, Outbreak, Public health
Research in context.
Evidence before this study
We searched PubMed and Scopus for any previous outbreaks of salmonellosis in people linked to contaminated pig ear pet treats or other food or treats for dogs. We used the following search terms: (“Salmonella” OR “salmonellosis”) AND (“outbreak”) AND (“pig ears” OR “pig ear pet treats” OR “pig ear treats” OR “dehydrated treats” OR “dry dog treats” OR “dry dog food” OR “dog food” OR “dog treats”). Our search was limited to publications before January 1, 2024. Our search identified eight reports of outbreaks linked to pig ear pet treats or other dog treats or foods or other relevant studies. Outbreaks caused by nontyphoidal Salmonella occur globally. In the United States, salmonellosis commonly results from contaminated food products; however, outbreaks are rarely linked to contaminated pet food or treats. Pet treats derived from the byproducts of animals such as pigs, chickens, and cattle have been associated with salmonellosis outbreaks. Before this investigation, pig ear pet treats had not been reported as linked to a multistate outbreak of Salmonella illnesses in the United States.
Added value of this study
We describe the first reported United States multistate outbreak of salmonellosis linked to pig ear pet treats and the associated antimicrobial resistance characteristics based on whole genome sequencing and antimicrobial susceptibility testing. Traceback investigation revealed that contaminated pig ear treats, some of which were labeled as irradiated, were imported by three pet treat companies from three countries in South America. Ultimately, six companies that supplied retail stores across the United States issued nationwide recalls of pig ear treats.
Implications of all the available evidence
This outbreak illustrates the potential for contaminated pig ear pet treats to present a risk to human and companion animal health in the United States. In this outbreak, pre- and post-processing pathogen reduction efforts were insufficient to mitigate this risk. Intensified surveillance of internationally traded pet food products for foodborne pathogens may be warranted, and international producers should consider bolstering strategies that reduce product contamination. Pet owners should be made aware of disease risk associated with pig ear pet treats and should take appropriate precautions when handling any pet foods or treats (e.g., handwashing after feeding pets) to avoid infection.
Introduction
Nontyphoidal Salmonella results in over one million infections and more than 26,000 hospitalizations in the United States annually.1, 2, 3 Illnesses can be linked to ingestion of contaminated food or water, contact with feces of infected humans, or contact with animals either directly or via fomites such as bedding or items kept within their habitats.2,3 Infection commonly leads to self-limiting diarrhea, abdominal pain, and fever. Severe illness can result if the bacteria spread from the intestinal tract to the bloodstream, necessitating antimicrobial therapy.4,5 Among human infections in the United States, Salmonella infections are increasingly resistant to antimicrobial drugs, with estimates indicating a 40% increase in the annual incidence of infections with clinically important resistance (i.e., resistance to ampicillin or ceftriaxone or nonsusceptibility to ciprofloxacin) from 2004 to 2016.2,6,7
International distribution of contaminated foods can be a source of Salmonella infections in people and can contribute to the spread of antimicrobial-resistant bacteria across countries. In the United States, surveillance studies of imported foods have most frequently isolated multidrug-resistant (MDR) Salmonella from frozen seafood, produce, and dried herbs and spices,8, 9, 10 some of which have been linked to multistate salmonellosis outbreaks.11, 12, 13, 14, 15
Companion animals, including dogs and cats, have also been identified as potential sources of human salmonellosis and can shed Salmonella in the feces even when they appear healthy.16, 17, 18 Studies have shown <1%–36% of dogs without diarrhea demonstrate Salmonella positive fecal cultures.16,19, 20, 21 Salmonella infection in dogs can result in signs such as diarrhea, vomiting, and lethargy; bacteremia and subsequent systemic infection can occur but is less common.22
Pet food and certain treats produced from animal byproducts have been linked to transmission of Salmonella to people and pets.17,23, 24, 25, 26, 27, 28 Pig ear pet treats produced in Canada were first identified as a source of a human Salmonella enterica serotype Infantis outbreak in Canada in 1999.27 The outbreak led to the United States Food and Drug Administration initiating an import alert in 1999 for violative products from impacted firms.29 Surveillance in the United States at the time of the outbreak in Canada found 41% (65/158) of domestically produced and imported pig ear treat samples tested were contaminated with one of 24 different Salmonella serotypes, and 36% (28/78) of isolates obtained demonstrated resistance to at least one antimicrobial drug.30 Salmonella has been detected, though at a lower prevalence, in pig ear treats in other countries, including Japan (7 positive of 303 tested, 2%), New Zealand (36/600, 6%), and the United Kingdom (184/2369, 8%).31, 32, 33 Imported pet treats potentially introduced novel strains of antimicrobial-resistant Salmonella to New Zealand.32 The first reported human illness outbreak in the United States linked to pet treats resulted from Salmonella Thompson contamination of dehydrated pet treats derived from beef and salmon byproducts produced at manufacturing plants in the United States and Canada in 2004.34
In May 2019, PulseNet, the national molecular subtyping network for foodborne disease surveillance at the United States Centers for Disease Control and Prevention (CDC), detected 23 human illnesses in nine states with S. enterica serotype I 4,[5],12:i:- isolates that demonstrated indistinguishable pulsed-field gel electrophoresis (PFGE) patterns and were within 0–8 single nucleotide polymorphism differences by whole genome sequencing (WGS) analysis. Here we report the subsequent investigation that was initiated to further characterize patient exposures, to identify potential sources of illness, and to implement prevention measures.
Methods
Case definitions
Cases were initially defined as infection with Salmonella I 4,[5],12:i:- with the outbreak strain (one of two PFGE patterns). PulseNet transitioned from PFGE to WGS analyzed by core-genome multi-locus sequence typing (cgMLST) as the primary pathogen subtyping approach during this outbreak investigation in the summer of 2019,35 and further cases were defined based on genetic relatedness to the outbreak strain as determined by either PFGE or cgMLST. The case definition was later expanded to include human infection with a Salmonella serotype Cerro, Derby, I 4,[5],12:i:-, London, Infantis, Newport, or Rissen strain genetically related by PFGE or cgMLST (see ranges for allele differences in Table 1) to isolates from pig ear pet treats, with isolation dates between June 2015 through September 2019. The case definition was expanded as testing of pig ear pet treats for Salmonella yielded strains that were genetically related to isolates obtained from ill people.
Table 1.
Descriptive analysis of patient epidemiologic information by serotype, including pig ear pet treats and dog Salmonella isolates.
| Serotype | Patient count n (%) | Median Age (y)a | Female n (%)a | Hospitalized n (%)a | Dog contact n (%)a | Pig ear treat contact n (%)a | Isolation date range | Pig ear treat Salmonella isolates n (%) | Dog Salmonella isolates n (%) | Allele differences rangeb |
|---|---|---|---|---|---|---|---|---|---|---|
| I 4,[5],12:i:- | 76 (49%) | 30 | 35/72 (49%) | 16/67 (24%) | 52/62 (84%) | 33/46 (72%) | Jun. 2015–Aug. 2019 | 7 (5%) | 0 (0%) | Clade 1: 2–14 Clade 2: 0–8 |
| Infantis | 40 (26%) | 44 | 19/37 (51%) | 13/37 (35%) | 29/32 (91%) | 19/29 (66%) | Oct. 2017–Sept. 2019 | 48 (35%) | 1 (25%) | 0–22 |
| London | 23 (15%) | 47 | 13/19 (68%) | 4/17 (24%) | 13/15 (87%) | 8/11 (73%) | May 2018–Aug. 2019 | 28 (20%) | 0 (0%) | Clade 1: 0–5 Clade 2: 0–3 Clade 3: 0–4 |
| Newport | 11 (7%) | 47 | 2/10 (20%) | 2/9 (22%) | 10/10 (100%) | 5/9 (56%) | Apr. 2019–Aug. 2019 | 21 (16%) | 0 (0%) | 0–6 |
| Rissen | 2 (1%) | 35 | 0/2 (0%) | 0/2 (0%) | 2/2 (100%) | 0/1 (0%) | Mar. 2019–Apr. 2019 | 1 (<1%) | 0 (0%) | 0 |
| Derby | 1 (<1%) | N/A | 1/1 (100%) | N/A | N/A | N/A | Feb. 2019 | 3 (2%) | 0 (0%) | 0–7 |
| Cerro | 1 (<1%) | N/A | 0/1 (0%) | 0/1 (0%) | 1/1 (100%) | 0/1 (0%) | Jul. 2019 | 2 (1%) | 2 (50%) | 0–1 |
| Total | 154 (100%) | 40 | 70/142 (49%) | 35/133 (26%) | 107/122 (88%) | 65/97 (67%) | Jun. 2015–Sept. 2019 | 137 (100%)c | 4 (100%)d | N/A |
Median and frequencies calculated based on total number of patients within serotype with information available. N/A = not applicable.
Genetic similarity of clinical and non-human isolates was determined based on core genome multi-locus sequence typing.
Total includes Salmonella serotypes Agona (n = 1), Anatum (n = 3), Give (n = 2), Senftenberg (n = 2), Uganda (n = 5), Worthington (n = 1), Brandenburg (n = 1), Livingstone (n = 5), and Panama (n = 6) isolated from pig ear treats, all of which were not genetically related to any human cases.
Total includes a Salmonella enterica subspecies arizonae isolate from a dog, which was not genetically related to any human cases.
In the United States, salmonellosis is a nationally notifiable disease.36 Salmonella isolates obtained from ill people are sequenced by state and local public health laboratories and analyzed for genetic relatedness through PulseNet to identify potential multistate outbreaks. Additionally, public health officials routinely interview people with laboratory-confirmed Salmonella infections (or their caregiver/proxy) with a standardized questionnaire designed to collect demographic information and general food, animal, and other exposures the week prior to illness onset.37 After interviews of initial patients included in this investigation indicated frequent exposure to dogs, CDC used a binomial probability analysis to compare the proportion of patients reporting contact with dogs within seven days of illness onset with the proportion of healthy individuals who reported contacting dogs in the seven days before interview as part of the 2018–2019 Foodborne Diseases Active Surveillance Network (FoodNet) population survey.37,38 This analysis prompted CDC to request further information from patients about the type of dog contact, pet illness, and pet food and pet treat purchase or exposure using a standardized questionnaire. Subsequent interviews indicated that most patients with dog contact also had contact with pig ear pet treats before illness onset, which prompted further investigation into pig ear pet treats.
The FDA Center for Veterinary Medicine Office of Surveillance and Compliance (OSC) reviewed adverse events reported either through an online public reporting system39 or through consumer complaints to FDA via email or phone call about pet illnesses associated with exposure to pig ear treats. A subset of complaints with pertinent information available were investigated by the FDA Veterinary Laboratory Investigation and Response Network (Vet-LIRN). Vet-LIRN reviewed medical records, interviewed pet owners, and conducted non-regulatory testing of pig ear treats and feces from ill dogs (fecal culture for enteric pathogens) who had consumed pig ear treats.
CDC and state partners collected pig ear treat receipts and purchase records from patients, when available. FDA’s Office of Regulatory Affairs and state public health and agriculture departments obtained records from retail locations regarding distributors and wholesalers of pig ear treats. State health and agriculture departments in collaboration with FDA collected pig ear treats for bacterial culture from retail locations in Arizona, Kansas, Michigan, Pennsylvania, and Rhode Island, where ill people reported buying the products. Pig ear treats were also tested from suppliers and distributors to those retail locations. One patient in Connecticut provided a sample from an opened bag of pig ear treats from their home. FDA traced a subset of Salmonella-positive pig ear treats to their most likely country of origin.
Laboratory tests and analysis
Salmonella culture of human specimens was performed by individual diagnostic laboratories according to their usual protocols.40 Salmonella culture of samples from dogs and pig ear treats was performed by state laboratories and the FDA and followed standard techniques.41 State laboratories performed PFGE on isolates from people and pig ear treat samples following the PulseNet protocol,42 analyzed patterns using BioNumerics 6.6 (Applied Maths, Sint-Martens-Latem, Belgium), and uploaded patterns to the national database for comparison and naming. WGS was performed on human clinical and pig ear treat isolates using the Nextera XT library preparation kit (Illumina, San Diego, CA) followed by sequencing on the Illumina MiSeq according to PulseNet protocols.43 Sequences were shared with the CDC for genomic analysis. Initially, high-quality single nucleotide polymorphism (hqSNP) analysis was performed44; however, cgMLST was adopted as the primary subtyping approach during the outbreak investigation35 and was performed using standard PulseNet protocol, with analysis done using BioNumerics 7.6.43
WGS of dog fecal and pig ear treat isolates obtained by Vet-LIRN was completed at one of two laboratories. Briefly, one laboratory extracted DNA using automated magnetic bead-based processing (MagMAX CORE; Thermo Fisher Scientific) and quantified with a Qubit 2.0 fluorometer (Thermo Fisher Scientific). Genomic libraries were prepared and barcoded using the Nextera XT DNA Library Preparation Kit (Illumina, Inc., San Diego, California, USA) and were then sequenced on the Illumina MiSeq platform using the MiSeq Reagent Kit version 3 (Illumina, Inc.) with 2 × 250 base pair chemistry. The second laboratory extracted DNA using a spin column method (DNeasy Blood and Tissue Kit, QIAGEN) and then quantified and sequenced the DNA using the same methods as the first laboratory. Sequencing of these isolates followed PulseNet protocols.43,45
Sequences from clinical isolates were deposited to the National Center for Biotechnology Information (NCBI) BioProject PRJNA230403 (Supplementary Table S1). Sequences from non-clinical isolates were deposited through NCBI under BioProject IDs PRJNA183851, PRJNA186035, and PRJNA292666 (Supplementary Table S1).
The National Antimicrobial Resistance Monitoring System (NARMS) laboratory at CDC performed antimicrobial susceptibility testing (AST) by broth microdilution on select clinical isolates using a custom Sensititre® panel (Trek Diagnostics, Westlake, OH; product number CMV4AGNF) with 14 drugs (Supplementary Table S2).46 Clinical and Laboratory Standards Institute (CLSI) breakpoints, if available, were used to define susceptible, intermediate, and resistant minimum inhibitory concentration (MIC) ranges; otherwise, we used NARMS-established breakpoints.46,47 Four Vet-LIRN laboratories performed AST using the same methods as above on dog and pig ear treat isolates collected following consumer complaints but tested isolates for resistance to either 19 (commercial Sensititre® panel; product number BOPO7F or COMPGN1F) or 22 drugs (commercial Sensititre® panel; product number COMPAN2F; Supplementary Table S2).
Resistance determinants were identified for clinical and pig ear treat isolates using standardized methods.48 Briefly, sequence data were assembled using shovill v.1.0.9 (https://github.com/tseemann/shovill) omitting contigs with less than 10% of the genome coverage, and assemblies were then screened for resistance determinants using staramr 0.4.0 (https://github.com/phac-nml/staramr) using the ResFinder database (updated 19 Feb 2021) and the PointFinder database for Salmonella (updated 01 Feb 2021).49,50 Isolates without AST results were assigned a predicted resistance pattern based on the presence of resistance determinants in genome assemblies.51 We considered isolates with intermediate interpretation to ciprofloxacin by AST or one quinolone resistance mechanism by WGS to be ciprofloxacin non-susceptible, and used nonsusceptibility rather than resistance in our definitions for MDR and clinically important resistance because Salmonella isolates with ciprofloxacin nonsusceptibility may be associated with clinical failure or delayed treatment response.47 We defined MDR as resistance or predicted resistance (or nonsusceptibility, for ciprofloxacin) to at least one antimicrobial in three or more CLSI drug classes47 and clinically important resistance as resistance or predicted resistance (or nonsusceptibility, for ciprofloxacin) to at least one antimicrobial commonly recommended for treatment (i.e., ampicillin, azithromycin, ceftriaxone, ciprofloxacin, or trimethoprim-sulfamethoxazole).7,52
Ethics and patient consent
This investigation, which spanned May 2019–November 2019, was reviewed by CDC and was conducted consistent with applicable federal law and CDC policy (See e.g., 45C F R. part 46, 21C F R. part 56; 42 U S C. §241(d); 5 U S C. §552a; 44 U S C. §3501 et seq.).
Role of the funding source
Funding sources for this outbreak investigation had no role in investigation design, data collection, data analysis, data interpretation, writing of the report, or the decision to publish.
Results
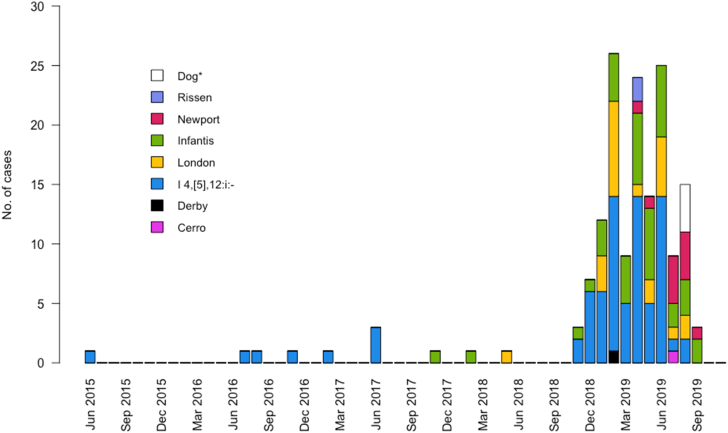
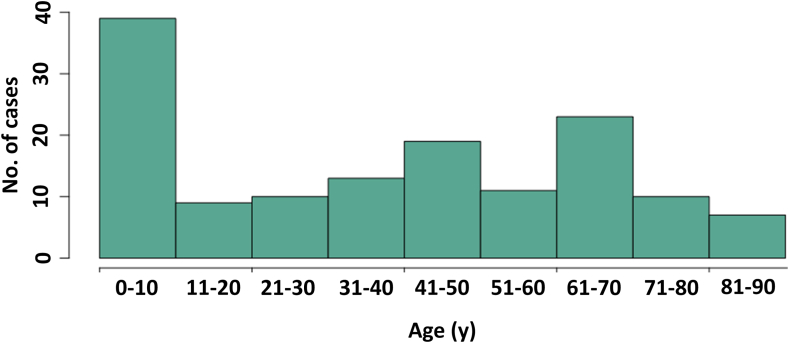
In 34 states, 154 human cases were identified (Fig. 1). Fifteen cases were identified based on PFGE pattern only, 33 were identified based on cgMLST analysis only, and 106 cases were identified by both PFGE and cgMLST. Isolation dates ranged from June 10, 2015, to September 15, 2019 (Fig. 2); 94% of cases (145/154) occurred during 2018–2019. Of 136 patients with information available, the median patient age was 40 years, with a range of <1–90 years (Fig. 3). Twenty-seven patients (20%) were children <5 years. Of 118 patients with race information available, 110 (93%) were White, six (5%) were African American/Black, and two (2%) were Asian and White. Of 107 patients with ethnicity information available, four (4%) were Hispanic. Thirty-five (26%) of 133 patients with information available were hospitalized; six hospitalized patients were <5 years of age, and six were ≥65 years of age. Salmonella (serotype I 4,[5],12:i:- or London) was isolated from the blood of two patients. No deaths were reported. For patients with information available, 107 (88%) of 122 reported contact with dogs before illness onset, and 65 (67%) of 97 reported handling pig ear dog treats. The number of patients reporting contact with dogs was significantly higher compared with the proportions of healthy people interviewed in the FoodNet Population survey reporting contact with a dog (68% of respondents, p-value = 0.010). A similar comparison could not be performed for the incidence of exposure to pig ear pet treats because this specific exposure is not captured in the FoodNet Population survey.38 The earliest patient to report contact with pig ear treats became ill in 2017. Fifteen (32%) of 47 patients reporting contact with pig ear treats said that they always washed their hands after handling pet food or treats, and 18 (38%) reported that they rarely or never washed their hands after handling pet food or treats. Five (11%) of 47 patients indicated that their pet dog exhibited signs consistent with bacterial infection, such as diarrhea, after consuming pig ear treats.
Fig. 1.
Human and dog cases of salmonellosis and pig ear pet treat isolates – United States, 2015–2019. Cases were defined as human infection with Salmonella serotypes Cerro, Derby, I 4,[5],12:i:-, London, Infantis, Newport, or Rissen genetically related to isolates from pig ear pet treats, with isolation dates from June 2015 to September 2019. Genetic relatedness of human and pig ear treat isolates was determined based on the identification of a matching pulsed field gel electrophoresis pattern or within a specific range of allele differences determined by whole genome sequencing analysis by core genome multi-locus sequence typing. A total of 34 states had human cases in this outbreak. The highest number of cases (24) occurred in Iowa. Black circles indicate states in which sampling of pig ear treats was conducted, and the number of positive samples is indicated. Dog icons indicate dogs testing positive for Salmonella.
Fig. 2.
Epidemiologic curve of isolation dates of Salmonella from humans and dog isolates, June 10, 2015, to September 13, 2019. Cases were defined as human infection with Salmonella serotypes Cerro, Derby, I 4,[5],12:i:-, London, Infantis, Newport, or Rissen genetically related to isolates from pig ear pet treats. Dogs were tested for Salmonella following consumer complaints of illness in their dogs after feeding pig ear pet treats. ∗ Isolates obtained from dogs include Salmonella serotypes Infantis (n = 1) and Cerro (n = 2) and Salmonella enterica subspecies arizonae (n = 1).
Fig. 3.
Age distribution of patients. Cases were defined as human infection with Salmonella serotypes Cerro, Derby, I 4,[5],12:i:-, London, Infantis, Newport, or Rissen genetically related to isolates identified from sampling of pig ear pet treats, with isolation dates from June 2015 through September 2019. Patient age was collected during routine interviews performed by state and local health officials. Median patient age was 40 years (range < 1–90 years).
Human infections resulted from seven Salmonella serotypes including: l 4,[5],12:i:- (49%), Infantis (26%), London (15%), Newport (7%), Rissen (1%), Derby (<1%), and Cerro (<1%) (Table 1). Eight of nine cases identified prior to 2018 were Salmonella I 4,[5],12:i:-. Across the four most common serotypes in this outbreak, patients reported similar frequencies of exposure to dogs (84–100%) or pig ear treats (56–73%) (Table 1).
A total of 137 clinical isolates had antimicrobial resistance information: two assessed by AST only, 24 with resistance predicted by WGS and confirmed by AST, and 111 with resistance predicted by WGS only (Table 2, Table 3); the 24 isolates analyzed by both methods showed concordant results. Of these, 92% (126/137) were resistant (or non-susceptible, for ciprofloxacin) to at least one antimicrobial, and 77% (105/137) were MDR. Ninety-one percent (125/137) demonstrated clinically important resistance: 105 (77%) isolates were resistant to ampicillin, 83 (61%) were non-susceptible to ciprofloxacin, three (2%) were resistant to trimethoprim-sulfamethoxazole, and one (1%) was resistant to azithromycin (Table 2, Table 3). No isolates were resistant to ceftriaxone or meropenem. Three serotype London isolates carried a qnrE1 gene (conferring ciprofloxacin nonsusceptibility), and one serotype I 4,[5],12:i:- isolate carried mef(C) and mph(G) genes (conferring azithromycin resistance, Table 2).
Table 2.
Predicted resistance of human and pig ear treat Salmonella isolates.
| Serotype | Clinical isolates screened for resistancea n = 135 n (%) | Resistance determinants present; No. with resistance mechanism/No. of isolates of that serotype with resistance information (%), Antimicrobial resistance predicted by mechanism | No. MDRb isolates n (%) | No. clinically important resistantc isolates n (%) | Pig ear treat isolates screened for resistance n = 135 n (%) | Resistance determinants present; No. with resistance mechanism/No. of isolates of that serotype with resistance information (%), Antimicrobial resistance predicted by mechanism | No. MDRb isolates n (%) | No. clinically important resistantc isolates n (%) |
|---|---|---|---|---|---|---|---|---|
| I 4,[5],12:i:- | 62 (46%) |
aac(3)-IId: 62/62 (100%), gentamicin aadA2: 62/62 (100%), streptomycin aph(3″)-Ib & aph(6)-Id: 61/62 (98%), streptomycin aph(3′)-IIa: 4/62 (6%), kanamycin aph(6)-Ic: 4/62 (6%), streptomycin blaTEM-1B: 61/62 (98%), ampicillin dfrA12: 55/62 (89%), trimethoprimd floR: 5/62 (8%), chloramphenicol gyrA(83): 62/62 (100%), nalidixic acid, ciprofloxacine mef(C): 1/62 (2%), azithromycin mph(G): 1/62 (2%), azithromycin sul2: 56/62 (90%), sulfisoxazole tet(A): 2/62 (3%), tetracycline tet(B): 61/62 (98%), tetracycline |
61/62 (98%) | 62/62 (100%) | 4 (3%) |
aac(3)-IId: 4/4 (100%), gentamicin aadA2: 4/4 (100%), streptomycin aph(3″)-Ib & aph(6)-Id: 4/4 (100%), streptomycin aph(3′)-IIa: 2/4 (50%), kanamycin aph(6)-Ic: 2/4 (50%), streptomycin blaTEM-1B: 4/4 (100%), ampicillin dfrA12: 2/4 (50%), trimethoprimd floR: 2/4 (50%), chloramphenicol gyrA(83): 4/4 (100%), nalidixic acid, ciprofloxacine mef(C): 0/4 (0%), azithromycin mph(G): 0/4 (0%), azithromycin sul2: 2/4 (50%), sulfisoxazole tet(A): 0/4 (0%), tetracycline tet(B): 4/4 (100%), tetracycline |
4/4 (100%) | 4/4 (100%) |
| Infantis | 39 (29%) |
blaTEM-1B: 39/39 (100%), ampicillin dfrA8: 36/39 (92%), trimethoprim floR: 39/39 (100%), chloramphenicol qnrB19: 0/39 (0%), ciprofloxacinf tet(A): 39/39 (93%), tetracycline No determinants detected: 0/39 (0%) |
39/39 (100%) | 39/39 (100%) | 47 (35%) |
blaTEM-1B: 40/47 (85%), ampicillin dfrA8: 41/47 (87%), trimethoprim floR: 41/47 (87%), chloramphenicol qnrB19: 3/47 (6%), ciprofloxacinf tet(A): 41/47 (87%), tetracycline No determinants detected: 6/47 (13%) |
41/47 (87%) | 40/47 (85%) |
| London | 21 (16%) |
aac(3)-IIa: 3/21 (14%), gentamicin aadA1: 3/21 (14%), streptomycin aph(3″)-Ib & aph(6)-Id: 3/21 (14%), streptomycin blaTEM-1B: 3/21 (14%), ampicillin dfrA1: 3/21 (14%), trimethoprim floR: 2/21 (10%), chloramphenicol qnrB19: 18/21 (86%), ciprofloxacinf qnrE1: 3/21 (14%), ciprofloxacinf sul1: 3/21 (14%), sulfisoxazole tet(A): 3/21 (14%), tetracycline |
3/21 (14%) | 21/21 (100%) | 28 (21%) |
aac(3)-IIa: 4/28 (14%), gentamicin aadA1: 4/28 (14%), streptomycin aph(3″)-Ib & aph(6)-Id: 4/28 (14%), streptomycin blaTEM-1B: 4/28 (14%), ampicillin dfrA1: 4/28 (14%), trimethoprim floR: 3/28 (11%), chloramphenicol qnrB19: 24/28 (86%), ciprofloxacinf qnrE1: 4/28 (14%), ciprofloxacinf sul1: 4/28 (14%), sulfisoxazole tet(A): 4/28 (14%), tetracycline |
4/28 (14%) | 28/28 (100%) |
| Newport | 10 (7%) |
qnrB19: 1/10 (10%), ciprofloxacinf No determinants detected: 9/10 (93%) |
0/10 (0%) | 1/10 (10%) | 20 (15%) |
qnrB19: 1/20 (5%), ciprofloxacinf No determinants detected: 19/20 (93%) |
0/20 (0%) | 1/20 (5%) |
| Rissen | 1 (<1%) | No determinants detected: 1/1 (100%) | 0/1 (0%) | 0/1 (0%) | 1 (<1%) | No determinants detected: 1/1 (100%) | 0/1 (0%) | 0/1 (0%) |
| Derby | 1 (<1%) | fos7: 1/1 (100%), fosfomycin | 0/1 (0%) | 0/1 (0%) | 3 (2%) |
fos7: 3/3 (100%), fosfomycin tet(A): 1/3 (33%), tetracycline |
0/3 (0%) | 0/3 (0%) |
| Cerro | 1 (<1%) | No determinants detected: 1/1 (100%) | 0/1 (0%) | 0/1 (0%) | 2 (1%) | No determinants detected: 2/2 (100%) | 0/2 (0%) | 0/2 (0%) |
| Agona | N/A | N/A | N/A | N/A | 1 (<1%) | fos7: 1/1 (100%), fosfomycin | 0/1 (0%) | 0/1 (0%) |
| Anatum | N/A | N/A | N/A | N/A | 3 (2%) | No determinants detected: 3/3 (100%) | 0/3 (0%) | 0/3 (0%) |
| Give | N/A | N/A | N/A | N/A | 2 (1%) | No determinants detected: 2/2 (100%) | 0/2 (0%) | 0/2 (0%) |
| Typhimurium | N/A | N/A | N/A | N/A | 4 (<1%) | No determinants detected: 4/4 (100%) | 0/4 (0%) | 0/4 (0%) |
| Senftenberg | N/A | N/A | N/A | N/A | 2 (1%) |
aph(3″)-Ib & aph(6)-Id: 1/2 (50%), streptomycin floR: 1/2 (50%), chloramphenicol qnrB19: 1/2 (50%), ciprofloxacinf sul2: 1/2 (50%), sulfisoxazole tet(A): 1/2 (50%), tetracycline No determinants detected: 1/2 (50%) |
1/2 (50%) | 1/2 (50%) |
| Uganda | N/A | N/A | N/A | N/A | 5 (4%) | No determinants detected: 5/5 (100%) | 0/5 (0%) | 0/5 (0%) |
| Worthington | N/A | N/A | N/A | N/A | 1 (<1%) |
aadA1: 1/1 (100%), streptomycin aph(3″)-Ib & aph(6)-Id: 1/1 (100%), streptomycin blaTEM-1A: 1/1 (100%), ampicillin dfrA1: 1/1 (100%), trimethoprim floR: 1/1 (100%), chloramphenicol qnrB19: 1/1 (100%), ciprofloxacinf sul1: 1/1 (100%), sulfisoxazole sul2: 1/1 (100%), sulfisoxazole tet(A): 1/1 (100%), tetracycline |
1/1 (100%) | 1/1 (100%) |
| Brandenburg | N/A | N/A | N/A | N/A | 1 (<1%) |
aph(3″)-Ib & aph(6)-Id: 1/1 (100%), streptomycin blaTEM-1A: 1/1 (100%), ampicillin gyrA(87): 1/1 (100%), nalidixic acid, ciprofloxacine sul2: 1/1 (100%), sulfisoxazole tet(A): 1/1 (100%), tetracycline |
1/1 (100%) | 1/1 (100%) |
| Livingstone | N/A | N/A | N/A | N/A | 5 (4%) | No determinants detected: 5/5 (100%) | 0/5 (0%) | 0/5 (0%) |
| Panama | N/A | N/A | N/A | N/A | 6 (4%) |
aadA5: 6/6 (100%), streptomycin aph(3″)-Ib & aph(6)-Id: 6/6 (100%), streptomycin dfrA17: 6/6 (100%), trimethoprim floR: 6/6 (100%), chloramphenicol qnrB19: 6/6 (100%), ciprofloxacinf sul2: 6/6 (100%), sulfisoxazole tet(B): 6/6 (100%), tetracycline |
6/6 (100%) | 6/6 (100%) |
Two hundred and seventy isolates (135 human, 135 pig ear treat samples) were screened for resistance determinants via whole genome sequencing with results as shown.
All sequenced clinical isolates have been deposited to the National Center for Biotechnology Information BioProject PRJNA230403.
MDR = Multidrug resistant, which was defined as resistance (or nonsusceptibility, for ciprofloxacin) to ≥3 antimicrobial classes.
Clinically important resistance was defined as resistance (or nonsusceptibility, for ciprofloxacin) to ≥1 antimicrobial recommended for treatment of salmonellosis (i.e., ampicillin, azithromycin, ceftriaxone, ciprofloxacin, or trimethoprim-sulfamethoxazole).
Although the dfrA12 gene was identified by ResFinder, the gene is interrupted, and the product does not appear functional. Therefore, these isolates are not expected to show phenotypic resistance to trimethoprim.
Single chromosomal mutations in the quinolone resistance-determining region (QRDR) of target enzyme genes such as gyrA typically confers resistance to nalidixic acid and intermediate interpretation to ciprofloxacin by phenotypic testing.
Single plasmid-mediated quinolone resistance genes (such as qnr genes) typically confer intermediate susceptibility to ciprofloxacin by phenotypic testing. No isolates harbored more than one quinolone resistance gene.
Table 3.
Antimicrobial resistance of Salmonella outbreak isolates from human and pig ear treat samples.
| Antimicrobial | Human N = 137 n (%) | Pig ear treats N = 135 n (%) | Dog N = 4 n (%) | Totala n (%) |
|---|---|---|---|---|
| Amikacin | NT | NT | 1 (25%) | 1/4 (25%) |
| Ampicillin | 105 (77%) | 50 (37%) | 4 (100%) | 159/276 (58%) |
| Amoxicillin-clavulanic acid | NT | NT | 4 (100%) | 4/4 (100%) |
| Azithromycin | 1 (1%) | 0 (0%) | NT | 1/272 (<1%) |
| Cefazolin | NT | NT | 2 (50%) | 2/4 (50%) |
| Cefpodoxime | NT | NT | 1 (25%) | 1/4 (25%) |
| Ceftriaxone | 0 (0%) | 0 (0%) | NT | 0/272 (0%) |
| Cephalexin | NT | NT | 2 (50%) | 2/4 (50%) |
| Chloramphenicol | 46 (34%) | 54 (40%) | 1 (25%) | 101/276 (37%) |
| Ciprofloxacinb | 84 (61%) | 45 (33%) | NT | 129/272 (47%) |
| Doxycycline | NT | NT | 1 (25%) | 1/4 (25%) |
| Fosfomycinc,d | 1 (1%) | 4 (3%) | NT | 5/272 (2%) |
| Gentamicin | 67 (49%) | 8 (6%) | 2 (50%) | 77/276 (28%) |
| Kanamycinc,d | 4 (3%) | 2 (2%) | NT | 6/272 (2%) |
| Meropenem | 0 (0%) | 0 (0%) | NT | 0/272 (0%) |
| Nalidixic acidc | 63 (53%) | 5 (5%) | NT | 68/272 (32%) |
| Streptomycinc | 65 (48%) | 17 (13%) | NT | 82/272 (30%) |
| Sulfisoxazole | 61 (45%) | 15 (11%) | NT | 76/272 (28%) |
| Tetracycline | 105 (77%) | 59 (44%) | 1 (25%) | 165/276 (60%) |
| Trimethoprimd | 39 (28%) | 52 (39%) | NT | 91/272 (33%) |
| Trimethoprim-sulfamethoxazole | 3 (2%) | 12 (9%) | 1 (25%) | 16/272 (6%) |
| Any resistance | 126 (92%) | 87 (64%) | 4 (100%) | 217/276 (79%) |
| Multidrug resistancee | 105 (77%) | 58 (43%) | 2 (50%) | 165/276 (60%) |
| Clinically important resistancef | 125 (90%) | 82 (61%) | 4 (100%) | 211/276 (76%) |
Antimicrobial resistance information was available for isolates from 137 ill people, including two assessed by standard antimicrobial susceptibility testing (AST) only, 24 with resistance predicted by whole genome sequencing (WGS) and confirmed by AST, and 111 with resistance predicted by WGS only. Pig ear treat isolates were only evaluated by WGS. Dog isolates were only evaluated by AST. Two pig ear treat isolates and 17 human isolates were not available for resistance screening.
Total in each row based on the number tested for that given antimicrobial. NT = Not tested.
Percentages reflect ciprofloxacin nonsusceptibilty (intermediate interpretation by AST or single quinolone resistance mechanism). No isolates showed resistance by AST or harbored multiple quinolone resistance mechanisms.
Antimicrobial resistance information was available for a subset of isolates: fosfomycin (n = 270), kanamycin (n = 270), nalidixic acid (n = 214), streptomycin (n = 270).
Resistance information was predicted based on WGS alone for these antimicrobials.
Multidrug resistance was defined as resistance (or nonsusceptibility, for ciprofloxacin) to ≥3 antimicrobial classes.
Clinically important resistance was defined as resistance (or nonsusceptibility, for ciprofloxacin) to ≥1 antimicrobial recommended for treatment of salmonellosis (i.e., ampicillin, azithromycin, ceftriaxone, ciprofloxacin, or trimethoprim-sulfamethoxazole).
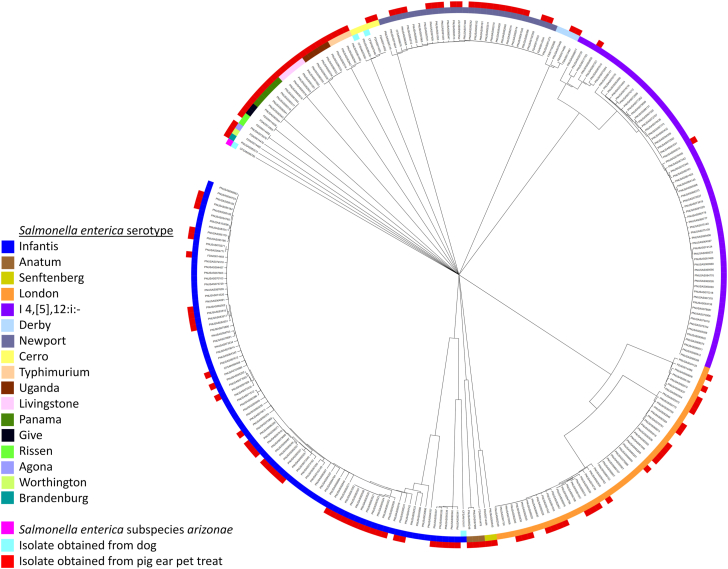
In total, 137 pig ear treat samples from 10 states yielded Salmonella (Fig. 1); 110 (80%) were closely genetically related to clinical isolates (Fig. 4, Table 1). Seventeen serotypes were detected (Table 1, Table 2). Resistance was predicted by WGS for all but two pig ear treat isolates; 64% (87/135) were resistant to at least one antimicrobial, and 43% (58/135) were MDR (Table 3). Four serotype London isolates from pig ear treats carried the qnrE1 gene (Table 2).
Fig. 4.
This dendrogram represents the genetic relatedness of Salmonella isolates included in this outbreak that were collected from 2015–2019 in the United States and reported to PulseNet, the national molecular subtyping network for foodborne disease surveillance at the Centers for Disease Control and Prevention. The shaded ring represents each Salmonella serotype (or Salmonella enterica subspecies arizonae) detected among isolates obtained from ill people, pig ear pet treats, or dogs. Red boxes indicate isolates obtained from pig ear treats. Light blue boxes indicate isolates obtained from dogs.
Testing by the Michigan Department of Agriculture and Rural Development (MDARD) and the Pennsylvania Department of Agriculture found that bulk pig ear treats stocked in open bins of retail stores owned by a United States pet treat retailer (Company A) yielded isolates with the outbreak strains (serotypes London, Newport, Infantis, and I 4,[5],12:i:-) as well as serotypes not associated with reported human illness in this outbreak (Typhimurium, Uganda, Brandenburg, Livingstone, Senftenberg, and Panama) (Fig. 5). Pig ear treats sampled at Company A’s distribution facility in Indiana yielded eight Salmonella isolates: four matching clinical isolates (serotypes Derby, Infantis, London, and Rissen) and four only detected in pig ear treats (serotypes Agona, Anatum, Senftenberg, and Worthington). Nineteen (32%) of 59 patients with information available reported purchasing pig ear treats from stores owned by Company A across 11 states. Four patients (7%) from different states reported purchasing pig ear treats sold as 8-pack pouches, individually shrink-wrapped, or in open bulk bins unwrapped from stores supplied by another company (Company B). Pig ear treats produced by Company B and sampled by FDA, MDARD, and Kansas, Washington, and Arizona state officials yielded outbreak strains (serotypes Newport, Infantis, London, and Cerro) and other Salmonella strains (serotypes Livingstone, Give, and Anatum). Company B also reported receiving two consumer complaints of illness in dogs that had ingested these pig ear treats. The Rhode Island Department of Health isolated Salmonella Infantis from one pig ear treat sample from Company C.
Fig. 5.
Traceback diagram of pig ear pet treats yielding Salmonella isolates. Traceback conducted by FDA determined the routes of supply of pig ear pet treats between the South America suppliers and specific retail store locations (unique routes of supply are indicated by different colored arrows). Black arrows represent pig ear treats in retail stores that could be traced to supplying company B but not the country of origin. Pig ear treats were sampled from seven retail stores and from one distribution facility; pig ear treats at all of these locations were positive for Salmonella (as indicated by a star icon).
A subset of pig ear treats from Company A was traced to Argentina and Colombia, Company B to Argentina and Brazil, and Company C to Brazil (Fig. 5). Eleven pig ear treat samples collected by FDA and traced to one specific supplier in each country were found to contain Salmonella matching clinical isolates (serotypes London, Newport, Rissen, and Infantis) and other Salmonella strains (serotypes Give and Senftenberg). In response, FDA issued import alerts for three firms that supplied Companies A, B, and C.29 Three individually wrapped pig ear treats labeled as irradiated were produced in Argentina, collected in Kansas, and yielded Salmonella.
On July 31, 2019, CDC and FDA issued a recommendation to the public not to buy pig ear treats or feed them to pets.53 FDA worked with companies to recall potentially contaminated products; recalls were conducted by six firms with two firms initiating and then expanding their recall (Table 4). CDC distributed educational information to pet owners via a CDC Outbreak Notice and social media, recommending hand washing after handling pet food or treats and after cleaning up pet feces to prevent additional illnesses.53 FDA distributed information about the investigation and amplified outreach through social media.
Table 4.
Summary of firm recalls.
| Firm Alias | Date of recall | Products recalled | States involved | Source | Comments |
|---|---|---|---|---|---|
| Company A | July 3, 2019 | Bulk pig ear treats provided in Company A retail stores | 33 states | Undetermineda | |
| Company B | July 26, 2019 | One specific brand of packaged or individually wrapped pig ear treats | Nationwide | Argentina and Brazil | |
| July 30, 2019 | The same brand of packaged or individually wrapped pig ear treats as well as pig ear treats sold in bulk unwrapped. | Nationwide | Argentina and Brazil | The recall was expanded to include bulk unwrapped pig ear treats as well as a wider date range during which the products were distributed. | |
| Company C | August 16, 2019 | Specified lots of bulk and packaged pig ear treats of one specific brand. | Nationwide | Brazil | |
| September 3, 2019 | Packaged pig ear treats of one additional brand sold by Company C to one retailer. | Unspecified | Brazil | Recall was expanded following Rhode Island Department of Health detecting Salmonella-positive pig ear treats of this specific brand, which was different than the brand specified in the August 16 recall. | |
| Company E | August 27, 2019 | Variously sized bags of pig ear treats distributed online and in one FL store | Nationwide | Colombia | Pig ear products tested positive for Salmonella, but none were linked to the outbreak. |
| Company F | September 20, 2019 | Variously sized bags of pig ear treats distributed online | Nationwide | USA | Michigan Department of Agriculture and Rural Development sampling found one positive bag of pig ear treats. The isolate and products from this company were not linked to the outbreak. |
| Company G | October 11, 2019 | Bulk pig ear treats | Nationwide | Unspecified South American country | Firm found positive isolates among pig ear treats in self-initiated audit. Company G reported it was supplied by Company C, but this was not confirmed by FDA. Positive isolates were not linked to the outbreak. |
This table provides information on the nature of voluntary pig ear treat recalls issued by six firms throughout the course of the outbreak and afterward. Company D did not issue a recall because this company was only a supplier for Company A and did not directly market pig ear treats to consumers.
While the source of all recalled Company A pig ear treats was undetermined, some were traced to Colombia and Argentina (Fig. 5).
FDA Vet-LIRN investigated nine of 18 consumer complaints of adverse events after exposure to pig ear treats. The reports were from eight states and involved ten dogs in nine households exposed to bulk and packaged pig ear treats (Supplementary Table S3). Four of ten dog fecal cultures yielded Salmonella, including serotypes Cerro and Infantis and S. enterica subspecies arizonae. Pig ear treats were tested from six households, three of which had pig ear treats yielding Salmonella. Contaminated pig ear treats from two households were purchased in bulk products from Company A, and one household purchased from a retail store chain supplied by Companies B and C.
Discussion
To our knowledge, this was the first documented multistate outbreak of MDR Salmonella in humans in the United States linked to pig ear pet treats. Canada previously reported an outbreak of Salmonella Infantis associated with pig ear pet treats,27 and surveillance efforts have since identified Salmonella from pig ear pet treats domestically and internationally without confirmed human cases.30,33,54,55 In our investigation, as in the Canadian outbreak, it was unclear whether human exposures occurred solely from direct contact with pig ear treats themselves or if zoonotic transmission from companion dogs contributed. This investigation identified human cases as far back in time as June 2015; limited exposure information was available from cases detected before 2018, though most were caused by Salmonella I 4,[5],12:i:-. Enteric illness outbreaks can occur over a wide timeframe particularly when animal or environmental reservoirs allow strains to persist.56 Our investigation documented multiple reports of ill dogs, some of which had been fed pig ear treats. Clinically ill and carrier dogs are considered potential sources of zoonotic transmission of salmonellosis via fecal-oral routes.16, 17, 18 Furthermore, more patients reported owning or having contact with dogs before illness onset (n = 107, 88%) than having direct contact with pig ear pet treats (n = 65, 67%). Therefore, both pig ear treats and dogs were considered sources of salmonellosis in this outbreak.
Pet treats like pig ears are regulated by FDA under the Federal Food, Drug, and Cosmetic Act, which requires any food for animals to be safe to eat, produced under sanitary conditions, and free of harmful substances. Finished pet treats that are found to be contaminated with Salmonella are considered adulterated.57,58 Epidemiologic, laboratory, and traceback evidence was unable to identify the exact sources of contaminated pig ear treats. Although country of origin and exporting firms could be determined for selected pig ear treat isolates, it was not possible to identify slaughterhouses or manufacturing plants where contamination occurred. In general, pig ears are removed from the carcass at the slaughterhouse, de-haired, and frozen for shipment to pet food and treat manufacturers.55 They are then thawed, dried, coated in flavouring, and cooked at a temperature sufficient to kill Salmonella, though there is evidence to support that the drying step may protect the pathogen during cooking.55,59,60 As evidence mounted in the early 2000s about the risks associated with pet treats, CDC and FDA provided additional guidance to the industry to encourage enhanced protective measures such as irradiation to reduce pathogen burden on treats before sale.34,55 Irradiation employs gamma, x-ray, or electron beam radiation as a means of reducing pathogen burden, including Salmonella, on food products without requiring heating.61,62 A subset of pig ear treats testing positive for Salmonella in this outbreak were in packages labeled as irradiated. However, to our knowledge, no studies have evaluated the effectiveness of irradiation controlling Salmonella contamination of pet treats derived from dried animal byproducts (e.g., pig ear treats, jerky-style treats, bully sticks, cattle hooves). We could not determine whether available guidelines for irradiation were followed, and it is possible that pig ear treats were too heavily contaminated before irradiation to allow for complete sterilization even by appropriate protocols. Furthermore, pet food and treat retailers associated with this outbreak and subsequent recalls were stocking pig ear treats unwrapped in bulk bins and, in some cases, were comingling pig ear treats from multiple sources, potentially negating any processing-level disease mitigation steps by introducing the risk of cross-contamination at stores.
Identification of contaminated pig ear treats originating from multiple companies and distributed to multiple states signifies the widespread risk to pets and pet owners. Pig ear treats tested in this investigation were contaminated with seventeen serotypes of Salmonella, seven of which were closely genetically related to Salmonella isolates obtained from ill people. Traceback of some contaminated products indicated that implicated Salmonella strains may have originated in Brazil, Colombia, and Argentina. This investigation identified some serotypes less commonly associated with human illness in the United States63; however, some (e.g., serotypes Brandenburg, Livingston, Agona, Derby, and Panama) have been documented in pork production chains in Brazil and Argentina and might be more common in these countries.64,65 This investigation revealed how a single internationally traded product can become contaminated with multiple strains of a pathogen, resulting in an outbreak-level incidence of illnesses linked to contact with this product and requiring coordinated mitigation efforts across state and federal governmental agencies.
Multidrug resistance presents another concern in this outbreak. MDR salmonellosis has been associated with worse clinical outcomes, including a higher risk of bloodstream infection or hospitalization.2,66,67 In this outbreak, over 90% of clinical isolates were resistant to at least one antimicrobial recommended for treatment.7,52 Fortunately, resistance to some recommended antimicrobials was uncommon; only one isolate was resistant to azithromycin, and none were resistant to ceftriaxone. Nonetheless, the presence of rare resistance determinants such as qnrE1, mef(C), and mph(G) serves as a reminder that imported products for pets can be a mechanism for spreading clinically important resistance globally. Among more than 67,000 clinical Salmonella isolates screened by CDC NARMS surveillance by the end of 2019 (CDC unpublished data, CDC 2021), only one isolate (accession number SAMN09636156) not related to this outbreak also carried the qnrE1 gene and two other isolates (accession numbers SAMN08159923 and SAMN13905329) carried mef(C) and mph(G) genes. Of these NARMS surveillance isolates, the isolate carrying qnrE1 was serotype Infantis closely genetically related to a strain with known ties to South America.68 This surveillance isolate was genetically unrelated to the Infantis strains in this outbreak. The two NARMS surveillance isolates carrying mef(C) and mph(G) were both serotype I 4,[5],12:i:-. One surveillance isolate is genetically related to the I 4,[5],12:i:- outbreak strain reported here, but it was not identified at the time of the outbreak investigation. These findings suggest that these genes likely have reservoirs in South America but are currently rare in the United States.
Other limitations of this outbreak investigation should be considered. First, not all patients were available or agreed to be interviewed, limiting the amount of data on potential Salmonella source exposures that could be explored and the representativeness of the underlying population. Second, while the proportion of patients included in this outbreak who reported contact with dogs was higher than what is reported among healthy people based on the FoodNet Population survey, the baseline incidence of healthy peoples’ exposure to pig ear pet treats in the United States is not captured in this survey and was not collected through interview of control cases during the outbreak investigation.38 This precludes our ability to perform statistical inference that might bolster our understanding of the epidemiologic information collected in this investigation. Collection of the baseline exposure rates to different types of pet foods and treats in future surveys of healthy people could improve investigative capabilities during illness outbreaks linked to these vehicles. Third, resistance was determined for most isolates based on WGS, and only a subset could have resistance confirmed phenotypically by AST. As such, resistance for most isolates reported here is predicted and subject to limitations of these methods.51 It is possible that some of those isolates carrying resistant genes do not express them or express them at levels that do not confer clinical resistance. Yet, isolates analyzed by AST and WGS demonstrated concordant resistance profiles consistent with other studies.51,69 Finally, the identification of patients as part of an outbreak necessitates those ill individuals to seek medical care, healthcare providers to order appropriate diagnostic testing, and positive test results to be reported to public health departments. Thus, our investigation is likely an underestimate of the true number of people that were affected by the outbreak strain and an overestimate of the severity of illness.
This marks the first reported multistate salmonellosis outbreak associated with exposure to pig ear pet treats in the United States. Multiple Salmonella serotypes and antimicrobial resistance profiles were identified through epidemiologic, laboratory, and traceback efforts coordinated across multiple state and federal agencies. The health of dogs was also impacted by contact with contaminated treats, and zoonotic transmission of Salmonella was also considered a potential contributor to human cases. This outbreak highlighted the risk of human illness linked to pig ear pet treats because of widespread contamination of this product with MDR strains of Salmonella that were not mitigated by processing practices such as heat treatment or irradiation. Imported products can be contaminated with strains of Salmonella not commonly found in the United States. Although Salmonella caused this illness outbreak, there is also concern for the importation of other pathogens from pet treats when pathogen reduction measures are inadequate. Intensified surveillance of internationally traded pet food products for enteric pathogens might be warranted, and international producers should consider bolstering strategies that reduce product contamination. Consumers should be aware of the potential risks to human and animal health from these products and take measures, such as handwashing after feeding pets treats and picking up pet feces, to protect their health.
Contributors
MN contributed to the investigation methodology/design, data collection, data interpretation, manuscript development including editing and revision. GSS analyzed outbreak data, contributed to interpretation, and developed the manuscript including writing the original draft and revision. DSR & SP coordinated Vet-LIRN testing and revised this manuscript. LG contributed to the investigation methodology/design, data collection, and revising this manuscript. JA contributed PulseNet data analyses, interpretation and creation of figures, and revising this manuscript. JCC and HC analyzed antimicrobial resistance data and assisted with writing and revising this manuscript. AH, MG, LP, SP, AN, SD, LP, and DSR coordinated consumer complaint review, sample collection, product traceback, inspections, recall actions, import activities, public messaging, and manuscript revision. AN and SD drafted, cleared, and published public health recommendations/communications related to this outbreak and contributed to manuscript revision. DD assisted with epidemiologic data collection and management for Michigan and revised this manuscript. SD and EB performed PFGE and WGS, analyzed this data, and revised this manuscript. SG contributed to the outbreak investigation and revised this manuscript. BH identified and serotyped Salmonella isolates and revised this manuscript. KM, KP, SB, and EF conducted laboratory testing of pig ear treats collected at retail locations in Michigan and revised this manuscript. SO performed data collection and analysis associated with the Kansas Department of Agriculture Laboratory and revised this manuscript. DN coordinated sample collection and epidemiologic data collection and management for Kansas and revised this manuscript. Pennsylvania (KEK, BT) collected epidemiologic data, revised the manuscript, and tested both pig ear treats collected at a retail store in Pennsylvania, as well as environmental swabs of pig ear treat bins. Rhode Island (GC, BV, and AM) contributed to acquisition of data through coordination of sample collection and product traceback information, coordination of product recalls, or laboratory testing of product samples and revised this manuscript. Connecticut (CT, KHT, CN, LM, TN) ensured interview of patients, collected pig ear dog treat samples for testing, performed testing of these samples, and revised this manuscript. SH performed data collection and analysis during state level outbreak investigation and revised this manuscript. LKFW performed the primary epidemiological analysis of antimicrobial resistance data and contributed to data interpretation, writing, and editing the manuscript.
Data sharing statement
Sequences from clinical isolates were deposited to the National Center for Biotechnology Information (NCBI) BioProject PRJNA230403. Sequences from non-clinical isolates were deposited through NCBI under BioProject IDs PRJNA183851, PRJNA186035, and PRJNA292666. Accession numbers for isolates available in NCBI are provided in Supplementary Table S1. Data presented in this manuscript are available from the corresponding author (gpg6@cdc.gov) upon reasonable request following publication. This includes data that underlie the results reported in this article, after de-identification (text, tables, figures, and Supplemental materials).
Editor note
The Lancet Group takes a neutral position with respect to territorial claims in published maps and institutional affiliations.
Declaration of interests
The authors have no conflicts of interest to disclose. The contents are those of the author(s) and do not necessarily represent the official views of, nor an endorsement, by CDC, FDA, HHS, or the United States Government. For more information, please visit FDA.gov.
Acknowledgements
The authors would like to acknowledge investigation contributions from all state and local governments and would like to acknowledge public health partners in Arizona, Indiana, Washington, and Tennessee for conducting sampling of pig ear treats. The authors would like to thank Meseret Birhane, Jason Folster, Beth Karp, Kaitlin Tagg, and Hattie Webb for assistance with antimicrobial resistance testing and interpretation during this outbreak. The authors would like to thank Patrick Adams for technical writing assistance with this manuscript. The authors would like to thank members of the FDA Center for Veterinary Medicine including Xin Li. We thank the following Vet-LIRN laboratories for their diagnostic contributions: Cornell University Animal Health Diagnostic Center, University of Illinois Veterinary Diagnostic Laboratory, Ohio Department of Agriculture Animal Disease Diagnostic Laboratory, Auburn University College of Veterinary Medicine Diagnostic Services, Oklahoma Animal Disease Diagnostic Laboratory, Washington Animal Disease Diagnostic Laboratory, Maryland Department of Agriculture Frederick Animal Health Laboratory, and Michigan State University Veterinary Diagnostic Laboratory.
Funding: Testing by these Vet-LIRN laboratories was funded by Vet-LIRN infrastructure grants. This project was supported by the FDA as part of an Animal Feed Regulatory Program Standards award totaling $450,000 with zero percentage financed with non-governmental sources.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.lana.2024.100769.
Appendix ASupplementary data
References
- 1.Scallan E., Hoekstra R.M., Angulo F.J., et al. Foodborne illness acquired in the United States--major pathogens. Emerg Infect Dis. 2011;17(1):7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CDC . U.S. Department of Health and Human Services, CDC; Atlanta, GA: 2019. Antibiotic resistance threats in the United States, 2019. [Google Scholar]
- 3.Collier S.A., Deng L., Adam E.A., et al. Estimate of burden and direct healthcare cost of infectious waterborne disease in the United States. Emerg Infect Dis. 2021;27(1):140–149. doi: 10.3201/eid2701.190676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pui C.F., Wong W.C., Chai L.C., et al. Salmonella: a foodborne pathogen. Int Food Res J. 2011;18:465–473. [Google Scholar]
- 5.CDC . 2015. Health pets, healthy people: Salmonella infection.https://www.cdc.gov/healthypets/diseases/salmonella.html [Google Scholar]
- 6.Medalla F., Gu W., Friedman C.R., Judd M., Folster J., Griffin P.M. Increased incidence of antimicrobial-resistant nontyphoidal Salmonella infections, United States, 2004-2016. Emerg Infect Dis. 2021;27(6):1662–1672. doi: 10.3201/eid2706.204486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shane A.L., Mody R.K., Crump J.A., et al. 2017 infectious diseases society of America clinical practice guidelines for the diagnosis and management of infectious diarrhea. Clin Infect Dis. 2017;65(12):e45–e80. doi: 10.1093/cid/cix669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bae D., Kweon O., Khan A.A. Isolation and characterization of antimicrobial-resistant nontyphoidal Salmonella enterica serovars from imported food products. J Food Protect. 2016;79(8):1348–1354. doi: 10.4315/0362-028X.JFP-15-564. [DOI] [PubMed] [Google Scholar]
- 9.Zhao S., McDermott P.F., Friedman S., et al. Characterization of antimicrobial-resistant Salmonella isolated from imported foods. J Food Protect. 2005;69(3):500–507. doi: 10.4315/0362-028x-69.3.500. [DOI] [PubMed] [Google Scholar]
- 10.Zhao S. In: Imported foods: microbiological issues and challenges. Doyle M.P., Erickson M.C., editors. ASM Press; Washington, DC: 2008. Antimicrobial-resistant food-borne pathogens in imported food; pp. 159–186. [Google Scholar]
- 11.CDC . 2013. Salmonella Saintpaul infections linked to imported cucumbers, 2013.https://www.cdc.gov/salmonella/saintpaul-04-13/index.html [Google Scholar]
- 12.Julian E., MacDonald K., Marsden-Haug N., et al. Salmonella Montevideo infections associated with salami products made with contaminated imported black and red pepper - United States, July 2009-April 2010. Morb Mortal Wkly Rep. 2010;59(50):1647–1650. [PubMed] [Google Scholar]
- 13.Hassan R., Rounds J., Sorenson A., et al. Multistate outbreak of Salmonella Anatum infections linked to imported hot peppers - United States, May-July 2016. Morb Mortal Wkly Rep. 2017;66(25):663–667. doi: 10.15585/mmwr.mm6625a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hassan R., Whitney B., Williams D.L., et al. Multistate outbreaks of Salmonella infections linked to imported Maradol papayas - United States, December 2016-September 2017. Epidemiol Infect. 2019;147 doi: 10.1017/S0950268819001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laughlin M., Bottichio L., Weiss J., et al. Multistate outbreak of Salmonella Poona infections associated with imported cucumbers, 2015-2016. Epidemiol Infect. 2019;147 doi: 10.1017/S0950268819001596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez S., Hofacre C.L., Lee M.D., Maurer J.J., Doyle M.P. Animal sources of salmonellosis in humans. J Am Vet Med Assoc. 2002;221(4):492–497. doi: 10.2460/javma.2002.221.492. [DOI] [PubMed] [Google Scholar]
- 17.Finley R., Reid-Smith R., Weese J.S. Human health implications of salmonella-contaminated natural pet treats and raw pet food. Clin Infect Dis. 2006;46:686–691. doi: 10.1086/500211. [DOI] [PubMed] [Google Scholar]
- 18.Ghasemzadeh I., Namazi S.H. Review of bacterial and viral zoonotic infections transmitted by dogs. J Med Life. 2015;8(4):1–5. [PMC free article] [PubMed] [Google Scholar]
- 19.Reimschuessel R., Grabenstein M., Guag J., et al. Multilaboratory survey to evaluate Salmonella prevalence in diarrheic and nondiarrheic dogs and cats in the United States between 2012 and 2014. J Clin Microbiol. 2017;55(5):1350–1368. doi: 10.1128/JCM.02137-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Botha W.J., Schoeman J.P., Marks S.L., Whitehead Z., Annandale C.H. Prevalence of Salmonella in juvenile dogs affected with parvoviral enteritis. J S Afr Vet Assoc. 2018;89:e1–e6. doi: 10.4102/jsava.v89i0.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viegas F.M., Ramos C.P., Xavier R.G.C., et al. Fecal shedding of Salmonella spp., Clostridium perfringens, and Clostridioides difficile in dogs fed raw meat-based diets in Brazil and their owners’ motivation. PLoS One. 2020;15(4) doi: 10.1371/journal.pone.0231275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sykes J.E., Marks S.L. Elsevier Inc.; St. Louis, MO: 2014. Salmonellosis. Canine and feline infectious diseases; pp. 437–444. [Google Scholar]
- 23.Behravesh C.B., Ferraro A., Deasy M., 3rd, et al. Human Salmonella infections linked to contaminated dry dog and cat food, 2006-2008. Pediatrics. 2010;126(3):477–483. doi: 10.1542/peds.2009-3273. [DOI] [PubMed] [Google Scholar]
- 24.Deasy M., 3rd, Moll M., Urdaneta V., et al. Update: recall of dry dog and cat food products associated with human Salmonella Schwarzengrund infections--United States, 2008. MMWR Morb Mortal Wkly Rep. 2008;57(44):1200–1202. [PubMed] [Google Scholar]
- 25.Imanishi M., Rotstein D.S., Reimschuessel R., et al. Outbreak of Salmonella enterica serotype Infantis infection in humans linked to dry dog food in the United States and Canada, 2012. J Am Vet Med Assoc. 2014;244(5):545–553. doi: 10.2460/javma.244.5.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavallo S.J., Daly E.R., Seiferth J., et al. Human outbreak of Salmonella Typhimurium associated with exposure to locally made chicken jerky pet treats, New Hampshire, 2013. Foodborne Pathog Dis. 2015;12(5):441–446. doi: 10.1089/fpd.2014.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark C., Cunningham J., Ahmed R., et al. Characterization of Salmonella associated with pig ear dog treats in Canada. J Clin Microbiol. 2001;39(11):3962–3968. doi: 10.1128/JCM.39.11.3962-3968.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kepinska-Pacelik J., Biel W. Microbiological hazards in dry dog chews and feeds. Animals (Basel) 2021;11(3):631. doi: 10.3390/ani11030631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.FDA . 2021. Import Alert 72-03: detention without physical examination of pig ears and other pet treats due to the presence of Salmonella.https://www.accessdata.fda.gov/cms_ia/importalert_218.html [Google Scholar]
- 30.White D.G., Datta A., McDermott P., et al. Antimicrobial susceptibility and genetic relatedness of Salmonella serovars isolated from animal-derived dog treats in the USA. J Antimicrob Chemother. 2003;52(5):860–863. doi: 10.1093/jac/dkg441. [DOI] [PubMed] [Google Scholar]
- 31.Willis C. Isolation of Salmonella species from imported dog chews. Vet Rec. 2001;149:426–427. doi: 10.1136/vr.149.14.426. [DOI] [PubMed] [Google Scholar]
- 32.Wong T.L., Thom K., Nicol C., Heffernan H., MacDiarmid S. Salmonella serotypes isolated from pet chews in New Zealand. J Appl Microbiol. 2007;103(4):803–810. doi: 10.1111/j.1365-2672.2007.03303.x. [DOI] [PubMed] [Google Scholar]
- 33.Yukawa S., Uchida I., Tamura Y., Ohshima S., Hasegawa T. Characterisation of antibiotic resistance of Salmonella isolated from dog treats in Japan. Epidemiol Infect. 2019;147:e102. doi: 10.1017/S0950268819000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crowe L., Chui L., Everett D., et al. Human salmonellosis associated with animal-derived pet treats --- United States and Canada, 2005. MMWR Morb Mortal Wkly Rep. 2006;55(25):702–705. [PubMed] [Google Scholar]
- 35.Kubota K.A., Wolfgang W.J., Baker D.J., et al. PulseNet and the changing paradigm of laboratory-based surveillance for foodborne diseases. Public Health Rep. 2019;134(2_suppl):22S–28S. doi: 10.1177/0033354919881650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.CDC National notifiable disease surveillance system (NNDSS) salmonellosis (Salmonella spp.) 2021. https://ndc.services.cdc.gov/conditions/salmonellosis/
- 37.Marshall K.E., Nguyen T.A., Ablan M., et al. Investigations of possible multistate outbreaks of Salmonella, shiga toxin-producing Escherichia coli, and Listeria monocytogenes infections - United States, 2016. MMWR Surveill Summ. 2020;69(6):1–14. doi: 10.15585/mmwr.ss6906a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.CDC . 2021. Foodnet Fast population survey.https://wwwn.cdc.gov/Foodnetfast/PopSurvey Retrieved from. [Google Scholar]
- 39.FDA Safety reporting portal. 2022. https://www.safetyreporting.hhs.gov/SRP2/en/Home.aspx?sid=b4aadf98-2ed3-460f-b6d6-e1073099f179
- 40.Dekker J.P., Faron M.L., Humphries R.M., Buchan B.W., Ledeboer N.A. In: Manual of clinical microbiology. Caroll K.C., Pfaller M.A., editors. American Society for Microbiology; Washington DC: 2022. Salmonella isolation procedures. [Google Scholar]
- 41.Andrews W.H., Wang H., Jacobson A., Ge B., Zhang G., Hammack T. 8th ed. United States Food and Drug Administration; 2021. Bacteriological analytical manual. [Google Scholar]
- 42.Ribot E.M., Fair M.A., Gautom R., et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Food Path Dis. 2006;3(1):59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 43.CDC . 2016. PulseNet methods and protocols: whole genome sequencing (WGS)https://www.cdc.gov/pulsenet/pathogens/wgs.html [Google Scholar]
- 44.Katz L.S., Griswold T., Williams-Newkirk A.J., et al. A comparative analysis of the lyve-SET phylogenomics pipeline for genomic epidemiology of foodborne pathogens. Front Microbiol. 2017;8:375. doi: 10.3389/fmicb.2017.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tolar B., Joseph L.A., Schroeder M.N., et al. An overview of PulseNet USA databases. Foodborne Pathog Dis. 2019;16(7):457–462. doi: 10.1089/fpd.2019.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.CDC . 2018. National antimicrobial resistance monitoring system for enteric bacteria (NARMS): human isolates surveillance report for 2015 (final report). Atlanta, GA. [Google Scholar]
- 47.CLSI . Clinical and Laboratory Standards Institute; 2021. Performance standards for antimicrobial susceptibility testing CLSI supplement M100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tagg K.A., Amir A., Ikram A., et al. Sequencing and characterization of five extensively drug resistant Salmonella enterica serotype typhi isolates implicated in human infections from Punjab, Pakistan. Microbiol Resour Announc. 2020;9(13) doi: 10.1128/MRA.01466-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zankari E., Allesøe R., Joensen K.G., Cavaco L.M., Lund O., Aarestrup F.M. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother. 2017;72(10):2764–2768. doi: 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zankari E., Hasman H., Cosentino S., et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McDermott P.F., Tyson G.H., Kabera C., et al. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother. 2016;60(9):5515–5520. doi: 10.1128/AAC.01030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pediatrics AAO . 32nd ed. 2021. Red book: report of the committee on infectious diseases. [DOI] [PubMed] [Google Scholar]
- 53.CDC Outbreak of multidrug-resistant Salmonella infections linked to contact with pig ear pet treats. 2019. https://www.cdc.gov/salmonella/pet-treats-07-19/index.html
- 54.Adley C., Dillon C., Morris C.P., Delappe N., Cormican M. Prevalence of Salmonella in pig ear pet treats. Food Res Int. 2011;44(1):193–197. [Google Scholar]
- 55.Finley R., Reid-Smith R., Ribble C., Popa M., Vandermeer M., Aramini J. The occurrence and anti-microbial susceptibility of Salmonellae isolated from commercially available pig ear pet treats. Zoonoses Public Health. 2008;55(8–10):455–461. doi: 10.1111/j.1863-2378.2008.01144.x. [DOI] [PubMed] [Google Scholar]
- 56.Gerner-Smidt P., Besser J., Concepcion-Acevedo J., et al. Whole genome sequencing: bridging one-health surveillance of foodborne diseases. Front Public Health. 2019;7:172. doi: 10.3389/fpubh.2019.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.United States code title 21 chapter 9. The federal food, drug, and cosmetic Act. 2023. [Google Scholar]
- 58.FDA FDA’s regulation of pet food. 2022. https://www.fda.gov/animal-veterinary/animal-health-literacy/fdas-regulation-pet-food
- 59.Chiewchan N., Pakdee W., Devahastin S. Effect of water activity on thermal resistance of Salmonella krefeld in liquid medium and on rawhide surface. Int J Food Microbiol. 2007;114(1):43–49. doi: 10.1016/j.ijfoodmicro.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 60.Taormina P.J. Survival rate of salmonella on cooked pig ear pet treats at refrigerated and ambient temperature storage. J Food Protect. 2014;77(1):50–56. doi: 10.4315/0362-028X.JFP-13-305. [DOI] [PubMed] [Google Scholar]
- 61.Sherry A.E., Patterson M.F., Madden R.H. Comparison of 40 Salmonella enterica serovars injured by thermal, high-pressure and irradiation stress. J Appl Microbiol. 2004;96(4):887–893. doi: 10.1111/j.1365-2672.2004.02211.x. [DOI] [PubMed] [Google Scholar]
- 62.Ravindran R., Jaiswal A.K. Wholesomeness and safety aspects of irradiated foods. Food Chem. 2019;285:363–368. doi: 10.1016/j.foodchem.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 63.CDC . 2013. An atlas of Salmonella in the United States, 1968-2011: laboratory-based enteric disease surveillance. [Google Scholar]
- 64.Colello R., Ruiz M.J., Padín V.M., et al. Detection and characterization of Salmonella serotypes in the production chain of two pig farms in Buenos Aires province, Argentina. Front Microbiol. 2018;9:1370. doi: 10.3389/fmicb.2018.01370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rodrigues G.L., Panzenhagen P., Ferrari R.G., Paschoalin V.M.F., Conte-Junior C.A. Antimicrobial resistance in nontyphoidal Salmonella isolates from human and swine sources in Brazil: a systematic review of the past three decades. Microb Drug Resist. 2020;26(10):1260–1270. doi: 10.1089/mdr.2019.0475. [DOI] [PubMed] [Google Scholar]
- 66.Parisi A., Crump J.A., Glass K., et al. Health outcomes from multidrug-resistant Salmonella infections in high-income countries: a systematic review and meta-analysis. Foodborne Pathog Dis. 2018;15(7):428–436. doi: 10.1089/fpd.2017.2403. [DOI] [PubMed] [Google Scholar]
- 67.Varma J.K., Mølbak K., Barrett T.J., et al. Antimicrobial-resistant nontyphoidal Salmonella is associated with excess bloodstream infections and hospitalizations. J Infect Dis. 2005;191:554–561. doi: 10.1086/427263. [DOI] [PubMed] [Google Scholar]
- 68.Brown A.C., Chen J.C., Watkins L.K.F., et al. CTX-M-65 extended-spectrum β-lactamase-producing Salmonella enterica serotype Infantis, United States(1) Emerg Infect Dis. 2018;24(12):2284–2291. doi: 10.3201/eid2412.180500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frey E., Stapleton G.S., Nichols M.C., et al. Antimicrobial resistance in multistate outbreaks of nontyphoidal Salmonella infections linked to animal contact—United States, 2015–2018. J Clin Microbiol. 2024;62(1) doi: 10.1128/jcm.00981-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.