Abstract
Background and objective
Circular RNAs (circRNAs) have garnered considerable attention in the study of various human diseases due to their ubiquitous expression and potential biological functions. This study conducts a bibliometric and visualization-based analysis of circRNA-related research in diseases, aiming to reveal the current status, hotspots and emerging trends within the field.
Methods
Literature published between 2013 and 2022 and indexed in the Web of Science core databases was retrieved. Visualizations of publication volume, countries, authors, institutions, journals, references, and keywords were performed. Microsoft Excel (2021) was used to analyze and graph publication volume and growth trends. Additionally, CiteSpace (version 6.1.R6) and VOSviewer (version 1.6.18) were employed to visualize the bibliographic information.
Results
Between 2013 and 2022, a total of 4195 relevant articles on circRNA in the context of diseases were identified. These articles covered 56 countries, 2528 institutions, 19,842 authors and 698 journals, citing 85,541 references. The annual publication volume showed an exponential growth trend, with rapid development post-2017. China, the United States and Germany emerged as the top three contributors, demonstrating high publication volume and total citations. Notably, Nanjing Medical University exhibited the highest publication volume, boasting 291 articles. Burton B. Yang and Li Yang consistently ranked among the top 10 authors in terms of publication volume and citations, emerging as core contributors in this research field. The journal Bioengineered ranked first in terms of published articles (160), with an impact factor of 6.832, while Molecular Cancer garnered the highest impact factor (41.4), solidifying its position as a top journal in this field. Furthermore, high-frequency keywords included “expression” “proliferation” “biomarker” “microRNA” “cancer”, signifying the prevailing research hotspots and principal themes of this field over the past decade. As of 2022, “biomarker”, “prostate cancer”,“drug resistance”,“papillary thyroid carcinoma”, etc. continued as keywords during the outbreak period. At present, the value of circRNA application is mainly reflected in the two aspects of biomarkers and therapeutic targets, and the prediction of accurate diagnosis and precise treatment based on big data analysis, especially in cancer, will become a hot spot of research in the future.
Conclusion
The trajectory of circRNA research from its biological origins to its applications in diseases has been delineated from 2013 to 2022. However, the transition to disease-specific applications and exploration of biological functions warrants further attention in future research endeavors.
Keywords: circRNA, Bibliometric analysis, Visualization, CiteSpace, VOSviewer
1. Introduction
Circular RNA (circRNA) represents a distinctive class of endogenous non-coding RNA molecules with a closed-loop structure. Initially discovered in viruses by Sanger et al., in 1976 [1], circRNAs have since been identified across various organisms, including prokaryotes, unicellular eukaryotes, and mammals [2]. However, circRNA has previously been regarded as a byproduct of splicing errors. The emergence of advanced RNA sequencing techniques and bioinformatics analysis has unveiled the multifaceted roles of circRNAs in cellular processes.
In contrast to linear RNAs, circRNAs exhibit remarkable stability due to their closed-loop structure, enabling their presence in diverse cellular compartments such as exosomes and plasma [3]. Moreover, circRNAs often demonstrate tissue-specific or developmental stage-specific expression patterns, underscoring their functional diversity and regulatory complexity [4]. Notably, circRNAs function as miRNA sponges, interact with proteins, and regulate gene expression at the post-transcriptional level [5]. The burgeoning interest in circRNAs stems from their implication in various human diseases, including cancer, kidney disorders, cardiovascular conditions, diabetes, preeclampsia, and cellular senescence [[6], [7], [8], [9]]. Systematic reviews and meta-analyses have further highlighted the significance of circRNAs in disease pathogenesis and progression, providing comprehensive overviews of their roles as diagnostic biomarkers and therapeutic targets. For example, the systematic review by Li et al. [10]provides insights into the role of circRNAs in cancer progression and their potential as diagnostic markers, while the meta-analysis by Zhang et al. [11]explores the diagnostic accuracy of circRNAs in cardiovascular diseases. These studies underscore the growing recognition of circRNAs as key players in human health and disease.
In light of the rapidly expanding body of circRNA literature, conducting a systematic and comprehensive bibliometric analysis becomes imperative. Bibliometric analysis serves as a mature methodology for quantitatively assessing research outputs, identifying key contributors, and delineating research trends within a specific field [12,13]. By elucidating bibliometric parameters such as core authors, institutions, countries, collaborative networks, and emerging topics, this analysis facilitates a deeper understanding of the research landscape surrounding circRNAs in disease contexts [14]. Cancer as examples, a scientific publications on cancer epidemiology in Indonesia through bibliometric analysis aiming to complement the national survey data [15]. Another study used bibliometric approach to identify key research areas in pharmaceutical fields [16].
Previous studies have collectively investigated global trends in circRNA research from various perspectives and time frames. They revealed a significant surge in circRNA-related publications, particularly post-2010, with China, the USA, and Germany emerging as major contributors. Notable circRNAs, such as circCDR1as and circHIPK3, have garnered extensive attention, particularly in cancer research [17]. Themes in circRNA studies evolved towards exploring biogenesis and functional aspects [18]. Additionally, the research increasingly focused on circRNA roles in diseases, notably cancer, with circFOXO3 and circITCH among the prominently studied circRNAs [11]. These findings underscore the dynamic nature of circRNA research and its potential implications for future investigations.
2. Materials and methods
2.1. Sources and search strategies
Web of Science, a renowned and authoritative citation database with robust indexing capabilities, is widely employed in bibliometric research [14,19]. In this study, the database was employed as the data source to ensure data accuracy and comprehensiveness. The selected citation indexes were Science Citation Index Expanded and Social Sciences Citation Index. The search strategy was outlined as follows: TI=(‘circRNA*’) OR TI=(‘circular RNA*’) OR TI=(‘closed circular RNA’) OR TI=(‘circular noncoding RNA*’) OR TI=(‘circular non coding RNA*’).
2.2. Inclusion and exclusion criteria
Inclusion and exclusion criteria were established to ensure the selection of relevant literature for this study. The time frame for inclusion ranged from January 1, 2013 to December 31, 2022, encompassing recent research within the defined period. Only articles categorized as "Article" or "Review" were considered for inclusion, reflecting the focus on primary research and comprehensive reviews. Furthermore, to maintain consistency and accessibility, only publications in the English language were included. Exclusion criteria were applied to refine the selection process. Firstly, articles from non-relevant disciplines, as determined by the Web of Science subject categories, were excluded following a thorough review of article titles, abstracts, and keywords to ensure alignment with the research theme. Secondly, retracted literature was excluded to uphold the integrity and validity of the selected publications. Lastly, literature lacking complete bibliographic information was excluded to maintain data accuracy and reliability.
2.3. Data extraction and cleaning
By applying the inclusion criteria, the search yielded a total of 5598 articles. Following the exclusion of 1403 articles, a final set of 4195 articles was obtained in plain text format. Each article contained comprehensive information such as title, authors, affiliations, journal name, publication date, abstract, keywords, research content, references and more. To ensure data reliability, the CiteSpace software was employed to eliminate erroneous and duplicate records. Keywords were standardized by merging synonymous terms; for example, “miRNA” and “microRNA” were consolidated as “microRNA”. Additionally, different institution names were unified, and relevant countries were combined, such as grouping “Northern Ireland,” “England,” “Scotland” and “Wales” under “United Kingdom”. The analysis of source publication influence relied on data from the 2022 Journal Citation Reports published by the American Institute for Scientific Information. Furthermore, supplementary bibliometric information, including citation frequency and the h-index, was obtained from the “Citation Reports” feature within the Web of Science database. In our study, keyword selectiona and data extraction were conducted independently by two authors to ensure accuracy and reliability. Each author performed the analysis separately, and any discrepancies or disagreements were resolved through discussion and consensus.
2.4. Research methodology
Microsoft Excel (2021) was employed for analyzing and graphing publication volume and growth trends. Additionally, CiteSpace (version 6.1.R6) and VOSviewer (version 1.6.18) were utilized for visualizing the bibliographic information.
CiteSpace, developed by Chaomei Chen's research team at Drexel University in the United States [20], constructs knowledge networks among documents based on temporal dimensions. It visually represents the structure, patterns and distribution of scientific knowledge within a specific discipline or between different disciplines. CiteSpace reveals research status, identifies popular research themes and predicts emerging trends [21]. The software's configuration parameters included: a time span from January 2013 to December 2022, with one year per time slice. It employed the "pathfinder" modification algorithm and set the initial g-index (k) threshold at 25, adjusted as required. Node types such as "Country," "Institution," "Author," "Keyword," and "Source" were represented, with node size reflecting their frequency. Thicker lines between nodes indicated stronger collaborations, while node color and thickness represented different years and their respective publication volumes [22]. Betweenness centrality, calculated by CiteSpace, measures the proportion of the shortest paths passing through a certain point to the total number of shortest paths between these points [23]. It identifies highly connected nodes in a network, reflecting their significance and influence. In the network graph, nodes with a centrality >0.1 were indicated with purple circles [24]. Cluster analysis employed the Log Likelihood Ratio (LLR) algorithm to cluster and name keywords, with modularity (Q) and average silhouette score (S) evaluating clustering quality. Q > 0.3 indicates significant clustering structures, S > 0.5 indicates reasonable clustering and S > 0.7 signifies reliable clustering [25,26]. The timeline graph depicted the chronological evolution of keywords categorized by clusters, facilitating the analysis of research evolution [20]. Keyword burst analysis predicted research frontiers [27], with red lines in the graph representing periods of keyword bursts and indicating burst strength. Keywords with high burst strengths and closer burst years represented research frontiers.
VOSviewer, developed by Eck and his team at Leiden University, Netherlands, visualizes knowledge unit networks in literature. It excels in clustering analysis of knowledge units based on association strength or similarity [28]. In this study, VOSviewer was utilized to conduct a co-citation analysis on references. Different colors represented distinct clusters, with the same colour indicating similar research directions.
3. Results
3.1. Publication trend analysis
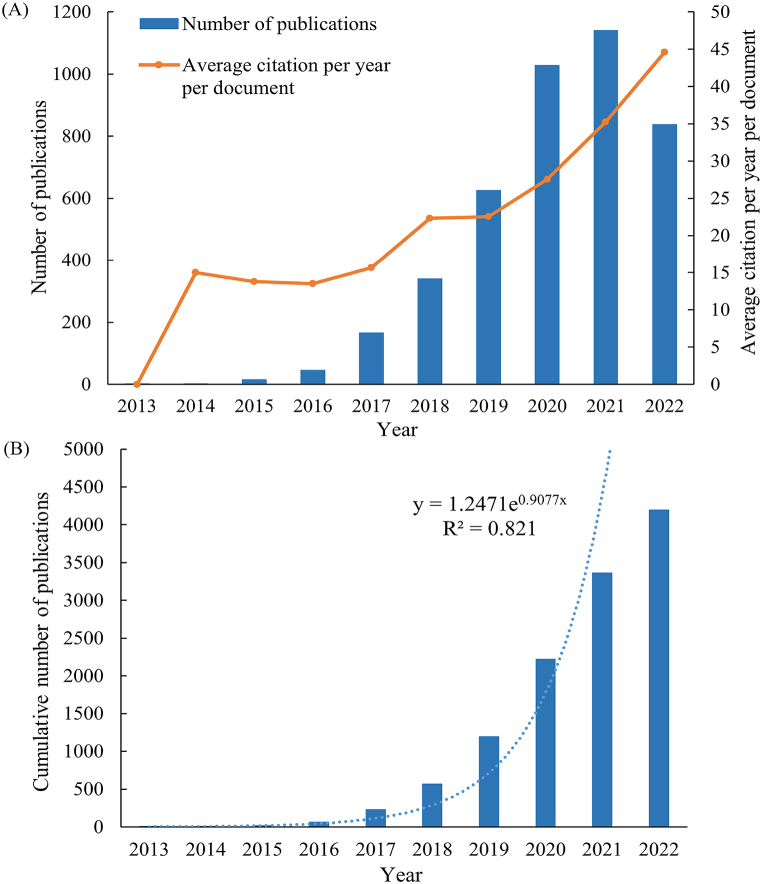
This study included 4195 relevant articles on circRNA within the field of disease research, from 2013 to 2022. Among these, 3393 constituted research papers (80.88 %), with the remaining 802 being reviews (19.12 %). Based on citation analysis, the total number of citations for the 4195 articles reached 131,119 (including 58,884 self-citations). The average citations per paper stood at 31.26, accompanied by an h-index of 153. Between 2013 and 2021, the annual publication volume of circRNA-related research in diseases exhibited a consistent upward trend. There was a rapid increase in publication volume post-2017, surging beyond 1000 articles in both 2020 and 2021. Notably, after the onset of the COVID-19 pandemic, relevant publications increased by approximately 64 %, with 625 papers in 2019 and 1027 papers in 2020.However, in 2022, a minor decline in publication volume was observed. The average citations per paper increased over the years, except for a minor dip in 2015–2016, ultimately reflecting an overall rising trend (Fig. 1A). The cumulative annual publication volume during this period demonstrated an exponential growth trend (y = 1.2471e0.9077x, R2 = 0.821; Fig. 1B).
Fig. 1.
Distribution of the number of publications related to circRNA research in disease areas.
3.2. National and institutional analysis of publications
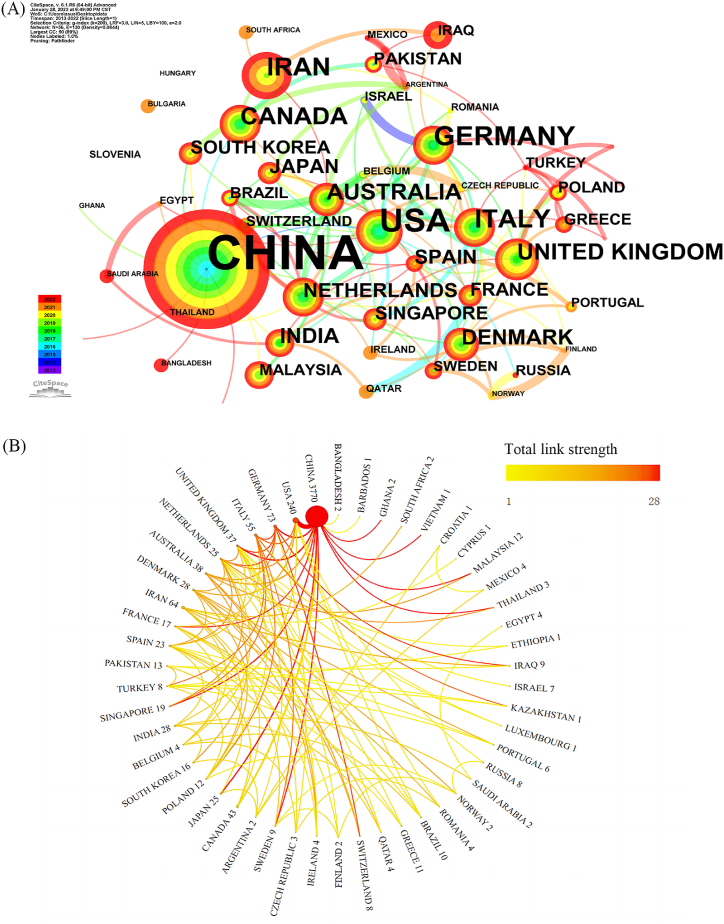
The 4195 articles originated from 56 different countries, as depicted in the co-authorship network analysis (Fig. 2A). Among these, five countries contributed over 50 articles, 16 countries contributed between 10 and 50 articles, and 35 countries contributed fewer than 10 articles. Moreover, the distribution of publication output across countries was disparate, showing a pronounced top-heavy effect, with a majority of articles concentrated within a limited number of countries. The top 10 countries by publication volume are listed in Table 1. The top three countries in both publication volume and total citations were China (3770 articles, constituting 80.8 %, with 104,318 citations), the United States (240 articles, constituting 5.1 %, with 19,559 citations), and Germany (73 articles, constituting 1.6 %, with 6103 citations). The collaboration relationships between countries are illustrated in Fig. 2B, with China exhibiting close collaboration with other countries, particularly the United States. The top three countries with the highest average citations per paper were Denmark (177.4 citations), followed by Germany (83.6 citations) and Canada (83 citations). The top three countries in terms of betweenness centrality were China (0.44), the United States (0.28) and Germany (0.21).
Fig. 2.
Country distribution of circRNA and disease research.(A)Co-occurrence network map, each node represents a country, the nodes are displayed in the form of a yearly wheel, the width of the wheel represents the number of papers published by the country in that year, and the size of the node represents the frequency of occurrence.(B) Cooperation network map, the connection between nodes represents cooperation, and the thickness of the line represents the strength of cooperation.
Table 1.
Top 10 countries with circRNA research publications in diseases.
| Rank | Country | Number of publications | proportion (%) | Total citation frequency | Average citation frequency per paper | Centrality |
|---|---|---|---|---|---|---|
| 1 | China | 3770 | 80.8 | 104,318 | 27.7 | 0.44 |
| 2 | USA | 240 | 5.1 | 19,559 | 81.5 | 0.24 |
| 3 | Germany | 73 | 1.6 | 6103 | 83.6 | 0.23 |
| 4 | Iran | 64 | 1.4 | 806 | 12.6 | 0.1 |
| 5 | Italy | 55 | 1.2 | 3440 | 62.5 | 0.13 |
| 6 | Canada | 43 | 0.9 | 3569 | 83.0 | 0.00 |
| 7 | Australia | 38 | 0.8 | 1475 | 38.8 | 0.07 |
| 8 | United Kingdom | 37 | 0.8 | 1503 | 40.6 | 0.15 |
| 9 | Denmark | 28 | 0.6 | 4966 | 177.4 | 0.09 |
| 10 | India | 28 | 0.6 | 1894 | 67.6 | 0.02 |
The 4195 articles originated from 2528 different institutions. As shown in Table 2, the institution with the highest publication output was Nanjing Medical University (291 articles), followed by Sun Yat-sen University (165 articles) and Fudan University (162 articles). Notably, the top 10 institutions in terms of publication output were all from China, contributing between 96 and 291 articles. Their betweenness centrality ranged from 0.03 to 0.12, with significantly high values for institutions such as Nanjing Medical University, Sun Yat-sen University, Fudan University, and Shanghai Jiao Tong University, all exceeding 0.1. Harvard Medical School ranked 65th in terms of publication volume (18 articles), but exhibited the highest betweenness centrality value (0.13).
Table 2.
Top 10 institutions with circRNA research publications in diseases.
| Rank | Institution | Country | Number of publications | Proportion (%) | Total citation frequency | Average citation frequency per paper | Centrality |
|---|---|---|---|---|---|---|---|
| 1 | Nanjing Med. Univ. | China | 291 | 11.5 | 10,344 | 35.5 | 0.12 |
| 2 | Sun Yat-sen Univ. | China | 165 | 6.5 | 5863 | 35.5 | 0.1 |
| 3 | Fudan Univ. | China | 162 | 6.4 | 6612 | 40.8 | 0.12 |
| 4 | Shanghai Jiao Tong Univ. | China | 142 | 5.6 | 6181 | 43.5 | 0.12 |
| 5 | China Med. Univ. | China | 126 | 5.0 | 2164 | 17.2 | 0.03 |
| 6 | Zhengzhou Univ. | China | 122 | 4.8 | 2397 | 19.6 | 0.06 |
| 7 | Southern Med. Univ. | China | 121 | 4.8 | 3216 | 26.6 | 0.09 |
| 8 | Cent. South Univ. | China | 113 | 4.5 | 3480 | 30.8 | 0.06 |
| 9 | Zhejiang Univ. | China | 100 | 4.0 | 2867 | 28.7 | 0.08 |
| 10 | Harbin Med. Univ. | China | 96 | 3.8 | 3211 | 33.4 | 0.05 |
3.3. Contribution of top journals
The 4195 articles were sourced from 698 different journals, primarily concentrated in the fields of medicine and biology. Table 3 displays the top 10 journals with the highest publication volume in this field, each with 50 articles or more, collectively constituing 1266 articles, which accounts for 30.2 % of the total publication output. Bioengineered (160 articles, 3.8 % of the total), with an impact factor of 6.832, emerged as the journal with the highest publication volume. It is a fully open-access interdisciplinary journal covering topics such as bioengineering and biotechnology, including research on biological processes, biomaterials, miRNA, other biomolecules and cancer. The second-highest publication volume was contributed by Frontiers in Oncology (104 articles, 2.5 %), which had an impact factor of 5.738. It primarily focuses on cancer research across all stages, from basic research to clinical studies. Frontiers in Genetics (97 articles, 2.3 %), with an impact factor of 4.772, was the third-highest publication volume journal. It primarily focuses on genes and genomics related to biological processes. Among the top 10 journals, Molecular Cancer had the highest impact factor (41.4), with 95 related articles published, contributing to 2.3 % of the total publication volume. It emerges as one of the top journals in this field.
Table 3.
Top 10 journals with circRNA research publications in diseases.
| Rank | Journal | Number of publications | Proportion (%) | Total citation frequency | Average citation frequency per paper | Impact Factor | JCRa |
|---|---|---|---|---|---|---|---|
| 1 | Bioengineered | 160 | 22.9 | 699 | 4.4 | 4.9 | Q1 |
| 2 | Frontiers in Oncology | 104 | 14.9 | 1034 | 9.9 | 4.7 | Q2 |
| 3 | Frontiers in Genetics | 97 | 13.9 | 863 | 8.9 | 3.7 | Q2 |
| 4 | Aging-US | 96 | 13.8 | 1863 | 19.4 | 5.2 | Q2 |
| 5 | Molecular Cancer | 95 | 13.6 | 10,350 | 108.9 | 37.3 | Q1 |
| 6 | Biochemical And Biophysical Research Communications | 91 | 13.0 | 4843 | 53.2 | 3.1 | Q2 |
| 7 | Cancer Management and Research | 79 | 11.3 | 1199 | 15.2 | 3.3 | Q3 |
| 8 | Molecular Therapy-Nucleic Acids | 77 | 11.0 | 2425 | 31.5 | 8.8 | Q1 |
| 9 | Cell Death & Disease | 72 | 10.3 | 2565 | 35.6 | 9.0 | Q1 |
| 10 | Cancer Cell International | 70 | 10.0 | 1120 | 16.0 | 5.8 | Q1 |
JCR: Journal Citation Reports.
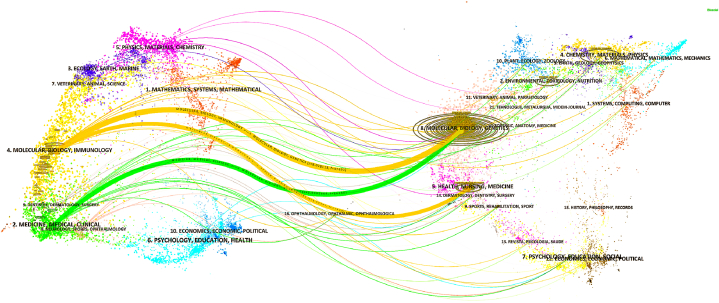
Fig. 3 illustrates the relationship between citing and cited journals through a dual map representation. The cited papers predominantly fall within two major fields: (1) Molecular Biology and Genetics, and (2) Health, Nursing, and Medicine. On the other hand, the citing papers are primarily clustered in two distinct areas: (1) Molecular Biology and Immunology, and (2) Medicine, Medical Sciences, and Clinical Research. These findings indicate the frontiers of knowledge exploration and innovation.
Fig. 3.
The dual-map overlay of journals. Publications on this field primarily appeared from two regions of the citing map: Molecular/Biology/Immunology in orange and Medicine/Medical/Clinical in green. The orange-colored curve indicated that the publications belonging to Molecular/Biology/Immunology journals primarily cited those in Molecular/Biology/Genetics journals and Health/Nursing/Medicine. The green-colored curve revealed that the publications from Medicine/Medical/Clinical journals mainly cited thuse in Molecular/Biology/Genetics journals.
Bradford's Law is used to quantitatively describe the concentration and dispersion distribution pattern of scientific papers in journals [29]. It involves arranging journals in descending order based on the number of papers published within a specific subject domain. This arrangement aids in delineating a core zone with the highest publication rate, followed by several zones (related zones and peripheral zones). The number of papers in each zone is approximately equal, and the number of journals in the core zone and subsequent zones follows a proportional relationship of 1: a:a2:... .Table 4 summarizes the distribution of circRNA-related research papers within the landscape of diseases, employing Bradford's Law. According to this law, the number of papers in each zone should be roughly equal, resulting in a journal ratio of 1:3.8:36.3. However, the 1:a:a2 proportionality is not adhered to, indicating the absence of a consolidated core zone for journals focused on circRNA-related research within the domain of diseases. Consequently, no pronounced concentration advantage in terms of publication output was discernible.
Table 4.
The distribution of journals.
| Partition | Number of articles in journals | Number of Journals | Number of papers | Proportion (%) |
|---|---|---|---|---|
| 1 | ≥54 | 17 | 1375 | 32.8 |
| 2 | 11–51 | 64 | 1405 | 33.5 |
| 3 | ≤10 | 617 | 1415 | 33.7 |
| SUM | – | 698 | 4195 | 100 |
3.4. Authors and their collaborations
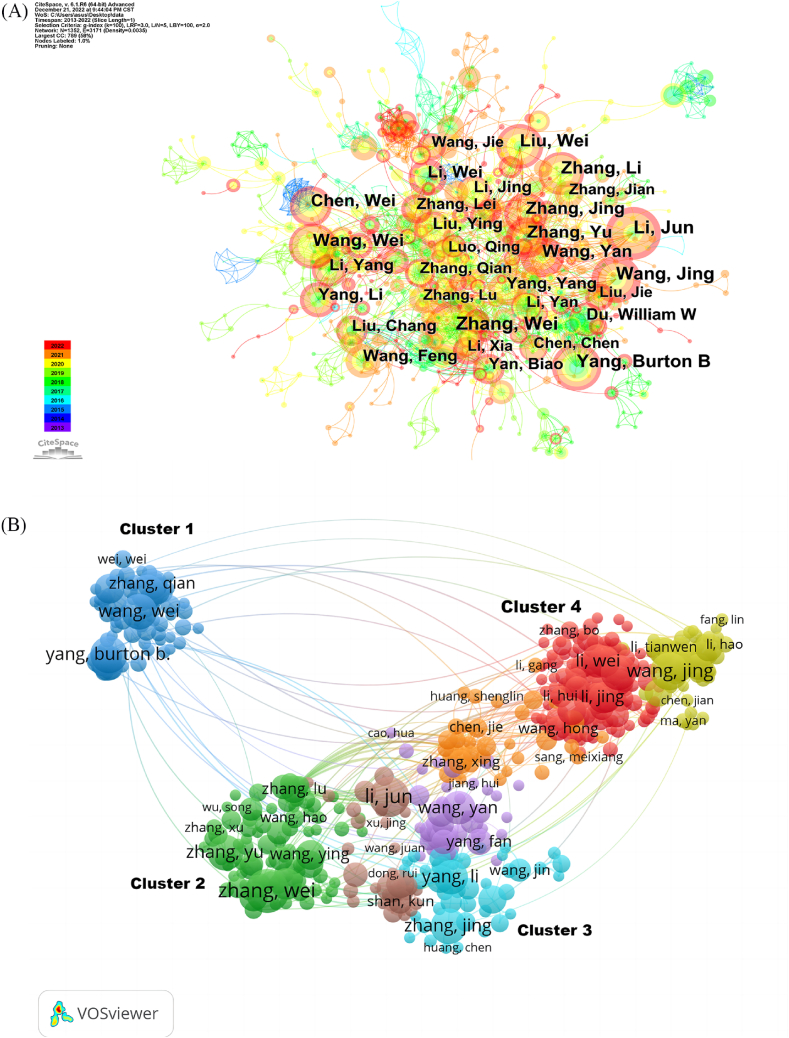
The 4195 articles were contributed by 19,842 authors (Fig. 4A). The distribution of authors was relatively concentrated, and a high degree of collaboration strength was observed. Among them, Professor Burton B. Yang from Sunnybrook Health Sciences Centre at the University of Toronto in Canada authored 24 papers, accumulating a total of 2555 citations and an average citation per paper of 106.5. His research primarily focuses on the regulatory roles and mechanisms of circRNA and miRNA in tumour initiation, progression and angiogenesis. Additionally, Researcher Yang Li (Yang, L) from the Institute of Biomedical Sciences at Fudan University authored 22 papers, earning a total of 3577 citations and an average citation per paper of 162.6. His work centers around employing cutting-edge techniques such as non-coding RNA and genome editing, to explore areas such as exonic circRNA processing, novel functional mechanisms, base editing and regulatory networks. Burton B. Yang and Yang Li consistently maintained their positions among the top 10 authors in both publication volume and citation frequency, establishing themselves as core authors in this research field (Table 5). Chen Lingling (Chen, LL), from the Centre of Excellence for Molecular Cell Science at the Chinese Academy of Sciences, holds the highest citation frequency among authors in this field. She has authored eight papers and garnered a total of 4385 citations, with an average citation per paper of 548. Her research primarily focuses on RNA processing, metabolism and functional mechanisms.
Fig. 4.
Author distribution of circRNA and disease research.(A)Co-occurrence of authors network map, nodes represent different authors, node size represents the number of author posts, the thickness of the connecting line between nodes represents the strength of cooperation. (B)Cooperation network map, the color represents different clusters, and the same color represents similar research directions.
Table 5.
Top 10 authors with circRNA research publications in diseases.
| Rank | Author | Number of publications | Total citation frequency | Average citation frequency per paper |
|---|---|---|---|---|
| 1 | Zhang, W | 30 | 898 | 29.9 |
| 2 | Li, J | 27 | 415 | 15.4 |
| 3 | Wang, J | 25 | 390 | 15.6 |
| 4 | Yang, BB | 24 | 2555 | 106.5 |
| 5 | Zhang, L | 23 | 278 | 12.1 |
| 6 | Zhang, J | 23 | 742 | 32.3 |
| 7 | Wang, W | 23 | 504 | 21.9 |
| 8 | Zhang, Y | 23 | 700 | 30.4 |
| 9 | Liu, W | 23 | 431 | 18.7 |
| 10 | Yang, L | 22 | 3577 | 162.6 |
Using VOSviewer, a collaborative network analysis was conducted on authors with publication volumes of 5 or more. The software categorizes authors with similar research content or directions into clusters (Fig. 4B). Table 6 summarizes key collaborative clusters among researchers in circRNA-related research, highlighting their main research areas and key partnerships, facilitating a better understanding of the interdisciplinary collaborations and thematic foci within the field.
Table 6.
Key collaborative clusters and research focus in circRNA-related studies.
| Cluster | Colour | Key partnership | Main research area |
|---|---|---|---|
| 1 | Anchored by Professor Burton B. Yang from Sunnybrook Health Sciences Centre at the University of Toronto, includes collaborations with colleagues such as William W Du, Yang WN and Awan Faryal Mehwish from the same institution. | To explore the biological functions of circRNA and its regulatory role in tumours (mainly breast cancer) and cardiovascular diseases. | |
| 2 | Composed of Luo Q, Zhang L, Huang ZK, Li JM and Guo Y from Nanchang University. | To investigate differentially expressed circRNA in rheumatoid arthritis, systemic lupus erythematosus and tuberculosis as potential biomarkers for diagnosis and treatment. | |
| 3 | Yang Li, Researcher, Institute of Biomedical Research, Fudan University, and Professor Chen Lingling, Center of Excellence for Molecular and Cellular Sciences, Chinese Academy of Sciences, are the center. | To explore the generation and processing of circRNA and their characteristics. | |
| 4 | Huang Jie from Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Lin Tiantian from Sun Yat-sen University, and Liu Huawei from Jinan University are the center authors. | To investigate the regulatory role of differentially expressed circRNA as miRNA sponges in bladder cancer. |
3.5. Analysis of funding institutions
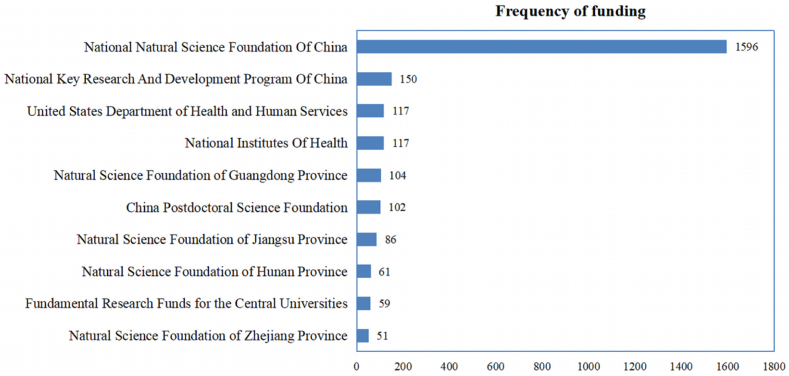
The analysis of funding institutions reveals important insights into the financial landscape surrounding research on the topic. Among the 4195 papers examined, a total of 3038 papers received funding support (Fig. 5). This indicates the significance of financial backing in facilitating research endeavors within the field. Leading the list of funding institutions is the National Natural Science Foundation Of China, which contributed to the largest number of papers, totaling 1596. Additionally, prominent Chinese institutions such as the National Key Research And Development Program Of China, as well as provincial-level natural science foundations such as those of Guangdong, Jiangsu, and Hunan, featured prominently in funding support. Interestingly, international funding bodies such as the National Institutes Of Health and the United States Department of Health and Human Services also played a role in supporting research, highlighting the global collaboration and exchange of resources in advancing knowledge on the topic. Overall, this analysis underscores the importance of funding institutions in driving scientific research forward and sheds light on the diverse sources of financial support contributing to the advancement of knowledge in this field.
Fig. 5.
Funding support landscape for research on circRNA in diseases.
3.6. Citation analysis and Co-citation analysis
A total of 4195 articles garnered a total of 131,119 citations, with self-citations accounting for 58,884 citations (44.9 %). The average citation per article stood at 31.26. Notably, 269 articles garnered over 100 citations, constituting 6.4 % of the total, while 522 articles remained uncited, comprising 12.4 % of the total. Table 7 presents the top 10 highly cited papers in the field, with citation counts ranging from 901 to 1864. Among these, eight papers surpassed 1000 citations. Among them, three were published in Molecular Cell, one in Nature Reviews Molecular Cell Biology, one in Nature Reviews Genetics, one in Molecular Cancer and one in Cancer Letters, which are prestigious journals within the medical and biological sciences domains. The most highly cited article, published in 2014 in Molecular Cell titled “circRNA Biogenesis Competes with Pre-mRNA Splicing” [30], delves into the biological genesis of circRNA. The article titled “The biogenesis, biology and characterization of circular RNAs” [31], published in 2019 in the journal Nature Reviews Molecular Cell Biology, ranks second in terms of citation frequency. This article primarily expounds on the biogenesis, biological characteristics and identification methods of circRNA, providing a comprehensive summary of the circRNA-related biological functions and the methodologies used to elucidate their functions. The article published in 2013 in the journal PLoS Genetics titled “Cell-Type Specific Features of Circular RNA Expression” [32] ranks third in terms of citation frequency. The article used refined bioinformatics and statistical methods to identify circRNAs, revealing that the regulation of splicing sites during RNA circularization displays both gene-specific and cell-type-specific features.
Table 7.
Top 10 studies with the most citation frequencies related to circRNA research in diseases.
| Rank | Article Title | First Author | Source Journal | Impact Factor |
Year | Citation Frequency |
|---|---|---|---|---|---|---|
| 1 | circRNA Biogenesis Competes with Pre-mRNA Splicing | Ashwal-Fluss, Reut | Molecular Cell | 16/Q1 | 2014 | 1864 |
| 2 | The Biogenesis, Biology and Characterization of Circular RNAs | Kristensen, Lasse S. | Nature Reviews Genetics |
42.7/Q1 | 2019 | 1718 |
| 3 | Cell-Type Specific Features of Circular RNA Expression | Salzman, Julia | PLoS Genetics | 4.5/Q1 | 2013 | 1319 |
| 4 | Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis | Legnini, Ivano | Molecular Cell | 16/Q1 | 2017 | 1251 |
| 5 | Circular RNA: A New Star of Noncoding RNAs | Qu, Shibin | Cancer Letters | 9.7/Q1 | 2015 | 1176 |
| 6 | Regulation of circRNA Biogenesis | Chen, Ling-Ling | RNA Biology | 4.1/Q2 | 2015 | 1169 |
| 7 | The Biogenesis and Emerging Roles of Circular RNAs | Chen, Ling-Ling | Nature Reviews Molecular Cell Biology |
112.7/Q1 | 2016 | 1049 |
| 8 | The Biogenesis, Functions, and Challenges of Circular RNAs | Li, Xiang | Molecular Cell | 16/Q1 | 2018 | 1028 |
| 9 | CircRNA: Functions and Properties of a Novel Potential Biomarker for Cancer | Meng, Shujuan | Molecular Cancer | 37.3/Q1 | 2017 | 970 |
| 10 | Circular RNAs in Cancer: Opportunities and Challenges in the Field | Kristensen, Lasse S. | Oncogene | 8.0/Q1 | 2018 | 901 |
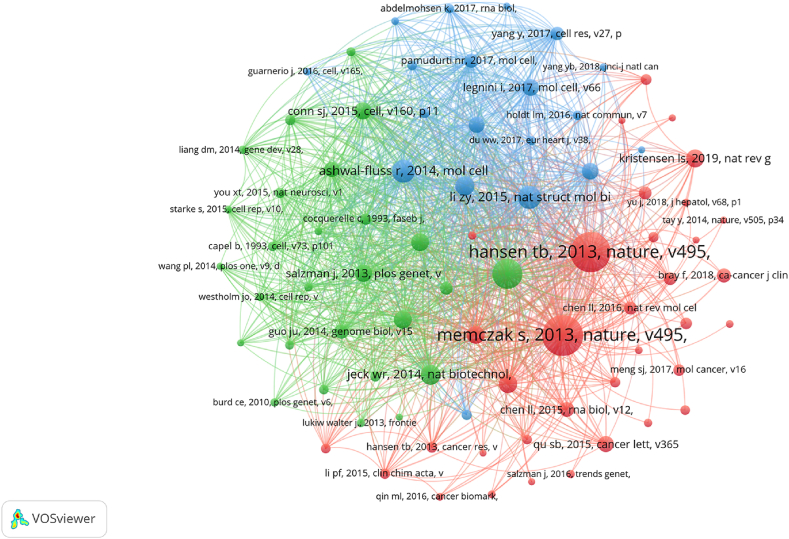
The 4195 articles collectively cited 85,541 references (Fig. 6). The top 10 most cited references primarily encompass the biological origin, characteristics, and functions of circRNAs, along with their significance as diagnostic biomarkers for cancer. The frequency of co-citations between articles reflects the degree of similarity in the concepts, theories, or methodologies they present [33]. Nodes of the same color in the graph indicate similar themes across publications. Furthermore, each cluster represents a distinct research domain: the red cluster focuses on the biological functions of circRNA; the green cluster emphasizes the biogenesis and biological characteristics of circRNA; and the blue cluster predominantly addresses the regulatory roles and diagnostic prognostic values of dysregulated circRNA in diseases.
Fig. 6.
Co-citation analysis of references used in publications on circRNA-related research in diseases, The color represents different clusters, and the same color represents similar research directions.
3.7. Analysis of research hotspots and frontiers
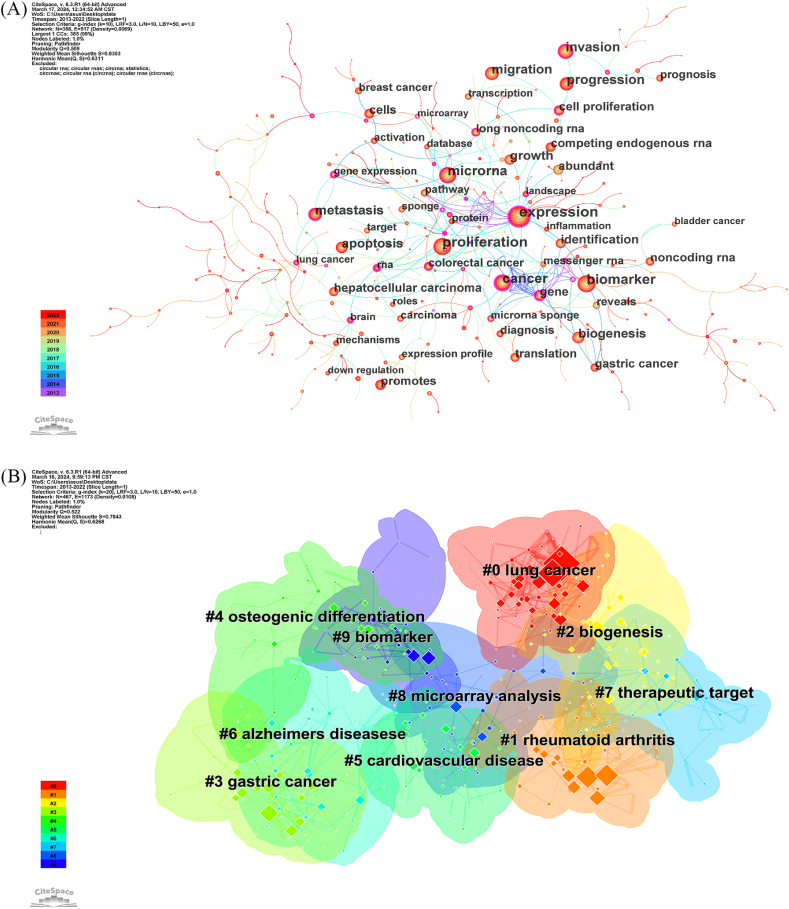
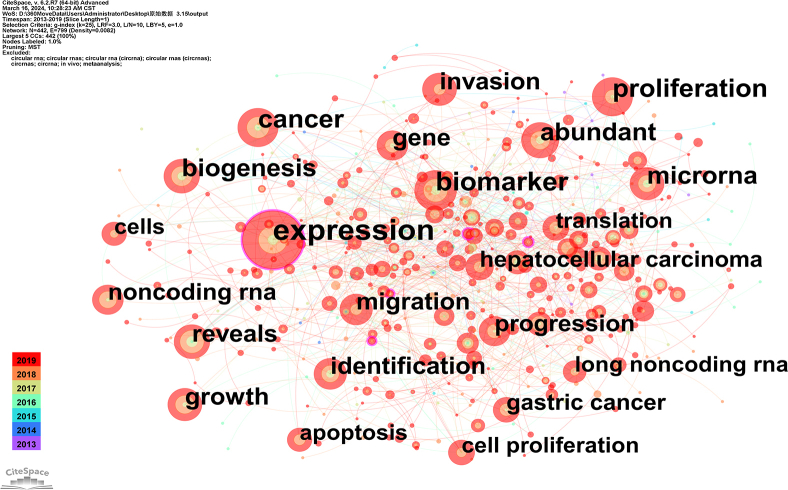
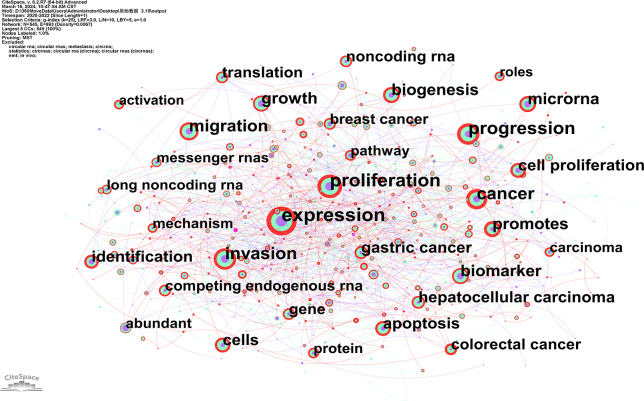
Utilizing CiteSpace for keyword analysis, synonym merging, co-occurrence, clustering and burst analysis, a co-occurrence knowledge map was constructed (Fig. 7A), containing 388 nodes and 317 edges. The frequently occurring keywords were as follows: expression (1243 occurrences), proliferation (938 occurrences), biomarker (908 occurrences), microRNA (775 occurrences), cancer (711 occurrences), invasion (654 occurrences), progression (613 occurrences), migration(516 occurrences), metastasis (483 occurrences), biogenesis (448 occurrences), and apoptosis (426 occurrences). These keywords collectively represent the major research themes of circRNAs in diseases from 2013 to 2022. Notably, expression (0.33), transcripts (0.3), cell(0.31), and regulation (0.28) were among the highest centrality nodes, indicating their pivotal role as “bridges” to various keywords in the field. The clustering analysis results are illustrated in Fig. 7, Fig. 8. The module value (Q value) was elucidated to be 0.522, and the average silhouette (S value) was 0.7842. A total of 10 distinct hotspot topic clusters were identified: Clusters #0, #1, #3, #4, #5 and #6 explored the primary disease types associated with circRNA research.Cluster #2 mainly centred on the biogenesis of circRNA. It revolves around the formation of circRNA through the reverse splicing of pre-mRNAs. Cluster #8, titled “microarray analysis” primarily concentrated on research methodologies. Microarray analysis is a key technique for identifying differentially expressed circRNA in tissues. Clusters #7 and #9 primarily focused on the application of circRNA. Characteristics such as specific expression, high conservation and stability highlight the potential of circRNAs as prognosis and diagnostic biomarkers, as well as their potential as novel therapeutic targets for diseases.
Fig. 7.
Keyword distribution of circRNA and disease research.(A)Co-occurrence of keywords, each node represents a keyword, and the size of the node represents the frequency of their occurrence, and the line between nodes represents the intensity of co-occurrence.(B) Keyword clustering map, CiteSpace uses Log-likelihood rate(LLR) to cluster closely related keywords. Different patterns represent a cluster. Tag # was assigned to the cluster, and the smaller the number is, the more keywords are in the cluster.
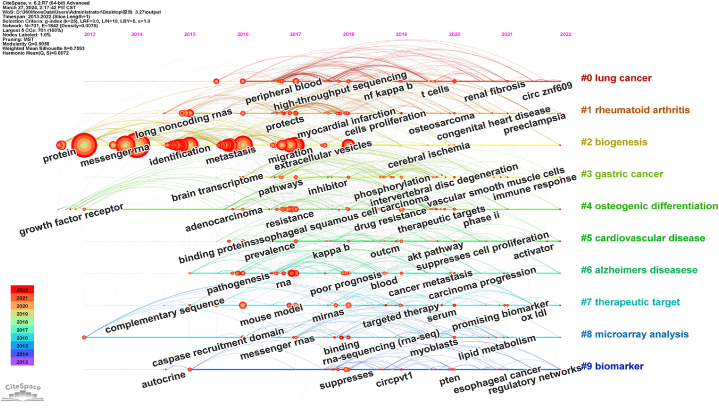
Fig. 8.
Timeline view of keyword clustering, in the timeline view, the keywords on the same horizontal line belong to the right cluster. The colors of lines and keywords in the view correspond to the colors of the time bar in the lower left corner.
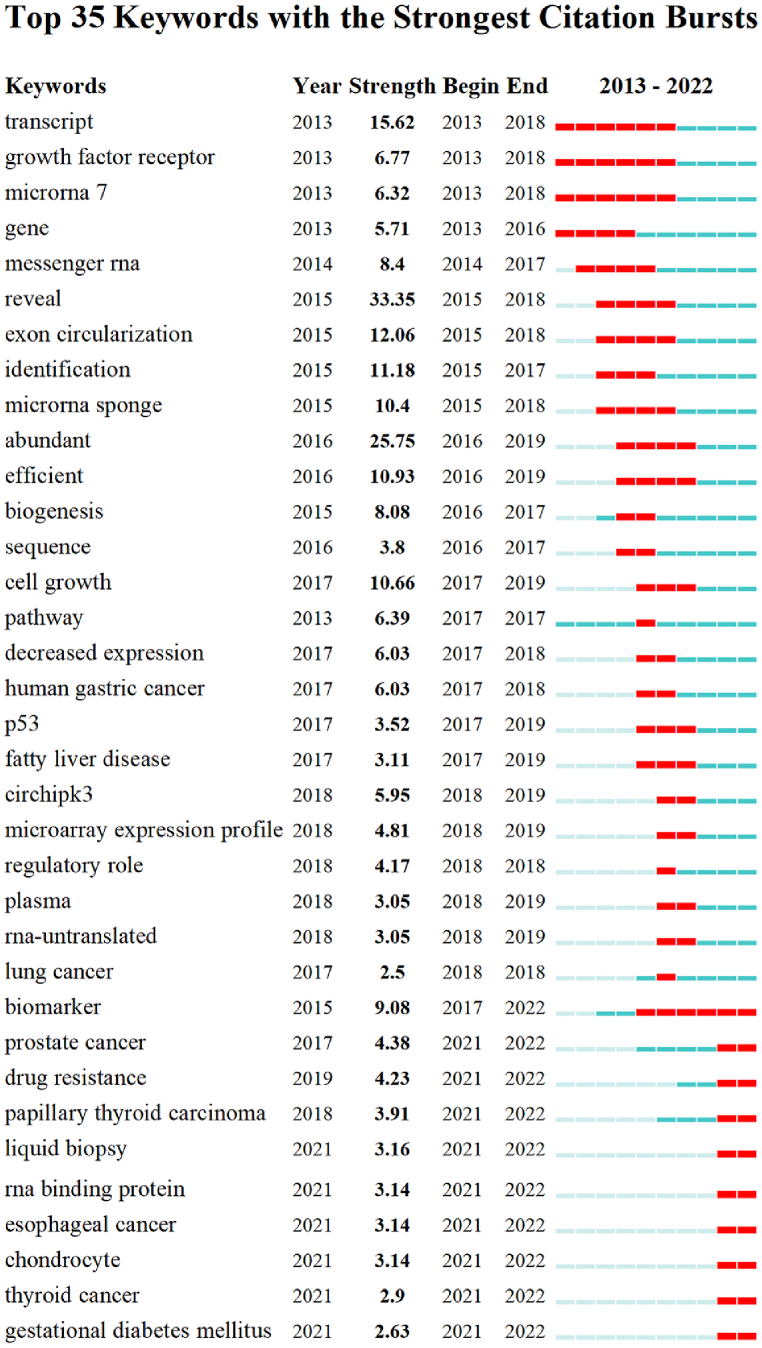
Fig. 9 presents the top 35 keywords with the highest burst strengths. Over the period from 2013 to 2022, keywords such as "transcript," "growth factor receptor," "microrna 7," and "biomarker" exhibited the longest burst durations, indicating sustained interest and research focus. Additionally, the keyword "reveal" showed the most intense burst strength during this timeframe. As of 2022, keywords such as "biomarker," "prostate cancer," "drug resistance," "papillary thyroid carcinoma," "liquid biopsy," "RNA binding protein," "esophageal cancer," "chondrocyte," "thyroid cancer," and "gestational diabetes mellitus" continue to experience bursts, indicating their ongoing significance and relevance in this field.
Fig. 9.
Top 35 keywords with the strongest citation bursts. From left to right, the first column represents the year in which this keyword first appeared. The second column represents the bursts' strength of the keyword. The third column represents the year in which this keyword begins and ends the burst. The fourth column in red represents the period during which the keyword is burst.
4. Discussion
Since 2013, research on circRNAs has witnessed remarkable growth, driven by its unique structure and biological characteristics, particularly in the field of human disease research. The quantity and trajectory of papers published within a specific academic field over time serve as crucial indicators for assessing the field's current status [26]. It is evident that circRNA has emerged as a hotspot in the field of disease research. From 2013 to 2022, the annual average citation frequency, which is a representative metric used in citation analysis for scientific evaluation, has consistently increased. This metric is also indicative of academic influence and research quality. Thus, based on bibliometric analyses, circRNA research in the context of diseases can be inferred to be continuously advancing, with the quality of papers in this field improving annually.
The countries with the highest publication volume are primarily China, the United States and Germany. China ranks first in terms of publication quantity, total citation frequency and intermediary centrality, highlighting its active and prominent roles in this field. However, the average citation frequency per paper is relatively low, at 28, reflecting a trend where many top Chinese authors have lower citation frequencies. This trend may be attributed to the large number of articles being published, which could dilute the proportion of high-quality papers. Additionally, Chinese research tends to focus more on the biological roles of circRNA in diseases rather than on investigating circRNA mechanisms and functions directly. Nonetheless, certain Chinese researchers have made significant contributions to the field of circRNA research. Professor Chen Lingling, for instance, ranks first in terms of citation frequency and represents a prominent figure in the field. Her team's work on circRNA biogenesis and mechanisms contributes significantly to this field [10,[34], [35], [36], [37]]. Furthermore, Germany consistently ranks within the top three in terms of publication quantity, total citation frequency, average citation frequency and intermediary centrality, highlighting its robust research capabilities and international influence. The top journals in this field are predominantly distributed within the domains of basic research and oncology. Notably, the top 10 institutions contributing to publication volume were all from China, indicating the pivotal role of collaborative efforts among institutions in driving circRNA-related research in the context of human diseases. Harvard Medical School, with the highest intermediary centrality, serves as a crucial contributor to fundamental circRNA research.
Co-citation analysis serves as a tool to elucidate the foundational knowledge within a specific research domain by examining cited references. It facilitates the exploration and analysis of the interconnectedness and developmental context among cited documents. Building upon the insights derived from co-citation analysis, research on circRNA within the context of diseases encompasses three key areas. Firstly, investigations into the biogenesis and biological characteristics of circRNA have revealed its formation through back-splicing of pre-mRNAs, regulated by cis-acting elements and trans-acting factors, as proposed by Salzman et al. [38]. Despite ongoing research, the complete mechanism of circRNA formation remains unclear, with reported models including intron-pairing-driven circularization, circularization mediated by RNA binding proteins (RBPs), and loop-driven circularization. Notably, circRNA exhibits highly conserved sequences and stable structures, with diverse expression patterns across cells and tissues [39]. Secondly, circRNA demonstrates various biological functions. In the cell nucleus, it influences RNA splicing and indirectly regulates parent gene transcription. In the cytoplasm, circRNA acts as microRNA (miRNA) sponges, modulating mRNA expression [40]. Additionally, it mediates translation via internal ribosome entry sites (IRES) and N6-methyladenosine (m6A) modifications [5], and interacts with proteins or protein complexes to exert specific function [41]. Lastly, circRNA plays crucial regulatory roles in diseases and holds diagnostic and prognostic significance, particularly in cancer. Noteworthy examples include CDR1as [[42], [43], [44], [45], [46]], CircFoxo3 [[47], [48], [49], [50]], CircPABPN1 [51,52], circHIPK3 [[53], [54], [55]] and circZNF609 [[56], [57], [58]], which are implicated in various tumor types. These insights underscore the multifaceted roles and clinical relevance of circRNA in disease contexts.
Keywords serve as concise summaries and encapsulations of the main topics covered in an article. Analyzing circRNA-related keywords in the context of disease research highlights "biomarker" as a consistent research focal point throughout the study period. CircRNA, due to its ubiquity, stability, specificity, and conservation, and circRNA can be detected and quantified through liquid biopsies in body fluids (including blood, saliva and urine), which are easy to repeat sampling, less invasive and more feasible, and more convenient and efficient than tissue biopsies. Therefore, it has potential as an vital biomarker for diagnosising and predicting disease progression and prognosis [59,60]. Over the past decade, circRNA research has transitioned from exploring its biogenesis, transcription, and functional mechanisms to investigating its application in disease. Dysregulation of circRNA expression has been linked to a variety of diseases, including cancer, cardiovascular disease, autoimmune disease, and Alzheimer's disease. Thus, endogenous circRNAs have been investigated as prospective therapeutic targets, particularly in tumor contexts. For instance, circRNAs function as competitive endogenous RNAs by competitively binding with miRNAs, thereby regulating the expression of their target mRNAs and participating in various disease pathways such as cell proliferation, apoptosis, invasion, metastasis and differentiation [61]. This regulatory mechanism has consistently been a central focus in research related to circRNA-mediated gene expression regulation [62]. In terms of research methodologies, microarray analysis has been the primary means for identifying and screening circRNAs. However, the circRNAs detected using microarrays are limited. With the continuous advancement of RNA sequencing technology and bioinformatics tools, the identification and in-depth exploration of circRNAs show promise. Foundational research aspects of circRNA, encompassing identification, biogenesis and biological characteristics, have been well-established. Future research is expected to focus on the specific applications of circRNA in particular diseases, emphasizing their biological functions. This includes exploring the pivotal roles of circRNA in cancer cells, including proliferation, apoptosis, and differentiation, as well as their function as miRNA sponges and diagnostic biomarkers.
While our study offers a comprehensive and objective bibliometric analysis of circRNA's relevance to diseases, it is important to acknowledge several limitations. Firstly, our analysis was confined to literature sourced exclusively from the Web of Science databases, namely the Science Citation Index Expanded and Social Sciences Citation Index, and was limited to publications in English. This approach may have overlooked relevant studies not indexed in these databases or published in languages other than English. Despite the Web of Science's reputation as a widely recognized citation database, it does not encompass all research outputs within the field, potentially resulting in the omission of pertinent publications [63]. However, given its prominence, the impact of such omissions on the overall findings is expected to be minimal. To mitigate these limitations, future research endeavors could incorporate data from additional sources and languages, facilitating a more comprehensive analysis of circRNA's relevance to diseases [64]. Nevertheless, future research on circRNA could explore its specific roles in disease pathways, particularly cancer, and validate circRNA biomarkers for various diseases to enhance early diagnosis and prognosis prediction. Investigating novel therapeutic strategies targeting circRNA, such as modulating circRNA expression or activity, could lead to innovative treatments. Integrating circRNA research with precision medicine initiatives and exploring its cross-talk with other regulatory networks can provide comprehensive insights into disease pathogenesis. Advancements in circRNA detection technologies and translation of research findings into clinical practice are also crucial for realizing the diagnostic and therapeutic potential of circRNA in disease management.
5. Conclusion
In summary, this study presents a comprehensive and objective analysis of circRNA in the field of disease research, facilitated by the use of CiteSpace and VOSviewer software. The advancement of high-throughput RNA sequencing technology and bioinformatics analysis has enabled extensive exploration of circRNA, spanning its biogenesis, biological characteristics, functional mechanisms, and applications in diseases. With its significant potential in clinical therapy, future research endeavors may focus on uncovering novel functions, elucidating formation processes, and understanding the mechanisms of action of circRNA in diseases.
Funding
Supported by Natural Science Foundation of Gansu Province(No.ZX-2020-040); Lanzhou Science and Technology Planing Project(No.2020-ZD-90); The Second Hospital of Lanzhou University's Cuiying Science and Technology Innovation ' Project(No.CY2020-BJ08) and Education Technology Innovation Research Project(No.2022B-122).
Data availability statement
Data will be made available on request.
Additional information
No additional information is available for this paper.
CRediT authorship contribution statement
Reyila Tuerdi: Writing – original draft, Visualization, Software, Methodology, Conceptualization. Hui Zhang: Writing – review & editing, Validation, Supervision. Wenxin Wang: Validation. Minghui Shen: Writing – review & editing, Validation, Supervision, Project administration, Funding acquisition. Xingmin Wei: Writing – review & editing, Methodology, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e31478.
Contributor Information
Minghui Shen, Email: shenmh21@lzu.edu.cn.
Xingmin Wei, Email: weixingmin@gszy.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
References
- 1.Abbaszadeh-Goudarzi K., Radbakhsh S., Pourhanifeh M.H., et al. Circular RNA and diabetes: epigenetic regulator with diagnostic role. Curr. Mol. Med. 2020;20(7):516–526. doi: 10.2174/1566524020666200129142106. [DOI] [PubMed] [Google Scholar]
- 2.Zhou W.Y., Cai Z.R., Liu J., et al. Circular RNA: metabolism, functions and interactions with proteins. Mol. Cancer. 2020;19(1) doi: 10.1186/s12943-020-01286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng Z., Xia L.X., Fan S.Y., et al. Circular RNA CircMAP3K5 acts as a MicroRNA-22-3p sponge to promote resolution of intimal hyperplasia via TET2-mediated smooth muscle cell differentiation. Circulation. 2021;143(4):354–371. doi: 10.1161/circulationaha.120.049715. [DOI] [PubMed] [Google Scholar]
- 4.Li J., Sun D., Pu W.C., et al. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020;6(4):319–336. doi: 10.1016/j.trecan.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 5.Lei M., Zheng G.T., Ning Q.Q., et al. Translation and functional roles of circular RNAs in human cancer. Mol. Cancer. 2020;19(1):9. doi: 10.1186/s12943-020-1135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu J.W., Xie D.L., Huang N.Y., et al. Circular RNAs as novel diagnostic biomarkers and therapeutic targets in kidney disease. Front. Med. 2021;8:16. doi: 10.3389/fmed.2021.714958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beilerli A., Gareev I., Beylerli O., et al. Circular RNAs as biomarkers and therapeutic targets. cancer. Semin. Cancer Biol. 2022;83:242–252. doi: 10.1016/j.semcancer.2020.12.026. [DOI] [PubMed] [Google Scholar]
- 8.Liu C., Li N., Dai G.F., et al. A narrative review of circular RNAs as potential biomarkers and therapeutic targets for cardiovascular diseases. Ann. Transl. Med. 2021;9(7) doi: 10.21037/atm-20-7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haddad G., Lorenzen J.M. Biogenesis and function of circular RNAs in health and in disease. Front. Pharmacol. 2019;10:10. doi: 10.3389/fphar.2019.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X., Yang L., Chen L.L. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z.X., Guo R.M., Wang Y.H., et al. Diagnostic value of circRNAs as effective biomarkers in human cardiovascular disease: an updated meta-analysis. Int. J. Med. Sci. 2022;19(3):446–459. doi: 10.7150/ijms.67094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper I.D. Bibliometrics basics. J. Med. Libr. Assoc. 2015;103(4):217–218. doi: 10.3163/1536-5050.103.4.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayr P., Scharnhorst A. Scientometrics and information retrieval: weak-links revitalized. Scientometrics. 2015;102(3):2193–2199. doi: 10.1007/s11192-014-1484-3. [DOI] [Google Scholar]
- 14.Cheng K.M., Guo Q., Yang W.G., et al. Mapping knowledge landscapes and emerging trends of the links between bone metabolism and diabetes mellitus: a bibliometric analysis from 2000 to 2021. Front. Public Health. 2022;10 doi: 10.3389/fpubh.2022.918483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iqhrammullah M.R.R., Ri Rasmi, Andika Ff, Hajjah H., Marlina M., Ningsih R. Cancer in Indonesia: a bibliometric surveillance. Narra X. 2023;1(2) doi: 10.52225/narrax.v1i2.86. [DOI] [Google Scholar]
- 16.Zulkifli B.F.F., Odigie J., Nnabuife L., Isitua Cc, Chiari W. Chemometric-empowered spectroscopic techniques in pharmaceutical fields: a bibliometric analysis and updated review. Narra X. 2023;1(1) doi: 10.52225/narrax.v1i1.80. [DOI] [Google Scholar]
- 17.Lv Y.H., Li Z.H., Tong F., et al. Bibliometric analysis of global research on circular RNA: current status and future directions. Mol. Biotechnol. 2023:14. doi: 10.1007/s12033-023-00830-y. [DOI] [PubMed] [Google Scholar]
- 18.Wu R., Guo F., Wang C., et al. Bibliometric analysis of global circular RNA research trends from 2007 to 2018. Cell J. 2021;23(2):238–246. doi: 10.22074/cellj.2021.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thelwall M. Bibliometrics to webometrics. J. Inf. Sci. 2008;34(4):605–621. doi: 10.1177/0165551507087238. [DOI] [Google Scholar]
- 20.Chen C.M. CiteSpace II: detecting and visualizing emerging trends and transient patterns in scientific literature. J. Am. Soc. Inf. Sci. Technol. 2006;57(3):359–377. doi: 10.1002/asi.20317. [DOI] [Google Scholar]
- 21.Song J.B., Zhang H.L., Dong W.L. A review of emerging trends in global PPP research: analysis and visualization. Scientometrics. 2016;107(3):1111–1147. doi: 10.1007/s11192-016-1918-1. [DOI] [Google Scholar]
- 22.Tu J.X., Lin X.T., Ye H.Q., et al. Global research trends of artificial intelligence applied in esophageal carcinoma: a bibliometric analysis (2000-2022) via CiteSpace and VOSviewer. Front. Oncol. 2022;12:18. doi: 10.3389/fonc.2022.972357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C.M., Ibekwe-Sanjuan F., Hou J.H. The structure and dynamics of cocitation clusters: a multiple-perspective cocitation analysis. J. Am. Soc. Inf. Sci. Technol. 2010;61(7):1386–1409. doi: 10.1002/asi.21309. [DOI] [Google Scholar]
- 24.Xu X., Wang Y.J., Li Y.M., et al. The future landscape of macrophage research in cardiovascular disease: a bibliometric analysis. Curr. Probl. Cardiol. 2022;47(10):31. doi: 10.1016/j.cpcardiol.2022.101311. [DOI] [PubMed] [Google Scholar]
- 25.Liu T.T., Yang L.P., Mao H.M., et al. Knowledge domain and emerging trends in podocyte injury research from 1994 to 2021: a bibliometric and visualized analysis. Front. Pharmacol. 2021;12:15. doi: 10.3389/fphar.2021.772386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin J.P., Ling F., Huang P., et al. The development of GABAergic network in depression in recent 17 Years: a visual analysis based on CiteSpace and VOSviewer. Front. Psychiatr. 2022;13:13. doi: 10.3389/fpsyt.2022.874137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C.M., Dubin R., Kim M.C. Emerging trends and new developments in regenerative medicine: a scientometric update (2000 - 2014) Expet Opin. Biol. Ther. 2014;14(9):1295–1317. doi: 10.1517/14712598.2014.920813. [DOI] [PubMed] [Google Scholar]
- 28.Van Eck N.J., Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics. 2010;84(2):523–538. doi: 10.1007/s11192-009-0146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin G.-X., Kotheeranurak V., Chen C.-M., et al. Global research hotspots and trends in the field of spine surgery during the COVID-19 pandemic: a bibliometric and visual analysis. Frontiers in Surgery. 2022;9 doi: 10.3389/fsurg.2022.976546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashwal-Fluss R., Meyer M., Pamudurti N.R., et al. circRNA biogenesis Competes with pre-mRNA splicing. Mol. Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 31.Kristensen L.S., Andersen M.S., Stagsted L.V.W., et al. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 32.Salzman J., Chen R.E., Olsen M.N., et al. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):15. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Nunen K., Li J., Reniers G., et al. Bibliometric analysis of safety culture research. Saf. Sci. 2018;108:248–258. doi: 10.1016/j.ssci.2017.08.011. [DOI] [Google Scholar]
- 34.Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17(4):205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 35.Chen L.L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020;21(8):475–490. doi: 10.1038/s41580-020-0243-y. [DOI] [PubMed] [Google Scholar]
- 36.Chen L.L., Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Xue W., Li X., et al. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15(3):611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 38.Salzman J., Gawad C., Wang P.L., et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):12. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Memczak S., Jens M., Elefsinioti A., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 40.Liu J., Liu T., Wang X.M., et al. Circles reshaping the RNA world: from waste to treasure. Mol. Cancer. 2017;16:12. doi: 10.1186/s12943-017-0630-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zang J.K., Lu D., Xu A.D. The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J. Neurosci. Res. 2020;98(1):87–97. doi: 10.1002/jnr.24356. [DOI] [PubMed] [Google Scholar]
- 42.Geng H.H., Li R., Su Y.M., et al. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One. 2016;11(3):17. doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanniford D., Ulloa-Morales A., Karz A., et al. Epigenetic silencing of CDR1as drives IGF2BP3-mediated melanoma invasion and metastasis. Cancer Cell. 2020;37(1):55. doi: 10.1016/j.ccell.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weng W.H., Wei Q., Toden S., et al. Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin. Cancer Res. 2017;23(14):3918–3928. doi: 10.1158/1078-0432.Ccr-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu H.Y., Guo S., Li W., et al. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci. Rep. 2015;5:12. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu L.L., Zhang M., Zheng X.B., et al. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2017;143(1):17–27. doi: 10.1007/s00432-016-2256-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du W.W., Fang L., Yang W.N., et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24(2):357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du W.W., Yang W.N., Chen Y., et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017;38(18):1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 49.Du W.W., Yang W.N., Liu E., et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen Z.Y., Zhou L., Zhang C., et al. Reduction of circular RNA Foxo3 promotes prostate cancer progression and chemoresistance to docetaxel. Cancer Lett. 2020;468:88–101. doi: 10.1016/j.canlet.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 51.Abdelmohsen K., Panda A.C., Munk R., et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14(3):361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X.X., Xiao L., Chung H.K., et al. Interaction between HuR and circPABPN1 modulates autophagy in the intestinal epithelium by altering ATG16L1 translation. Mol. Cell Biol. 2020;40(6):15. doi: 10.1128/mcb.00492-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y.W., Zheng F.X., Xiao X.Y., et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18(9):1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng Q.P., Bao C.Y., Guo W.J., et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:13. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeng K.X., Chen X.X., Xu M., et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:15. doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56.Luo J.Y., Wang S.C., Zhang L., et al. Research advance and clinical implication of circZNF609 in human diseases. Biotechnol. Biotechnol. Equip. 2022;36(1):668–683. doi: 10.1080/13102818.2022.2118076. [DOI] [Google Scholar]
- 57.Wang S.T., Xue X.K., Wang R., et al. CircZNF609 promotes breast cancer cell growth, migration, and invasion by elevating p70S6KI via sponging miR-145-5p. Cancer Manag. Res. 2018;10:3881–3890. doi: 10.2147/cmar.S174778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu W.D., Wei N.X., Shao G., et al. circZNF609 promotes the proliferation and migration of gastric cancer by sponging miR-483-3p and regulating CDK6. OncoTargets Ther. 2019;12:8197–8205. doi: 10.2147/ott.S193031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siede D., Rapti K., Gorska A.A., et al. Identification of circular RNAs with host gene-independent expression in human model systems for cardiac differentiation and disease. J. Mol. Cell. Cardiol. 2017;109:48–56. doi: 10.1016/j.yjmcc.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 60.Gupta S.K., Garg A., Bar C., et al. Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ. Res. 2018;122(2):246–254. doi: 10.1161/circresaha.117.311335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rong D.W., Sun H.D., Li Z.X., et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8(42):73271–73281. doi: 10.18632/oncotarget.19154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Z.R., Yang T.T., Xiao J.J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274. doi: 10.1016/j.ebiom.2018.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang L., Chen Z.L., Liu T., et al. Global trends of solid waste research from 1997 to 2011 by using bibliometric analysis. Scientometrics. 2013;96(1):133–146. doi: 10.1007/s11192-012-0911-6. [DOI] [Google Scholar]
- 64.Ginting B., Chiari W., Duta T.F., et al. COVID-19 pandemic sheds a new research spotlight on antiviral potential of essential oils - a bibliometric study. Heliyon. 2023;9(7) doi: 10.1016/j.heliyon.2023.e17703. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.