Abstract
RhaS activates transcription of the Escherichia coli rhaBAD and rhaT operons in response to l-rhamnose and is a member of the AraC/XylS family of transcription activators. We wished to determine whether ς70 might be an activation target for RhaS. We found that ς70 K593 and R599 appear to be important for RhaS activation at both rhaBAD and rhaT, but only at truncated promoters lacking the binding site for the second activator, CRP. To determine whether these positively charged ς70 residues might contact RhaS, we constructed alanine substitutions at negatively charged residues in the C-terminal domain of RhaS. Substitutions at four RhaS residues, E181A, D182A, D186A, and D241A, were defective at both truncated promoters. Finally, we assayed combinations of the RhaS and ς70 substitutions and found that RhaS D241 and ς70 R599 met the criteria for interacting residues at both promoters. Molecular modeling suggests that ς70 R599 is located in very close proximity to RhaS D241; hence, this work provides the first evidence for a specific residue within an AraC/XylS family protein that may contact ς70. More than 50% of AraC/XylS family members have Asp or Glu at the position of RhaS D241, suggesting that this interaction with ς70 may be conserved.
The RhaS protein is the l-rhamnose-responsive transcription activator of the Escherichia coli l-rhamnose catabolic and transport operons rhaBAD and rhaT, respectively (12, 13, 52, 53, 57), and is a member of the AraC/XylS family of transcription activators (17, 18, 44, 53). Full activation of both the rhaBAD and rhaT promoters requires activation by CRP binding immediately upstream of RhaS (13, 57). RhaS alone is able to activate rhaBAD expression by about 1,000-fold (13). In the presence of RhaS, CRP activates rhaBAD an additional 30- to 50-fold; however, CRP is unable to activate to any significant extent in the absence of RhaS (13).
The AraC/XylS family of transcription activators is named for its most well-studied member, AraC. The AraC protein consists of two functionally separable domains (7). The N-terminal AraC domain is responsible for both dimerization and l-arabinose binding, while the C-terminal domain is responsible for both DNA binding and transcription activation. In RhaS, the C-terminal domain is also responsible for DNA binding (4), and it is likely that the N-terminal domain functions in dimerization and l-rhamnose binding. The AraC/XylS family consists of more than 130 proteins that are identified by a 99-amino-acid region of sequence similarity within the DNA-binding domain of AraC (17, 18, 44, 53). One subset of AraC/XylS family proteins regulates expression of genes involved in carbon metabolism. This group includes AraC, RhaS, RhaR, and MelR from E. coli and XylS from Pseudomonas putida, which are among the most well characterized of the AraC/XylS family proteins (4, 5, 8, 12, 13, 16, 19, 29, 30, 38, 39, 53–55). Another large and important subset of AraC/XylS family proteins are those that regulate expression of virulence factors in bacterial pathogens (18). A few examples of this large group include CfaD from enterotoxigenic E. coli, SprA from Salmonella enterica serovar Typhimurium, and UreR from a variety of enteric pathogens (9, 14, 28, 48).
While DNA binding has been well characterized in a number of AraC/XylS family members (4–6, 12, 43, 45, 54), transcription activation by AraC/XylS family proteins is less well understood. It has been shown that activation of several promoters dependent upon AraC/XylS family activators requires the C-terminal domain (CTD) of the α subunit of RNA polymerase (RNAP). The α-CTD is the most well-characterized activation target and is required by a large number of activator proteins (reviewed in references 11 and 24). Perhaps the most direct evidence for an interaction between an AraC/XylS family activator and α-CTD has been found with the Ada protein at the alkA promoter. In this case mobility shift assays showed that a substitution in α-CTD eliminated the ability of purified α subunit to supershift the DNA-bound form of either Ada or meAda (34). Strong evidence also exists for an interaction between α-CTD and the MarA, SoxS, and Rob proteins in cases where these activators bind to DNA upstream but not overlapping the −35 region of the promoter (25–27). Finally, at a truncated rhaBAD promoter where RhaS was the only activator, deletion of α-CTD led to a 180-fold defect, and alanine substitutions identified eight residues in α-CTD that were candidates for making contacts with RhaS (23).
There is also evidence that the mechanism of transcription activation by some AraC/XylS family proteins may involve interactions with the ς70 subunit of RNAP, usually in cases where the binding site for the activator overlaps the −35 region of the promoter. In fact, the very first substitution isolated in ς70 (originally named alt and with the substitution R596H) involved an interaction with AraC. This substitution increased the ability of AraC to activate transcription in the absence of CRP, such that cya mutant cells regained the ability to use arabinose as the sole carbon source (50, 56). The more recent finding that other ς70 substitutions at positions near R596, especially K593A, significantly reduce activation by AraC in the absence of CRP supports the hypothesis that wild-type AraC and ς70 make an interaction that contributes to transcription activation (36).
Biochemical evidence for an interaction between ς70 and Ada also exists. Ada differs from many other AraC/XylS family proteins in that it can activate transcription from either a site that overlaps the −35 region (at alkA) or from a site that is 5 to 7 bp upstream of the −35 region (at ada and aidB) (1, 15, 35, 47). The N-terminal half of Ada, which includes the AraC/XylS family domain, is capable of binding to DNA and activating transcription at the alkA promoter but is not sufficient for transcription activation at promoters where Ada binds upstream of the −35 region (1). At the alkA promoter, a set of positively charged amino acids in ς70 was important for activation by Ada (33). A heparin-resistant ternary complex could be formed between DNA, Ada, and RNAP containing wild-type ς70, but not with RNAP containing ς70 substitutions K593A, K597A, or R603A (33), indicating that these ς70 residues might be directly involved in an interaction with Ada.
The focus of our work has been the mechanism of transcription activation by the RhaS protein. The binding site for RhaS overlaps the −35 region of both the rhaBAD and rhaT promoters by 4 bp, and hence it seemed likely that a target of transcription activation by RhaS might be ς70. To test this possibility, we first tested activation by RhaS in strains expressing a library of ς70 derivatives with single alanine substitutions in region 4.2 and at the very C-terminal end of ς70. We found that activation by RhaS was defective in the presence of several ς70 derivatives, most notably K593A and R599A, but only at truncated promoters that lacked the binding sites for the second activator, CRP. In an effort to identify RhaS amino acids that might contact these positively charged ς70 residues, we constructed alanine substitutions in nearly all of the negatively charged residues in the C-terminal domain of RhaS. A number of the RhaS derivatives were defective for activation in combination with wild-type ς70. Finally, we combined the RhaS and ς70 derivatives and found one combination, RhaS D241A plus ς70 R599A, which showed no greater defect than the individual derivatives at both the truncated rhaBAD and rhaT promoters. This phenotype suggests that the two substitutions may define an interaction between the RhaS and ς70 proteins that is important for transcription activation.
MATERIALS AND METHODS
Culture media and growth conditions.
Cultures for β-galactosidase assay were grown in 1× MOPS buffered medium (42); 1× MOPS consisted of 40 mM 3-(N-morpholino)propanesulfonic acid (MOPS); 4 mM tricine, 0.01 mM FeSO4, 9.5 mM NH4Cl, 0.276 mM K2SO4, 0.5 μM CaCl2, 0.528 mM MgCl2, 50 mM NaCl, 3 × 10−9 M Na2Mo4, 4 × 10−7 M H3BO3, 3 × 10−8 M CoCl2, 10−8 M CuSO4, 8 × 10−8 M MnCl2, 10−8 M ZnSO4, 1.32 mM K2HPO4, 10 mM NaHCO3, 0.2% Casamino Acids, and 0.002% thiamine. For other experiments (cloning, strain construction, Ter test, etc.), cells were grown in tryptone-yeast extract medium (37), with or without antibiotic, or TB maltose (0.8% Bacto-Tryptone, 0.5% NaCl, 0.2% maltose). Ampicillin was used at 125 or 200 μg/ml, as indicated.
General methods.
Standard methods were used for restriction endonuclease digestion, ligation, transformation, and purification of plasmid DNA. Primers for automated DNA sequencing were IRD41 dye labeled (Table 1) and custom synthesized by LI-COR, Inc. (Lincoln, Nebr.). DNA sequences were verified by automated dideoxy sequencing on a LI-COR 4000L sequencer. Sequencing reactions were performed using the Thermo Sequenase fluorescence-labeled-primer cycle sequencing kit from Amersham Pharmacia Biotech (Piscataway, N.J.). All DNA sequences were confirmed on both strands.
TABLE 1.
Oligos used in this studya
| Oligo no. | Oligo sequence (5′–3′) | Use |
|---|---|---|
| 744 | CGC GGA TCC CCA CTG GAT GCG CCG AGA TCG | Hybridizes within rhaB; used to amplify recombined rhaS alleles for diagnostic PCR and sequencing |
| 880 | CTA ACA TCG TCG GCA TCG | Hybridizes within rhaT; used to amplify recombined rhaS alleles for sequencing |
| 898 | TGA GTA AAG CTT TTA TTG CAG AAA GCC ATC CCG | Downstream end of rhaS; used to amplify rhaS alleles for chromosomal replacements |
| 1170 | CCG GAA TTC TTG TGG TGA TGT GAT GCT CAC | Upstream of rhaS; used to amplify rhaS alleles for chromosomal replacements |
| 2068 | ATG ACC GTA TTA CAT AGT GTG GATb | rhaS sequencing |
| 2069 | TTA TTG CAG AAA GCC ATC CCG TCCb | rhaS sequencing |
| 2074 | TGG TTG CAC AGA TGG AAC AGCb | rhaS sequencing |
| 2075 | GTT GAG ACG TGA TGC GCT GTTb | rhaS sequencing |
| 2083 | GTG GGA TCC ATG ACC GTA TTA CAT AGT | Upstream for diagnostic PCR on plasmid clones of all rhaS alleles |
| 2096 | GCG GGA TCC GCG TTA CTC ATC TTC TTA | Downstream Φ(rhaT-lacZ)Δ84 and Δ133 |
| 2097 | CGC GAA TTC AAG GGT ATG GTT TTG CAG | Upstream Φ(rhaT-lacZ)Δ133 |
| 2130 | GGC CTG GCT GGC AGA CCA TTT TG | SDM; RhaS E181Ala |
| 2131 | CAT CGG CAA AAT GGT CTG | Diagnostic PCR; RhaS E181Ala |
| 2134 | CCA TTT TGC CGC AGA GGT GAA TTG | SDM; RhaS E186Ala |
| 2135 | CAT CCC AAT TCA CCT CTG | Diagnostic PCR; RhaS E186Ala |
| 2136 | TTG CCG ATGCAG TGA ATT GG | SDM; RhaS E187Ala |
| 2137 | CGG CAT CCC AAT TCA CTG | Diagnostic PCR; RhaS E187Ala |
| 2138 | CCG TGG CGGCAC AAT TTT CT | SDM; RhaS D195Ala |
| 2139 | CGC AGT GAA AGA GAA AAT TGT G | Diagnostic PCR; RhaS D195Ala |
| 2141 | TGT CAG TAA CGC TGG CTG | Diagnostic PCR; RhaS E236Ala |
| 2142 | CGT TAC TGC AAT CGC CTA TC | SDM; RhaS D241Ala |
| 2143 | CAC AGC GAT AGG CGA TTG | Diagnostic PCR; RhaS D241Ala |
| 2146 | TCA CCG CGT GCA ATT CGC CA | SDM; RhaS D268Ala |
| 2147 | CCG TCC CTG GCG AAT TG | Diagnostic PCR; RhaS D268Ala |
| 2148 | AGG GAC GGGCAG GCT TTC T | SDM; RhaS D274Ala |
| 2149 | TTA TTG CAG AAA GCC TG | Diagnostic PCR; RhaS D274Ala |
| 2152 | CCG GAA TTC ACT TAA TGC CGT GAT TG | Upstream Φ(rhaT-lacZ)Δ84 |
| 2154 | TGG CTG GAG GCT CAT TTT GCC | SDM; RhaS D182Ala |
| 2155 | ACG CCA CAG CGC AGC CAG CGT TA | SDM; RhaS E236Ala |
| 2156 | CCT CAT CGG CAA AAT GAG | Diagnostic PCR; RhaS D182Ala |
| 2161 | TTT GTT TGC GTT TAC TGG CAG ATA | Downstream Plac-bet exo kan |
| 2162 | ACG GCA ACG GCC TTG AAC TGA AAT | Upstream Plac-bet exo kan |
| 2185 | TTC GCC GAG CAT TTA ACT GGT C | SDM; RhaS E261Ala |
| 2186 | GCG GTG ACC AGT TAA ATG | Diagnostic PCR; RhaS E261Ala |
Oligos were used for cloning, diagnostic or regular PCR, and site-directed mutagenesis (SDM). Regions not complementary to wild-type rha genes are underlined.
Oligos IRD41 dye labeled for use in a LI-COR automated sequencer.
Strains, plasmids, and phage.
The E. coli strains, λ phage, and plasmids used in this study are described in Table 2. All assays were performed using cultures of strains derived from ECL116 (2). In all cases, lacZ translational fusions were assayed as single-copy lysogens integrated into the E. coli chromosome at attλ. A library encoding wild-type ς70 and alanine substitution derivatives of ς70 were a gift from C. Gross and were carried on the plasmid pGEX2T (10, 36).
TABLE 2.
Strains used in this study
| Strain, phage, or plasmid | Genotype | Source or reference |
|---|---|---|
| E. coli strains | ||
| KM22 | Δ(recC ptr recB recD)::Plac-bet exo kan | 41 |
| ECL116 | F− ΔlacU169 endA hsdR thi | 2 |
| SME1035 | ECL116 recA::cat λ SME103 | 13 |
| SME1036 | ECL116 recA::cat λ SME104 | 13 |
| SME1082 | ECL116 ΔrhaSa | Laboratory collection |
| SME1087 | ECL116 ΔrhaS recA::cat λ SME101 | Laboratory collection |
| SME1088 | ECL116 ΔrhaS recA::cat λ SME104 | Laboratory collection |
| SME1222 | SME1082 λ SME103 | 4 |
| SME1851 | ECL116 λ SME104 | Laboratory collection |
| SME2186 | ECL116 λ SME107 | This study |
| SME2187 | ECL116 λ SME108 | This study |
| SME2341 | SME2186 ΔrhaS zih-35::Tn10 | This study |
| SME2342 | SME2187 ΔrhaS zih-35::Tn10 | This study |
| SME2393 | SME1222 ΔrhaS zih-35::Tn10 | This study |
| SME2394 | SME2393 Plac-bet exo kan | This study |
| SME2603 | SME1851 rhaS(E181A) recA::kan | This study |
| SME2604 | SME1851 rhaS(D182A) recA::kan | This study |
| SME2605 | SME1851 rhaS(D186A) recA::kan | This study |
| SME2606 | SME1851 rhaS(E187A) recA::kan | This study |
| SME2607 | SME1851 rhaS(D241A) recA::kan | This study |
| SME2608 | SME1851 rhaS(wt) recA::kanb | This study |
| SME2609 | SME2187 rhaS(E181A) recA::kan | This study |
| SME2610 | SME2187 rhaS(D182A) recA::kan | This study |
| SME2611 | SME2187 rhaS(D186A) recA::kan | This study |
| SME2612 | SME2187 rhaS(E187A) recA::kan | This study |
| SME2613 | SME2187 rhaS(D241A) recA::kan | This study |
| SME2614 | SME2187 rhaS(wt) recA::kan | This study |
| Phage | ||
| λRS45 | bla′-lacZscatt+ imm21ind+ | 51 |
| λRS74 | bla′-placUV5-lacZ+att+imm21ind+ | 51 |
| λ SME101 | λ RS45 Φ(rhaB-lacZ)Δ226 | 13 |
| λ SME103 | λ RS45 Φ(rhaB-lacZ)Δ110 | 13 |
| λ SME104 | λ RS45 Φ(rhaB-lacZ)Δ84 | 13 |
| λ SME107 | λ RS45 Φ(rhaT-lacZ)Δ133 | This study |
| λ SME108 | λ RS74 Φ(rhaT-lacZ)Δ84 | This study |
| Plasmids | ||
| pALTER-1 | Aps Tetr; lacZ, f1 ori | Promega Corp. |
| pSE159 | Apr pALTER-1 rhaS (wt) | 4 |
| pSE193 | pSE159 (RhaS E181A) | This study |
| pSE194 | pSE159 (RhaS D182A) | This study |
| pSE195 | pSE159 (RhaS D186A) | This study |
| pSE196 | pSE159 (RhaS E187A) | This study |
| pSE197 | pSE159 (RhaS D195A) | This study |
| pSE198 | pSE159 (RhaS E236A) | This study |
| pSE199 | pSE159 (RhaS D241A) | This study |
| pSE200 | pSE159 (RhaS E261A) | This study |
| pSE201 | pSE159 (RhaS D268A) | This study |
| pSE202 | pSE159 (RhaS D274A) | This study |
| pRS414 | Apr ′lacZ lacY lacA | 51 |
| pSE203 | pRS414 Φ(rhaT-lacZ)Δ133 | This study |
| pSE204 | pRS414 Φ(rhaT-lacZ)Δ84 | This study |
Construction of this ΔrhaS allele is described elsewhere (13).
wt, wild type.
Alanine substitutions of negatively charged amino acids in the DNA-binding domain of RhaS were constructed by site-directed mutagenesis of rhaS (Promega GeneEditor In Vitro Mutagenesis System) with plasmid pSE159 as the template. The recommended protocol was followed, except that we found that lengthening the expression period after transformation into the mutS strain from 1 to 2 h greatly increased the success of the procedure. Single-stranded plasmid template was used to construct all substitutions. Oligos, Etc., and Integrated DNA Technologies synthesized oligonucleotide primers for site-directed mutagenesis and identification of mutants (Table 1). Mutations were initially identified by a diagnostic PCR procedure using oligonucleotide (oligo) 744 and a second diagnostic oligo for each mutation. In the diagnostic oligos, two nucleotides at the 3′ end were complementary to the mutant allele and therefore not to the wild-type allele (Table 1). No PCR product was generated in any case from the wild-type allele; however, templates carrying the mutant alleles yielded a product in all cases. DNA sequencing of the entire rhaS gene on both strands confirmed all mutations and ensured that there were no additional mutations.
Construction of rhaT-lacZ fusions.
The full-length rhaT promoter (including both the CRP and RhaS-binding sites) was amplified by PCR using primers 2097 and 2096 and whole cells of E. coli ECL116 as the source of template DNA. The truncated rhaT promoter (with only the RhaS-binding site) was amplified by PCR using primers 2096 and 2152 and whole cells of E. coli DH5α as the source of template DNA. The PCR products were digested at the EcoRI site in 2097 and 2152 and the BamHI site in 2096 and cloned between the EcoRI and BamHI sites of pRS414, yielding plasmids carrying full-length (pSE203) and truncated (pSE204) fusions, respectively. The DNA sequence of the promoter regions and fusion junctions were sequenced on both strands. The translational fusions thus constructed with full-length and truncated rhaT promoters were transferred to λRS45 and λRS74 (both λimm21), respectively, by in vivo recombination (51) to generate recombinant phages λSME107 and λSME108 (Table 2). Strains SME2186 and SME2187 carrying a single-copy lysogen of the recombinant λ phage were obtained by transducing ECL116 with phage carrying the full-length and the truncated fusions, respectively. Lysogens carrying the full-length fusion were identified as pinpoint blue colonies amid a white lawn on a nutrient agar plate containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and l-rhamnose. Lysogens of the truncated fusion were selected by spreading the transduction mixture on a plate carrying a lawn of λgt30 (λimm21). In this case, lysogens were differentiated from λ-resistant cells by their sensitivity to the heteroimmune phage λCh6 (λimm434) when cross-streaked. For both the full-length and the truncated fusions, single lysogens were identified by the Ter test (22) and confirmed by β-galactosidase assay. P1 phage-mediated generalized transduction (40) was used to introduce an in-frame deletion of approximately two-thirds of rhaS (13) linked to Tn10 into SME2186 and SME2187 to generate SME2341 and SME2342, respectively. The presence of the rhaS deletion was confirmed by PCR analysis.
Recombination of rhaS alleles into the chromosome.
Chromosomal replacements by mutant rhaS alleles were constructed using an E. coli strain carrying bacteriophage λ recombination functions resulting in increased homologous recombination frequencies (41). SME2394 was constructed by P1 transduction of the λ bet exo operon under the control of the lac promoter from KM22 into SME 2393 with selection for kanamycin resistance (41). The presence of Plac-bet exo was confirmed by PCR with oligos 2161 and 2162. Alleles of rhaS to be recombined were amplified by PCR using oligos 898 and 1170 with the corresponding rhaS clone in pALTER-1 (pSE193-196 and pSE199) as a template. Then, 100 μl of CaCl2-treated SME2394 competent cells were transformed with approximately 500 ng of unpurified PCR product. The transformation mixtures were plated onto nutrient agar plates containing X-Gal, IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM), and l-rhamnose and incubated at 37°C for 72 to 96 h. Tiny blue colonies picked from amid the white lawn were patched onto nutrient agar plates containing X-Gal and l-rhamnose with and without ampicillin. Blue, ampicillin-resistant colonies had been transformed with plasmid DNA that had served as template in the PCR reaction, while blue, ampicillin-sensitive colonies had been transformed with the PCR-generated DNA fragments and had undergone the desired chromosomal replacement. Performing PCR on blue, ampicillin-sensitive colonies with oligo 898 downstream and oligo 744 upstream identified replacements of the in-frame rhaS deletion in SME2394 with full-length rhaS alleles. The presence of the mutant rhaS allele was tested by the same diagnostic PCR used to identify the mutations after the initial site-directed mutagenesis. For DNA sequencing, the rhaS alleles were amplified by PCR using oligos 880 and 744 with whole cells as the source of template DNA. The PCR products were purified using QIAquick PCR purification kit (Qiagen, Inc.). Both strands of the PCR products were sequenced using the IRD44-labeled oligos listed in Table 1. Once confirmed, the wild-type and mutant rhaS alleles were introduced into SME1851 and SME2187 by phage P1-mediated generalized transduction (40) using selection for the linked Tn10. Finally, recA::kan was moved into each of the strains by P1 transduction with selection for kanamycin resistance.
β-Galactosidase assay.
Strains to be assayed for β-galactosidase activity were grown and assayed as previously described (4). Briefly, they were first grown in TY broth containing ampicillin and then transferred to 1× MOPS minimal medium with ampicillin (200 μg/ml) and limiting carbon source (0.04% glycerol) for overnight growth. The overnight culture was diluted 1:100 into 1× MOPS medium containing 0.4% glycerol as the carbon source; 0.2% l-rhamnose was added as the inducer along with ampicillin (200 μg/ml), and the culture was grown to an A600 of approximately 0.4. Assays were performed as described by Miller (40) except that in assays of the truncated rhaT fusion [Φ(rhaT-lacZ)Δ84], cultures were concentrated 20-fold (ς70 derivatives) or 114-fold (RhaS derivatives and RhaS-ς70 derivative combinations) upon addition of Z buffer. This is much greater than the 2.5-fold concentration in the standard assay. These assays were allowed to incubate for up to 3 days. Assays of RhaS derivatives and combinations of RhaS and ς70 derivatives at the truncated rhaT fusion were performed in a total volume of 0.1 ml rather than in the standard 1 ml so that very large culture volumes did not need to be grown. Under these conditions, the vast majority of the optical-density-at-420-nm readings were greater than 0.1, while the very lowest readings were greater than 0.05. Specific activities were averaged from at least three independent assays, with two replicates in each assay.
RESULTS
Sigma70 substitutions at rhaBAD.
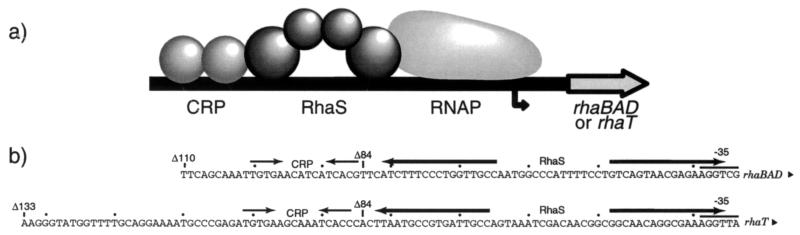
Lonetto et al. (36) constructed a library of 17 single alanine substitutions near the C-terminal end of the ς70 subunit of RNA polymerase. They found that ς70 residues in this region were required for activation of a variety of promoters in which an activator protein binds to a site that overlaps the −35 region of the promoter. To determine whether contacts with ς70 were important for activation by RhaS, we first tested the library of alanine substitutions in ς70 at the rhaBAD and rhaT promoters. The strains that we assayed had a gene encoding wild-type ς70 in the chromosome and carried the gene encoding the ς70 derivatives on a plasmid. Lonetto et al. (36) showed that in the absence of IPTG induction the ς70 derivatives were produced from these plasmids at a level that is only slightly higher than that of wild-type ς70; hence, only about 50% of the RNAP is expected to contain non-wild-type ς70. The strains also carried a wild-type rha locus at the normal chromosomal location and a single-copy λ specialized transducing phage carrying a translational fusion of the rhaBAD or rhaT promoter with lacZ. The promoter fusions were either full length and included the binding sites for both the CRP and RhaS activators or truncated and included only the binding site for RhaS (Fig. 1). Deletion of the CRP-binding site from the fusions was preferable to deletion of the crp gene has been shown to decrease rhaBAD expression both due to the direct loss of CRP activation and to decreased rhaS expression from the CRP-dependent rhaSR promoter.
FIG. 1.
(a) Schematic representation of the rhaBAD and rhaT promoter regions. RNA polymerase and the two activator proteins CRP and RhaS are shown bound to DNA in their respective positions. (b) DNA sequences of the rhaBAD and rhaT promoter regions, extending from the −35 regions to the most upstream endpoint of the promoter fusions used in this work. The positions of the RhaS-binding sites are shown by everted arrows, and the positions of the CRP-binding sites are shown by inverted arrows. The −35 regions of each promoter are marked, and the upstream endpoints of promoter fusions with lacZ are identified by a “Δ.”
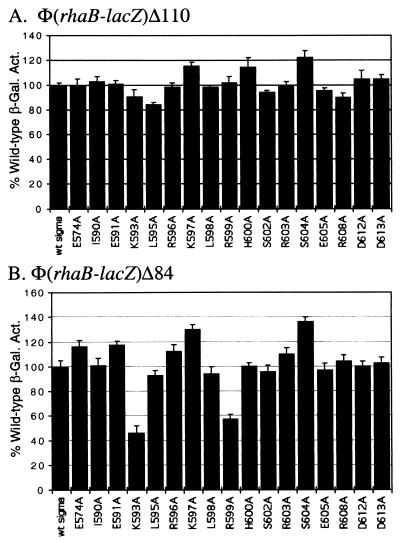
We first tested the ς70 substitution library at the full-length rhaBAD promoter fusion [Φ(rhaB-lacZ)Δ110] and found that there were no significant defects with any of the ς70 substitutions at this promoter (Fig. 2A). We next assayed the library at the truncated rhaBAD promoter fusion that included only the RhaS-binding site [Φ(rhaB-lacZ)Δ84] (Fig. 2B). At this fusion, two of the ς70 substitutions, K593A and R599A, allowed activation to only 46 and 58% of the wild type, respectively. Given that only 50% of the RNAP was likely to contain the ς70 substitution at K593 or R599 in each case, these defects are reasonably large. Residue K593 was also found to be important for activation by AraC, but R599 was not (36). Also, similar to the findings at araBAD, the ς70 substitutions only had a significant effect when the CRP-binding site was not present upstream of rhaBAD.
FIG. 2.
Alanine substitutions in the ς70 subunit of E. coli RNA polymerase analyzed at a full-length fusion of the rhaBAD promoter with lacZ [Φ(rhaB-lacZ)Δ110] (A) and at a truncated fusion of the rhaBAD promoter with lacZ [Φ(rhaB-lacZ)Δ84] (B). In each case, a strain carrying the indicated translational fusion as a single-copy λ lysogen was transformed with a plasmid encoding either wild-type ς70 or a derivative with a single alanine substitution at the positions indicated. β-Galactosidase activity was measured from cultures grown in minimal medium with glycerol, l-rhamnose, and ampicillin. The x axis represents the ς70 derivative. The y axis represents the β-galactosidase specific activity for each ς70 derivative as a percentage of the activity for wild-type ς70. The wild-type activity in panel A was 402 Miller units, while the wild-type activity in panel B was 9 Miller units. Both were set to 100%.
Although the ς70 K593A and R599A derivatives only showed defects at non-native, truncated promoters, we would argue that this information is likely to be biologically relevant. It is possible that these residues are also important for RhaS activation in the full-length promoter, but for reasons described in the Discussion, such as redundancy, they did not show any detectable defect in the presence of CRP activation. Further, if these ς70 residues are important for activation by RhaS in the absence of CRP, it is possible that other AraC/XylS family proteins that activate transcription without the aid of a second activator, such as CRP, may also require these residues.
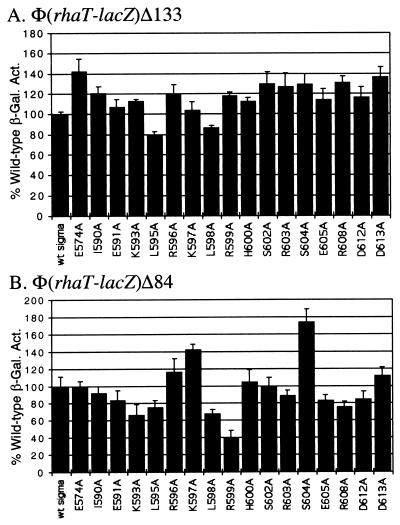
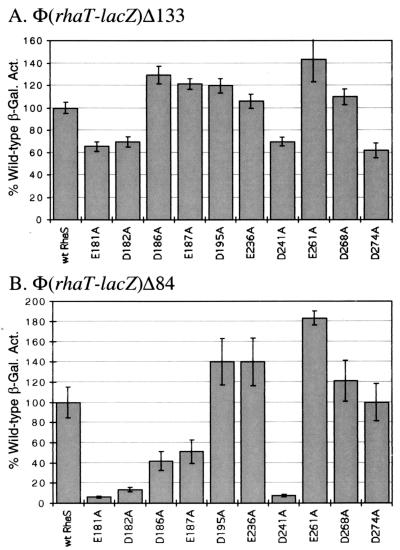
ς70 substitutions at rhaT.
To develop a more general picture of the role of the C-terminal end of ς70 in activation by RhaS, we tested the ς70 library at rhaT promoter fusions. As shown in Fig. 1, the location of the RhaS and CRP-binding sites relative to the core promoter at rhaT is the same as that at rhaBAD. Assays of β-galactosidase activity were modified as described in Materials and Methods to allow accurate measurement of the low activities expressed from the truncated rhaT fusion. At the full-length rhaT fusion that included both the RhaS and CRP-binding sites [Φ(rhaT-lacZ)Δ133], L595A was slightly defective, but none of the other substitutions were defective (Fig. 3A). When tested at a truncated rhaT promoter where only the RhaS-binding site was present [Φ(rhaT-lacZ)Δ84], five of the ς70 substitutions gave a value that was less than 80% of the wild-type activation (Fig. 3B). The largest defects were found with K593A and R599A, the same two substitutions that were defective at Φ(rhaB-lacZ)Δ84, while smaller defects were also seen with L595A, L598A, and R608A. One of the ς70 substitutions, S604A, activated to more than 170% of the level of the wild type. This finding is similar to the increased activation observed with two other ς70 substitutions at the araBAD promoter, although the magnitudes of the effects were greater at araBAD (36).
FIG. 3.
Alanine substitutions in the ς70 subunit of E. coli RNA polymerase analyzed at a full-length fusion of the rhaT promoter with lacZ [Φ(rhaT-lacZ)Δ133] (A) and at a truncated fusion of the rhaT promoter with lacZ [Φ(rhaT-lacZ)Δ84] (B). In each case, a strain carrying the indicated translational fusion as a single-copy λ lysogen was transformed with a plasmid encoding either wild-type ς70 or a derivative with a single alanine substitution at the positions indicated. β-Galactosidase activity was measured in cultures grown in minimal medium with glycerol, l-rhamnose, and ampicillin. The x axis represents the ς70 derivative; the y axis represents the β-galactosidase specific activity for each ς70 derivative as a percentage of the activity for wild-type ς70. The wild-type activity in panel A was 2.5 Miller units; in panel B it was 0.079 Miller units. Both were set to 100%.
Substitution of negatively charged amino acids in RhaS.
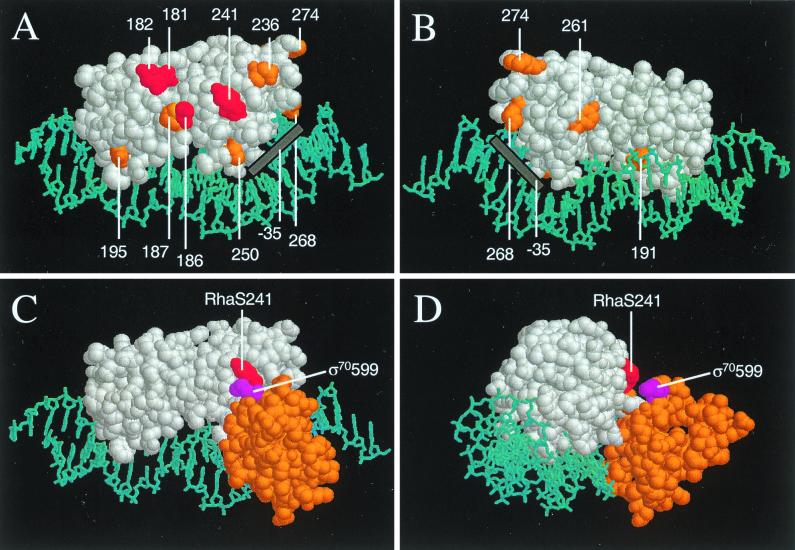
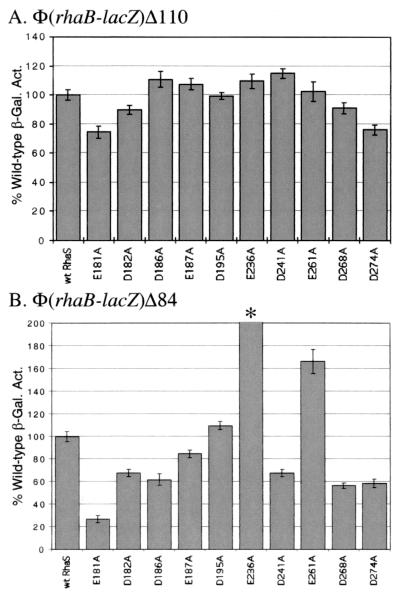
We noticed that the two ς70 residues that were defective at rhaBAD were positively charged amino acids (K593A and R599A). While additional substitutions were also found at rhaT, the same two positively charged amino acids were defective at this promoter as well. It has been proposed that ς70 residues identified using this library define interactions with the activator protein that overlaps the −35 region of that promoter (32, 33, 36). It has previously been shown that an overexpressed DNA-binding domain of AraC could weakly activate transcription (7), indicating that this domain of AraC is capable of transcription activation. As this is the conserved domain of AraC/XylS family proteins, it is likely that many other family members utilized this same domain for transcription activation. We hypothesized that if the ς70 residues that were defective at rhaBAD indeed defined an interaction with RhaS, then the RhaS amino acids involved in this interaction would be among the 12 negatively charged amino acids located within the DNA-binding domain of RhaS (Fig. 4A and B). We previously determined that one of these, D250, was involved in base-specific DNA contacts at rhaBAD (4). We therefore constructed alanine substitutions by site-directed mutagenesis at 10 of the remaining 11 positions. Based on the position of residue 191 on the crystal structure of MarA (46) (Fig. 4B), it seemed very unlikely to contact ς70, so when technical difficulties were encountered in this construction it was not further pursued.
FIG. 4.
Model of the C-terminal domain of RhaS bound to DNA based on the crystal structure of a MarA-DNA complex (44). (A) “Front” view of RhaS C-terminal domain (white) in a space-filling model with the negatively charged residues highlighted and numbered. DNA is shown in a stick model and is colored cyan. RhaS residues (in red) were defective at both the rhaBAD and the rhaT promoters, while residues in orange were either not defective, were defective at only one promoter, or were not tested (D250 and D191). In this view the N-terminal subdomain of RhaS is on the left and the C-terminal subdomain is on the right. The approximate position of the −35 region of the promoter is shown as a gray bar. (B) Same as panel A, except rotated around the vertical axis by approximately 180° to give the “back” view (i.e., the N-terminal subdomain is on the right, and the C-terminal subdomain is on the left). (C) A model of the C-terminal region of ς70 (residues 550 to 613, orange, based on the DNA-binding domain of NarL) has been added to the RhaS C-terminal domain model. RhaS is in the same view as in panel A, but only the RhaS residue 241 is highlighted in red. The ς70 residue 599 is highlighted in violet. (D) Same as panel C, but rotated by somewhat less than 90° around the vertical axis. The modeling of ς70 onto the MarA-DNA complex was performed using the program Insight II, and panels A through D were drawn using RasMol version 2.6 for the Macintosh.
RhaS substitutions at rhaBAD.
We first tested the substitutions in negatively charged amino acids of RhaS at the rhaBAD promoter. At Φ(rhaB-lacZ)Δ110, two of the RhaS substitutions (E181A and D274A) showed slight defects of about 75% of the level of wild-type activation (Fig. 5A). At the truncated rhaBAD promoter fusion, Φ(rhaB-lacZ)Δ84, six of the alanine substitutions in RhaS were defective (Fig. 5B). E181A showed the greatest defect at 28% of the level of wild-type activation, while the other five defective substitutions activated to 56 to 68% of the wild-type level. It is also interesting to notice that the substitution at E236 resulted in a level of 279% of the wild-type activation at this truncated promoter but was not significantly different than wild-type at the full-length Φ(rhaB-lacZ)Δ110 fusion. This is similar to the increased activation by two of the ς70 substitutions (E591A and R596A) when tested at araBAD in the absence of CRP (36). E261A also resulted in greater than wild-type activation of Φ(rhaB-lacZ)Δ84, in this case to 166% of the wild-type level.
FIG. 5.
Alanine substitutions in RhaS analyzed at a full-length fusion of the rhaBAD promoter with lacZ [Φ(rhaB-lacZ)Δ110] (A) and at a truncated fusion of the rhaBAD promoter with lacZ [Φ(rhaB-lacZ)Δ84] (B). In each case, a strain carrying the indicated translational fusion as a single-copy λ lysogen was transformed with a plasmid encoding either wild-type RhaS or a derivative with a single alanine substitution at the positions indicated. β-Galactosidase activity was measured from cultures grown in minimal medium with glycerol, l-rhamnose, and ampicillin. The x axis represents the RhaS derivative. The y axis represents the β-galactosidase specific activity for each RhaS derivative as a percentage of the activity for wild-type RhaS. The wild-type activity in panel A was 453 Miller units for all of the assays except for E261A, where the wild-type activity was 204 Miller units, and in panel B it was 9.4 Miller units for all of the assays except for D241A, where the wild-type activity was 9.3 Miller units, and E261A, where wild-type activity was 3.9 Miller units. The activity in the case of E236A in panel B (marked with an asterisk) was 279%, but is drawn off the scale to avoid compression of the other values. The wild-type activity was set to 100%.
RhaS substitutions at rhaT.
The same substitutions of negatively charged residues of RhaS were also tested for activation of rhaT. At the full-length rhaT promoter fusion [Φ(rhaT-lacZ)Δ133], four of the substituted RhaS proteins were slightly defective for activation (Fig. 6A). Each of the substitutions at positions E181, D182, D241, and D274 activated to 62 to 70% of the wild-type RhaS. Three of those four substitutions (E181, D182, and D241) were also defective at the truncated rhaT fusion that lacked the CRP-binding site [Φ(rhaT-lacZ)Δ84] (Fig. 6B). The defects of these substitutions at the truncated promoter were much more severe and resulted in only about 10% of the wild-type activation. Interestingly, the substitution at D274 was not defective at the truncated promoter. Two additional substitutions were somewhat defective at the truncated promoter but not at the full-length promoter (D186A and E187A).
FIG. 6.
Alanine substitutions in RhaS analyzed at a full-length fusion of the rhaT promoter with lacZ [Φ(rhaT-lacZ)Δ133] (A) and a truncated fusion of the rhaT promoter with lacZ [Φ(rhaT-lacZ)Δ84] (B). In each case, a strain carrying the appropriate translational fusion as a single-copy λ lysogen was transformed with a plasmid encoding either wild-type RhaS or a derivative with a single alanine substitution at the positions indicated. β-Galactosidase activity was measured from cultures grown in minimal medium with glycerol, l-rhamnose, and ampicillin. The x axis represents the RhaS derivative. The y axis represents the β-galactosidase specific activity for each RhaS derivative as a percentage of the activity for wild-type RhaS. The wild-type activity in panel A was 1.38 Miller units for all of the assays except for with E261A, where the wild-type activity was 0.34 Miller units, and in panel B was 0.048 Miller units for all of the assays except for with E261A, where the wild-type activity was 0.027 Miller units. The wild-type activity was set to 100%.
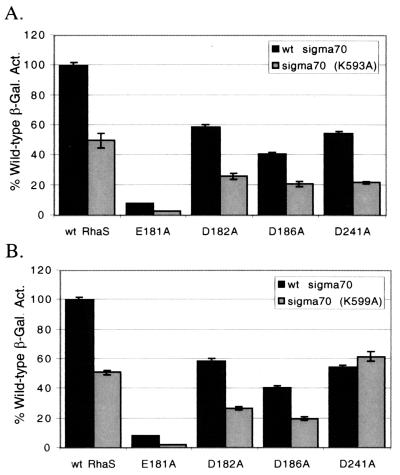
Combination of RhaS and ς70 substitutions.
We next wished to combine the RhaS and ς70 substitutions to test for evidence of interactions between combinations of alleles. We recombined the defective rhaS alleles into the normal chromosomal rhaS locus using the gene replacement strategy of Murphy (41). In this procedure an E. coli strain carries phage λ recombination genes and, as a result, is capable of high-frequency replacement of chromosomal genes with alleles carried on PCR-generated DNA fragments. In the original description of this method, the recombined alleles could be identified by positive selection (for example lacZ::kan). Using a rhaB-lacZ fusion strain background, we were able to identify replacements of an in-frame deletion of rhaS with our partially functional rhaS alleles by screening for tiny blue colonies amid a lawn and so did not require positive selection (see Materials and Methods).
The goal of our analysis was to determine genetically whether any of the combinations of defective substitutions in RhaS and ς70 might identify specific amino acid contacts between the two proteins. The logic behind our analysis was that the combination of any two substitutions that do not identify specific amino acid contacts should result in a greater defect than either of the individual substitutions. On the other hand, the combination of two substitutions that do identify specific amino acid contacts would be expected to result in a defect that is no greater than the more defective individual substitution. In this case, each of the individual substitutions would have already lost the contact, so a substitution in the second residue involved in that contact would be expected to result in no further defect. We tested the RhaS E181A, D182A, D186A, and D241A substitutions in combination with the ς70 K593A and R599A substitutions at each of the truncated rhaB-lacZ and rhaT-lacZ fusions.
The combinations of RhaS and ς70 substitutions were first tested for activation of the truncated Φ(rhaB-lacZ)Δ84 fusion. In most cases, the combinations of substitutions gave percent activation values that were less than the values for either of the substitutions alone (Fig. 7). In fact, in all but one case the percent activation for the combination of two substitutions was approximately equal to the product of the values for each of the substitutions alone in the same assay. Since each of the two ς70 substitutions (K593A and R599A) alone activated to approximately 50%, one can easily see that most of the combinations of RhaS and ς70 substitutions activated to very nearly half of the percent activation by the RhaS substitution alone. In contrast, the combination of RhaS D241A and ς70 R599A resulted in a percent activation that was no less (and in fact was somewhat greater) than the percent activation of the RhaS D241A and ς70 R599A substitutions individually. These results are consistent with the conclusion that RhaS D241A and ς70 R599A may define an interaction between RhaS and ς70 and that none of the other combinations of substitutions tested define an interaction at Φ(rhaB-lacZ)Δ84.
FIG. 7.
Combinations of RhaS and ς70 alanine substitutions at Φ(rhaB-lacZ)Δ84. aRhaS substitutions were tested in combination with either ς70 K593A (A) or ς70 R599A (B) at the Φ(rhaB-lacZ)Δ84 fusion. The β-galactosidase specific activity for each combination is represented as a percentage of the activity found for the combination of wild-type RhaS and wild-type ς70, which was 9.1 Miller units and was set to 100% for both graphs.
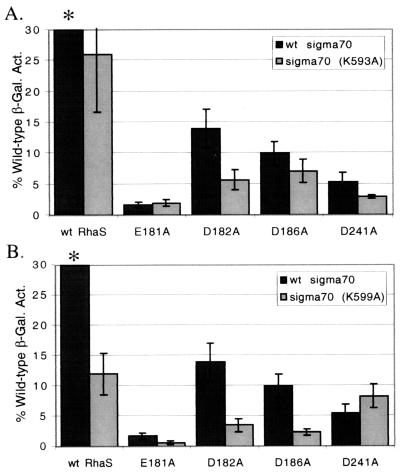
The RhaS and ς70 combinations were also tested for activation at the truncated Φ(rhaT-lacZ)Δ84 fusion (Fig. 8). Again the combination of RhaS D241A and ς70 R599A resulted in a percent activation that was no worse than that of the each of the two substitutions alone and was somewhat greater than the RhaS substitution alone. This result further strengthens the hypothesis that RhaS D241A and ς70 R599 define an interaction between the wild-type RhaS and ς70 proteins.
FIG. 8.
Combinations of RhaS and ς70 alanine substitutions at Φ(rhaT-lacZ)Δ84. aRhaS substitutions were tested in combination with either ς70 K593A (A) or ς70 R599A (B) at the Φ(rhaT-lacZ)Δ84 fusion. The β-galactosidase specific activity for each combination is represented as the percentage of the activity found for the combination of wild-type RhaS and wild-type ς70, which was 0.18 Miller units and was set to 100% for both graphs.
One additional combination of RhaS and ς70 substitutions, RhaS E181A and ς70 R593A, was no worse than the individual substitutions at the Φ(rhaT-lacZ)Δ84 fusion (Fig. 8). In this case, the value for the β-galactosidase expression with RhaS 181A alone was extremely low (in the range of background levels); therefore, we are not confident that we could reproducibly measure a lower level from the combination of the RhaS and ς70 derivatives. This combined with the fact that this combination was only identified at rhaT and not at rhaBAD suggests that this may not represent a real interaction. This hypothesis is further supported by our molecular modeling (see below) which does not place RhaS 181 and ς70 593 in close proximity (not shown).
Modeling of RhaS interaction with ς70.
There are currently structures available for the DNA-binding domain of two AraC/XylS family proteins, MarA and Rob (31, 46). The C-terminal domain of RhaS shares 24% identity and 46% similarity with MarA and 31% identity and 45% similarity with Rob. According to Kwon et al., the main chain atoms of the conserved portions of the MarA and Rob structures are extremely similar, with a root mean square deviation of 0.9 Å (31), suggesting that modeling of RhaS residues onto either structure would give nearly the same result. The only major difference between the MarA and Rob structures is that MarA makes base-specific contacts with DNA using both of its helix-turn-helix motifs, while Rob only makes base-specific contacts with its N-terminal helix-turn-helix motif (31, 46). As we have evidence that both helix-turn-helix motifs of RhaS make base-specific contacts with DNA (4), RhaS was modeled based on the structure of the MarA-DNA complex (Brookhaven Data Bank file 1BLO) (Fig. 4) (46).
We know (based on specific amino-acid–base-pair contacts [4]) that RhaS is oriented with its C-terminal subdomain overlapping the −35 region of the promoter by 4 bp, thereby defining the position of ς70 relative to the RhaS model. We modeled ς70 residues 550 to 613 based on the DNA-binding domain of NarL (Brookhaven Data Bank file 1RNL) as previously proposed by Lonetto et al. (36) and substituted the residues of ς70 for the NarL residues (Fig. 4C and D). This region of NarL was modeled onto DNA exactly as described earlier (3). The DNAs in the NarL-DNA complex and the MarA-DNA complex were manually superimposed, and ς70 residues 584 and 588 were aligned as closely as possible with the fifth and third positions of the −35 hexamer, respectively, based on previously identified contacts (20, 49). Once the C-terminal region of ς70 was modeled onto the MarA-DNA complex, the DNA onto which NarL was initially modeled was deleted and ς70 residue 599 was highlighted. Finally, RhaS residue 241, which our results indicate interacts with ς70 residue 599, was also highlighted. As is shown in Fig. 4C and D, RhaS 241 and ς70 599 are very near one another on the model and are therefore in an excellent position to participate in a contact between RhaS and ς70.
DISCUSSION
Activation by RhaS requires amino acids near the C-terminal end of ς70.
Two residues near the C-terminal end of ς70, K593A and R599A, were found to be important for activation at lacZ fusions with both the truncated rhaBAD and rhaT promoters (Fig. 2B and Fig. 3B). Other work has shown that none of these ς70 substitutions are generally defective for transcription (32, 33, 36). These truncated promoters have binding sites for only one activator protein, RhaS. Hence, residues K593 and R599 in ς70 are apparently required for transcription activation by RhaS and might be involved in direct contacts with RhaS.
It is very interesting that none of the ς70 substitutions resulted in defects worse than 79% of wild-type at full-length rhaBAD and rhaT promoter fusions (Fig. 2A and Fig. 3A). The full-length fusions include the binding site for CRP in addition to that for RhaS, indicating that in the presence of CRP the contribution of these residues to rhaBAD and rhaT activation was either decreased or eliminated. Very similar results were found at the araBAD promoter where residues in this region of ς70 were only important in a cya mutant strain (36). In the cya mutant strain, CRP would not be bound to its site, and AraC would be the only activator of araBAD expression. These results may indicate that CRP has an influence on transcription activation of rhaBAD, rhaT, and araBAD that is redundant with the role of these ς70 residues. Alternatively, in the presence of CRP the total number of interactions at this promoter may be large enough that the loss of any one interaction does not result in a significant defect. Finally, it is also possible that the proposed contacts between RhaS and ς70 only occur in the absence of CRP binding. CRP might alter the geometry of the transcription activation complex such that RhaS and ς70 are no longer in precisely the correct position to interact.
Negatively charged residues in RhaS important for activation.
We reasoned that ς70 K593A and R599A might define interactions with RhaS and, if so, that the partner residues in RhaS would probably be negatively charged. Upon substitution of most of the Asp and Glu residues in the C-terminal domain of RhaS to Ala, we found that E181A, D182A, D186A, and D241A were defective at both the truncated rhaBAD and rhaT promoters (Fig. 5B and Fig. 6B). When modeled on the structure of MarA (46), the positions of these residues of RhaS suggest a possible face of RhaS that could interact with ς70 (Fig. 4A). RhaS E181, however, aligns with a residue on MarA, where alanine substitution resulted in a severe defect both at promoters where MarA binds overlapping the −35 region and at promoters where MarA binds further upstream (21), suggesting that this residue may have a role other than interaction with ς70. We cannot rule out that some of these RhaS residues are defective due to DNA-binding defects. We would argue, however, that the evidence for residue D241, in particular when in combination with substitutions in ς70 (Fig. 7 and 8 and see below), argues that the defect caused by at least this substitution is not due to a DNA-binding defect.
Genetic evidence for contacts between RhaS and ς70.
If residues within two proteins are involved in direct protein-protein contacts with each other, than one would expect that substitution of either one or both of the residues might have the same phenotype (in this case, the same defect in transcription activation). It is also possible, however, that one or both of the residues will have secondary effects on protein folding or stability and therefore would have a larger overall effect on transcription activation. In this case, substitution of both of the residues involved in a contact would be expected to have the same defect as that of the single residue with the greater defect. On the other hand, the combination of two substitutions that do not define a direct protein-protein contact would be expected to have a defect that was greater than either of the individual substitutions. We have used this reasoning to analyze the combination of substitutions in ς70 and RhaS to determine whether any of the residues might define a protein-protein contact that might contribute to transcription activation.
The combination of the RhaS D241A and ς70 R599A substitutions showed a pattern of defects that was consistent with the wild-type RhaS and ς70 proteins making protein-protein contacts at these positions at both the truncated rhaBAD and rhaT promoters (Fig. 7 and 8). We do not yet have direct biochemical evidence to support the existence of a contact between these residues; however, several arguments can be made to support the hypothesis that these genetic results may indicate a real interaction. First, the same combination of residues showed genetic evidence for an interaction at both the truncated rhaBAD and rhaT promoters. Second, considering that D241 is located within the first helix of H-T-H 2 of RhaS (helix-5 of the MarA structure) (4) and that H-T-H 2 binds to a major groove that overlaps the −35 region of the promoter (12), D241 appears to be ideally positioned on the surface of RhaS to make contact with ς70 (Fig. 4). Third, we have modeled the C-terminal region of ς70 onto the model of the RhaS-DNA complex and found that RhaS D241 and ς70 R599 lie in very close proximity to one another (Fig. 4C and D). Finally, more than half of the AraC/XylS family proteins aligned by Gallegos et al. (18) have an Asp or Glu that aligns with RhaS D241, indicating that this residue is conserved among family members, perhaps for a role in transcription activation. Consistent with this, neither AraC nor Ada have a negatively charged residue that aligns with RhaS D241, and in both of these cases ς70 R599A was not defective for activation (33, 36). Further, RhaR does have an Asp at the position that aligns with RhaS D241, and R599A was found to be defective for activation by RhaR (V. Rao and S. M. Egan, unpublished results). RhaS D241 represents the first residue of an AraC/XylS family protein that has been implicated in a direct role in transcription activation through a contact with ς70.
ACKNOWLEDGMENTS
We are very grateful to Carol Gross for providing the ς70 alanine substitution library, Jeffrey Urbauer for assistance with the modeling of ς70 onto the MarA-DNA complex, and Keenan Murphy for providing strain KM22. We thank the members of our laboratory for critical discussions and Carolyn Holcroft for comments on the manuscript. We also thank Susan Bear for constructing pSE159; Patrick Angell for construction of pSE204 and λ SME108; and Jessica Kueker, Vydehi Rao, and Patrick Angell for technical assistance with strain construction and β-galactosidase assays. We thank an anonymous reviewer for suggesting the modeling of ς70 on the MarA-DNA complex and James Therrien and other members of the University of Kansas Biochemical Research Service Laboratory for help with automated DNA sequencing.
This work was supported by Public Health Service grant GM55099 from the National Institute of General Medical Sciences and the Franklin Murphy Molecular Biology Endowment, both to S.M.E.
REFERENCES
- 1.Akimuru H, Sakumi K, Yoshikai T, Anai M, Sekiguchi M. Positive and negative regulation of transcription by a cleavage product of Ada protein. J Mol Biol. 1990;216:261–273. doi: 10.1016/S0022-2836(05)80318-3. [DOI] [PubMed] [Google Scholar]
- 2.Backman K, Chen Y-M, Magasanik B. Physical and genetic characterization of the gln A-glnG region of the Escherichia coli chromosome. Proc Natl Acad Sci USA. 1981;78:3743–3747. doi: 10.1073/pnas.78.6.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baikalov I, Schroder I, Kaczor-Grzeskowiak M, Grzeskowaik K, Gunsalus R P, Dickerson R E. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- 4.Bhende P M, Egan S M. Amino acid-DNA contacts by RhaS: an AraC family transcription activator. J Bacteriol. 1999;181:5185–5192. doi: 10.1128/jb.181.17.5185-5192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourgerie S J, Michan C M, Thomas M S, Busby S J W, Hyde E I. DNA binding and DNA bending by the MelR transcription activator protein from Escherichia coli. Nucleic Acids Res. 1997;25:1685–1693. doi: 10.1093/nar/25.9.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunelle A, Schleif R. Determining residue-base interactions between AraC protein and araI DNA. J Mol Biol. 1989;209:607–622. doi: 10.1016/0022-2836(89)90598-6. [DOI] [PubMed] [Google Scholar]
- 7.Bustos S A, Schleif R F. Functional domains of the AraC protein. Proc Natl Acad Sci USA. 1993;90:5638–5642. doi: 10.1073/pnas.90.12.5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caswell R, Williams J, Lyddiatt A, Busby S. Overexpression, purification and characterization of the Escherichia coli MelR transcription activator protein. Biochem J. 1992;287:493–499. doi: 10.1042/bj2870493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Orazio S E F, Collins C M. The plasmid-encoded urease gene cluster of the family Enterobacteriaceae is positively regulated by UreR, a member of the AraC family of transcriptional activators. J Bacteriol. 1993;175:3459–3467. doi: 10.1128/jb.175.11.3459-3467.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dombroski A J, Walter W A, Record M T J, Siegele D A, Gross C A. Polypeptides containing highly conserved regions of transcription initiation factor ς70 exhibit specificity of binding to promoter DNA. Cell. 1992;70:501–512. doi: 10.1016/0092-8674(92)90174-b. [DOI] [PubMed] [Google Scholar]
- 11.Ebright R H, Busby S. The Escherichia coli RNA polymerase α subunit: structure and function. Curr Opin Genet Dev. 1995;5:197–203. doi: 10.1016/0959-437x(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 12.Egan S M, Schleif R F. DNA-dependent renaturation of an insoluble DNA binding protein. Identification of the RhaS binding site at rhaBAD. J Mol Biol. 1994;243:821–829. doi: 10.1006/jmbi.1994.1684. [DOI] [PubMed] [Google Scholar]
- 13.Egan S M, Schleif R F. A regulatory cascade in the induction of rhaBAD. J Mol Biol. 1993;234:87–98. doi: 10.1006/jmbi.1993.1565. [DOI] [PubMed] [Google Scholar]
- 14.Eichelberg K, Hardt W-D, Galan J E. Characterization of SprA, an AraC-like transcriptional regulator encoded within the Salmonella typhimurium pathogenicity island 1. Mol Microbiol. 1999;33:139–152. doi: 10.1046/j.1365-2958.1999.01458.x. [DOI] [PubMed] [Google Scholar]
- 15.Furuichi M, Yu C G, Anai M, Sakumi L, Sekiguchi M. Regulatory elements for expression of the alkA gene in response to alkylating agents. Mol Gen Genet. 1992;236:25–32. doi: 10.1007/BF00279639. [DOI] [PubMed] [Google Scholar]
- 16.Gallegos M-T, Marqués S, Ramos J L. Expression of the TOL plasmid xylS gene in Pseudomonas putida occurs from a ς70-dependent promoter or from ς70- and ς54-dependent tandem promoters according to the compound used for growth. J Bacteriol. 1996;178:2356–2361. doi: 10.1128/jb.178.8.2356-2361.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallegos M-T, Michán C, Ramos J L. The XylS/AraC family of regulators. Nucleic Acids Res. 1993;21:807–810. doi: 10.1093/nar/21.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallegos M-T, Schleif R, Bairoch A, Hofmann K, Ramos J L. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallegos M-T, Marqués S, Ramos J L. The TACAN4TGCA motif upstream from the −35 region in the ς70-ςS-dependent Pm promoter of the TOL plasmid is the minimum DNA segment required for transcription stimulation by XylS regulators. J Bacteriol. 1996;178:6427–6434. doi: 10.1128/jb.178.22.6427-6434.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardella T, Moyle H, Susskind M M. A mutant Escherichia coli ς70 subunit of RNA polymerase with altered promoter specificity. J Mol Biol. 1989;206:579–590. doi: 10.1016/0022-2836(89)90567-6. [DOI] [PubMed] [Google Scholar]
- 21.Gillette W K, Martin R G, Rosner J L. Probing the Escherichia coli transcriptional activator MarA using alanine-scanning mutagenesis: residues important for DNA binding and activation. J Mol Biol. 2000;299:1245–1255. doi: 10.1006/jmbi.2000.3827. [DOI] [PubMed] [Google Scholar]
- 22.Gottesman M E, Yarmolinsky M B. The integration and excision of the bacteriophage lambda genome. Cold Spring Harbor Symp Quant Biol. 1968;33:735–747. doi: 10.1101/sqb.1968.033.01.084. [DOI] [PubMed] [Google Scholar]
- 23.Holcroft C C, Egan S M. Roles of cyclic AMP receptor protein and the carboxyl-terminal domain of the α subunit in transcription activation of the Escherichia coli rhaBAD operon. J Bacteriol. 2000;182:3529–3535. doi: 10.1128/jb.182.12.3529-3535.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishihama A. Role of the RNA polymerase α subunit in transcription activation. Mol Microbiol. 1992;6:3283–3288. doi: 10.1111/j.1365-2958.1992.tb02196.x. [DOI] [PubMed] [Google Scholar]
- 25.Jair K, Martin R G, Rosner J L, Fujita N, Ishihama A, Wolf R E J. Purification and regulatory properties of MarA protein, a transcriptional activator of Escherichia coli multiple antibiotic and superoxide resistance promoters. J Bacteriol. 1995;177:7100–7104. doi: 10.1128/jb.177.24.7100-7104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jair K-M, Yu X, Skarstad K, Thony B, Fujita N, Ishihama A, Wolf R E J. Transcriptional activation of promoters of the superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J Bacteriol. 1996;178:2507–2513. doi: 10.1128/jb.178.9.2507-2513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jair K-W, Fawcett W P, Fujita N, Ishihama A, Wolf R E., Jr Ambidextrous transcriptional activation by SoxS: requirement for the C-terminal domain of the RNA polymerase alpha subunit in a subset of Escherichia coli superoxide-inducible genes. Mol Microbiol. 1996;19:307–317. doi: 10.1046/j.1365-2958.1996.368893.x. [DOI] [PubMed] [Google Scholar]
- 28.Jordi B J A M, van der Zeijst B A M, Gaastra W. Regions of the CFA/I promoter involved in the activation by the transcriptional activator CfaD and repression by the histone-like protein H-NS. Biochimie. 1994;76:1052–1054. doi: 10.1016/0300-9084(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 29.Kaldalu N, Mandel T, Ustav M. TOL plasmid transcription factor XylS binds specifically to the Pm operator sequence. Mol Microbiol. 1996;20:569–579. doi: 10.1046/j.1365-2958.1996.5381060.x. [DOI] [PubMed] [Google Scholar]
- 30.Kessler B, Herrero M, Timmis K N, Lorenzo V DE. Genetic evidence that the XylS regulator of the Pseudomonas TOL meta operon controls the Pm promoter through weak DNA-protein interactions. J Bacteriol. 1994;176:3171–3176. doi: 10.1128/jb.176.11.3171-3176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwon H J, Bennik M H J, Demple B, Ellenberger T. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat Struct Biol. 2000;7:424–430. doi: 10.1038/75213. [DOI] [PubMed] [Google Scholar]
- 32.Landini P, Brown J A, Volkert M R, Busby S J W. Ada protein-RNA polymerase ς subunit interaction and α subunit-promoter DNA interactions are necessary at different steps in transcription activation at the Escherichia coli ada and aidB promoters. J Biol Chem. 1998;273:13307–13312. doi: 10.1074/jbc.273.21.13307. [DOI] [PubMed] [Google Scholar]
- 33.Landini P, Busby S J. The Escherichia coli Ada protein can interact with two distinct determinants in the ς70 subunit of RNA polymerase according to promoter architecture: identification of the target of Ada activation at the alkA promoter. J Bacteriol. 1999;181:1524–1529. doi: 10.1128/jb.181.5.1524-1529.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landini P, Gaal T, Ross W, Volkert M R. The RNA polymerase α subunit carboxyl-terminal domain is required for both basal and activated transcription from the alkA promoter. J Biol Chem. 1997;272:15914–15919. doi: 10.1074/jbc.272.25.15914. [DOI] [PubMed] [Google Scholar]
- 35.Landini P, Volkert M R. Transcriptional activation of the Escherichia coli adaptive response gene aidB is mediated by binding of methylated Ada protein. Evidence for a new consensus sequence for Ada-binding sites. J Biol Chem. 1995;270:8285–8289. doi: 10.1074/jbc.270.14.8285. [DOI] [PubMed] [Google Scholar]
- 36.Lonetto M A, Rhodius V, Lamberg K, Kiley P, Busby S, Gross C. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase ς70 subunit. J Mol Biol. 1998;284:1353–1365. doi: 10.1006/jmbi.1998.2268. [DOI] [PubMed] [Google Scholar]
- 37.Maloy S R, Stewart V J, Taylor R K. Genetic analysis of pathogenic bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 38.Marqués S, Gallegos M-T, Manzanera M, Holtel A, Timmis K N, Ramos J L. Activation and repression of transcription at the double tandem divergent promoters for the xylR and xylS genes of the TOL plasmid of Pseudomonas putida. J Bacteriol. 1998;180:2889–2894. doi: 10.1128/jb.180.11.2889-2894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michán C M, Busby S J W, Hyde E I. The Escherichia coli MelR transcription activator: production of a stable fragment containing the DNA-binding domain. Nucleic Acids Res. 1995;23:1518–1523. doi: 10.1093/nar/23.9.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 41.Murphy K C. Use of bacteriophage λ recombination functions to promote gene replacement in Escherichia coli. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neidhardt F C, Bloch P L, Smith D F. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niland P, Huhne R, Muller-Hill B. How AraC interacts specifically with its target DNAs. J Mol Biol. 1996;254:667–674. doi: 10.1006/jmbi.1996.0668. [DOI] [PubMed] [Google Scholar]
- 44.Ramos J L, Rojo F, Zhou L, Timmis K N. A family of positive regulators related to the Pseudomonas putida TOL plasmid XylS and the Escherichia coli AraC activators. Nucleic Acids Res. 1990;18:2149–2152. doi: 10.1093/nar/18.8.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramos J L, Marqués S, Timmis K N. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through interplay of host factors and plasmid-encoded regulators. Annu Rev Microbiol. 1997;51:341–373. doi: 10.1146/annurev.micro.51.1.341. [DOI] [PubMed] [Google Scholar]
- 46.Rhee S, Martin R G, Rosner J L, Davies D R. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc Natl Acad Sci USA. 1998;95:10413–10418. doi: 10.1073/pnas.95.18.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakumi K, Sekiguchi M. Regulation of the expression of the ada gene controlling the adaptive response: interactions with the ada promoter and RNA polymerase. J Mol Biol. 1989;205:373–385. doi: 10.1016/0022-2836(89)90348-3. [DOI] [PubMed] [Google Scholar]
- 48.Savelkoul P H M, Willshaw G A, McConnell M M, Smith H R, Hamers A M, van der Zeijst B A M, Gaastra W. Expression of CFA/I fimbriae is positively regulated. Microb Pathog. 1990;8:91–99. doi: 10.1016/0882-4010(90)90073-y. [DOI] [PubMed] [Google Scholar]
- 49.Siegele D A, Hu J C, Walter W A, Gross C A. Altered promoter recognition by mutant forms of the sigma 70 subunit of Escherichia coli RNA polymerase. J Mol Biol. 1989;206:591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- 50.Silverstone A E, Goman M, Scaife J G. ALT: a new factor involved in the synthesis of RNA by Escherichia coli. Mol Gen Genet. 1972;118:223–234. doi: 10.1007/BF00333459. [DOI] [PubMed] [Google Scholar]
- 51.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 52.Tate C G, Muiry J A R, Henderson P J F. Mapping, cloning, expression, and sequencing of the rhaT gene which encodes a novel l-rhamnose-H+ transport protein in Salmonella typhimurium and Escherichia coli. J Biol Chem. 1992;287:6923–6932. [PubMed] [Google Scholar]
- 53.Tobin J F, Schleif R F. Positive regulation of the Escherichia colil-rhamnose operon is mediated by the products of tandemly repeated regulatory genes. J Mol Biol. 1987;196:789–799. doi: 10.1016/0022-2836(87)90405-0. [DOI] [PubMed] [Google Scholar]
- 54.Tobin J F, Schleif R F. Purification and properties of RhaR, the positive regulator of the l-rhamnose operons of Escherichia coli. J Mol Biol. 1990;211:75–89. doi: 10.1016/0022-2836(90)90012-B. [DOI] [PubMed] [Google Scholar]
- 55.Tobin J F, Schleif R F. Transcription from the rha operon psr promoter. J Mol Biol. 1990;211:1–4. doi: 10.1016/0022-2836(90)90003-5. [DOI] [PubMed] [Google Scholar]
- 56.Travers A A, Buckland R, Goman M, LeGrice S S G, Scaife J G. A mutation affecting the ς subunit of RNA polymerase changes transcriptional specificity. Nature. 1978;273:354–358. doi: 10.1038/273354a0. [DOI] [PubMed] [Google Scholar]
- 57.Via P, Badia J, Baldoma L, Obradors N, Aguilar J. Transcriptional regulation of the Escherichia coli rhaT gene. Microbiology. 1996;142:1833–1840. doi: 10.1099/13500872-142-7-1833. [DOI] [PubMed] [Google Scholar]