Abstract
Stress-induced regulatory networks coordinated with a procaryotic developmental program were revealed by two-dimensional gel analyses of global gene expression. Four developmental stages were identified by their distinctive protein synthesis patterns using principal component analysis. Statistical analyses focused on five stress stimulons (induced by heat, cold, salt, ethanol, or antibiotic shock) and their synthesis during development. Unlike other bacteria, for which various stresses induce expression of similar sets of protein spots, in Streptomyces coelicolor heat, salt, and ethanol stimulons were composed of independent sets of proteins. This suggested independent control by different physiological stress signals and their corresponding regulatory systems. These stress proteins were also under developmental control. Cluster analysis of stress protein synthesis profiles identified 10 different developmental patterns or “synexpression groups.” Proteins induced by cold, heat, or salt shock were enriched in three developmental synexpression groups. In addition, certain proteins belonging to the heat and salt shock stimulons were coregulated during development. Thus, stress regulatory systems controlling these stimulons were implicated as integral parts of the developmental program. This correlation suggested that thermal shock and salt shock stress response regulatory systems either allow the cell to adapt to stresses associated with development or directly control the developmental program.
With the availability of complete microbial genome sequences that define the components of numerous forms of life, integrative methods must be developed to understand the complex networks that coordinate expression of these genes. Methods of recording global patterns of gene expression, including two-dimensional (2D) gels and microarray hybridization, have allowed snapshots of various static patterns of gene expression and have identified gene products that change in response to certain stimuli. Genes having similar functions can often be identified by their similar temporal expression profiles in eucaryotic developmental processes (“synexpression groups”) (24). Synexpression groups have been identified by applying standard methods of cluster analysis to Saccharomyces cerevisiae gene expression data recorded by microarray hybridization (8). While synexpression groups are thought to provide for coordinated gene expression in eucaryotic cells comparable to procaryotic operons (24), the concept has not been explored in bacterial systems.
Image analysis of 2D gel protein spot intensity has provided a quantitative “proteome” database for Escherichia coli (29) and Bacillus subtilis (28) describing how sets of proteins change in response to physiological conditions. We use the term proteome here to describe the complex state of an organism under defined conditions rather than its complete protein repertoire. Methods of data analysis must be developed to compare proteomes in order to simplify and thus extract relevant information from these complex databases.
Stress proteins, initially studied as adaptive components induced by heat shock and providing for repair of denatured proteins, have proven to play essential and diverse roles in bacterial physiology. In E. coli, the heat shock regulon is induced in response to carbon, nitrogen, or phosphate starvation (19). The stringent response appears to be induced by heat shock and be repressed by cold shock (4). While global regulators of the cold shock response have not been defined, the heat shock response is controlled by sigma 32 (RpoH) and sigma 24 (RpoE) (12). Sigma 38 (RpoS), first identified for its role in adaptation to the stationary phase, provides a general response (15, 19) to starvation, osmotic, oxidative, and heat stresses (22). In B. subtilis, a diverse range of stresses, including heat, salt, ethanol, and starvation, all induce synthesis of a similar set of proteins (13). These so-called “general stress proteins” are under the direct control of a single specialized sigma factor, SigB.
Here we report global changes in gene expression in response to stress conditions that relate to developmental stages in a simple differentiating prokaryote, Streptomyces coelicolor (5). As a group, Streptomyces is best known for its ability to produce thousands of diverse antibiotics, triggered in response to undefined stress conditions (including elevated temperature [7]; our unpublished observations]). Studies of the heat shock response in S. coelicolor have suggested that developmental and thermal induction of a stress regulon might have common control elements (25). Many genes have been identified which control the Streptomyces heat shock regulon (2, 3, 10, 11, 27). At least one of these, a Clp protease, is required for morphological development on solid medium (6). In chemically defined liquid medium, S. coelicolor undergoes at least four stages of development (25). After an initial phase of rapid growth (RG1 phase), a transitory slowdown in growth provides a transition phase (T phase) to a second period of rapid growth (RG2 phase) and finally to stationary phase (S phase). Similar growth kinetics have been observed in liquid cultures of various Streptomyces species. The T phase has been associated with the activation of antibiotic biosynthetic genes (16) and regulatory elements needed for antibiotic-induced expression of a multidrug resistance gene (26), a starvation response indicated by the accumulation of ppGpp (16), and decreases in the rates of synthesis of ribosomal proteins (1). When thermal stress was applied to these cultures at different times during growth, the levels of thermal induction attained were closely related to growth stage, supporting the notion that developmental and thermal induction of this stress stimulon has common control elements (25). Here we demonstrate that five stress stimulons are developmentally controlled, and we suggest that certain stress response regulatory systems either allow the cell to adapt to developmental stresses that occur during development or directly control the developmental program.
MATERIALS AND METHODS
Cultivation conditions and sample preparation.
Cultures of S. coelicolor J1501 (hisA1 uraA1 strA1 pgl SCP1− SCP2−) were grown in liquid minimal medium at 30°C (25). Samples (1 ml) of 17-h cultures were radiolabeled for 60 min or were stress induced, i.e., either treated with 100 μg of pristinamycin I (P1) per ml, 0.5 M NaCl, and 4% ethanol or temperature shocked (heat shift from 30 to 37°C, cold shift from 30 to 12°C), and simultaneously radiolabeled with 100 μCi of 35S-labeled methionine-cysteine labeling mix (ICN Biomedicals) for 60 min. The mycelia were collected by centrifugation, washed twice with TA disruption buffer containing Tris (pH 7.5), 100 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol (DTT), 0.1% Triton, 10% glycerol, and protease inhibitor cocktail (Complete; Boehringer), resuspended in 500 μl of TA buffer, and disrupted in a Bead Beater using two 30-s treatments in the presence of 500 μg of zirconium beads. Cell debris was removed by centrifugation in a Microfuge (20 min, 13,000 rpm, 4°C). Proteins were precipitated by the addition of 4 volumes of acetone, incubated overnight at 80°C, and centrifuged in a Microfuge (30 min, 13,000 rpm, 4°C). The pellet was resuspended in sample solubilization buffer containing 3 g of urea, 0.2 g of CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 80 mg of DTT, 275 μl of the carrier ampholytes 3-10 (Oxford GlycoSystems, Oxford, United Kingdom), and 2.3 ml of H2O.
2D gel electrophoresis.
Radiolabeled proteins (ca. 106 dpm of trichloroacetic acid-precipitable protein) were separated on high-resolution 2D gels (Investigator System; Oxford GlycoSystems). Isoelectric focusing was carried out using Millipore tubes (1 mm, inside diameter; 26 cm in length). The solutions used to pour the isoelectric focusing gel contained 6 ml of Millipore isoelectric focusing gel stock solution, 360 μl of 3-10 ampholytes, and 40 μl of 8.5-10 ampholytes (Pharmalyte; Pharmacia Biotech). Isoelectric focusing was carried out at 16,000 V · h. In the second dimension, proteins were separated on vertical 12.5% gels (Duracryl; 30% acrylamide, 0.8% Bis; Oxford GlycoSystems). The gels were fixed in 40% methanol–10% acetic acid and dried without staining.
Image analysis.
Images were recorded on X-ray films (Curix; AGFA) using two different exposure times. The film density scale was calibrated by a set of step wedges containing known numbers of disintegrations per minute (dpm) per square centimeter and exposed together with gels to be analyzed. The sensitivity range of the film was expanded by merging two different exposures of a single gel. The films were scanned using a Molecular Dynamics laser scanner and processed with pdQuest gel analysis software (30). Under these conditions, the electropherogram was able to reliably resolve more than 1,000 protein spots.
Normalization.
The radioactive spot intensity was expressed in parts-per-million (ppm) units defined as the ratio of its integrated volume dpm in a spot to the total dpm applied to the gel multiplied by 106. Stress inducers caused a decrease in the overall protein biosynthesis. Therefore, induction or repression of synthesis of an individual protein was not absolute but was relative to the overall rates of protein biosynthesis.
Molecular mass and isoelectric point calibration.
2D gels were calibrated by comigrating radiolabeled S. coelicolor extracts with standards of known Mr and pI (Bio-Rad). The pI and Mr values of individual radiolabeled spots were interpolated using pdQuest software.
Database organization.
The developmental database recorded changes in a culture radiolabeled at 16 different time points (12, 16, 20, 24, 26, 28, 30, 32, 36, 40, 44, 48, 54, 60, 66, and 72 h; Fig. 1) (14). Each of these samples was run in triplicate (“gelsets”), thus generating 48 gels (16 gelsets). The final comprehensive reference gel contained 1,238 spots, each represented in a developmental database which recorded its rates of synthesis at the 16 time points.
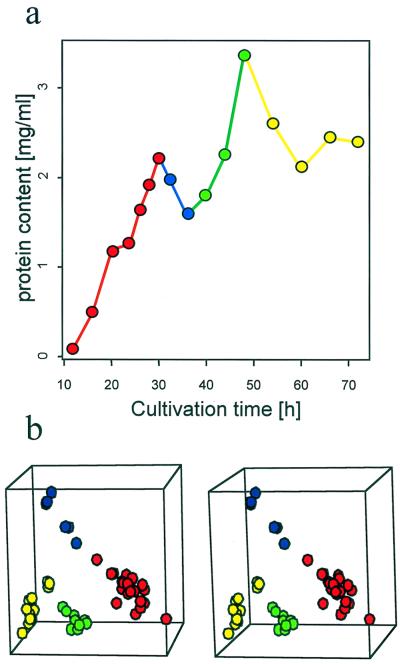
FIG. 1.
Principal component analysis identified four distinct patterns of expression corresponding to developmental stages. (a) S. coelicolor J1501 was cultured in a defined liquid minimal medium (MG) (14). Samples were taken at short intervals (circles), pulse-radiolabeled for 60 min, and analyzed by using 2D gels. Colors indicate time points representing different developmental phases (see panel b): RG1 (red), T (blue), RG2 (green), or S (yellow). (b) PCA of gelsets which record gene expression during the S. coelicolor growth curve (i.e., panel a). This 3D representation of the first three principal components reflected 56% of the variability contained in the experimental set. Gelsets corresponding to the time points in panel a are indicated using the same colors. PCA was done using a representative set of 150 “high-quality spots,” as defined by Garrels (9), so that the gel pattern correlation matrix of the selected set was the same as that of the set including all spots. The number of spots was minimized until the correlation matrix of the selected set diverged significantly from the correlation matrix of the full set. This method allowed us to define the minimal set of spots capable of characterizing the features of the whole set. Some readers may require a stereoviewer to fuse the images. The S script used for drawing the stereoplot was obtained from B. Carr (http://www.galaxy.gmu.edu/∼dcarr/).
The stress database recorded the effects of five different stress inducers applied at 17 h, a time when developmentally programmed changes were minimal. Gels from each series of experiments describing development or the five stress responses (before and after induction) were grouped to form individual “matchsets.” An artificial “reference gel” was created composed of all spots which appeared in the gels of a matchset. Each spot in the reference gel was represented in a list indicating the identification number, apparent molecular weight, pI, and radioactive intensity in each of the member gels of the matchset. At least three different cultures were analyzed in a control or stress set for each shock induction.
To identify proteins whose synthesis was stress induced (“stress proteins) in the developmental database, all five stress matchsets and one developmental matchset were combined to one “high-level” matchset, connecting all spots recorded in all reference gels of all matchsets. Each spot in the reference gel of the high-level matchset thus represented its ppm value at 16 points of the developmental database together with the ppm values before and after stress treatment. Stress-induced proteins were thus automatically identified in the developmental set.
Cluster analysis.
Similarity among the developmental profiles of 136 stress protein spots was analyzed by clustering using a dissimilarity matrix calculated from correlation coefficients (ri,j) among all of the developmental patterns and slopes among adjacent points in the profile. The pairwise inverted correlations (1 − ri,j) served as input for an agglomerative nesting clustering algorithm (17), using the group average method of clustering. The limits of the clusters were identified by visual inspection. The structure of clusters was confirmed, and the level of cluster separation was defined by comparison with the results of divisive clustering (using an algorithm for partitioning around medoids).
A second, more-inclusive distance measure employed the concept of mutual information (18). This approach quantifies the reduction of uncertainty of one random variable given knowledge about another random variable. In our case, normalized mutual information was used as a measure of the extent to which knowledge about the rate of synthesis of one protein reduced the uncertainty about the rate of synthesis of another given protein. Mutual information between two profiles was calculated from the information entropy of the individual profile entropies as described previously (20). In order to calculate the information entropy of each spot profile, the series were transformed by binning the normalized expression levels into 10 discrete equidistant levels. The mutual information M(i,j) of each pair was normalized to the maximal entropy of each of the contributing spot developmental profiles [M(i,j)nrm]. The equation 1 − M(i,j)nrm was used to calculate the distance matrix used for clustering. Algorithms for agglomerative nesting clustering and partitioning around medoids were applied.
RESULTS
S. coelicolor developmental and stress-induced protein databases.
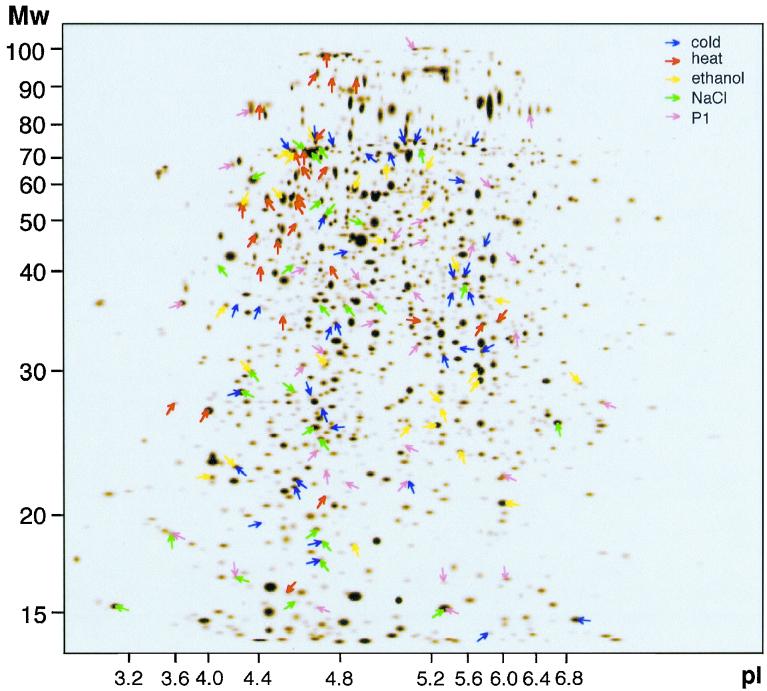
We have previously presented a quantitative database that records changes in the rates of synthesis of 1,238 S. coelicolor proteins during development (30). The database describes 2D gel profiles of a culture pulse-labeled with [35S]methionine-[35S]cysteine at 16 different time points (gelsets) representing the developmental program (Fig. 1). An artificial reference gel (Fig. 2) containing all of the proteins detected (1,238 spots) was linked to a database that recorded their relative rates of synthesis throughout development.
FIG. 2.
An artificial reference gel of 1,238 protein spots in the S. coelicolor proteome database. Many protein spots synthesized during development were induced by one of five different stress treatments (marked with different colors). Each spot in the gel is reflected in the proteome database as its relative levels of incorporation at 16 different time points during development. Altogether, 136 stress-induced (defined as statistically significant and induced more than 2.5-fold) proteins could be identified in the developmental database.
Developmental stages.
The S. coelicolor developmental system was characterized by multiphasic growth kinetics in liquid medium (RG1, T, RG2, and S; Fig. 1). Developmental stages could be identified by their characteristic 2D gel spot density patterns using principal component analysis (PCA). This standard statistical method begins by representing the combination of all (n) spot intensities defining one gel pattern as a point in a conceptual n-dimensional space (i.e., one dimension is assigned to each spot). PCA is designed to reduce this mathematically defined multidimensional space into two to three visually perceptible dimensions, thereby providing a model representing a maximal amount of database information in a form that can be intuitively understood. PCA can thus be viewed as the best projection of this multidimensional space to the visually perceptible 3D space, preserving the original distribution of points and therefore the original similarity among corresponding gelsets. Thus, the information content in each of the gelsets representing 16 time points (48 gels) was assigned to a point in a 3D space (i.e., three principal components) (Fig. 1b).
The spatial arrangement of the points representing gelsets in the derived space indicated how the corresponding patterns of gene expression related to each other. From a simple inspection of the plot (Fig. 1b) it was obvious that the gel patterns were arranged into four distinct groups. Thus, PCA documented the patterns of gene expression defining four developmental stages. The significance of this statistical deduction was independently confirmed by the fact that these stages corresponded perfectly to the four phases of the growth curves: RG1, T, RG2, and S. The degree of separation depends on the level of dissimilarity among the labeled protein intensity patterns. Therefore, the distance between groups of points in the 3D view of the first three principal components indicates rapid change in the average protein pattern during the transition from one stage to another (Fig. 1). This implies switches in regulatory systems that occur at the beginning and/or end of each phase.
Stress stimulons.
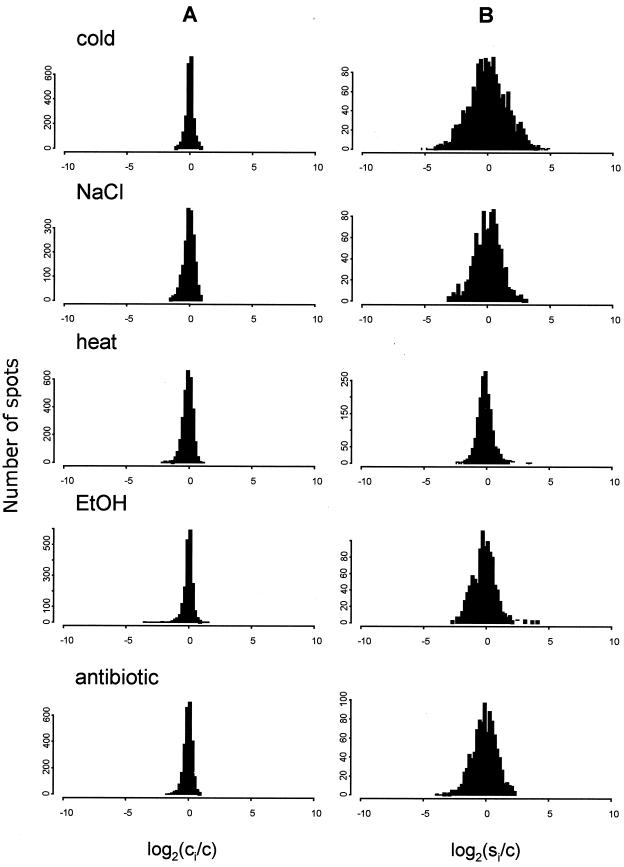
S. coelicolor cultures were subjected to five different stress treatments: cold, heat, ethanol, salt, and P1, a protein synthesis inhibitor. Changes in the rates of synthesis of proteins in response to a certain stress (note that this labeling time was chosen based on kinetic studies showing that most changes in heat shock gene expression in S. coelicolor take place in the first hour [25]) were demonstrated as histograms comparing the distribution of ratios between individual spot intensity before (Fig. 3A) and after (Fig. 3B) stress application. The width of the distribution of control samples (Fig. 3A) represented the random experimental error. In response to stress, the dispersion in both directions increased symmetrically to reflect roughly equivalent numbers of proteins similarly induced or repressed. Synthesis of the majority of proteins was significantly increased or decreased by stress (defined by the t test; P = 0.05): 53% by cold shock, 13% by heat shock, 30% by ethanol shock, 30% by NaCl shock, and 22% by P1 shock. Synthesis of 17 proteins was induced by two different stress conditions. Only one was induced by three different stresses. None were induced by more than three stresses.
FIG. 3.
Stress-induced changes in gene expression. Cultures were subjected to stress during RG1 phase (17 h) preceding major developmental changes (Fig. 1a). Sets of control and induced samples were resolved by 2D gel electrophoresis. Images of electropherograms were processed, data sets representing relative rates of synthesis (in ppm) of each protein spot in the control and stress samples were created for each of the inductions, and the data sets were then analyzed statistically. Histograms of the distribution of logarithmic ratios between the spot ppm average in control samples (A) and the spot ppm after different stress treatments (B) are shown. For each protein spot the base 2 logarithm of the ratio between its induced or repressed PPM and the average ppm value in control samples was calculated. The logarithm was used to numerically equalize rates of induction and repression. c, average spot ppm in control samples; ci, spot ppm in control samples; si, spot ppm in stress samples.
Since this study did not indicate discrete subpopulations of proteins whose synthesis was significantly induced or repressed, an arbitrary decision was made to focus on a subgroup which displayed increases in expression that were both statistically significant and greater than 2.5-fold. To analyze stress protein expression during development, 136 protein spots representing the combined stress stimulon (35 cold, 27 heat, 26 ethanol, 28 NaCl, and 35 P1 shocks) were identified in the S. coelicolor database (Fig. 2).
Correlation of specific stress stimulons with developmental patterns of expression.
Hierarchical cluster analysis is a standard statistical method used to visualize similarities within groups of objects, a finding most familiar to molecular geneticists as a tool commonly employed to represent sequence relationships within gene families as a tree. Pairwise comparisons of the objects are used to define the length and thereby the arrangement of branches. The combined branch lengths separating two objects reflect their degree of similarity; clustering of branch points in the tree indicates that they have a shared feature.
We have applied hierarchical clustering algorithms to visualize possible relationships in patterns of developmental gene expression as branches of a tree. Groups of proteins with similar patterns of developmental expression would be identified as those that are closely linked and branching from a localized region of the tree. Coregulated proteins identified in this manner are therefore likely to have related functions or be under the control of a common regulatory mechanism.
Cluster analysis identified 10 different branch clusters (CL1 to CL10; Fig. 4A), each defining an average developmental expression profile (Fig. 4C). Visual inspection suggested synexpression groups were synthesized primarily during one of the four stages: early growth (CL3 and CL5), transition phase (CL4, CL6, and CL7), both early and transition phases (CL10), or stationary phase (CL1 and CL9) (CL8 had few members and was not analyzed). Some of the stress shock stimulons were enriched in certain developmental expression clusters (Fig. 4D). This indicated that particular stress regulatory systems were an integral part of the developmental program.
FIG. 4.
Stress proteins classified into developmentally coregulated families. Two kinds of cluster analysis were used to analyze a “combined-stress stimulon” that included 136 spots whose relative intensity increased in response to one of the five stresses. Correlation (A) and mutual-information-based (B) clustering were used to compare the developmental expression profiles of individual proteins, thus identifying synexpression groups. (A) Correlation-based cluster analysis identified 10 clusters of stress proteins (CL1 to CL10) with similar expression patterns. The partitioning was confirmed by divisive clustering showing that the kinetic patterns were best separated into 10 groups having a similar spot membership (not shown). (B) Mutual-information-based cluster analysis identified four clusters of stress proteins (CL1* to CL4*) potentially in the same regulon that may have different expression profiles (see text) yet be controlled by the same regulatory protein. (C) Patterns of expression in developmentally coregulated families. The bar graphs represent the average normalized spot profiles of clusters corresponding to those in panel A. The bars are arranged from left to right in the order in which the samples were pulse-labeled at the following times postinoculation: 12, 16, 20, 24, 26, 28, 30, 32, 36, 40, 44, 48, 54, 60, 66, and 72 h. Each row in the box plot above the average profile represents the spot profile of one particular protein. The relative protein dpm value is expressed as shades of gray. (D) Relative abundance of different stress regulons in clusters CL1 to CL10 and clusters CL2* to CL4*. The bars are arranged from left to right in the following order: cold, heat, ethanol, salt, and antibiotic P1. Cold (C), heat (H), and salt (S) shock proteins were enriched in certain developmental clusters. The heights of the bars represent the percentages of the proteins in the given stimulon present in each cluster. Notable is the correlation in percentages between salt and heat, especially in clusters CL5 and CL7, which are associated with the initiation of growth at the beginning and in the hiatus of the T phase. Cold shock proteins, on the other hand, were associated with the phases of retarded growth later in the T and S phases (CL1 and CL4). CL2*, defined by mutual-information-based clustering, which groups coordinately regulated patterns, is also enriched in proteins induced by salt and heat stress. CL3* and CL4* are nonspecific with respect to individual stress conditions. CL1* was not considered as it was too small.
Almost half of all heat or salt shock proteins were distributed in one of two branches of the tree representing synexpression groups CL5 and CL7. CL5 contained proteins synthesized during an early stage of development. CL7 was characterized by a peak of synthesis at the beginning of the T phase; 48% of all heat shock proteins exhibited maximal levels of synthesis at that time. Maximal rates of cold shock protein synthesis (CL4) occurred 2 h later.
Cold shock proteins were distributed primarily in CL1 and CL4. CL1 proteins were mostly synthesized during stationary phase; their levels of synthesis during exponential growth were very low. Synthesis levels of CL4 proteins were strongly associated with the phase of retarded growth (T phase), at which 30% of all cold shock proteins exhibited maximal rates of synthesis. They were not synthesized during the other phases of growth. A large subgroup of cold shock proteins was therefore associated with periods of retarded growth (T or S phase). The remaining clusters either had few members or did not show any apparent association with one of the stresses.
Networks of developmentally regulated stress stimulons identified by “mutual-information”-based cluster analysis.
Due to the complex, interactive requirements of microbial regulatory systems, genes belonging to the same regulon are often subject to multiple control systems. As a result, genes partially controlled by the regulatory protein that defines the regulon often display different developmental or stimulus-response patterns (23). For example, genes in the same regulon may display differences in the onsets or rates of change and, in the extreme, the same regulatory protein may have opposite effects on the expression of different genes. The cluster analysis algorithm described earlier (Fig. 4A) can only detect similar patterns of expression and therefore cannot identify all proteins belonging to the same regulon. Such variable response patterns in proteins controlled by the same regulatory element (therefore in the same regulon) can be identified by “mutual-information”-based cluster analysis (18).
Mutual-information-based analysis of the database confirmed that heat shock and salt shock proteins had similar developmental patterns of synthesis. Proteins fell into two major developmental clusters (CL2* and CL3*; Fig. 4B) that included 104 protein profiles (76%). This suggested the influence of two principal regulatory mechanisms (networks) during development. Cluster 2* (24%) was dominated by salt and heat shock proteins (Fig. 4D, bottom), thus confirming their similar regulatory patterns (Fig. 4D, top) detected by cluster analysis. Cluster 3* (52%) contained all stress stimulons in proportional amounts; other clusters contained too few members for further analysis.
Inspection of the developmental profiles of the members of clusters did not reveal any predominant patterns. This illustrated that these mutual information analyses were, as expected, able to identify proteins having different developmental profiles as belonging to a smaller number of coregulated families.
DISCUSSION
Our study of a series of developmental stages and stress responses allowed us to identify coordinately synthesized gene products and utilize statistical approaches that can be generally applied to biological response systems or developmental processes. While most global studies of gene expression initially focus on individual gene products, the limited resolution, identification, quantification, and reproducibility of spots on 2D gels continue to pose serious technical problems. To avoid these limitations in quantifying individual spot density, we analyzed representative trends within populations of proteins. PCA was used to compare global expression patterns during development, and cluster analysis identified groups of cosynthesized proteins.
Developmental stages identified by PCA.
The complex patterns of global gene expression associated with developmental stages was mathematically defined as four stages by PCA using a statistically representative set of 150 spots (Fig. 1). The significance of this statistical deduction was independently confirmed by the fact that these stages correspond perfectly to the four phases of the growth curve (25): RG1, T, RG2, and S (Fig. 1a). In principle, similar analysis of developmentally blocked mutants might define their stage of arrest. PCA will undoubtedly be useful in many other biological systems to define expression patterns associated with disease, nutritional conditions, responses to stimuli, genetic variation, etc.
Similarities in patterns of gene expression: stress modulons.
As in numerous other bacteria, stress treatments induced synthesis of many S. coelicolor proteins. However, unlike descriptions of other bacteria, statistical analysis of our data showed that stress stimulons could not be identified as discrete groups of highly induced proteins. Diverse shock treatments, including heat, cold, ethanol, antibiotic, or salt produced global changes involving both induction and repression of the majority of cellular gene products. The normalized rates of changes of all proteins defined a symmetric bell-shaped curve that did not identify any discrete subgroups of proteins with either decreased or increased rates of synthesis. This indicated that most stress-induced changes were under the control of promoters having diverse activities and/or under the control of multiple regulatory genes. Since these groups of proteins were apparently induced or repressed by multiple systems of regulation which transmit environmental changes, they will be referred to as “stimulons” or “inhibulons.”
We define stress inhibulons here as groups of protein spots whose relative rates of synthesis decreased in response to a particular stress. It is curious that these proteins form a group that is comparable in size to stress stimulons. The developmental regulation of such global “inhibulons” has been largely ignored, except in the case of the stringent response induced by starvation in E. coli (4). The molecular mechanisms that effect repression of the diverse inhibulons we observed will be the subject of future studies. An arbitrary decision was made to study only “stress stimulons,” i.e., proteins having statistically significant increases after induction.
It is commonly believed that stress regulons are largely overlapping and reflect the limited number of basic signaling and response systems (16). In B. subtilis, a “general stress response” elicited by heat, salt, and ethanol is controlled by one sigma factor (13, 14). However, in S. coelicolor, independent groups of proteins were induced in response to these three treatments. Of the 105 proteins belonging to one of these stimulons, very few were induced by more than one stress. Transcriptional regulation of large groups of genes is commonly achieved by reprogramming the RNA polymerase using alternative sigma subunits or global transcriptional regulatory proteins. The fact that multiple SigB paralogs have been identified in the S. coelicolor genome database (Sanger Centre) lends support to the idea that, in contrast to B. subtilis and E. coli, at least some individual stress responses in S. coelicolor may be independently controlled by specific sigma factors belonging to the same family. Further specificity of the response is suggested by the observation that there are at least three SigB-like sigma factors that respond to salt induction under the conditions described here (P. H. Viollier, G. Kelemen, M. J. Buttner, and C. J. Thompson, unpublished data). The existence of multiple stress response regulatory systems implies different molecular signals sensed by these regulons.
In E. coli and B. subtilis, denatured proteins are sometimes considered to be the common stress signal generated by diverse physical stress effectors, including heat, salt, and ethanol (12, 21). Nevertheless, the fact that these treatments induced different sets of proteins in S. coelicolor might result from unique primary signals. This could reflect subsets of proteins differentially susceptible to heat, osmotic, or ethanol denaturation because of their intracellular location or inherent stability characteristics. Alternatively, these stress treatments may generate different low-molecular-weight alarmones. The specialization of the S. coelicolor stress response systems may reflect the need to specifically control expression of individual stress stimulons during certain stages of the developmental program.
Stress stimulons form synexpression groups during development.
Most of these stress proteins were developmentally controlled. In general, the highest levels of synthesis of individual proteins were associated with one of the four developmental phases of the culture. Ten basic patterns of stress protein synthesis were identified by cluster analysis. Some of these clusters were enriched for proteins also associated in the same stress stimulons.
Cluster analysis proved that salt and heat shock stress stimulons were developmentally coregulated, notably during the transition phase. Both salt and heat shock stress regulators may be required for construction or repair during this stage.
We suggest that evolutionary pressures could have adopted these stress regulatory systems as parts of a developmental program associated with abrupt changes in physiology. To test this hypothesis, we are exploring the prediction that mutants defective in heat, salt, and cold shock stress responses may be developmental mutants (bald). These concepts have focused our interest on SigB-like sigma factors as possible global transcriptional regulators of development in Streptomyces spp. The other testable prediction, that developmental mutations affect one or more of these stress response systems, has now been verified in experiments demonstrating altered expression of a sigB-like gene in the bldD mutant (Viollier et al., unpublished data).
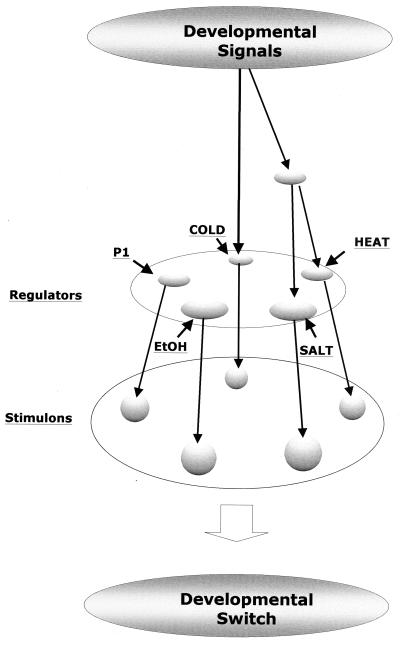
In conclusion, cluster analysis suggested potential interactions among developmental genes and stress stimulons that gave an initial glimpse of a regulatory network (Fig. 5). The network's structure could help to define the hierarchy of underlying regulatory mechanisms and corresponding genes and thus ultimately determine whether different patterns of gene expression have functional significance (23). Future work will focus on inactivating genetic elements such as sigma factors that might control critical branch points within this network. Completion of the S. coelicolor genome sequence will facilitate identification of proteins having conserved developmental patterns of expression that should establish biologically meaningful correlations between functional and kinetic groups.
FIG. 5.
An integrated network of developmental and stress-induced proteins in S. coelicolor. 2D gel patterns indicated that S. coelicolor cultures underwent a developmental program involving four stages (Fig. 1). Developmental signals controlled the family of proteins (see Fig. 4; indicated here by arrows) which were also induced by stress treatments (heat, NaCl, cold, ethanol [EtoH], or P1 shock) (short arrows). Stress stimulons contained unique nonoverlapping groups of proteins (Stimulons; see Fig. 2), presumably reflecting the role of independent regulators. Subsets of heat, salt, and cold shock proteins had similar patterns of synthesis as a function of development (see Fig. 4), suggesting that corresponding stress regulators were functional during growth. In addition, the observation that many members of two different stress stimulons (heat and salt shock) had related patterns of synthesis during development (Fig. 4C and D) suggested that their corresponding stress-specific regulators were under the control of a common signal from a higher-level regulator. The existence of this regulatory cascade involving stress stimulons suggests that corresponding regulatory systems may play a role in mediating developmental switches. There may also be alternative developmental pathways that bypass these stress circuits (not shown).
ACKNOWLEDGMENTS
We thank Urs Jenal, Kien Nguyen, and Harley McAdams for comments on the manuscript.
This work was supported by grants from the Swiss National Science Foundation Grant (NF31000039699) and the Czech Ministry of Education (no. 310/96/k102).
REFERENCES
- 1.Blanco G, Rodicio M R, Puglia A M, Méndez C, Thompson C J, Salas J A. Synthesis of ribosomal proteins during growth of Streptomyces coelicolor. Mol Microbiol. 1994;12:375–385. doi: 10.1111/j.1365-2958.1994.tb01027.x. [DOI] [PubMed] [Google Scholar]
- 2.Bucca G, Ferina G, Puglia A M, Smith C P. The dnaK operon of Streptomyces coelicolor encodes a novel heat-shock protein which binds to the promoter region of the operon. Mol Microbiol. 1995;17:663–674. doi: 10.1111/j.1365-2958.1995.mmi_17040663.x. [DOI] [PubMed] [Google Scholar]
- 3.Bucca G, Hindle Z, Smith C P. Regulation of the dnaK operon of Streptomyces coelicolor A3(2) is governed by HspR, an autoregulatory repressor protein. J Bacteriol. 1997;179:5999–6004. doi: 10.1128/jb.179.19.5999-6004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 5.Chater K F. Taking a genetic scalpel to the Streptomyces colony. Microbiology. 1998;144:1465–1478. doi: 10.1099/00221287-144-6-1465. [DOI] [PubMed] [Google Scholar]
- 6.De Crecy-Lagard V, Servant-Moisson P, Viala J, Grandvalet C, Mazodier P. Alteration of the synthesis of the clp ATP-dependent protease affects morphological and physiological differentiation in streptomyces. Mol Microbiol. 1999;32:505–517. doi: 10.1046/j.1365-2958.1999.01364.x. [DOI] [PubMed] [Google Scholar]
- 7.Doull J L, Ayer S W, Singh A K, Thibault P. Production of a novel polyketide antibiotic, jadomycin B, by Streptomyces venezuelae following heat shock. J Antibiot. 1993;46:869–871. doi: 10.7164/antibiotics.46.869. [DOI] [PubMed] [Google Scholar]
- 8.Eisen M B, Spellman P T, Brown P O, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrels J. The QUEST system for quantitative analysis of two-dimensional gels. J Biol Chem. 1989;264:5269–5282. [PubMed] [Google Scholar]
- 10.Grandvalet C, de Crecy-Lagard V, Mazodier P. The ClpB ATPase of Streptomyces albus G belongs to the HspR heat shock regulon. Mol Microbiol. 1999;31:521–532. doi: 10.1046/j.1365-2958.1999.01193.x. [DOI] [PubMed] [Google Scholar]
- 11.Grandvalet C, Rapoport G, Mazodier P. hrcA, encoding the repressor of the groEL genes in Streptomyces albus G, is associated with a second dnaJ gene. J Bacteriol. 1998;180:5129–5134. doi: 10.1128/jb.180.19.5129-5134.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross C. Function and regulation of the heat shock proteins. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1382–1399. [Google Scholar]
- 13.Hecker M, Schumann W, Völker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 14.Hecker M, Volker U. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the sigmaB regulon. Mol Microbiol. 1998;29:1129–1136. doi: 10.1046/j.1365-2958.1998.00977.x. [DOI] [PubMed] [Google Scholar]
- 15.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1497–1512. [Google Scholar]
- 16.Holt T G, Chang C, Laurent-Winter C, Murakami T, Garrels J I, Davies J E, Thompson C J. Global changes in gene expression related to antibiotic biosynthesis in Streptomyces hygroscopicus. Mol Microbiol. 1992;6:969–980. doi: 10.1111/j.1365-2958.1992.tb02163.x. [DOI] [PubMed] [Google Scholar]
- 17.Kaufman L, Rousseeuw P. Finding groups in data: an introduction to cluster analysis. New York, N.Y: John Wiley & Sons, Inc.; 1990. [Google Scholar]
- 18.Liang S. REVEAL, a general reveres engineering algorithm for inference of genetic network architectures. Pacific Symp Biocomputing. 1998;3:18–29. [PubMed] [Google Scholar]
- 19.Matin A. The molecular basis of carbon-starvation-induced general resistance in Escherichia coli. Mol Microbiol. 1991;5:19–22. doi: 10.1111/j.1365-2958.1991.tb01819.x. [DOI] [PubMed] [Google Scholar]
- 20.Michaels G, Carr D, Askenazi M. Cluster analysis and data visualization of large scale gene expression data. Pacific Symp Biocomputing. 1998;3:42–53. [PubMed] [Google Scholar]
- 21.Mogk A, Volker A, Engelmann S, Hecker M, Schumann W, Volker U. Nonnative proteins induce expression of the Bacillus subtilis CIRCE regulon. J Bacteriol. 1998;180:2895–2900. doi: 10.1128/jb.180.11.2895-2900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muffler A, Barth M, Marschall C, Hengge-Aronis R. Heat shock regulation of ςS turnover: a role for DnaK and relationship between stress responses mediated by ςS and ς32 in Escherichia coli. J Bacteriol. 1997;179:445–452. doi: 10.1128/jb.179.2.445-452.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neidhardt F C, Savageau M A. Regulation beyond the operon. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1310–1324. [Google Scholar]
- 24.Niehrs C, Pollet N. Synexpression groups in eukaryotes. Nature. 1999;402:483–487. doi: 10.1038/990025. [DOI] [PubMed] [Google Scholar]
- 25.Puglia A M, Vohradsky J, Thompson C J. Developmental control of the heat-shock stress regulon in Streptomyces coelicolor. Mol Microbiol. 1995;17:737–746. doi: 10.1111/j.1365-2958.1995.mmi_17040737.x. [DOI] [PubMed] [Google Scholar]
- 26.Salah-Bey K, Blanc V, Thompson C J. Stress-activated expression of a Streptomyces pristinaespiralis multidrug resistance gene (ptr) in various Streptomyces and Escherichia coli. Mol Microbiol. 1995;17:1001–1012. doi: 10.1111/j.1365-2958.1995.mmi_17051001.x. [DOI] [PubMed] [Google Scholar]
- 27.Servant P, Mazodier P. Heat induction of hsp18 gene expression in Streptomyces albus G: transcriptional and posttranscriptional regulation. J Bacteriol. 1996;178:7031–7036. doi: 10.1128/jb.178.24.7031-7036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tobisch S, Zuhlke D, Bernhardt J, Stulke J, Hecker M. Role of CcpA in regulation of the central pathways of carbon catabolism in Bacillus subtilis. J Bacteriol. 1999;181:6996–7004. doi: 10.1128/jb.181.22.6996-7004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.VanBogelen R A, Abshire K Z, Pertsemlidis A, Clark R L, Neidhardt F C. Gene-protein database of Escherichia coli K-12. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: ASM Press; 1996. pp. 2067–2202. [Google Scholar]
- 30.Vohradsky J, Li X M, Thompson C J. Identification of procaryotic developmental stages by statistical analyses of two-dimensional gel patterns. Electrophoresis. 1997;18:1418–1428. doi: 10.1002/elps.1150180817. [DOI] [PubMed] [Google Scholar]