Abstract
The stringent response in Staphylococcus aureus is mediated by the nucleotide guanosine pentaphosphate, whose synthesis is catalyzed by the product of the rel gene. We report here that the rel gene is essential for the in vitro growth of S. aureus, distinguishing it from all other bacteria tested for this requirement.
The stringent response is a pleiotropic physiological reaction exhibited by bacteria in response to amino acid deprivation or to inhibition of tRNA amino acylation. The hallmark of the stringent response is an abrupt cessation of stable RNA synthesis, but it also involves the stimulation of some genes involved in various anabolic functions and of some stationary-phase-specific genes (3). Limitation for other nutrients elicits a similar response, so that the stringent response has come to be referred to as a response to all nutrient limitations. The stringent response is usually mediated by the nucleotides guanosine 3′,5′-bis(diphosphate) (ppGpp) and guanosine 3′-diphosphate,5′-triphosphate (pppGpp). In Staphylococcus aureus, only pppGpp accumulates (4). The two nucleotides appear to be functionally interchangeable and are collectively known as (p)ppGpp.
Most work on the stringent response has been performed with Escherichia coli, which possesses two (p)ppGpp synthetases encoded by the relA and spoT genes (7, 17). The RelA enzyme is required for the rapid increase in (p)ppGpp synthesis following amino acid starvation (or inhibition of amino acylation of tRNA). The homologous SpoT enzyme is required for maintenance of basal levels of (p)ppGpp and is responsible for (p)ppGpp accumulation following nutrient limitations that do not involve amino acid starvation. SpoT is a bifunctional enzyme that also catalyzes the degradation of (p)ppGpp (1, 12). E. coli strains with relA and spoT deleted lack detectable (p)ppGpp and exhibit a pleiotropic phenotype (7, 17) that includes attenuation in pathogenic isolates (D. R. Gentry, A. P. Bryant, I. Critchley, and A. Marra, submitted for publication).
Genomic sequencing results reveal a trend in which gram-negative organisms possess two synthetases similar to what is seen in E. coli while most gram-positive organisms appear to have a single gene, generally called rel, which encodes a bifunctional enzyme responsible for both (p)ppGpp synthesis and degradation (2, 9, 10, 14). Studies have shown that a Bacillus subtilis rel deletion mutant lacks detectable (p)ppGpp and has difficulty in responding to stress (15). Also, a Streptococcus pneumoniae rel mutant exhibits attenuation in a murine respiratory tract infection model and a gerbil otitis media model (R. Greenwood, A. P. Bryant, A. Marra, K. A. Ingraham, D. Holmes, and D. R. Gentry, submitted for publication). These observations predict that similar rel deletions in gram-positive human pathogens may result in decreased virulence and poor stress responsiveness. This paper reports our attempts to isolate a rel deletion mutant of the important human pathogen S. aureus. In contrast to other gram-positive organisms studied to date, in S. aureus rel was found to be essential for bacterial viability.
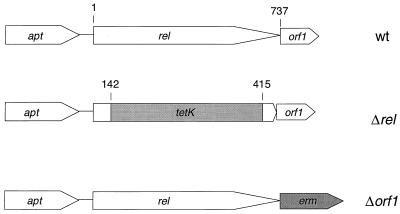
The S. aureus rel gene was identified from sequence data generated during an effort to sequence the S. aureus genome. As with most gram-positive bacteria, upstream of rel is the apt gene, encoding adenine phosphoribosyltransferase, while downstream is a highly conserved gene of unknown function called orf1. The rel gene product is identical to the predicted Rel sequence from the previously published S. aureus rel gene (5). The Rel enzyme is bifunctional, as indicated by the ability of a plasmid carrying the rel gene to complement the growth defects of E. coli mutants defective in (p)ppGpp synthesis or degradation (Greenwood et al., submitted). A plasmid-borne rel insertion-deletion construct was isolated in the following manner. A 2.6-kb SalI-EcoRI fragment containing the rel locus was inserted into the SalI-EcoRI sites of pBluescript. The internal Mlu-NdeI fragment (1 kb) in the rel gene was replaced by the S. aureus tetK gene from plasmid pCW59 (16). Thus, the final construct contained upstream sequence-tetK-resistance marker-downstream sequence (3.9 kb) cloned into pBluescript (shown schematically in Fig. 1). So that a counterscreen could be used to detect double-recombination allelic replacement events following transformation, the 3.9-kb SalI-EcoRI rel deletion-insertion fragment was cloned into pBluescriptErm, generating the plasmid pEKerm. Plasmid pBluescriptErm is pBluescript with an erythromycin resistance cassette inserted into the NaeI site.
FIG. 1.
Schematic representation of the rel region on the S. aureus chromosome. (Top) Wild-type (wt) orientation of the apt, rel, and orf1 genes. The numbers indicate the first and last codons of the rel gene. (Middle) Orientation of the rel gene in the deletion construct. The numbers indicate the codons on the flanks of the tetK insertion; thus, codons 143 through 414 of rel are deleted in the construct. (Bottom) Orientation of the orf1 deletion. This drawing is not to scale.
Plasmid pEKerm was transformed into S. aureus RN4220. A total of five transformants were obtained from tryptic soy agar (TSA; Difco) plates containing 5 μg of tetracycline per ml. Both PCR and Southern hybridization analysis confirmed that all five of the transformants were cointegrants. Because cointegrants contain the insertion-deletion mutation with sufficient flanking sequences to recombine at the rel locus, the deletion mutation should be obtainable via generalized phage transduction if the rel gene is nonessential. An attempt to resolve a cointegrant by φ11 transduction yielded 2,200 Tcr transductants, none of which were Ems, suggesting that it was impossible to resolve the cointegrant and that rel was probably essential for in vitro growth. For nonessential genes, the frequency of cointegrant resolution is typically between 0.5 and 5%. We then sought to determine if we could resolve the cointegrant into a strain with the wild-type gene provided in trans on a plasmid. The rel gene, on a 5.3-kb EcoRI fragment, was cloned into the Cmr plasmid pSK265 to generate plasmid pSKrel. In addition to rel, pSKrel contains part of the downstream gene, orf1, as well as about 2.0 kb of sequence upstream of rel. This plasmid was introduced into S. aureus 8325-4. In this strain it was possible to resolve the cointegrant (26 of 2,000 transductants [1.3%] were Ems Cmr) within the expected frequency range. For 2 of the 26 resolved mutants, the pSKrel plasmid had integrated illegitimately into the chromosome (Cmr). For these two mutants, the natural rel locus was clearly disrupted, as predicted. All the remaining 24 resolved mutants possessed extrachromosomal plasmid pSKrel. PCR (10 of 10 mutants) and sequencing analysis (2 of 2 mutants) confirmed the correct structure of those resolved mutants.
A phage lysate was prepared from one of the resolved mutants and used to transduce the Tcr marker. Transductants were isolated; however, 100% of these (70 of 70) had also inherited the pSKrel Cmr plasmid. Transfer of plasmids via transduction is commonly observed in S. aureus. Thus, it was not possible to dissociate the rel deletion mutation and the rel complementing plasmid. This result further indicated that rel is essential for in vitro growth.
As mentioned above, plasmid pSKrel carries a fragment of orf1. To ensure that the lethality of the rel deletion is not due to polar effects on the expression of this gene, a deletion strain was constructed in which the entire gene was replaced with an erythromycin resistance cassette. Results show that orf1 is not essential, as indicated by the ability to readily isolate allelic replacement mutants. This mutant exhibits no obvious phenotype and has not been studied any further.
S. aureus stands out among the gram-positive organisms by its dependence on rel for growth. This puts S. aureus on the extreme end of the variability of the effect of mutations on (p)ppGpp metabolism. The reason for this is open to speculation. While Rel-related enzymes are the most broadly distributed (p)ppGpp synthetases, other, unrelated enzymes that can catalyze (p)ppGpp synthesis are known. For example, Streptoverticillium morookaensis and some of its relatives produce an extracellular nucleotide 3′-pyrophosphokinase (11). Additionally, polynucleotide phosphorylase from Streptomyces antibioticus catalyzes the synthesis of (p)ppGpp in vitro (8). If another source of (p)ppGpp synthesis were present in S. aureus and if rel is the sole source of (p)ppGpp degradation, the rel gene would be essential because its absence would lead to prohibitively high intracellular levels of (p)ppGpp. A homolog of the Streptoverticillium morookaensis nucleotide 3′-pyrophosphokinase is not present in the several S. aureus genomic sequence databases available, and only Streptomyces antibioticus polynucleotide phosphorylase has been shown to synthesize (p)ppGpp in vitro [and its role in in vivo (p)ppGpp synthesis is unclear]. It therefore seems that if another (p)ppGpp synthetic enzyme exists in S. aureus, it is not related to any enzyme known to have the activity. It is formally possible that the N-terminal 142 amino acids of the S. aureus Rel protein, which is still intact in the deletion construct made, has (p)ppGpp synthetic activity but no degradative activity. Given the lack of residual activity of similar peptides of E. coli SpoT (6), this seems very unlikely.
Based on what is known of the E. coli system (7, 17), a likely cause of the lethality of the rel deletion is some defect in amino acid biosynthesis. Such a defect is not likely to be due to a straightforward amino acid auxotrophy, given that the medium, TSA, used in the experiments described here is likely to contain a full complement of amino acids provided for by the predominant ingredients, tryptone and soytone. This is best shown by the ability of an E. coli ΔrelA ΔspoT strain to grow well in this medium (data not shown). Also, TSA, in our experience, is one of the better media for propagating S. aureus in terms of both growth rate and growth yield. A more complicated amino acid defect, such as amino acid sensitivities, is more likely. An example of an amino acid sensitivity is the inability of E. coli relA mutants to grow in the presence of serine, methionine, and glycine because isoleucine biosynthesis is inhibited under such conditions (13).
TSA could be deficient for some other nutrient whose biosynthesis is under tight (p)ppGpp control. In addition to tryptone and soytone, TSA contains glucose, NaCl, and K2HPO4. It is therefore possible that it is deficient in a key vitamin or nucleobase, given the lack of added vitamins or yeast extract. A defect in the ability to use the carbon sources in TSA is less likely, given the carbohydrates provided by soytone and the added glucose. Finally, a defect in the transport of any nutrient cannot be excluded. It is also possible that rel has a function other than its role in (p)ppGpp metabolism and that it is this function that is essential in S. aureus.
Further work is clearly needed in this area, with a key requirement for a conditional rel mutant. We have constructed a strain in which the rel gene is under control of a regulatable promoter, and our initial finding, i.e., that the strain is not viable in the absence of rel expression, supports our conclusions reported here. Unfortunately, the strain has proven to be unstable, and further refinement is needed before it can be effectively used to address this problem.
Acknowledgments
The work was funded in part by DARPA grant N65236-97-1-5810.
REFERENCES
- 1.An G, Justesen J, Watson R J, Friesen J D. Cloning the spoT gene of Escherichia coli: identification of the spoT gene product. J Bacteriol. 1979;137:1100–1110. doi: 10.1128/jb.137.3.1100-1110.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avarbock D, Salem J, Li L, Wang Z, Rubin H. Cloning and characterization of a bifunctional RelA/SpoT homologue from Mycobacterium tuberculosis. Gene. 1999;233:261–269. doi: 10.1016/s0378-1119(99)00114-6. [DOI] [PubMed] [Google Scholar]
- 3.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W, Riley M, Shaechter M, Umbarger A E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 4.Cassels R, Oliva B, Knowles D. Occurrence of the regulatory nucleotides ppGpp and pppGpp following induction of the stringent response in staphylococci. J Bacteriol. 1995;177:5161–5165. doi: 10.1128/jb.177.17.5161-5165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujimura T, Murakami K. Increase of methicillin resistance in Staphylococcus aureus caused by deletion of a gene whose product is homologous to lytic enzymes. J Bacteriol. 1997;179:6294–6301. doi: 10.1128/jb.179.20.6294-6301.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gentry D R, Cashel M. Mutational analysis of the Escherichia coli spoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol Microbiol. 1996;19:1373–1384. doi: 10.1111/j.1365-2958.1996.tb02480.x. [DOI] [PubMed] [Google Scholar]
- 7.Hernandez V J, Bremer H. Escherichia coli ppGpp synthetase II activity requires spoT. J Biol Chem. 1991;266:5991–5999. [PubMed] [Google Scholar]
- 8.Jones G H, Bibb M J. Guanosine pentaphosphate synthetase from Streptomyces antibioticus is also a polynucleotide phosphorylase. J Bacteriol. 1996;178:4281–4288. doi: 10.1128/jb.178.14.4281-4288.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez-Costa O H, Fernandez-Moreno M A, Malpartida F. The relA/spoT-homologous gene in Streptomyces coelicolor encodes both ribosome-dependent (p)ppGpp-synthesizing and -degrading activities. J Bacteriol. 1998;180:4123–4132. doi: 10.1128/jb.180.16.4123-4132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mechold U, Cashel M, Gentry D, Malke H. Functional analysis of a relA/spoT gene homolog from Streptococcus equisimilis. J Bacteriol. 1996;178:1401–1411. doi: 10.1128/jb.178.5.1401-1411.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muta S, Osoegawa K, Ezaki S, Zubair M, Kuhara S, Mukai J, Dixon R. Streptomyces ATP nucleotide 3′-pyrophosphokinase and its gene. Nucleic Acids Symp Ser. 1992;422:165–166. [PubMed] [Google Scholar]
- 12.Sy J. In vitro degradation of guanosine 5′-diphosphate, 3′-diphosphate. Proc Natl Acad Sci USA. 1977;74:5529–5533. doi: 10.1073/pnas.74.12.5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uzan M, Danchin A. A rapid test for the relA mutation in E. coli. Biochem Biophys Res Commun. 1976;69:751–758. doi: 10.1016/0006-291x(76)90939-6. [DOI] [PubMed] [Google Scholar]
- 14.Wehmeier L, Schafer A, Burkovski A, Kramer R, Mechold U, Malke H, Puhler A, Kalinowski J. The role of the Corynebacterium glutamicum rel gene in (p)ppGpp metabolism. Microbiology. 1998;144:1853–1862. doi: 10.1099/00221287-144-7-1853. [DOI] [PubMed] [Google Scholar]
- 15.Wendrich T M, Marahiel M A. Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol Microbiol. 1997;26:65–67. doi: 10.1046/j.1365-2958.1997.5511919.x. [DOI] [PubMed] [Google Scholar]
- 16.Wilson C R, Skinner S E, Shaw W V. Analysis of two chloramphenicol resistance plasmids from S. aureus: insertional inactivation of Cm resistance, mapping of restriction sites, and constructiont of cloning vectors. Plasmid. 1981;5:245–258. doi: 10.1016/0147-619x(81)90002-0. [DOI] [PubMed] [Google Scholar]
- 17.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]