Abstract
Two putative Methanococcus jannaschii isocitrate dehydrogenase genes, MJ1596 and MJ0720, were cloned and overexpressed in Escherichia coli, and their gene products were tested for the ability to catalyze the NAD- and NADP-dependent oxidative decarboxylation of dl-threo-3-isopropylmalic acid, threo-isocitrate, erythro-isocitrate, and homologs of threo-isocitrate. Neither enzyme was found to use any of the isomers of isocitrate as a substrate. The protein product of the MJ1596 gene, designated AksF, catalyzed the NAD-dependent decarboxylation of intermediates in the biosynthesis of 7-mercaptoheptanoic acid, a moiety of methanoarchaeal coenzyme B (7-mercaptoheptanylthreonine phosphate). These intermediates included (−)-threo-isohomocitrate [(−)-threo-1-hydroxy-1,2,4-butanetricarboxylic acid], (−)-threo-iso(homo)2citrate [(−)-threo-1-hydroxy-1,2,5-pentanetricarboxylic acid], and (−)-threo-iso(homo)3citrate [(−)-threo-1-hydroxy-1,2,6-hexanetricarboxylic acid]. The protein product of MJ0720 was found to be α-isopropylmalate dehydrogenase (LeuB) and was found to catalyze the NAD-dependent decarboxylation of one isomer of dl-threo-isopropylmalate to 2-ketoisocaproate; thus, it is involved in the biosynthesis of leucine. The AksF enzyme proved to be thermostable, losing only 10% of its enzymatic activity after heating at 100°C for 10 min, whereas the LeuB enzyme lost 50% of its enzymatic activity after heating at 80°C for 10 min.
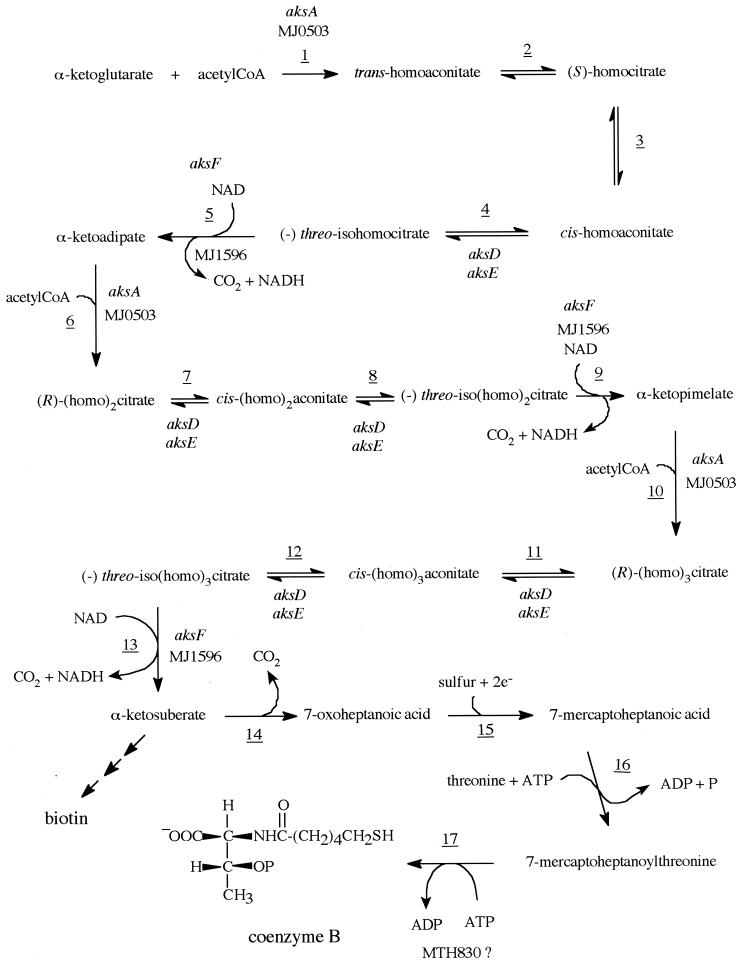
Recent work has established the biochemistry of the thirteen steps involved in the conversion of α-ketoglutarate and acetyl coenzyme A (CoA) to α-ketosuberate (aks) a precursor to coenzyme B (7-mercaptoheptanoylthreonine phosphate) and biotin, in the methanoarchaea (Fig. 1) (4). The gist of these steps consists of the repeated application of the α-ketoacid chain elongation series of reactions to increase the number of methylenes from two, as found in α-ketoglutaric acid, to five, as found in α-ketosuberic acid, the final product of this α-ketoacid chain elongation series of reactions. The α-ketosuberic acid then serves as a precursor to the biosynthesis of the 7-mercaptoheptanoic acid moiety of coenzyme B, as shown in Fig. 1 (15).
FIG. 1.
Biosynthetic pathway for coenzyme B.
Previous work has shown that the protein product of the Methanococcus jannaschii MJ0503 gene aksA, AksA, catalyzes the condensation of α-ketoglutarate and acetyl-CoA to form trans-homoaconitate and the condensation of α-ketoadipate or α-ketopimelate with acetyl-CoA to form, respectively, the (R)-(homo)2citrate[(R)-2-hydroxy-1,2,5-pentanetricarboxylic acid] or (homo)3citrate [(R)-2-hydroxy-1,2,6-hexanetricarboxylic acid] (4). These steps are shown in Fig. 1 as steps 1, 6, and 10. In this biosynthetic pathway there are also three reactions in which homologs of (−)-threo-isocitrate undergo reactions mechanistically like the NAD-dependent isocitrate dehydrogenase reactions to form α-ketoacids (Fig. 1, steps 5, 9, and 13). Based on the expected mechanism of these reactions and the observed stereochemistry of the reaction products, it is very likely that genes homologous to NAD-dependent isocitrate dehydrogenase may be responsible for producing the enzymes that catalyze these reactions. M. jannaschii has two open reading frames (ORFs) that can produce proteins homologous to isocitrate dehydrogenase, the NADP-dependent isocitrate dehydrogenase gene MJ0720 (37.3% identity in 193 amino acids to the Escherichia coli isocitrate dehydrogenase), and the NAD-dependent isocitrate dehydrogenase gene MJ1596 (32.0% identity in 219 amino acids to the E. coli isocitrate dehydrogenase) (1). These ORFs correspond, respectively, to the MTH0184 and MTH1388 genes in Methanobacterium thermoautotrophicum (11). We have cloned the genes for these proteins from M. jannaschii, overproduced their enzymes in E. coli, isolated the enzymes, and determined the enzymatic reactions that each catalyzes. We can now report that the gene product of the MJ1596 gene is the AksF enzyme involved in the biosynthesis of coenzyme B that carries out the NAD-dependent decarboxylation of the homologs of (−)-threo-isocitrate and that the MJ0720 gene encodes a NAD-dependent 3-isopropylmalate dehydrogenase (LeuB).
Identification, cloning, and high-level expression of the gene product.
Expression of the MJ1596 and MJ0720 genes in E. coli was accomplished by the following procedure. The MJ1596 and MJ0720 genes were amplified by PCR using genomic DNA from M. jannaschii (David E. Graham, Urbana, Ill.) as the template. The synthetic oligonucleotide primers 5′CATGCATATGATGAAGGTGTGTGTTATAGAA3′ and 5′CAGTGGATCCTTAATATCCCTTTAACTTCTTTCT3′ (derived from the MJ1596 DNA sequence) and the primers 5′CATGCATATGGTGATAAGTATGGATAAA3′ and 5′GATCGGATCCTTATTCTTCTCTTACTCTTTT3′ (derived from the MJ0720 DNA sequence) were used. Each set of primers was used to insert the NdeI and BamHI restriction sites at the 5′ and 3′ ends. PCR was performed, with 1 μg of genomic DNA as the template, with 20 μmol of each primer, 3.75 U of AmpliTaq DNA polymerase, and 10 μl of 10× PCR buffer (Perkin Elmer, Branchburg, N.J.) in a final volume of 100 μl. Each cycle was set for 1 min of denaturation at 95°C, 2 min of annealing at 60°C, and 3 min of extention at 72°C, and 35 reaction cycles were carried out in a DNA thermal cycler. After purification of the PCR products via absorption and desorption with a QIAquick spin column (QIAGEN, Valencia, Calif.), the products were digested with NdeI and BamHI and were cloned into a NdeI-BamHI-digested pT7-7 plasmid vector to obtain the constructs pT7-7-MJ1596 and pT7-7-MJ0720. The constructs were transformed into E. coli TB1 for plasmid preparation and into E. coli BL21(DE3) for protein expression. The cells of E. coli BL21(DE3) transformed with pT7-7-MJ1596 or pT7-7-MJ0720 plasmid were grown in Luria-Bertani medium (200 ml), supplemented with 100 mg of ampicillin/liter at 37°C to an absorbance at 600 nm of 1.0.
Protein production was then induced by the addition of isopropylthio-β-d-galactoside (IPTG) to a final concentration of 0.1 mM, and the cells were grown for an additional 4 h at 37°C. The cells were harvested by centrifugation (5 min, 4,000 × g) and frozen at −20°C until used. High-level expression of the desired protein was confirmed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (12% polyacrylamide) of the SDS-soluble cellular proteins.
Preparation and analysis of cell extracts.
Cell extracts were prepared by sonication of the cell pellets (∼300 mg [wet weight]) suspended in 2 to 3 ml of buffer [50 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES) (pH 7.0), 10 mM MgCl2, and 20 mM mercaptoethanol], followed by centrifugation (10 min, 14,000 × g). SDS-PAGE analysis of the pellets and the cell extracts showed that most (>90%) of the overexpressed proteins were present in a soluble form. Heating the extracts at 60°C for 10 min, followed by centrifugation (10 min, 14,000 × g), removed most of the E. coli proteins and left essentially pure solutions of the overexpressed proteins (>95% pure by SDS-PAGE). These solutions were used for the analyses reported here. The protein concentrations were determined using the Bio-Rad protein assay.
Measurement of enzymatic activities.
The activities of the enzymes were measured spectrophotometrically in 1-ml quartz cuvettes at 60°C by following NAD reduction at 340 nm. A molar absorptivity of 6.2 × 103 M−1 cm−1 was used to determine the concentrations of NADH and NADPH. Thus, into a quartz cell at 60°C we added 500 μl of TES buffer (50 mM TES [pH 7.4], 10 mM MgCl2 and 20 mM mercaptoethanol), 6 μl of a 0.1 M solution of the desired substrate, 2 μl of a 0.1 M solution of NAD or NADP, and water to 1 ml. After equilibration to 60°C for ∼5 min, 10 μl of the enzyme solution was added with mixing. For determination of Km and Vmax values, concentrations of the various substrates were varied over a range of 0 to 1.0 mM in this 1.0-ml assay mixture. One unit of enzyme activity was defined as the reduction of 1 μmol NAD or NADP per min. dl-(threo)Isopropylmalic acid was obtained from Waco Pure Chemical Industries, Ltd., Osaka, Japan. threo-ds(+)Isocitrate, threo-isocitrate, and dl-isocitrate were obtained from Sigma Chemical Co. threo-Isohomocitrate (1-hydroxy-1,2,4-butanetricarboxylic acid), threo-iso(homo)2citrate (1-hydroxy-1,2,5-pentanetricarboxylic acid), and threo-iso(homo)3citrate (1-hydroxy-1,2,6-hexanetricarboxylic acid), their (−) isomers, and a mixture of threo-isocitrate and erythro-isocitrate were synthesized as previously described (4).
Product identification.
The products of the NAD-dependent oxidations of each of the substrates were identified by gas chromatography-mass spectrometry (GC-MS). The general procedure in each case was to incubate the enzyme with the substrates, isolate the products, convert the products into suitable methyl ester derivatives, and analyze them by GC-MS using a known sample as a reference. Incubations were conducted as described above except that an excess of NAD and enzyme was used to drive the reaction to completion. The procedures for the preparation, isolation, and subsequent GC-MS analyses of the methyl esters have been described previously (4).
Reactions catalyzed by cloned enzymes.
As reported in Table 1, the MJ1596-encoded enzyme was able to use only the homologs of threo-isocitrate as substrates. The enzyme was also unable to use any isomer of isocitrate or isopropylmalate as a substrate. This behavior is to be contrasted with the MJ0720-encoded enzyme, which was found to use only threo-isopropylmalate as a substrate. Neither enzyme was found to use NADP as an oxidant. This coenzyme specificity is the same as that observed for the isopropylmalate dehydrogenase isolated from Salmonella enterica serovar Typhimurium (10).
TABLE 1.
Kinetic data for the overproduced NAD-dependent dehydrogenases from M. jannaschii
| Substrate | MJ1596
|
MJ0720
|
||||
|---|---|---|---|---|---|---|
| Km (mM) | Vmaxa | Vmax/Kmb | Km (mM) | Vmax | Vmax/Km | |
| threo-isocitratec | nrd | nr | ||||
| threo-isohomocitrate | 0.037 | 2.6 | 69 | nr | ||
| threo-iso(homo)2citrate | 0.016 | 8.7 | 530 | nr | ||
| threo-iso(homo)3citrate | 0.021 | 7.1 | 130 | nr | ||
| threo-isopropylmalate | nr | 0.021 | 0.43 | 20 | ||
U/mg of protein.
Values are in milliliters per minute per milligram.
None of the diastereomers of isocitrate served as a substrate for either enzyme.
nr, no reaction.
The MJ1596-encoded enzyme is therefore proposed to be involved in the biosynthesis of the α-ketosuberate used for coenzyme B biosynthesis, and the MJ0720-encoded enzyme is proposed to be the isopropylmalate dehydrogenase involved in leucine biosynthesis. The Km and Vmax/Km values for these enzymes are also consistent with the metabolic functions for those enzymes having Km values in the range of 20 to 40 μM. These values are typical for related or identical enzymes from other organisms. Thus, the isocitrate dehydrogenase from Sulfolobus solfataricus has a Km for isocitrate of 130 μM and a Vmax of 68 μmol/min/mg of protein (2), whereas the same enzyme from Archaeoglobus fulgidus has a Km for isocitrate of 118 μM and a Vmax of 141 μmol/min/mg of protein (12). The Km and Vmax values for threo-isopropylmalate for the isopropylmalate dehydrogenases from S. enterica serovar Typhimurium (10) and Saccharomyces cerevisiae (5) have been measured and are, respectively, 20 μM and 54 μmol/min/mg of protein and 23 μM and 19 μmol/min/mg of protein for the respective organisms. The Km for this substrate for the isopropylmalate dehydrogenase from Sulfolobus sp. strain 7 has been determined to be 1.2 μM (13).
Thermostability of cloned enzymes.
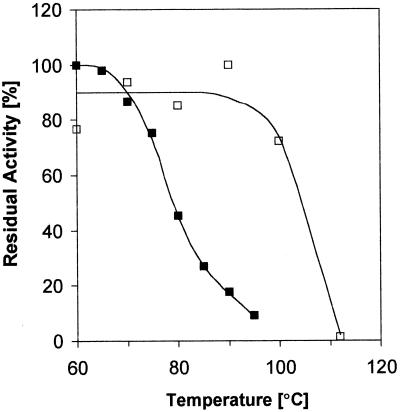
The crude extracts containing the cloned enzymes were heated for 10 min at different temperatures, and after the precipitated proteins were removed by centrifugation, the remaining activity was measured at 60°C using either threo-isohomocitrate, threo-iso(homo)2citrate, threo-iso(homo)3citrate, or threo-isopropylmalate and NAD as substrates. As can be seen from Fig. 2, the MJ1596-encoded enzyme was stable at 100°C for 10 min, whereas the LeuB enzyme lost 50% of its enzymatic activity when heated at 80°C for 10 min. Thus, the enzyme is not quite as thermostable as the LeuB enzyme isolated from Thermus thermophilus (6).
FIG. 2.
Heat stabilities of the iso(homo)1→3citrate dehydrogenase (□) and isopropylmalate dehydrogenase (■). The crude extracts containing the overexpressed enzymes were heated for 10 min at the indicated temperatures, and after the precipitated proteins were removed by centrifigation, the remaining activity was measured at 60°C using NAD plus threo-isohomocitrate or threo-isopropylmalate. Identical inactivation curves were obtained for the iso(homo)1→3citrate dehydrogenase with threo-iso(homo)2citrate and threo-iso(homo)3citrate.
While it seems unusual for a single enzyme to catalyze more than one step in a biochemical pathway, exactly the same reaction is catalyzed in each case, but with homologs of compounds containing the same structural groups. There are many biochemical examples where one enzyme carries out the same reaction on several different compounds. Among these are hexokinase (3), transaminases (8), fatty acid synthases (8), acetohydroxy acid synthases (14), and nucleoside mono- and diphosphate kinases (9). Even in the case of coenzyme B, it has already been demonstrated that the AksA gene product (4) as well as the AksD and AksE gene products can catalyze the same reaction using different homologs as substrates (M. Graupner, H. Xu, and R. H. White, unpublished results).
Having identified the M. jannaschii MJ1596 gene as one of the genes involved in the biosynthesis of coenzyme B, it is both important and useful to establish whether or not this gene, or any of its orthologs in the other Archaea, is present in gene clusters, for its presence would indicate additional genes that could be involved in coenzyme B biosynthesis. Unfortunately, neither the M. jannaschii MJ1596 gene nor its M. thermoautotrophicum ortholog, MTH0184, appears to reside within a gene cluster containing coenzyme B biosynthetic-relevant genes. The other known gene in coenzyme B biosynthesis is the aksA gene MJ0503 that encodes for the (homo)1→3citrate synthase (4). The MJ0503 gene does not appear to be in a cluster of genes related to coenzyme B biosynthesis in M. jannaschii, but its ortholog in M. thermoautotrophicum (MTH1630) is downstream of MTH1631, an ortholog to the M. jannaschii gene MJ1003 (aksD). The MJ1003 encodes a gene for the large subunit of an aconitase that is specific for the dehydration and hydration reactions involved in coenzyme B biosynthesis (Graupner et al., unpublished results). As expected, the genes MJ0504 and MTH1630 for the (homo)1→3citrate synthases, which are involved in the biosynthesis of coenzyme B, which is only needed for methane formation, are exclusively found in the methanoarchaea.
The aconitase small subunit for coenzyme B biosynthesis MJ1271 (aksE) is an ortholog of MTH0829, which has next to it a putative kinase MTH830, which could be the enzyme responsible for the addition of the phosphate in the last step of coenzyme B biosynthesis (4).
The MJ0720-encoded isopropylmalate dehydrogenase has clear orthologs in all four of the published archaea genomes (M. jannaschii, M. thermoautotrophicum, A. fulgidus, and Pyrococcus horikoshii) (7). Although the M. jannaschii MJ0720 gene is not in a gene cluster containing other genes related to leucine biosynthesis, it clearly is in a cluster of leucine-related genes in all the other archaea. The ortholog for this gene in M. thermoautotrophicum is MHT1388, in a cluster with three other genes that are related to leucine biosynthesis, MTH1387, MTH1386, and MTH1385. This cluster has been established since the first two of these genes correspond, respectively, to MJ1277 and MJ0499, genes now known to function as the small and large subunits of 3-isopropylmalate dehydratase (Graupner et al., unpublished results). The MTH1385 gene putatively encodes an aminotransferase (11). This grouping of genes is analogous to that seen for the leuABCD operon in E. coli (14).
Acknowledgments
We thank Kim Harich for the GC-MS analyses of enzymatic products, Jonathan Lau for assistance in the measurement of enzymatic activities, and Walter G. Niehaus for reviewing the manuscript prior to submission.
This work was supported in part by the National Science Foundation Grant MCB963086.
REFERENCES
- 1.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1997;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 2.Camacho M L, Brown R A, Bonete M-J, Danson M J, Hough D W. Isocitrate dehydrogenase from Haloferax volcanii and Sulfolobus solfataricus: enzyme purification, characterization and N-terminal sequence. FEMS Microbiol Lett. 1995;134:85–90. doi: 10.1111/j.1574-6968.1995.tb07919.x. [DOI] [PubMed] [Google Scholar]
- 3.Crane R K. Hexokinases and pentokinases. In: Boyer P D, Lardy H, Myrback K, editors. The enzymes. Vol. 6. New York, N.Y: Academic Press; 1962. pp. 47–66. [Google Scholar]
- 4.Howell D M, Harich K, Xu H, White R H. The α-keto acid chain elongation reactions involved in the biosynthesis of coenzyme B (7-mercaptoheptanoylthreonine phosphate) in methanogenic Archaea. Biochemistry. 1998;37:10108–10117. doi: 10.1021/bi980662p. [DOI] [PubMed] [Google Scholar]
- 5.Hsu Y-P, Kohlhaw G B. Leucine biosynthesis in Saccharomyces cerevisiae. J Biol Chem. 1980;255:7255–7260. [PubMed] [Google Scholar]
- 6.Kirino H, Aoki M, Aoshima M, Hayashi Y, Ohba M, Yamagishi A, Wakagi T, Oshima T. Hydrophobic interaction at the subunit interface contributes to the thermostability of 3-isopropylmalate dehydrogenase from the extreme thermophile, Thermus thermophilus. Eur J Biochem. 1994;220:275–281. doi: 10.1111/j.1432-1033.1994.tb18623.x. [DOI] [PubMed] [Google Scholar]
- 7.Makarova K S, Aravind L, Galperin M Y, Grishin N V, Tatusov R L, Wolf Y I, Koonin E V. Comparative genomics of the archaea (Euryarchaeota): evolution of conserved protein families, the stable core, and the variable shell. Genome Res. 1999;9:608–628. [PubMed] [Google Scholar]
- 8.Pittard A J. Biosynthesis of the aromatic amino acids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 458–484. [Google Scholar]
- 9.Parks R E, Jr, Agarwal R P. Nucleoside diphosphokinases. In: Boyer P D, editor. The enzymes. 3rd ed. Vol. 8. New York, N.Y: Academic Press; 1973. pp. 307–333. [Google Scholar]
- 10.Parsons S J, Burns R O. Purification and properties of β-isopropylmalate dehydrogenase. J Biol Chem. 1969;244:996–1003. [PubMed] [Google Scholar]
- 11.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J, Rice P, Nölling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautophicum. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steen I H, Lien T, Birkeland N-K. Biochemical and phylogenetic characterization of isocitrate dehydrogenase from a hyperthermophilic archaeon, Archaeoglobus fulgidus. Arch Microbiol. 1997;168:412–420. doi: 10.1007/s002030050516. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki T, Inoki Y, Yamagisi A, Iwasaki T, Wakagi T, Oshima T. Molecular and phylogenetic characterization of isopropylmalate dehydrogenase from a thermoacidophilic archaeon, Sulfolobus sp. strain 7. J Bacteriol. 1997;179:1174–1179. doi: 10.1128/jb.179.4.1174-1179.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Umbarger H E. Biosynthesis of branched-chain amino acids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 442–457. [Google Scholar]
- 15.White R H. Steps in the conversion of α-ketosuberate to 7-mercaptoheptanoic acid in methanogenic bacteria. Biochemistry. 1989;28:9417–9423. doi: 10.1021/bi00450a026. [DOI] [PubMed] [Google Scholar]