Alkylation damage to DNA occurs when cells encounter alkylating agents in the environment or when cellular metabolism produces active alkylators. To cope with DNA alkylation, cells have evolved genes that encode proteins with alkylation-specific DNA repair activities. In Escherichia coli, the main response specific for alkylation damage has been called the adaptive response (53). The adaptive response genes are induced upon exposure to exogenous alkylators by Ada-dependent induction, and also during stationary phase by rpoS-dependent gene expression, possibly to prevent accumulation of DNA damage due to increased endogenous production of alkylating agents. Recent studies of the regulatory mechanisms of Ada protein and the various responses of the individual promoters regulated by this protein has revealed a complexity of regulation not initially recognized. In this review we describe the roles of the Ada-regulated genes and the regulatory mechanisms that activate gene expression from the three Ada-dependent promoters. We will focus on Ada-dependent induction of the adaptive response genes, fine tuning of individual gene expression according to the growth phase, and the role played by Ada in shutting off the adaptive response.
ADA-DEPENDENT REGULATION OF THE ADAPTIVE RESPONSE GENES
The adaptive response set of genes is comprised of the ada, alkA, alkB, and aidB genes. Expression of these genes is regulated by Ada, and their induction provides protection against alkylation damage to DNA. The ada gene product has both repair and regulatory activities. These two activities are closely tied to one another, as the Ada protein must be activated to perform its regulatory function and activation is a consequence of its DNA repair activity. Ada has two active methyl acceptor cysteine residues, Cys-69 and Cys-321, that are required for demethylation of DNA. Both sites can become methylated when Ada protein transfers the methyl group from the appropriate substrate DNA lesions to itself. This reaction is irreversible, and methylated Ada (meAda) is the terminal end product of the demethylation reaction (31). The two methyl acceptor sites present in Ada differ with respect to the lesions repaired. Cys-321 is the methyl acceptor site required for the removal of methyl groups from either O6-methylguanine or O4-methylthymine, two highly mutagenic lesions (10, 11). Cys-69 is required for demethylation of phosphomethyltriesters in the sugar-phosphate backbone. This lesion is apparently innocuous, since Ada repairs only one of two stereoisomers (16, 36, 37, 72), leaving the other to remain in DNA with no apparent deleterious consequences (28, 40). Although methylated phosphates are innocuous, this lesion is readily produced by methylating agents (37) and provides a sensitive regulatory signal that leads to induction of the Ada regulon. Once Ada protein transfers a methyl group from the methyl-phosphate to the Cys-69 residue, it becomes a transcriptional activator. Thus, the methylated phosphates in DNA serve as the signal that converts Ada to its transcriptionally active form, which, in turn, induces the Ada regulon, resulting in increased alkylation repair activities. The adaptive response genes are induced most effectively by methylating agents and are either not induced or induced only weakly by larger alkyl groups (67), presumably because alkyl lesions larger than methyl groups are efficiently repaired by the uvrABCD-dependent nucleotide excision repair pathway and are poor substrates for the adaptive response repair genes (65, 70).
ROLES OF ADA-REGULATED GENES
The functions of the ada-regulated genes have been the subject of several reviews (32, 56, 58, 66) and will be only briefly discussed here. The ada gene, described above, is in an operon with the alkB gene, and transcription of both genes is directed by the ada promoter. The enzymatic function of alkB remains elusive despite numerous efforts to determine its biochemical function (8, 71). alkB mutants are hypersensitive to the methylating agent methyl methanesulfonate (MMS) and dimethyl sulfate, showing only modest sensitivity to N-methyl-N′-nitro-N-nitrosoguanidine and methyl nitrosourea (5, 17, 68). Because alkB mutants were deficient in their ability to reactivate MMS-treated λ phage, which implies that AlkB is able to repair lesions introduced into phage DNA prior to infection, AlkB has been implicated as a DNA repair protein (17). More recently it has been demonstrated that alkB is required for reactivation of MMS-treated single-stranded phage. Since no lesions appear to be removed in this process, it has been suggested that alkB is involved in replication of damaged template DNA (9). Regardless of its precise function, the fact that alkB expression can confer MMS resistance when expressed in mammalian cells suggests it functions by itself (5). The alkA gene encodes a glycosylase that repairs a variety of lesions including N7-methylguanine and N3-methyl purines and O2-methyl pyrimidines (32). The AlkA protein removes a damaged base from the sugar-phosphate backbone by cleaving the glycosylic bond attaching the base to the sugar, producing an abasic site. Further processing of the abasic site by AP endonucleases, polymerase I, and ligase then completes the repair (32, 66). The function of the aidB gene has not been conclusively established, but it is homologous to the mammalian isovaleryl coenzyme A dehydrogenase (IVD), has IVD activity, and appears to function to inactivate nitrosoguanidines or their reactive intermediates produced during metabolic detoxification (25).
MECHANISM OF TRANSCRIPTION ACTIVATION BY MEADA
According to the model for meAda activation proposed by Sakumi et al. (52), meAda contacts RNA polymerase through protein-protein interaction with the C-terminal domain (CTD) of its α subunit (αCTD) and recruits RNA polymerase to the Ada-dependent promoters. The evidence for this model was based on the observation that truncations in αCTD abolish transcription from the ada promoter in the presence of meAda. However, more recent data indicate that RNA polymerase binds to the −60 to −40 region of the ada and aidB promoters via its αCTD, regardless of the presence of Ada (26). This region serves as the meAda binding site, but it also closely resembles, in A+T content, location, and function, the UP element transcription enhancer sequence identified in the rrnBP1 promoter (49). At this promoter, truncation of RNA polymerase αCTD, or substitution of its R265 residue by alanine, abolishes α binding to the UP element (14). These mutations also affect RNA polymerase binding to the −60 to −40 region of ada and aidB and impair transcription initiation (26). Based on these observations, the −60 to −40 regions of ada and aidB are functionally similar to the rrnB UP element and they are sufficient to recruit RNA polymerase. However, only upon binding of meAda does the formation of a ternary complex that is proficient in transcription initiation take place (26). Thus, substitutions in αCTD result in reduced transcription from ada-dependent promoters because they prevent α from binding the promoter −60 to −40 transcription enhancer sequence, rather than impairing its direct interaction with meAda.
Several lines of evidence clearly indicate that the target in RNA polymerase for activation by the Ada protein is the ς70 subunit. Gel retardation experiments (20) show that direct protein-protein interaction takes place between meAda and ς70 and that the determinants for such interaction reside in the C-terminal 39 amino acids of ς70. Substitutions of several amino acids in the C-terminal region of ς70 impair Ada-dependent transcription both in vivo and in vitro (20, 21), indicating that meAda-ς70 interaction is indeed necessary for transcription activation.
MEADA-ς70 INTERACTION AT THE ADA AND AIDB PROMOTERS
At the ada and aidB promoters, Ada interacts with a negatively charged patch in ς70: with the single exception of residue I590, substitutions affecting Ada-dependent transcription are in negatively charged amino acids (E574, E575, E591, E605, and D612). Two of these residues (E575 and E591) are also involved in the interaction with the activator proteins PhoB and cI, respectively (19, 35), suggesting that these amino acids might be surface exposed and accessible to different activators. In the absence of meAda, RNA polymerase can bind to the promoters via its αCTD, but it fails to establish any strong interaction with the core promoter. meAda does not recruit RNA polymerase to the ada and aidB promoters, since binding of Ada and α to the −60 to −40 region is noncooperative (26). Instead, upon binding of α to the UP element, meAda interacts with ς70, activating transcription; therefore, α subunit-promoter and meAda-ς70 interactions act at separate but interdependent steps of transcription initiation. At the ada and aidB promoters, meAda appears either to increase binding to the core promoter region or to favor the formation of the open complex, functions that are indeed typical for activators that interact with the ς subunit of RNA polymerase (29, 48).
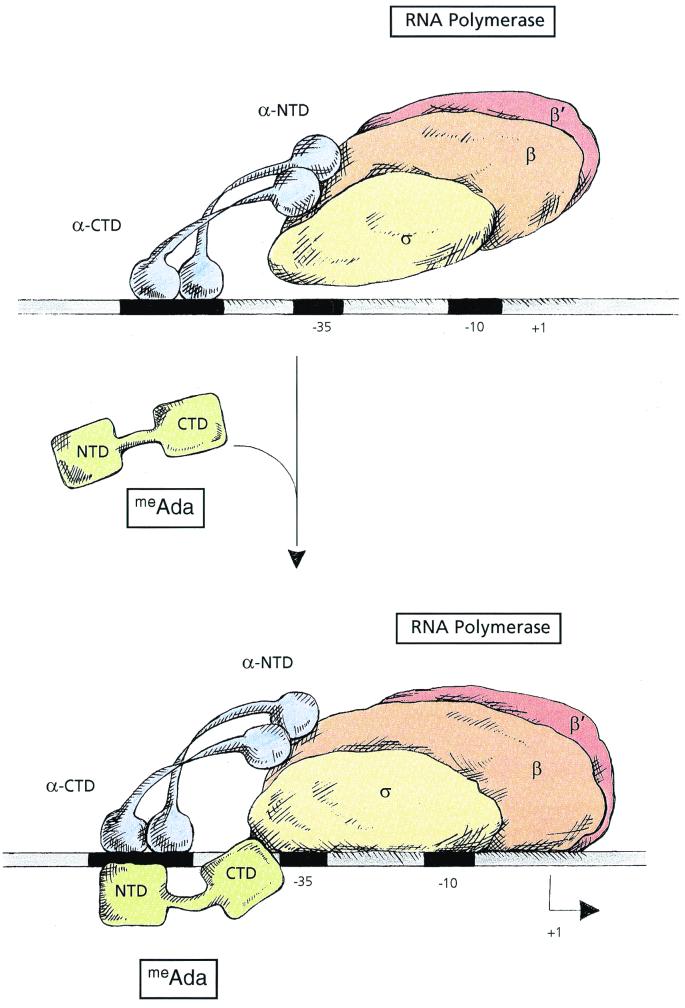
As shown in Fig. 1, the Ada protein is structured in two independent domains, linked by a hinge region that is highly susceptible to proteolytic cleavage (7, 63). The N-terminal domain of the Ada protein (AdaNTD) carries the determinants for specific DNA binding: methylation of cysteine-69, the methyl acceptor site for methyl-phosphotriesters, allows AdaNTD to specifically bind DNA. meAdaNTD binds to the Ada binding site with an affinity similar to that of the full-length protein and protects the same bases in DNase I protection assays. However, meAdaNTD is not able to activate transcription at the ada and aidB promoters. Thus, the determinants for interaction with RNA polymerase (i.e., the “activating region” of the Ada protein for ada and aidB) must reside in the CTD of the Ada protein (1). This is further substantiated by the extensive mutational studies of the AdaCTD performed by Shevell and Walker (57, 59).
FIG. 1.
Model for transcription activation by meAda at the ada and aidB promoters. The upper panel illustrates the specific interactions established between RNA polymerase and promoter DNA in the absence of meAda; the RNA polymerase-promoter complex results from protein-DNA interactions between αCTD and the UP elements. The lower panel shows the RNA polymerase-promoter-meAda ternary complex. meAda binds to its DNA site via its NTD and stimulates transcription initiation (black arrow) via protein-protein interaction between its own CTD and the CTD of ς70. Additional evidence suggests that RNA polymerase may also make DNA contacts farther upstream, most likely by bending the DNA. However, the nature of these contacts and their possible functions remain to be determined.
The results of recent studies (20) suggest the possibility that the activating region of the Ada protein at ada and aidB might be a positively charged patch in AdaCTD. Indeed, interaction between surface-exposed patches of opposite electrical charges is a common feature for transcription activation and for protein-protein interactions at large (29, 46–48). Interestingly, the methylation acceptor sites of Ada are part of two distinct positively charged patches: cysteine-69, (AdaNTD methylation site) is part of a PCKR amino acid sequence, while cysteine-321 (AdaCTD methylation site) is located in the similar sequence PCHR. The presence of positively charged amino acids in the proximity of the cysteine residues that function as methyl acceptor sites was proposed to be important for interaction with DNA and DNA repair activity of the Ada protein (42).
Structural data show that cysteine-321 and the flanking amino acids are buried inside the protein and are not accessible to solvents in the unmethylated Ada protein. However, upon DNA binding, the PCHR patch becomes exposed at the surface of the protein; methylation of cysteine-321 stabilizes this conformation (42). Methylation of cysteine-321 is necessary for optimal activation of ada transcription (61, 64), which is consistent with a possible involvement of the PCHR motif in Ada-ς70 interaction. Interestingly, substitution of cysteine-321 to an alanine results in the exposure of the histidine and arginine residues on the surface of the Ada protein, thus mimicking the effects of cysteine-321 methylation. The C321A mutation of Ada protein results in constitutive activation of the ada promoter (60, 61), again suggesting a direct role of the methylation site in the AdaCTD in transcription activation. Alternatively, it is possible that the role of AdaCTD methylation is indirect, triggering a conformational change required to expose an activating region. Such a role would be consistent with the results of Shevell and Walker (57), who reported that truncations of the terminal 20 to 30% of the AdaCTD result in constitutive activation of the ada promoter. The truncated proteins are missing the Cys-321 region, and its deletion may expose the interaction domain, allowing it to contact the ς70 subunit. However, further deletions in the AdaCTD abolish activation at the ada promoter, providing additional evidence that the determinants for Ada interaction with RNA polymerase reside in the CTD of the protein. Thus, conversion of Cys-321 to A, deletion of part of the CTD including the C321 region, and methylation of Cys-321 may all have similar consequences for Ada activation. The truncated Ada proteins that constitutively activate ada transcription require additional activation in order to induce alkA transcription, again demonstrating that the regulatory regions required for alkA and ada induction are distinct and separable by mutation.
ADA-ς70 INTERACTION AT THE ALKA PROMOTER
Although the C-terminal region of ς70 is also a target for Ada activation at the alkA promoter, a different set of amino acids (K593, K597, and R603) is involved (21). In contrast to the ς70 residues necessary for meAda-dependent transcription at ada and aidB, the amino acids involved in transcription activation by Ada at the alkA promoter are positively charged. The K593, K597, and R603 residues, as well as other neighboring positively charged residues, are also targeted by other activator proteins. Substitution to alanine of any of these residues severely affects transcription activation by the Fnr protein and by cyclic AMP receptor protein (CRP), at the dmsA and pmelRcon promoters, respectively, while R596 appears to be the target site for cI (30, 33). Although the three-dimensional structure of ς70 has not yet been solved, two alternative model structures were proposed, based on the highly similar DNA binding regions of the NarL and Cro proteins (3, 41). According to both models, residues K593 and K597 belong to a surface-exposed patch, thus providing an accessible target for activator proteins such as Ada, while R603 is removed from the protein surface and in closer contact with the helix-turn-helix DNA binding motif. The location of the R603 residue would suggest that the RA603 substitution can affect alkA transcription by altering the general conformation of the C-terminal region of ς70, consistent with the effects of the RA603 mutation on factor-independent transcription at some promoters (33).
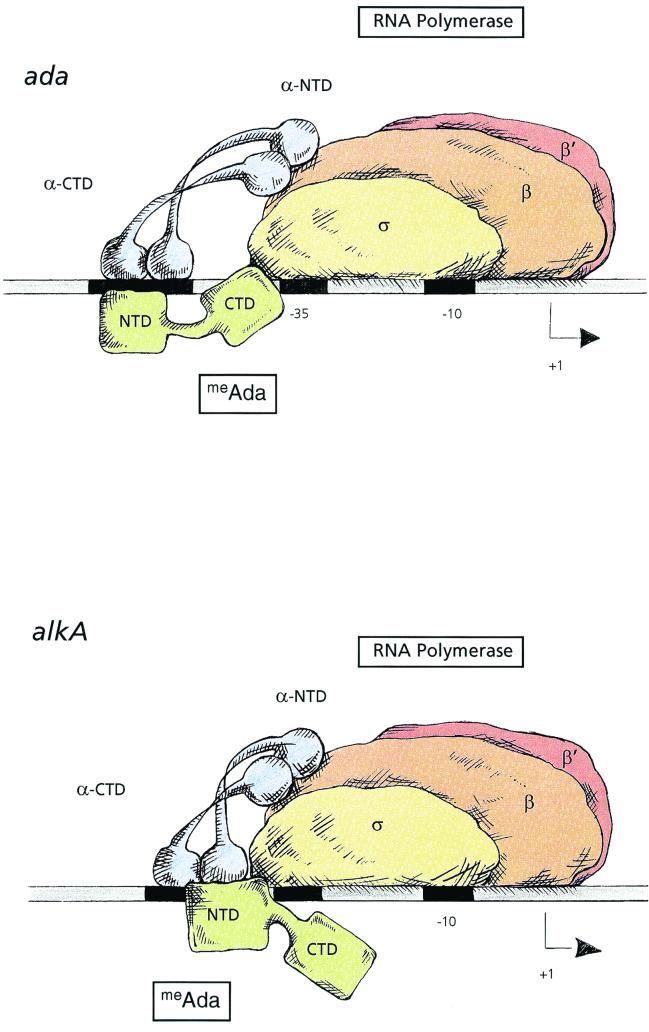
The Ada protein is the first example of a transcription activator able to contact two distinct determinants in the ς70 subunit of RNA polymerase in a promoter-specific fashion. Due to the limited flexibility in where the C-terminal region of ς70 can be positioned, an activator that interacts with ς70 is able to contact its target only if precisely placed. This is in contrast to the situation with activators that interact with αCTD, which, due to its flexible linker, can establish the same kind of interactions with activators at different locations (15, 38). This argues that a ς70-contacting activator that binds at different positions in different promoters must contact alternative determinants in ς70. This is indeed the case for the Ada protein, which binds the alkA promoter between −47 and −35, i.e., one helical turn downstream compared to the location of the Ada binding site in ada (between −57 and −45) and aidB (between −55 and −43). Thus, the use of different targets in ς70 appears to depend upon the location of the Ada binding site. The different position of the Ada binding site and the use of a different activation target at the amino acid level would strongly suggest that a different activating region of the Ada protein is responsible for transcription activation at alkA. Indeed, in contrast with the ada and aidB promoters, the unmethylated form of the Ada protein, as well as the methylated form of the AdaNTD, is able to activate transcription at alkA, although with a lower efficiency than the full-length methylated Ada protein (1, 43). These observations indicate that methylation of the Ada protein is not required to expose the activating region responsible for alkA induction and that these determinants might be located in the AdaNTD. A model contrasting Ada-RNA polymerase promoter interactions at ada and aidB with the interaction at alkA is shown in Fig. 2. Since the CTD of Ada is dispensable for activation at alkA, only the NTD is shown to make contact with both α and ς70. However, we cannot rule out the possibility that either α or ς70 might establish additional contacts with AdaCTD. Unlike at the ada and aidB promoters, where meAda does not stimulate binding of the αCTD, interaction between the α subunit of RNA polymerase and the Ada protein appears to make an important contribution to transcription activation of the alkA promoter (23). It is noteworthy that other transcription activators, such as CRP, Fnr, and the Mor protein, whose binding site is centered around −41, also interact simultaneously with the α and ς70 subunits and contact similar activation targets at the amino acid level (2, 33). These proteins are often referred to as ambidextrous activators (45).
FIG. 2.
Model for transcription activation by meAda at ada and alkA. At ada, RNA polymerase docks to the promoter region by interactions between the α subunit and DNA, and meAda-ς interaction triggers transcription initiation. At alkA, meAda interacts with both the α and ς subunits, recruiting RNA polymerase to the promoter. Since the CTD of Ada is dispensable for activation at alkA, the NTD is shown to be involved in both meAda-α and meAda-ς contacts.
EXPRESSION OF THE ADAPTIVE RESPONSE GENES IN STATIONARY PHASE AND ADA-EςS INTERACTION
A role for adaptive response genes in stationary phase was suggested first by the fact that cells lacking methyltransferase activity are spontaneous, stationary-phase-specific mutators (34, 44). Recent results indicate that the Ada regulon is indeed induced during stationary phase and protects against active alkylators produced by nitrosation of amino acids in nongrowing cells (55, 62). This form of regulation requires the rpoS gene product, which encodes the stationary-phase-specific sigma factor ςS, the key regulatory element required for expression of stationary-phase genes. ςS plays an important role in several stress responses, such as cellular responses to oxidative damage and osmotic shock (4, 27, 39).
meAda is able to activate transcription by EςS as well as Eς70 at both the ada and aidB promoters (24, 62). These observations are consistent with the fact that the ς70 amino acids important for activation by meAda at ada and aidB are also conserved in ςS (20). meAda activates ada transcription by either Eς70 or EςS with roughly the same efficiency, while the lack of a functional rpoS gene results in lower expression of the aidB gene even in the presence of meAda (22, 69), showing that the aidB promoter is dependent on both Ada and ςS for optimal expression. In contrast, not only does meAda fail to stimulate alkA transcription by EςS, consistent with the lack of conservation of K593, K597, and R603 in ςS, but it negatively affects EςS-dependent transcription both in vivo and in vitro (22). Gel retardation experiments have shown that meAda inhibits initial binding of EςS to the alkA promoter, possibly by competition with EςS for the same binding site. It has been shown that, although Eς70 and EςS can recognize the same promoter, they differ in the nature of their interaction with the promoter DNA, in particular in the degree of protein-induced DNA bending and possibly in the location of the αCTD in the EςS-DNA binary complex (6, 18). meAda might bind to a region of the alkA promoter important for recognition by EςS, but not by Eς70; alternatively, binding of meAda might alter the alkA conformation to make it less favorable for interaction with EςS.
The negative effect of meAda on EςS-dependent transcription of alkA is likely to have important physiological consequences for shutting off this component of the Ada regulon. Since methylation of the Ada protein is irreversible, the cells need a specific mechanism to turn off transcription of the Ada-dependent genes after the methylation damage of DNA has been repaired. The methylated NTD of Ada as well as the unmethylated form of the protein have been shown to negatively regulate the ada promoter and were proposed to be involved in shutting off the adaptive response. However, both methylated AdaNTD and unmethylated Ada protein can activate transcription at alkA, so that neither mechanism would result in down-regulation of this promoter. Thus, it was postulated that return to low levels of expression from alkA occurs by simple dilution of the Ada protein over several growth cycles. The function of meAda as a negative regulator of EςS-dependent transcription suggests a specific mechanism for turning off high-level expression of the alkA gene: alkA expression is activated as long as transcription in the cell is mostly dependent on Eς70. When cells reach stationary phase, intracellular concentrations of ςS increase; meAda prevents EςS from binding to alkA, thus reducing the amount of RNA polymerase available for alkA transcription and, in turn, its expression. Similar negative regulation by a functional rpoS gene has been observed for another ς70-dependent gene, uspA (12), suggesting that ς factors might indeed compete for a limiting amount of RNA polymerase during stationary phase.
Low levels of expression of alkA in stationary phase might be tolerated even upon exposure to alkylating agents: the main function of the AlkA protein is removal of 3-methyladenine from DNA. Methylation of this base is toxic because it blocks DNA replication (32). During stationary phase, very little DNA replication takes place, and the need to rapidly repair replication-blocking lesions might be less critical. Thus, the basal levels of alkA expression, together with the constitutive expression of the other 3-methyladenine-DNA glycosylase, the Tag protein (44), and with low levels of nucleotide excision repair (65) may be sufficient to repair these lesions, making high-level expression of alkA unnecessary.
Interestingly, the adaptive response genes, including alkA, are transcribed more efficiently by EςS than by Eς70 both in vitro and in vivo in the absence of meAda protein (22, 24). It has been proposed that low concentrations of alkylating agents such as methyl nitrosourea are generated during stationary phase through amino acid nitrosation (62). The observation that strains totally devoid of methyltransferase activity are spontaneous stationary-phase mutators (34, 44) indicates that these proteins are necessary to prevent alkylation mutagenesis. Therefore, an increase in expression of the adaptive response genes in parallel with expression of the genes producing active alkylators during stationary phase prevents alkylation damage to DNA and mutagenesis. It seems more efficient for Escherichia coli to counteract methylation damage by endogenously produced alkylating agents by preventing their accumulation, rather than by repairing DNA damage. Indeed, aidB, encoding a protein responsible for detoxification of some methylating agents (25, 71), is expressed at an increased rate during stationary phase (23, 67).
CONCLUSIONS
In this report, we have reviewed the mechanisms of transcription activation by the Ada protein. Although only three promoters (ada, aidB, and alkA) are the target of the Ada protein, major differences exist in the mechanisms for their activation. These differences allow fine regulation of the adaptive response genes, which can be differentially expressed according to the specific needs and physiological state of the cell. The main features of transcription activation by Ada are summarized in Table 1.
TABLE 1.
Properties of meAda-inducible promoters
| Promoter | Location of the Ada binding site | Target subunit in RNA polymerase | Effect of meAda on transcription by EςS | Activating region in Ada | Regulation by unmethylated Ada |
|---|---|---|---|---|---|
| ada | −57 to −45 | ς70 (negatively charged patch) | Activation | CTD | Repression |
| aidB | −55 to −43 | ς70 (negatively charged patch) | Activation (promoter is dependent on EςS for optimal expression) | CTD | ??? |
| alkA | −47 to −35 | ς70 (positively charged patch) and αCTD | Repression | NTD | Activation |
ACKNOWLEDGMENTS
We thank Tony Poteete and Martin Marinus for critical review of the manuscript.
Work in the laboratory of M.R.V. was supported by funds from NIH grant GM56420.
REFERENCES
- 1.Akimaru H, Sakumi K, Yoshikai T, Anai M, Sekiguchi M. Positive and negative regulation of transcription by a cleavage product of Ada protein. J Mol Biol. 1990;216:261–273. doi: 10.1016/S0022-2836(05)80318-3. [DOI] [PubMed] [Google Scholar]
- 2.Artsimovitch I, Murakami K, Ishihama A, Howe M M. Transcription activation by the bacteriophage Mu Mor protein requires the C-terminal regions of both α and ς70 subunits of Escherichia coli RNA polymerase. J Biol Chem. 1996;271:32343–32348. doi: 10.1074/jbc.271.50.32343. [DOI] [PubMed] [Google Scholar]
- 3.Baikalov I, Schroder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus R P, Dickerson R E. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- 4.Bordes P, Repoila F, Kolb A, Gutierrez C. Involvement of differential efficiency of transcription by EςS and Eς70 RNA polymerase holoenzymes in growth phase regulation of the Escherichia coli osmE promoter. Mol Microbiol. 2000;35:845–853. doi: 10.1046/j.1365-2958.2000.01758.x. [DOI] [PubMed] [Google Scholar]
- 5.Chen J B, Carroll P, Samson L. The Escherichia coli AlkB protein protects human cells against alkylation-induced toxicity. J Bacteriol. 1994;176:6255–6261. doi: 10.1128/jb.176.20.6255-6261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colland F, Fujita N, Kotlarz D, Bown J A, Meares C F, Ishihama A, Kolb A. Positioning of ςS, the stationary phase ς factor, in Escherichia coli RNA polymerase-promoter open complexes. EMBO J. 1999;18:4049–4056. doi: 10.1093/emboj/18.14.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demple B, Sedgwick B, Robins P, Totty N, Waterfield M D, Lindahl T. Active site and complete sequence of the suicidal methyltransferase that counters alkylation mutagenesis. Proc Natl Acad Sci USA. 1985;82:2288–2292. doi: 10.1073/pnas.82.9.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinglay S, Gold G, Sedgwick B. Repair in Escherichia coli alkB mutants of abasic sites and 3-methyladenine residues in DNA. Mutat Res. 1998;407:109–116. doi: 10.1016/s0921-8777(97)00065-7. [DOI] [PubMed] [Google Scholar]
- 9.Dinglay S, Trewick S C, Lindahl T, Sedgwick B. Defective processing of methylated single-stranded DNA by E. coli alkB mutants. Genes Dev. 2000;14:2097–2105. [PMC free article] [PubMed] [Google Scholar]
- 10.Dosanjh M K, Singer B, Essigmann J M. Comparative mutagenesis of O6-methylguanine and O4-methylthymine in Escherichia coli. Biochemistry. 1991;30:7027–7033. doi: 10.1021/bi00242a031. [DOI] [PubMed] [Google Scholar]
- 11.Eadie J S, Conrad M, Toorchen D, Topal M D. Mechanism of mutagenesis by O6-methylguanine. Nature. 1984;308:8–14. doi: 10.1038/308201a0. [DOI] [PubMed] [Google Scholar]
- 12.Farewell A, Kvint K, Nystrom T. Negative regulation by RpoS: a case of sigma factor competition. Mol Microbiol. 1998;29:1039–1051. doi: 10.1046/j.1365-2958.1998.00990.x. [DOI] [PubMed] [Google Scholar]
- 13.Furuichi M, Yu C G, Anai M, Sakumi K, Sekiguchi M. Regulatory elements for expression of the alkA gene in response to alkylating agents. Mol Gen Genet. 1992;236:25–32. doi: 10.1007/BF00279639. [DOI] [PubMed] [Google Scholar]
- 14.Gaal T, Ross W, Blatter E E, Tang H, Jia X, Krishnan V V, Assa-Munt N, Ebright R H, Gourse R L. DNA-binding determinants of the α subunit of RNA polymerase: novel DNA-binding domain architecture. Genes Dev. 1996;10:16–26. doi: 10.1101/gad.10.1.16. [DOI] [PubMed] [Google Scholar]
- 15.Gaston K, Bell A, Kolb A, Buc H, Busby S J W. Stringent spacing requirements for transcription activation by CRP. Cell. 1990;62:733–743. doi: 10.1016/0092-8674(90)90118-x. [DOI] [PubMed] [Google Scholar]
- 16.Hamblin M R, Potter B V L. E. coli Ada regulatory protein repairs the Sp diastereoisomer of alkylated DNA. FEBS Lett. 1985;189:315–317. doi: 10.1016/0014-5793(85)81047-4. [DOI] [PubMed] [Google Scholar]
- 17.Kataoka H, Yamamoto Y, Sekiguchi M. A new gene (alkB) of Escherichia coli that controls sensitivity to methyl methane sulfonate. J Bacteriol. 1983;153:1301–1307. doi: 10.1128/jb.153.3.1301-1307.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolb A, Kotlarz D, Kusano S, Ishihama A. Selectivity of the Escherichia coli RNA polymerase Eς38 for overlapping promoters and ability to support CRP activation. Nucleic Acids Res. 1995;23:819–826. doi: 10.1093/nar/23.5.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuldell N, Hochschild A. Amino acid substitutions in the −35 recognition motif of ς70 that result in defects in phage λ repressor-stimulated transcription. J Bacteriol. 1994;176:2991–2998. doi: 10.1128/jb.176.10.2991-2998.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landini P, Bown J A, Volkert M R, Busby S J W. Ada protein-RNA polymerase sigma subunit interaction and alpha subunit promoter DNA interaction are necessary at different steps in transcription initiation at the Escherichia coli ada and aidB promoters. J Biol Chem. 1998;273:13307–13312. doi: 10.1074/jbc.273.21.13307. [DOI] [PubMed] [Google Scholar]
- 21.Landini P, Busby S J W. The Escherichia coli Ada protein can interact with two distinct determinants in the ς70 subunit of RNA polymerase according to promoter architecture: identification of the target of Ada activation at the alkA promoter. J Bacteriol. 1999;181:1524–1529. doi: 10.1128/jb.181.5.1524-1529.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landini P, Busby S J W. Expression of the Escherichia coli ada regulon in stationary phase: evidence for rpoS-dependent negative regulation of alkA transcription. J Bacteriol. 1999;181:6836–6839. doi: 10.1128/jb.181.21.6836-6839.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landini P, Gaal T, Ross W, Volkert M R. The RNA polymerase a subunit carboxyl-terminal domain is required for both basal and activated transcription from the alkA promoter. J Biol Chem. 1997;272:15914–15919. doi: 10.1074/jbc.272.25.15914. [DOI] [PubMed] [Google Scholar]
- 24.Landini P, Hajec L I, Nguyen L H, Burgess R R, Volkert M R. The leucine-responsive protein (Lrp) acts as a specific repressor for ςS-dependent transcription of the Escherichia coli aidB gene. Mol Microbiol. 1996;20:947–955. doi: 10.1111/j.1365-2958.1996.tb02536.x. [DOI] [PubMed] [Google Scholar]
- 25.Landini P, Hajec L I, Volkert M R. Structure and transcriptional regulation of the Escherichia coli adaptive response gene aidB. J Bacteriol. 1994;176:6583–6589. doi: 10.1128/jb.176.21.6583-6589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landini P, Volkert M R. RNA polymerase α subunit binding site in positively controlled promoters: a new model for RNA polymerase/promoter interaction and transcriptional activation in the E. coli ada and aidB genes. EMBO J. 1995;14:4329–4335. doi: 10.1002/j.1460-2075.1995.tb00107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 28.Larson K, Sahm J, Shenkar R, Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutat Res. 1985;150:77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- 29.Li M, McClure W R, Susskind M M. Changing the mechanism of transcriptional activation by phage λ repressor. Proc Natl Acad Sci USA. 1997;94:3691–3696. doi: 10.1073/pnas.94.8.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, Moyle H, Susskind M M. Target of the transcriptional activation function of phage λ cI protein. Science. 1994;263:75–77. doi: 10.1126/science.8272867. [DOI] [PubMed] [Google Scholar]
- 31.Lindahl T, Demple B, Robins P. Suicide inactivation of the E. coli O6-methylguanine-DNA methyltransferase. EMBO J. 1982;1:1359–1363. doi: 10.1002/j.1460-2075.1982.tb01323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindahl T, Sedgwick B, Sekiguchi M, Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- 33.Lonetto M, Rhodius V, Lamberg K, Kiley P, Busby S, Gross C. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase ς70 subunit. J Mol Biol. 1998;284:1353–1365. doi: 10.1006/jmbi.1998.2268. [DOI] [PubMed] [Google Scholar]
- 34.Mackay W J, Han S, Samson L D. DNA alkylation repair limits spontaneous base substitution mutations in Escherichia coli. J Bacteriol. 1994;176:3224–3230. doi: 10.1128/jb.176.11.3224-3230.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makino K, Amemura M, Kim S K, Nakata A, Shinagawa H. Role of the ς70 subunit of RNA polymerase in transcriptional activation by activator protein PhoB in Escherichia coli. Genes Dev. 1993;7:149–160. doi: 10.1101/gad.7.1.149. [DOI] [PubMed] [Google Scholar]
- 36.Margison G P, Cooper D P, Brennand J. Cloning of the E. coli O6-methylguanine and methylphosphotriester methyltransferase gene using a functional DNA repair assay. Nucleic Acids Res. 1985;13:1939–1952. doi: 10.1093/nar/13.6.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCarthy T V, Lindahl T. Methyl phosphotriesters in alkylated DNA are repaired by the Ada regulatory protein of E. coli. Nucleic Acids Res. 1985;13:2683–2698. doi: 10.1093/nar/13.8.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meng W, Savery N J, Busby S J W, Thomas M S. The Escherichia coli RNA polymerase α subunit linker: length requirements for transcription activation at CRP-dependent promoters. EMBO J. 2000;19:1555–1566. doi: 10.1093/emboj/19.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michán C, Manchado M, Dorado G, Pueyo C. In vivo transcription of the Escherichia coli oxyR regulon as a function of growth phase and in response to oxidative stress. J Bacteriol. 1999;181:2759–2764. doi: 10.1128/jb.181.9.2759-2764.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller P S, Chandrasegaran S, Dow D L, Pulford S M, Kan L S. Synthesis and template properties of an ethyl phosphotriester modified decadeoxyribonucleotide. Biochemistry. 1982;21:5468–5474. doi: 10.1021/bi00265a014. [DOI] [PubMed] [Google Scholar]
- 41.Mondragon A, Harrison S. The phage 434 Cro/OR1 complex at a 2.5 angstrom resolution. J Mol Biol. 1991;2199:321–334. doi: 10.1016/0022-2836(91)90568-q. [DOI] [PubMed] [Google Scholar]
- 42.Moore M H, Gulbis J M, Dodson E J, Demple B, Moody P C E. Crystal structure of a suicidal DNA repair protein: the Ada O6-methylguanine-DNA methyltransferase from E. coli. EMBO J. 1994;13:1495–1501. doi: 10.1002/j.1460-2075.1994.tb06410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakabeppu Y, Sekiguchi M. Regulatory mechanisms for induction of synthesis of repair enzymes in response to alkylating agents: Ada protein acts as a transcriptional regulator. Proc Natl Acad Sci USA. 1986;83:6297–6301. doi: 10.1073/pnas.83.17.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rebeck G W, Samson L. Increased spontaneous mutation and alkylation sensitivity of Escherichia coli strains lacking the ogt O6-methylguanine DNA repair methyltransferase. J Bacteriol. 1991;173:2068–2076. doi: 10.1128/jb.173.6.2068-2076.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rhodius V A, Busby S J. Positive activation of gene expression. Curr Opin Microbiol. 1998;1:152–159. doi: 10.1016/s1369-5274(98)80005-2. [DOI] [PubMed] [Google Scholar]
- 46.Rhodius V A, Busby S J W. Interactions between activating region 3 of the Escherichia coli cyclic AMP receptor protein and region 4 of the RNA polymerase ς70 subunit: application of suppression genetics. J Mol Biol. 2000;299:311–324. doi: 10.1006/jmbi.2000.3737. [DOI] [PubMed] [Google Scholar]
- 47.Rhodius V A, Busby S J W. Transcription activation by the Escherichia coli cyclic AMP receptor protein: determinants within activating region 3. J Mol Biol. 2000;299:295–310. doi: 10.1006/jmbi.2000.3736. [DOI] [PubMed] [Google Scholar]
- 48.Rhodius V A, West D M, Webster C L, Busby S J W, Savery N J. Transcription activation at class II CRP-dependent promoters: the role of different activating regions. Nucleic Acids Res. 1997;25:326–332. doi: 10.1093/nar/25.2.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross W, Gosink K K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R L. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 50.Saget B, Walker G C. The Ada protein acts as both a positive and a negative modulator of Escherichia coli's response to methylating agents. Proc Natl Acad Sci USA. 1994;91:9730–9734. doi: 10.1073/pnas.91.21.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saget B M, Shevell D E, Walker G C. Alteration of lysine 178 in the hinge region of the Escherichia coli Ada protein interferes with activation of ada, but not alkA, transcription. J Bacteriol. 1995;177:1268–1274. doi: 10.1128/jb.177.5.1268-1274.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakumi K, Igarashi K, Sekiguchi M, Ishihama A. The Ada protein is a class I transcription factor of Escherichia coli. J Bacteriol. 1993;175:2455–2457. doi: 10.1128/jb.175.8.2455-2457.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Samson L, Cairns J. A new pathway for DNA repair in Escherichia coli. Nature. 1977;267:281–283. doi: 10.1038/267281a0. [DOI] [PubMed] [Google Scholar]
- 54.Sedgwick B. Genetic mapping of ada and adc mutations affecting the adaptive response of Escherichia coli to alkylating agents. J Bacteriol. 1982;150:984–988. doi: 10.1128/jb.150.2.984-988.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sedgwick B. Nitrosated peptides and polyamines as endogenous mutagens in O6-alkylguanine-DNA alkyltransferase deficient cells. Carcinogenesis. 1997;18:1561–1567. doi: 10.1093/carcin/18.8.1561. [DOI] [PubMed] [Google Scholar]
- 56.Sedgwick B, Vaughan P. Widespread adaptive response against environmental methylating agents in microorganisms. Mutat Res. 1991;250:211–221. doi: 10.1016/0027-5107(91)90178-q. [DOI] [PubMed] [Google Scholar]
- 57.Shevell D, Walker G C. A region of the Ada DNA repair protein required for the activation of ada transcription is not necessary for activation of alkA. Proc Natl Acad Sci USA. 1991;88:9001–9005. doi: 10.1073/pnas.88.20.9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shevell D E, Friedman B M, Walker G C. Resistance to alkylation damage in Escherichia coli: role of the Ada protein in induction of the adaptive response. Mutat Res. 1990;233:53–72. doi: 10.1016/0027-5107(90)90151-s. [DOI] [PubMed] [Google Scholar]
- 59.Shevell D E, LeMotte P K, Walker G C. Alteration of the carboxyl-terminal domain of Ada protein influences its inducibility, specificity, and strength as a transcriptional activator. J Bacteriol. 1988;170:5263–5271. doi: 10.1128/jb.170.11.5263-5271.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takano K, Nakkabeppu Y, Sekiguchi M. Functional sites of the Ada regulatory protein of Escherichia coli: analysis by amino acid substitutions. J Mol Biol. 1988;201:261–271. doi: 10.1016/0022-2836(88)90137-4. [DOI] [PubMed] [Google Scholar]
- 61.Taketomi A, Nakabeppu Y, Ihara K, Hart D J, Furuichi M, Sekiguchi M. Requirement for two conserved cysteine residues in the Ada protein of Escherichia coli for transactivation of the ada promoter. Mol Gen Genet. 1996;250:523–532. doi: 10.1007/BF02174440. [DOI] [PubMed] [Google Scholar]
- 62.Taverna P, Sedgwick B. Generation of edogenous methylating agents by nitrosation in Escherichia coli. J Bacteriol. 1996;178:5105–5111. doi: 10.1128/jb.178.17.5105-5111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teo I, Sedgwick B, Demple B, Li B, Lindahl T. Induction of resistance to alkylating agents in E. coli: the ada+ gene product serves both as a regulatory protein and as an enzyme for repair of mutagenic damage. EMBO J. 1984;3:2151–2157. doi: 10.1002/j.1460-2075.1984.tb02105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Teo I, Sedgwick B, Kilpatrick M W, McCarthy T V, Lindahl T. The intracellular signal for induction of resistance to alkylating agents in E. coli. Cell. 1986;45:315–324. doi: 10.1016/0092-8674(86)90396-x. [DOI] [PubMed] [Google Scholar]
- 65.Van Houten B, Sancar A. Repair of N-methyl-N′-nitro-N-nitrosoguanidine-induced DNA damage by ABC excinuclease. J Bacteriol. 1987;169:540–545. doi: 10.1128/jb.169.2.540-545.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Volkert M R. Adaptive response of Escherichia coli to alkylation damage. Environ Mol Mutagen. 1988;11:241–255. doi: 10.1002/em.2850110210. [DOI] [PubMed] [Google Scholar]
- 67.Volkert M R, Gately F H, Hajec L I. Expression of DNA damage-inducible genes of Escherichia coli upon treatment with methylating, ethylating and propylating agents. Mutat Res. 1989;217:109–115. doi: 10.1016/0921-8777(89)90062-1. [DOI] [PubMed] [Google Scholar]
- 68.Volkert M R, Hajec L I. Molecular analysis of the aidD6::MudI(bla lac) fusion mutation of Escherichia coli. Mol Gen Genet. 1991;229:319–323. doi: 10.1007/BF00272173. [DOI] [PubMed] [Google Scholar]
- 69.Volkert M R, Hajec L I, Matijasevic Z, Fang F C, Prince R. Induction of the Escherichia coli aidB gene under oxygen-limiting conditions requires a functional rpoS (katF) gene. J Bacteriol. 1994;176:7638–7645. doi: 10.1128/jb.176.24.7638-7645.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Warren W, Lawley P D. The removal of alkylation products from the DNA of Escherichia coli cells treated with the carcinogens N-ethyl-N-nitrosourea and N-methyl-nitrosourea: influence of growth conditions and DNA repair defects. Carcinogenesis. 1980;1:67–78. doi: 10.1093/carcin/1.1.67. [DOI] [PubMed] [Google Scholar]
- 71.Wei Y-F, Chen B J, Samson L. Suppression of Escherichia coli alkB mutants by Saccharomyces cerevisiae genes. J Bacteriol. 1995;177:5009–5015. doi: 10.1128/jb.177.17.5009-5015.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weinfeld M, Drake A F, Saunders J K, Paterson M C. Stereospecific removal of methylphosphotriesters from DNA by an E. coli Ada extract. Nucleic Acids Res. 1985;13:7067–7077. doi: 10.1093/nar/13.19.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]