Abstract
An arylketone monooxygenase was purified from Pseudomonas putida JD1 by ion exchange and affinity chromatography. It had the characteristics of a Baeyer-Villiger-type monooxygenase and converted its substrate, 4-hydroxyacetophenone, into 4-hydroxyphenyl acetate with the consumption of one molecule of oxygen and oxidation of one molecule of NADPH per molecule of substrate. The enzyme was a monomer with an Mr of about 70,000 and contained one molecule of flavin adenine dinucleotide (FAD). The enzyme was specific for NADPH as the electron donor, and spectral studies showed rapid reduction of the FAD by NADPH but not by NADH. Other arylketones were substrates, including acetophenone and 4-hydroxypropiophenone, which were converted into phenyl acetate and 4-hydroxyphenyl propionate, respectively. The enzyme displayed Michaelis-Menten kinetics with apparent Km values of 47 μM for 4-hydroxyacetophenone, 384 μM for acetophenone, and 23 μM for 4-hydroxypropiophenone. The apparent Km value for NADPH with 4-hydroxyacetophenone as substrate was 17.5 μM. The N-terminal sequence did not show any similarity to other proteins, but an internal sequence was very similar to part of the proposed NADPH binding site in the Baeyer-Villiger monooxygenase cyclohexanone monooxygenase from an Acinetobacter sp.
One of the strategies used by microorganisms for the aerobic metabolism of ketones in degradative processes is to perform the biological equivalent of the Baeyer-Villiger reaction. This involves the insertion of an oxygen between the keto carbon and an adjacent carbon to form an ester, which can then be readily hydrolyzed. This sequence has been described for the degradation of a number of cyclic ketones, for example cyclohexanone and camphor, which yield lactones on oxygenation (6, 20), and for the splitting of aliphatic ketones such as 2-tridecanone (1) or the removal of the side chain of progesterone (13). Several of the enzymes involved in the oxygenating reaction have been purified, and the cyclohexanone monooxygenase from an Acinetobacter species has been studied in great detail and particularly well characterized (2, 7, 21). Generally they have the properties of flavoprotein monooxygenases with a requirement for reduced NAD or NADP, although the enzymes that lactonize the enantiomers of camphor require an additional protein to transfer electrons from the NADH to the monooxygenase component (20). Some of these enzymes have broad substrate specificities, including the ability to oxidize sulfur or selenium atoms in organic compounds, and they are capable of forming chiral products in high enantiomeric excess. Consequently, there has been considerable interest in this class of enzyme because of the potential use for the biological production of chiral synthons, and some of these enzymes are now produced commercially (14, 15).
In addition to the metabolism of cyclic and aliphatic ketones, the degradation of aryl ketones can also proceed by formation of an ester, apparently by the action of a Baeyer-Villiger type of monooxygenase. In the degradative pathway for acetophenone by an Arthrobacter sp. and by a Nocardia sp., the substrate is oxidized to phenyl acetate (3, 4), and a similar route has been proposed for degradation of chlorinated acetophenones by an Alcaligenes sp. and Pseudomonas fluorescens (9). However, the enzymes concerned have generally been unstable, and none have been purified and characterized to see if they resemble other Baeyer-Villiger monooxygenases. Another example of an aryl ketone is 4-hydroxyacetophenone, which is an intermediate in the degradation of 4-ethylphenol by Pseudomonas putida JD1, and, in the proposed pathway, it too is converted into an ester, 4-hydroxyphenyl acetate, followed by hydrolysis to give 1,4-dihydroxybenzene (hydroquinone) and acetate (Fig. 1) (5). Here we describe the purification and some of the characteristics of the enzyme involved in the Baeyer-Villiger oxygenation of this aryl ketone.
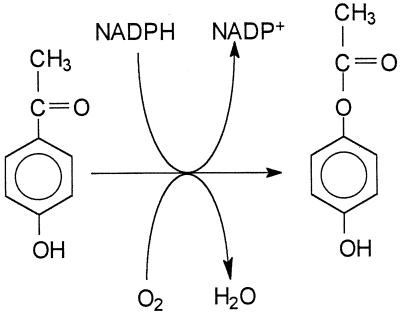
FIG. 1.
The reaction catalyzed by 4-hydroxyacetophenone monooxygenase to convert 4-hydroxyacetophenone into 4-phenyl acetate.
MATERIALS AND METHODS
Maintenance and growth of organism.
Pseudomonas putida JD1 was maintained on nutrient agar slopes. It was grown in liquid medium containing, per liter, 2.4 g of KH2PO4, 4.8 g of Na2HPO4, 2.0 g of NH4Cl, 4 ml of salts solution (16), and 0.4 g of 4-hydroxyacetophenone. A large crop of bacteria for enzyme purification was grown by inoculating 10 liters of medium in a New Brunswick laboratory fermentor, stirred at 300 rpm and aerated at a rate of 4 liters/min, with 1 liter of growing culture grown in a conical flask on an orbital incubator at 150 rpm and 30°C. When growth slowed due to depletion of the carbon source, as indicated by a rise in the oxygen concentration in the medium, further additions of 4-hydroxyacetophenone (0.4 g/liter) were made. Bacteria were harvested using an Alfa Laval continuous-flow centrifuge, and the cell paste was stored at −20°C.
Enzyme assay.
The enzyme was assayed spectrophotometrically by monitoring the oxidation of NADPH at 370 nm. This wavelength was chosen because of the high absorbance of 4-hydroxyacetophenone at 340 nm, the λmax for NADPH. A value of 2.86 mM−1 cm−1 was used for the ɛmM of NADPH at 370 nm. The reaction mixture at 30°C contained, in 1.0 ml of 50 mM Tris-HCl buffer (pH 8.0), 0.3 μmol of NADPH, 1.0 μmol of 4-hydroxyacetophenone, and enzyme. A unit of activity is defined as that amount of enzyme required to oxidize 1 μmol of NADPH/min.
This assay was used for kinetic studies with a range of substrate concentrations of 15 μM to 1 mM for 4-hydroxyacetophenone, 15 to 125 μM for 4-hydroxypropiophenone, and 225 to 900 μM for acetophenone, all at an NADPH concentration of 300 μM. For NADPH, a range from 12.8 to 64 μM was used with 1 mM 4-hydroxyacetophenone as the fixed substrate. The assays were performed in triplicate, and the kinetic constants were calculated using the ENZFITTER nonlinear regression analysis computer program (12).
For measurements of reaction stoichiometry, the spectrophotometric assay was used but with 0.17 U of enzyme and amounts of substrate that would give incomplete oxidation of the NADPH. Measurements of oxygen uptake in reaction stoichiometry were made in an oxygen monitor with a Clark-type oxygen electrode at 30°C (Yellow Springs Instrument, Yellow Springs, Ohio). The 3-ml reaction volume contained 50 mM Tris-HCl buffer (pH 8.0), 1 μmol of NADPH, and 0.425 U of enzyme. Reaction was started by the addition of substrate.
Purification of enzyme.
Frozen bacteria, grown on 4-hydroxyacetophenone, were thawed and resuspended in two volumes of 20 mM K–Na phosphate buffer (pH 7.0). Bacteria were disrupted by ultrasonic disintegration using eight 30-s bursts interspersed with cooling on ice. The suspension was then centrifuged at 28,000 × g for 20 min at 4°C, and the supernatant was taken as crude extract. All purification procedures were performed at 4°C. The crude extract was loaded on a column of Q Sepharose FF (6- by 2.5-cm diameter) equilibrated with 20 mM K–Na phosphate buffer (pH 7.0). The column was washed with 300 ml of buffer, and then the enzyme was eluted with a gradient from 0 to 0.4 M KCl in 500 ml of the phosphate buffer at a flow rate of 2 ml/min. Fractions of 7.7 ml were collected, and the enzyme was eluted at about 0.22 M KCl. Pooled fractions were dialyzed against 2 liters of 20 mM K–Na phosphate buffer (pH 7.0) for 3 h with two changes of buffer. The enzyme was then loaded onto a Reactive Red column (2- by 2.5-cm diameter) equilibrated with the phosphate buffer. The column was washed with buffer until the absorbance at 280 nm (A280) of the effluent returned to a low value (160 ml), and then the enzyme was eluted with phosphate buffer containing 1 mg of NADP+/ml at a flow rate of 0.8 ml/min. Fractions of 2.5 ml were collected.
UV/visible spectra.
Spectra of the enzyme and flavin were recorded using a Hewlett-Packard HP8452A diode array spectrophotometer. For spectra of the anaerobic reduction of the enzyme by NADPH, a cuvette contained 1.5 mg of enzyme in 0.8 ml of 20 mM Na–K phosphate buffer (pH 7.0). The cuvette contents were made anaerobic by blowing nitrogen over the liquid surface for several minutes and then sealing the cuvette with a rubber cap. Small volumes of NADPH solution were injected through the cap.
Sequencing of peptides.
The N-terminal analysis of the enzyme and sequencing of internal peptides produced by digestion with Lys-c was done by Edman degradation using a model 473A automated protein sequencer (Applied Biosystems Inc.) at the sequencing service of the University of Nottingham. Peptides, obtained by incubation of enzyme dissolved in 250 mM ammonium bicarbonate with sequencing grade Lys-c (Promega Ltd.) for 16 h at 37°C, were separated by high-pressure liquid chromatography (HPLC) on a Jupiter C18 (250 by 2.0 mm) column (Phenomex Ltd.). The peptides were eluted with solvent A (0.1% trifluoroacetic acid in water) and a gradient of solvent B (0.07% trifluoroacetic acid and 70% [vol/vol] acetonitrile in water) run over 80 min at a flow rate of 0.15 ml per min.
Chromatography.
Gas chromatography-mass spectroscopy (GC-MS) was performed on a Hewlett-Packard 5890 instrument with a 5971 mass-selective detector. This was fitted with an HP5 (cross-linked 5% phenylmethylsilicone) column (25 m by 0.2 mm; 0.33-μm film thickness) with helium as the carrier gas and using a temperature program of 2 min at 70°C rising at 20°C/min to 270°C.
Thin-layer chromatography (TLC) of reaction products was performed on precoated silica-gel GHLF plates (Analtech, Newark, N.J.) using a solvent of toluene,1,4-dioxan, and acetic acid (22.5:5:1, by volume). Compounds were detected by the quenching of background fluorescence when plates were viewed under UV light and by the color given by phenolic compounds after spraying with diazotized p-nitroaniline (18). TLC of flavins was performed on precoated cellulose plates (Merck, Darmstadt, Germany) using a solvent of tert-amyl alcohol, formic acid, and water (3:1:1 by volume). Flavins were detected by their yellow color and by their fluorescence under UV light.
Preparation of apoenzyme and reconstitution.
Apoenzyme was prepared from the purified enzyme preparation by the gradual addition of finely powdered ammonium sulfate to give 90% saturation followed by acidification with HCl to pH 2. The precipitated protein was collected by centrifuging, which left most of the yellow color in the supernatant. The precipitate was washed with cold 90% saturated ammonium sulfate solution (pH 3) and then redissolved in the original volume of 50 mM Tris-HCl buffer (pH 8.0).
Electrophoresis.
BioRad Mini Protean II Ready Gels (4 to 20% gradient) were used for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (SDS-PAGE). The gels were calibrated with Sigma 4,000- to 70,000- and 30,000- to 200,000-molecular-mass markers. Proteins were detected by staining with Coomassie brilliant blue R-250.
Mr determination by gel filtration.
The Mr of the native enzyme was determined by gel filtration on a Sephacryl S-200 column (77- by 2.5-cm diameter) that was run with 0.1 M Tris-HCl buffer (pH 8.0) at a flow rate of 17 ml/h and on a BioRad Bio-Sil TSK-125 (30- by 0.75-cm diameter) column by HPLC, using a solvent of 42 mM K–Na phosphate buffer (pH 7.0), containing 0.15 M NaCl at a flow rate of 0.5 ml/min. Proteins used to calibrate the columns were alcohol dehydrogenase (from Saccharomyces cerevisiae [Mr 150,000]), lactate dehydrogenase (from rabbit muscle [Mr 140,000]), 4-ethylphenol methylenehydroxylase (Mr 120,000), bovine serum albumin (Mr 66,000), ovalbumin (Mr 45,000), carbonic anhydrase (Mr 29,000), trypsin inhibitor (Mr 20,100), myoglobin (Mr 17,000), and cytochrome c (Mr 12,500).
RESULTS
Purification of 4-hydroxyacetophenone monooxygenase.
This enzyme catalyzes one of the steps in the catabolism of 4-ethylphenol, the carbon source on which P. putida JD1 was originally isolated. However, the organism will also grow on 4-hydroxyacetophenone, which is an intermediate in the pathway and the substrate for this enzyme. As this is an easier compound to handle, the enzyme was purified from bacteria grown on this substrate. The enzyme has a requirement for a reducing agent (NADPH) and O2, as is typical for a monooxygenase, and it was assayed spectrophotometrically by monitoring the oxidation of NADPH. In preliminary experiments to develop an assay, the enzyme was assayed at pH values ranging from 6.8 to 9.6 in phosphate, Tris-HCl, and glycine buffers. The optimum pH was 8.0 in Tris-HCl buffer, and this was used for all assays.
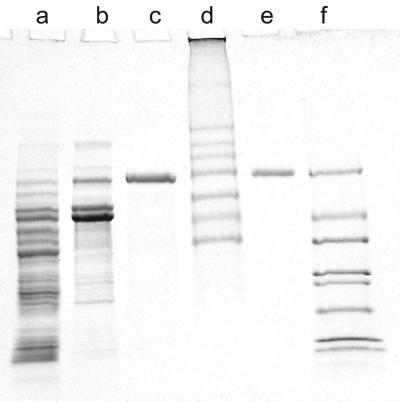
Just two steps, ion-exchange chromatography followed by chromatography on an affinity column, from which the enzyme was eluted with NADP+ resulted in apparently pure enzyme with a good recovery of 79% (Table 1). The purified enzyme gave a single band on an SDS-polyacrylamide gel after electrophoresis and staining for protein (Fig. 2). The purification of 98-fold indicated that the enzyme constituted about 1% of the soluble cell protein, which is typical for an inducible degradative enzyme.
TABLE 1.
Summary of the purification of 4-hydroxyacetophenone monooxygenase from P. putida JD1
| Purification step | Vol (ml) | Total protein (mg) | Total activity (U) | Sp. act. (U/mg of protein) | Purification (fold) | Recovery (%) |
|---|---|---|---|---|---|---|
| Crude extracta | 82 | 3300 | 271 | 0.082 | 100 | |
| Q-Sepharose FF chromatography | 116 | 565 | 260 | 0.46 | 5.6 | 97.5 |
| Reactive red chromatography | 30 | 26.5 | 215 | 8.1 | 98 | 79 |
From 39 g (wet weight) of P. putida JD1 grown on 4-hydroxyacetophenone.
FIG. 2.
SDS-PAGE of samples from the stages in purification of 4-hydroxyacetophenone monooxygenase. Lane a, crude extract (20 μg); lane b, protein after Q-Sepharose FF chromatography (10 μg); lane c, pure enzyme (7 μg); lane d, high-molecular-weight markers; lane e, pure enzyme (3.5 μg); lane f, low-molecular-weight markers.
Mr of enzyme.
Gel filtration of the enzyme on a calibrated Sephacryl S200 column gave a value for the native Mr of 70,000, and HPLC on a Bio-sil TSK-125 column gave a value of 63,000. The value from SDS-PAGE of the purified enzyme was 71,000 (Fig. 2). This indicates that the enzyme consists of a single polypeptide chain.
Spectrum and flavin content.
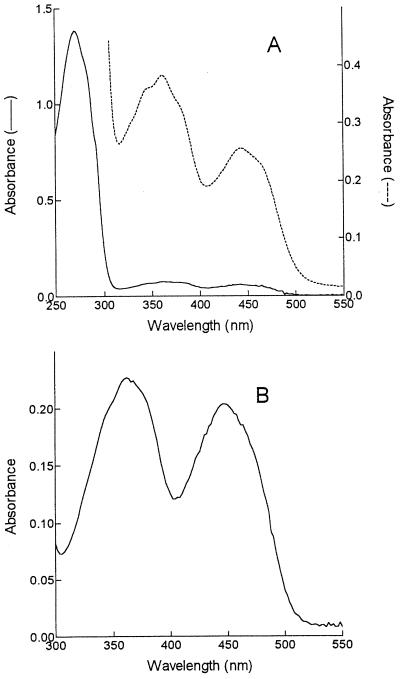
The purified enzyme was colored yellow, indicative of a flavoprotein. However, to obtain a spectrum, the NADP+ used in the final elution from the affinity column had to be removed. Dialysis of the protein failed to remove all the NADP+, and residual nucleotide was removed by fast protein liquid chromatography (FPLC) of the protein on a Superose 12 HR 10/30 column (Pharmacia Biotech, St. Albans, United Kingdom). The spectrum of the enzyme (Fig. 3A) shows peaks at 362 nm and 442 nm, indicative of a flavoprotein, although there is some perturbation of the typical flavin spectrum as shown by the shoulders on each side of the 362-nm peak and the fact that the A362 is much higher than the A442.
FIG. 3.
UV/visible absorbance spectra of enzyme (A) and extracted flavin (B). The spectra in A are of concentrated enzyme (2.3 mg/ml) (–––) to show the features between 300 and 550 nm and of diluted enzyme (0.56 mg/ml) (——) to show the UV peak.
Flavin was extracted by precipitation of the protein with trichloroacetic acid (TCA), followed by centrifugation. TLC of the extracted flavin gave a spot that corresponded with flavin adenine dinucleotide (FAD) (Rf, 0.13) and not flavin mononucleotide (FMN) (Rf, 0.35). The spectrum of the yellow supernatant is shown in Fig. 3B and was now more typical of FAD.
The ratio of FAD to enzyme was calculated from the A442 of the enzyme, assuming that the associated flavin has the same millimolar absorbance coefficient of 11.3 as free FAD (22). Using a value of 70,000 for the Mr of the protein, ratios for different preparations ranged from 0.64:1 to 0.77:1.
The flavin in the enzyme was reduced specifically by NADPH and not by NADH. This was shown by recording the spectrum from 400 to 500 nm of a 27-μM solution of enzyme in an anaerobic cuvette after successive additions of 5 μM NADPH. This led to a stepwise bleaching of the 442-nm peak with reduction for each addition, complete within the time taken to make the addition, mix the contents, and record the spectrum (about 5 s). Admittance of air to the cuvette and mixing of the contents resulted in the gradual reappearance of the oxidized spectrum over a period of 1 to 2 min. No reduction of the flavin was seen when NADH replaced NADPH in the experiment.
Some flavoprotein monooxygenases show a perturbation of the flavin spectrum on addition of the organic substrate, and this can be used to study the stoichiometry of binding and the dissociation constant. However, no perturbation of the flavin spectrum was seen when 4-hydroxyacetophenone was added to this enzyme in small portions to a final concentration of 0.125 mM.
Reconstitution of apoenzyme with FAD.
The activity of the purified enzyme preparation was not stimulated by incubation with either FMN or FAD at a concentration of 0.5 mM. However, apoenzyme could be produced by precipitation of purified enzyme with acid ammonium sulfate. This treatment released most of the flavin, leaving a very pale yellow precipitate. When redissolved, this precipitate had only 21% of the original activity. Incubation of this redissolved enzyme with 0.25 mM FMN gave no increase in activity. However, incubation with 0.25 mM FAD gave a twofold stimulation to 43% of the original activity. No further stimulation was seen with higher concentrations.
Stoichiometry of reaction.
The reaction was run using purified enzyme with a limiting amount of 4-hydroxyacetophenone, and it was monitored either by measuring NADPH oxidation spectrophotometrically at 370 nm or by using an oxygen monitor to measure O2 uptake. In the spectrophotometric assay, 0.1 and 0.05 μmol of substrate resulted in the oxidation of 0.117 and 0.057 μmol of NADPH, respectively. In the oxygen monitored, there was an uptake of 0.21 and 0.1 μmol of O2 on addition of 0.2 and 0.1 μmol of substrate, respectively. Thus, in each case the ratio is close to the 1:1 for NADPH or O2 used to 4-hydroxyacetophenone added as would be expected for a monooxygenase reaction.
Identification of reaction product.
A reaction mixture containing 3 μmol of 4-hydroxyacetophenone, 5 μmol of NADPH, and enzyme in 5 ml of 50 mM Tris-HCl buffer (pH 8.0) was incubated at 30°C with shaking for 1.5 h. The reaction mixture was then extracted with an equal volume of diethyl ether. This was dried over anhydrous sodium sulfate, evaporated to a small volume, and examined by GC-MS. It gave one major peak with a retention time of 8.4 min. The mass spectrum was identical with that for authentic 4-hydroxyphenyl acetate with a base peak at m/z 110 and an M+ peak of m/z 152, equivalent to the addition of one atom of oxygen (16) to 4-hydroxyacetophenone (M+, m/z 136). This was confirmed by TLC, in which the product corresponded to authentic 4-hydroxyphenyl acetate (Rf, 0.58) and not 4-hydroxyacetophenone (Rf, 0.5). Control reaction mixtures without enzyme or with boiled enzyme showed no change in the substrate.
Other substrates.
The products from other substrates were also analyzed by GC-MS. Acetophenone yielded a product giving a single peak with a retention time of 5.5 min and a mass spectrum with an M+ of m/z 136 and a base peak of m/z 94. This was identified from the Wiley library of spectra as phenyl acetate. 4-Hydroxypropiophenone also gave a product with a single peak on the GC and a mass spectrum with a similar pattern to that for 4-hydroxyphenyl acetate with a base peak at m/z 110 but with an M+ of m/z 166, as expected for the additional methylene group in 4-hydroxyphenyl propionate. No oxygen uptake or NADPH oxidation was seen when benzophenone or cyclohexylacetone was tested as the substrate, and no products were obtained from these compounds.
Steady-state kinetic experiments with the positive substrates gave Michaelis-Menten kinetics with apparent Km values (at 300 μM NADPH) of 47 ± 5 μM for 4-hydroxyacetophenone, 23 ± 4 μM for 4-hydroxypropiophenone, and 384 ± 45 μM for acetophenone. The highest Vmax value from these experiments was for 4-hydroxyacetophenone (ΔA370/min of 0.31). The values for 4-hydroxypropiophenone and acetophenone were 85 and 59% of this, respectively. The apparent Km for NADPH with 4-hydroxyacetophenone (1 mM) as the aromatic substrate was 17.5 ± 2.5 μM, and there was no reaction when NADPH was replaced by NADH in the reaction mixture.
Amino acid sequences.
The N-terminal sequence for the first 20 amino acids was found by automated Edman degradation of the purified enzyme. This gave the following result starting at the N terminal: MRTYNTTLASLEXDDETLRA. Two internal peptides generated by digestion of the enzyme with Lys-c were sequenced in the same way and gave, for peptide A, RVAVIGTGASAAQFIPQLAK, and, for peptide B, RIIRDDGTWISTLK.
DISCUSSION
The proposal that 4-hydroxyacetophenone is converted into 4-hydroxyphenyl acetate in the catabolism of 4-ethylphenol by P. putida JD1 was confirmed here by purification of the enzyme concerned and demonstration of the proposed reaction (Fig. 1). The enzyme had a requirement for oxygen and reduced pyridine nucleotide and showed a stoichiometry of 1:1:1 for substrate, oxygen, and NADPH, all characteristic of a monooxygenase-catalyzed reaction. In this case the monooxygenase inserted one atom of the oxygen molecule between a keto group and an adjacent carbon, and the product was isolated and identified as 4-hydroxyphenyl acetate. Such a reaction resembles a Baeyer-Villiger oxidation, making this enzyme a further example of a Baeyer-Villiger type of monooxygenase. Several of these have been described that attack alicyclic or aliphatic ketones (1, 7, 8), but in contrast we have here an example of oxygen insertion into an aryl ketone. Other aryl ketones were substrates, including acetophenone and 4-hydroxypropiophenone, the apparent Km for the latter being even lower than for 4-hydroxyacetophenone. For some monooxygenases, there are compounds that appear to be substrates but act only to stimulate NAD(P)H oxidase activity and are therefore classed as pseudosubstrates (19). Here, however, the products from the two additional substrates were identified as phenyl acetate and 4-hydroxyphenyl propionate, respectively, demonstrating that these are true substrates for the enzyme.
The enzyme was purified to homogeneity and gave a single band after electrophoresis on nondenaturing gels and by SDS-PAGE (Fig. 2). The Mr of the enzyme from the SDS-PAGE was about the same as that found by gel filtration, showing that the enzyme was a single polypeptide chain of Mr 70,000.
The purified enzyme was yellow in color with a spectrum indicative of a flavoprotein (Fig. 3A). The flavin was not covalently bound and could be extracted by denaturation of the protein. Its identity as an FAD rather than an FMN was demonstrated by its correspondence to FAD on TLC. In some instances the activity of purified flavoproteins could be stimulated by addition of the appropriate flavin, probably a reflection of the enzyme-flavin dissociation constant. For example, the activity of tridecanone monooxygenase from Pseudomonas cepacia was improved by 50% by the addition of FAD (1). In contrast, there was no such stimulation of purified 4-hydroxyacetophenone monooxygenase. It was, however, possible to prepare apoenzyme which was stimulated by FAD but not by FMN, providing further evidence for FAD as the flavin in this enzyme. The FAD did not restore complete activity to the apoenzyme and possibly removal of the cofactor lowers enzyme stability, resulting in some denaturation. This might also account for the inability of FAD to stimulate purified enzyme, some of which might be in a denatured form. The ratio of protein to FAD, calculated from the absorbance spectrum, suggests that the enzyme contains one molecule of FAD.
The Baeyer-Villiger monooxygenases as a group show considerable variation in complexity of their structures. For example camphor monooxygenase from P. putida C1B consists of two proteins, one of which acts to transfer electrons from NADH to the oxygenating protein (20). Others are single proteins capable of oxidizing reduced pyridine nucleotide directly, although these range from monomeric proteins such as cyclohexanone monooxygenase from Acinetobacter sp. strain NCIMB 9871 and Nocardia globerula CL1 (7) to the multimeric enzyme cyclopentanone monooxygenase from Pseudomonas sp. strain NCIMB 9872 (8). All are flavoproteins, and it has been shown that, as for flavin hydroxylases, the reactive oxygen species in cyclohexanone monooxygenase from Acinetobacter is 4a-hydroperoxyflavin (17). The 4-hydroxyacetophenone monooxygenase described here shows similarities with cyclohexanone monooxygenase. It is a monomeric protein, although slightly larger (Mr 70,000) than the cyclohexanone enzyme (Mr 59,000), and it contains one molecule of FAD. The FAD can be reduced rapidly by the electron donor NADPH under anaerobic conditions in the absence of substrate. There was no reduction by NADH, a further demonstration of the strict specificity for an electron donor. Unlike cyclohexanone monooxygenase and other flavin monooxygenases, there was no perturbation of the flavin spectrum by substrate. This precluded a study of substrate binding from difference spectra as has been done with other enzymes. Also the N-terminal sequence showed no similarity with cyclohexanone monooxygenase or indeed with any other protein. However, one of the internal sequences did show considerable similarity to part of the potential ADP-binding site recognized in the sequence of cyclohexanone monooxygenase from Acinetobacter sp. strain NCIMB 9871 (Fig. 4), which is suggested as being involved in binding the nicotinamide cofactor (2). Even greater similarity is seen with the sequence derived from a putative cyclohexanone monooxygenase gene in Pseudomonas fluorescens DSM 50106 (11), and the presence of such a site in the enzyme is in accord with its use of NADPH as a cofactor. Further comparisons will be made when the gene for this enzyme has been cloned and sequenced. This report on the purification and characteristics of the enzyme lays the foundations for such studies and for a more extensive study of specificity.
FIG. 4.
A comparison of an internal amino acid sequence from 4-hydroxyacetophenone with sequences from a putative cyclohexanone monooxygenase from P. fluorescens DSM 50106 (11) and the proposed NADP+-binding site in cyclohexanone monooxygenase from Acinetobacter sp. strain NCIMB 9871 (2).
ACKNOWLEDGMENTS
We thank K. Bailey at the sequencing service of the University of Nottingham for help with peptide sequencing and P. W. Trudgill for helpful discussions.
REFERENCES
- 1.Britton L N, Markovetz A J. A novel ketone monooxygenase from Pseudomonas cepacia. J Biol Chem. 1977;252:8561–8566. [PubMed] [Google Scholar]
- 2.Chen Y-C J, Peoples O P, Walsh C T. Acinetobacter cyclohexanone monooxygenase: gene cloning and sequence determination. J Bacteriol. 1988;170:781–789. doi: 10.1128/jb.170.2.781-789.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cripps R E. The microbial metabolism of acetophenone and some chloroacetophenones by an Arthrobacter species. Biochem J. 1975;152:233–241. doi: 10.1042/bj1520233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cripps R E, Trudgill P W, Whateley J G. The metabolism of 1-phenylethanol and acetophenone by Nocardia T5 and an Arthrobacter species. Eur J Biochem. 1978;86:175–186. doi: 10.1111/j.1432-1033.1978.tb12297.x. [DOI] [PubMed] [Google Scholar]
- 5.Darby J M, Taylor D G, Hopper D J. Hydroquinone as the ring-fission substrate in the catabolism of 4-ethylphenol and 4-hydroxyacetophenone by Pseudomonas putida JD1. J Gen Microbiol. 1987;133:2137–2146. [Google Scholar]
- 6.Donoghue N A, Trudgill P W. The metabolism of cyclohexanol by Acinetobacter NCIB 9871. Eur J Biochem. 1975;60:1–7. doi: 10.1111/j.1432-1033.1975.tb20968.x. [DOI] [PubMed] [Google Scholar]
- 7.Donoghue N A, Norris D B, Trudgill P W. The purification and properties of cyclohexanone oxygenase from Nocardia globerula CL1 and Acinetobacter NCIB 9871. Eur J Biochem. 1976;63:175–192. doi: 10.1111/j.1432-1033.1976.tb10220.x. [DOI] [PubMed] [Google Scholar]
- 8.Griffin M, Trudgill P W. Purification and properties of cyclopentanone oxygenase from Pseudomonas NCIB 9872. Eur J Biochem. 1976;63:199–209. doi: 10.1111/j.1432-1033.1976.tb10222.x. [DOI] [PubMed] [Google Scholar]
- 9.Higson F K, Focht D D. Bacterial degradation of ring-chlorinated acetophenones. Appl Environ Microbiol. 1990;56:3678–3685. doi: 10.1128/aem.56.12.3678-3685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Itagaki E. Studies on steroid monooxygenase from Cylindrosporon radicicola ATCC 11011. Purification and characterization. J Biochem (Tokyo) 1986;99:815–824. doi: 10.1093/oxfordjournals.jbchem.a135541. [DOI] [PubMed] [Google Scholar]
- 11.Khalameyzer, V., I. Fischer, U. T. Bornscheuer, and J. Altenbuchner. 1999. Screening, nucleotide sequence, and biochemical characterization of an esterase from Pseudomonas fluorescens with high activity towards lactones. 65:477–482. [DOI] [PMC free article] [PubMed]
- 12.Leatherbarrow R J. ENZFITTER. Cambridge, United Kingdom: Elsevier-Biosoft; 1987. [Google Scholar]
- 13.Rahim M A, Sih C J. Mechanisms of steroid oxidation by micoorganisms. XI. Enzymatic cleavage of the pregnane side chain. J Biol Chem. 1966;241:3615–3623. [PubMed] [Google Scholar]
- 14.Roberts S M, Wan P W H. Enzyme-catalysed Baeyer-Villiger oxidations. J Mol Catal B-Enzymat. 1998;4:111–136. [Google Scholar]
- 15.Roberts S M, Willetts A J. Development of the enzyme-catalysed Baeyer-Villiger reaction as a useful technique in organic synthesis. Chirality. 1993;5:334–337. [Google Scholar]
- 16.Rosenberger R F, Elsden S R. The yields of Streptococcus faecalis grown in continuous culture. J Gen Microbiol. 1960;22:726–739. doi: 10.1099/00221287-22-3-726. [DOI] [PubMed] [Google Scholar]
- 17.Ryerson C C, Ballou D P, Walsh C. Mechanistic studies on cyclohexanone oxygenase. Biochemistry. 1982;21:2644–2655. doi: 10.1021/bi00540a011. [DOI] [PubMed] [Google Scholar]
- 18.Smith I. Chromatographic and electrophoretic techniques. 2nd ed. Vol. 1. London: W. Heinemann (Medical Books) Ltd.; 1960. pp. 291–298. [Google Scholar]
- 19.Spector T, Massey V. Interactions of substrate and non-substrate effectors with p-hydroxybenzoate hydroxylase from Pseudomonas fluorescens. Biochem Biophys Res Commun. 1971;45:1219–1226. doi: 10.1016/0006-291x(71)90148-3. [DOI] [PubMed] [Google Scholar]
- 20.Taylor D G, Trudgill P W. Camphor revisited: studies of 2,5-diketocamphane, 1,2-monooxygenase from Pseudomonas ATCC 17453. J Bacteriol. 1986;165:489–497. doi: 10.1128/jb.165.2.489-497.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsh C T, Chen Y-C J. Enzymic Baeyer-Villiger oxidations by flavin-dependent monooxygenases. Angew Chem Int Ed. 1988;27:333–343. [Google Scholar]
- 22.Whitby L G. A new method for preparing flavin-adenine dinucleotide. Biochem J. 1953;54:437–442. doi: 10.1042/bj0540437. [DOI] [PMC free article] [PubMed] [Google Scholar]