Abstract
Purpose of review
Ribosomal RNAs (rRNAs) are transcribed within nucleoli from rDNA repeats by RNA Polymerase I (Pol I). There is variation in rRNA transcription rates across the hematopoietic tree, and leukemic blast cells have prominent nucleoli, indicating abundant ribosome biogenesis. The mechanisms underlying these variations are poorly understood. The purpose of this review is to summarize findings of rDNA binding and Pol I regulation by hematopoietic transcription factors.
Recent findings
Our group recently used custom genome assemblies optimized for human and mouse rDNA mapping to map nearly 2200 ChIP-Seq datasets for nearly 250 factors to rDNA, allowing us to identify conserved occupancy patterns for multiple transcription factors. We confirmed known rDNA occupancy of MYC and RUNX factors, and identified new binding sites for CEBP factors, IRF factors, and SPI1 at canonical motif sequences. We also showed that CEBPA degradation rapidly leads to reduced Pol I occupancy and nascent rRNA in mouse myeloid cells.
Summary
We propose that a number of hematopoietic transcription factors bind rDNA and potentially regulate rRNA transcription. Our model has implications for normal and malignant hematopoiesis. This review summarizes the literature, and outlines experimental considerations to bear in mind while dissecting transcription factor roles on rDNA.
Keywords: CEBPA, hematopoiesis, leukemia, ribosome, rRNA, transcription factor
INTRODUCTION
The hematopoietic system, like any major organ system, is composed of cells of varying sizes, functions, and proliferation rates [1,2■■,3■]. Each cell type has a tightly regulated number of ribosomes, and it is theorized that ribosome concentration is a crucial determinant of how a cell’s complex transcriptome, comprising mRNAs with a range of translation initiation efficiencies, is translated into its appropriate proteome [4]. Ribosomes are ribonucleoprotein machines composed of four ribosomal RNAs (rRNAs) and nearly 80 ribosomal proteins; altered ribosome abundance can result from altered levels of rRNAs, ribosomal proteins, or over 200 additional proteins that are required for appropriate ribosomal folding, packaging, and quality control [5]. The relevance of ribosomal proteins to hematopoietic biology and disease is widely appreciated; Diamond-Blackfan anemia is caused by germline haploinsufficiency of over 20 ribosomal protein proteins, and ribosomal protein somatic deletions or mutations are observed in myeloid and lymphoid malignancies [6]. Less well appreciated is the relevance of rRNAs and the regulation of their transcription, despite the fact that rRNAs form over half of the mass of the ribosome and account for over 80% of total cellular RNA.
The four rRNAs, in descending size, are 28S, 18S, 5.8S, and 5S rRNA, the first three of which are transcribed as a single 13.4-kb nascent 47S pre-rRNA by RNA Polymerase I (Pol I), while 5S is transcribed by RNA Pol III [1]. 47S pre-rRNA is transcribed from specialized repeats known as rDNA genes, which are present in 200–600 copies in tandem arrays across 5 chromosomes (Fig. 1). These copies are near-identical, each nearly 45 kb in length, and each containing promoters, a 13.4-kb transcribed region and an nearly 30-kb intergenic spacer (IGS). This short review will focus only on recent advances in our understanding of 47S transcription in hematopoiesis, and not on processing or modification of rRNAs, the role of ribosomal proteins, or factors controlling the translational roles of mature subunits, all of which merit their own detailed consideration beyond our current scope.
FIGURE 1.
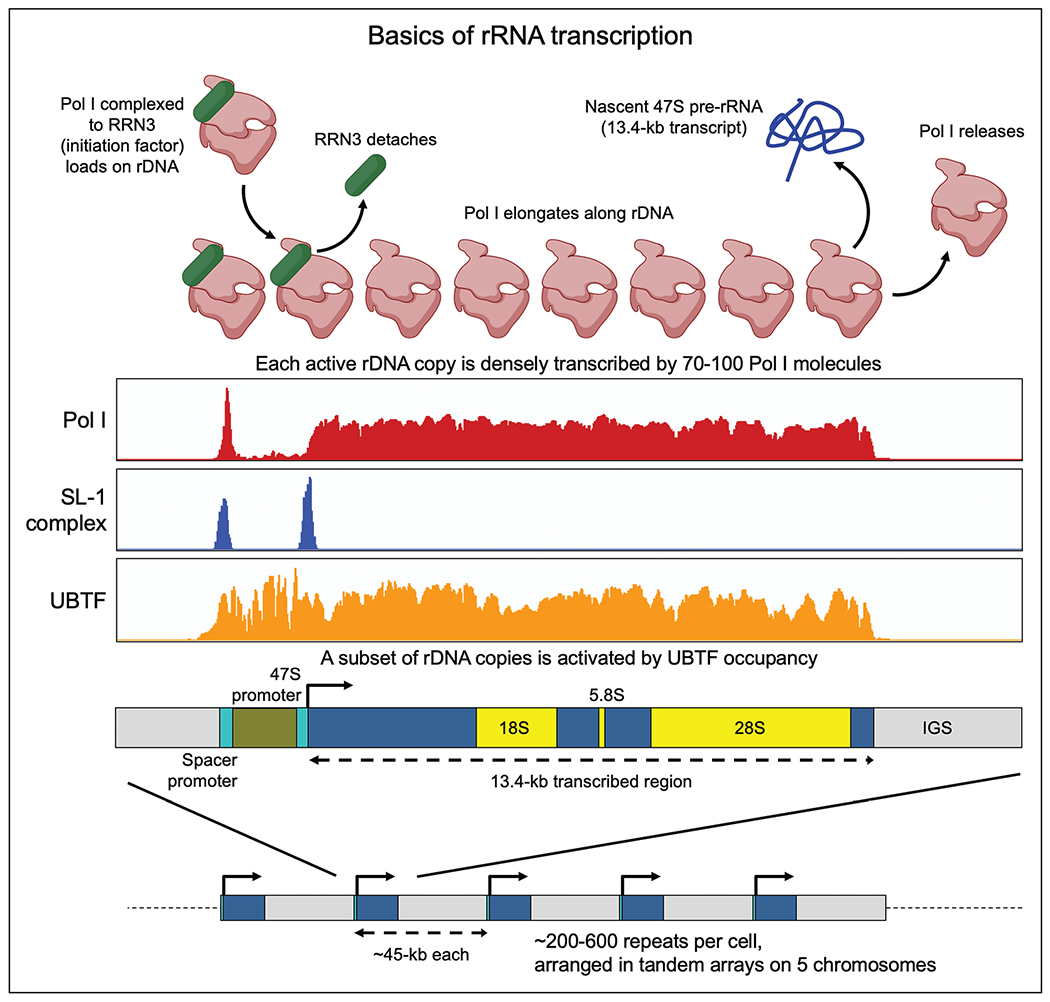
Basics of rRNA transcription. rDNA repeats are present in hundreds of near-identical copies per cell in tandem arrays. A subset of repeats is activated and transcribed in any given cell. Note: Pol I and SL-1 occupancy is seen both at the Spacer promoter and the 47S promoter, but their functions at the former are unknown. Adapted from [3■].
Mammalian rDNA transcription machinery
Each rDNA repeat has two promoters spaced nearly 800nt apart in humans and nearly 2 kb apart in mouse [3■,7] (Fig. 1). The downstream promoter (47S promoter) is the one from which 47S pre-rRNA transcription begins, and the upstream promoter (Spacer promoter), though conserved, has unknown functions. In any given cell, a subset of rDNA repeats is activated by occupancy of the promoters and the 13.4-kb transcribed region by the HMG box protein UBTF (aka UBF), accompanied by depletion of nucleosomes across the stretch [1,8]. UBTF recruits the TATA-binding protein complex SL-1 (five subunits) to both promoters. Pol I, a 13-subunit complex, can occupy rDNA only when bound to its initiation factor RRN3. Once the Pol I-RRN3 complex is loaded, and transcription of 47S pre-rRNA has begun, RRN3 detaches and Pol I elongates along the rDNA gene. A notable difference between Pol I and Pol II is that while Pol II has a well established pause-release mechanism, it is currently controversial whether Pol I pauses, and whether its pausing could be a potential node for regulation [1,9]. TTF1 (Transcription Termination Factor 1) binds to the promoters of rDNA as well as to the 3′ end of the transcribed region [10], is required for termination of rRNA transcription, and is believed to produce a looped structure between the promoter and termination site of each rDNA repeat [11]. All core machinery components (UBTF, SL-1, Pol I, RRN3, TTF1) have multiple regulatory PTMs (mostly phosphorylation) that are controlled by mTOR, AKT, MAPK, and other signaling kinases. See recent reviews for a more detailed overview of rRNA transcription [1,8,9].
Variations in rRNA transcription in the hematopoietic tree
It has long been known that terminal erythropoiesis involves shutdown of rRNA transcription and degradation of mature ribosomes [12,13]. Beyond erythropoiesis, only a handful of studies have quantified rRNA transcription in hematopoiesis. Hayashi et al. [14] and Jarzebowski et al. [15] have reported that mouse hematopoietic stem cells (HSCs) and multiple mature cell types have lower rRNA transcription compared to myeloid progenitor populations, with many-fold variation that is not explained by differences in proliferation. We recently published a protocol for FISH-Flow of nascent and mature rRNA [16■], and our group and others are currently in the process of assembling a detailed profile of rRNA dynamics in normal and disordered hematopoiesis.
Challenges in rDNA studies
Bioinformatic challenges
The repetitive nature of rDNA poses nontrivial challenges in high-throughput mapping. Standard genome assemblies historically did not contain intact rDNA sequences, and though reference sequences are available for human/mouse rDNA [17–19], they are typically not used by most investigators for routine mapping. Consequently, though ATAC-Seq, ChIP-Seq, RNA-Seq datasets and others contain rDNA-mapping reads, these reads are discarded or improperly mapped by standard pipelines. Inspired by past approaches [20,21], our group recently generated custom human and mouse genome assemblies optimized for rDNA mapping - these assemblies contain a single reference sequence inserted as an extra “chromosome R” [3■]; high-throughput datasets can be mapped to it using standard tools, enabling rDNA signal to be easily visualized (Fig. 1). We have made these custom genomes, annotation files, and positive and negative control datasets available on GitHub, hoping to make rDNA mapping more accessible to non-aficionados (https://github.com/vikramparalkar/rDNA-Mapping-Genomes). We recently used this approach to map nearly 2200 human and mouse ChIP-Seq datasets for nearly 250 transcription factors and chromatin proteins of relevance to normal and malignant hematopoiesis, and identified previously unknown rDNA-occupancy patterns for several important factors [2■■], some of which we discuss in this review.
Experimental challenges
A primary challenge in rDNA studies is determining whether a factor in question regulates transcription directly through rDNA binding, or indirectly through its Pol II mediated effects on the coding transcriptome. This question often arises in studies of gene regulation more broadly; it is increasingly appreciated that TFs (including oncogenic transcription factor fusions) regulate the transcription of only a minority of the genomic loci to which they bind [22–24]. Dissecting the direct targets of a transcription factor has historically been performed using a combination of approaches: (i) Inducible expression or degron studies to rapidly manipulate transcription factor levels and assay early effects on nascent transcription, (ii) Chromatin-tethering to selectively restore transcription factor binding only to a specific site, (iii) Editing a transcription factor motif to specifically abolish binding to a particular locus, and (iv) Reporter assays assessing a transcription factor’s ability to boost expression of a reporter transgene. Approaches (ii) and (iii) have been challenging at rDNA arrays due to their repetitive nature, and, to our knowledge, have not yet been reported in the literature. Approach (iv) has the weakness of being divorced from the native chromatin context of the locus in question, a weakness that is particularly salient for rDNA, given its specialized epigenetic features. With advancements in CRISPR base editing technology, we expect some of these challenges to soon be overcome.
Transcription factors that bind rDNA
Multiple transcription factors bind and potentially regulate rRNA transcription, but there have historically been only a small number of studies experimentally structured to uncouple direct transcription factor effects from indirect ones. We summarize below data for factors of hematopoietic relevance (Fig. 2). For factors like MYC and RUNX, we include findings from nonhematopoietic cell types that may inform future studies in hematopoietic cells. This list is not comprehensive, and our group and others have identified additional factors of interest that are worthy of investigation [2■■].
FIGURE 2.
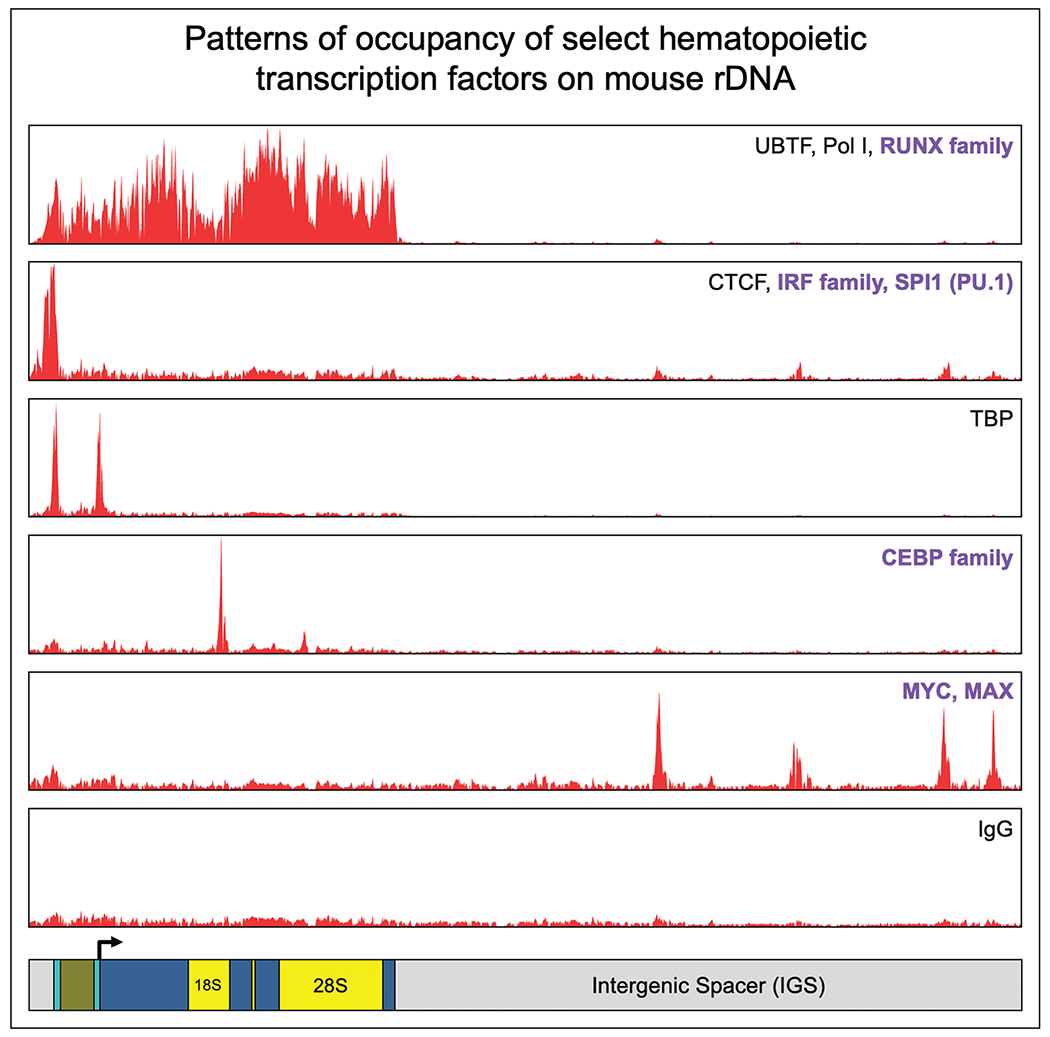
Occupancy patterns of selected hematopoietic transcription factors on mouse rDNA. Patterns of ChIP-Seq occupancy on mouse rDNA, shown as averaged consensus track signals for multiple factors per pattern. Select transcription factors of relevance to normal and malignant hematopoiesis are listed in bold for each binding pattern. Adapted from [2■■].
Universal transcription factors
TBP (TATA-binding protein), part of the SL-1 complex mentioned earlier, binds to the Spacer promoter and 47S promoter of rDNA, and is essential to recruit Pol I [25]. CTCF binds immediately upstream of the Spacer promoter, and its knockout leads to reduced rRNA transcription and altered nucleolar number [26], Notably, the entire cohesin complex shows rDNA occupancy immediately upstream to the CTCF peak [2■■,7], but it is unknown whether CTCF/cohesin regulate rDNA topology.
MYC
MYC is widely considered the master regulator of ribosome biogenesis, and is believed to coordinate transcription of rRNA by Pol I, transcription of genes coding for ribosomal proteins and other ribosome biogenesis factors by Pol II, and transcription of 5S rRNA by Pol III [27]. MYC directly binds human and mouse rDNA repeats, but our mapping of ChIP-Seq datasets for MYC and its dimerization partner MAX unexpectedly found that its rDNA binding sites fell in different regions of rDNA in the two species: human MYC binds within the transcribed region of rDNA, while mouse MYC shows multiple peaks in the IGS [2■■]. The strongest evidence of direct rRNA regulation by MYC comes from studies in human fibroblasts expressing MYC-ER fusion protein, in which activation of MYC by tamoxifen treatment led to increased nascent rRNA levels within 3–4 h, pointing to direct control [28,29]. However, it has also been reported that MYC promotes transcription of genes encoding UBTF and Pol I complex proteins, pointing to potential indirect mechanisms in addition to direct ones [30,31]. In contrast to our relatively detailed understanding of MYC effects on Pol II [32], there is limited mechanistic understanding of how MYC controls Pol I.
CEBP family
The transcription factor family that we found in our recent mapping to show the most striking and consistent binding to rDNA was the CEBP (CCAAT/enhancer-binding protein) family, with 19 human and 13 mouse ChIP-seq tracks showing a sharp conserved peak for four of six members of the CEBP family (CEBPA, CEBPB, CEBPG, CEBPD) [2■■]. The peak was at a canonical motif sequence nearly 5-kb downstream of the 47S transcription start site, within the region that is transcribed into 18S rRNA. CEBP factors are a family of leucine zipper TFs that bind DNA as homo or heterodimers [33], and CEBPA is a master myeloid TF whose hematopoietic deletion is known to cause loss of the myeloid lineage [34,35]. Using an FKBP degron system in a mouse myeloid cell line, we found that CEBPA degradation led to reduced Pol I occupancy on rDNA and reduced 47S rRNA levels within 4 h, without any change in the cellular abundance of core rDNA machinery [2■■]. Occupancy of RRN3 was also proportionately reduced, while occupancy of upstream complexes was not, indicating that CEBPA controls the loading of the Pol I-RRN3 complex on rDNA. CEBPA is a single exonic gene, translated into a p42 and p30 isoform from two start codons. An extended isoform of CEBPA with nucleolar localization was previously reported to be translated from an upstream alternate start codon [36]; however, we could not identify such an extended isoform in our cells, indicating that one or both of the “usual” CEBPA isoforms likely bind rDNA. This has implications for AML, in which N-terminal mutations in the CEBPA gene cause selective loss of the p42 isoform [37], and it is reasonable to speculate that p30 and p42 may play divergent roles on rDNA. Overall, our work suggests that one of the normal functions of CEBPA is to promote loading of the Pol I-RRN3 complex on rDNA in a cell-type specific manner (Fig. 3). The CEBP motif on rDNA provides an attractive site for motif editing to rigorously uncouple the Pol I vs. Pol II roles of CEBPA and other CEBP family members.
FIGURE 3.
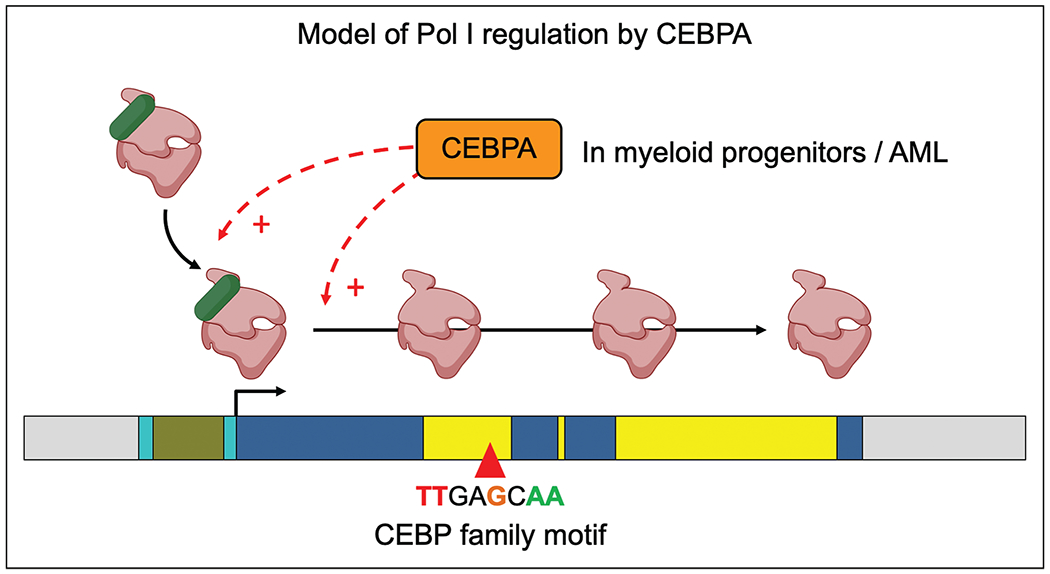
Model of Pol I regulation by CEBPA. We propose that in myeloid progenitors and in AML, CEBPA promotes the loading of Pol I-RRN3 complex on rDNA and/or the elongation of Pol I along the transcribed region. We speculate that this mechanism may be shared by other rDNA-binding TFs.
IRF family and SPI1
Another prominent set of transcription factors that we identified binding to rDNA was the IRF family (Interferon regulatory factor) and SPI1 (PU.1), which are known to combinatorially control important aspects of hematopoiesis, inflammation, and immunity [38,39]. We identified peaks for multiple IRF family members on both human and mice rDNA at a conserved IRF motif immediately upstream of the Spacer promoter [2■■]. SPI1 showed divergence in binding sites between the two species, with human binding in the IGS, and mouse binding upstream of the Spacer Promoter. To our knowledge, the roles of these factors on rDNA have not been explored, but their consistent binding across a large number of independent datasets provides compelling reason for further investigation.
RUNX family
RUNX factors, as well as RUNX1-RUNX1T1 fusion protein, have been reported to bind rDNA, with persistent binding during mitosis that was visualized as striking co-localization with UBTF on rDNA arrays of mitotic chromosomes in a number of human and mouse cell types [40–42]. In our ChIP-Seq mapping, we found broad occupancy of RUNX factors to promoters as well as to the entire 13-kb transcribed region of rDNA, without a discrete peak that would be expected if binding were driven by a TF motif [2■■]. Such a pattern of ChIP-Seq signal indicates that RUNX rDNA occupancy is likely mediated through interactions with core machinery, potentially UBTF. In nonhematopoietic cell types, RUNX2 and RUNX3 have been reported to repress rRNA transcription [40,41], while conditional hematopoietic deletion of RUNX1 leads to HSCs with smaller size, reduced ribosome content, and reduced expression of several ribosomal protein genes [43]. This effect is restricted to HSCs, indicating that RUNX1 may promote rRNA transcription and associated gene networks in a context-specific manner. Further studies will be required to illuminate the extent to which RUNX effects on rDNA/rRNA are mediated via its occupancy on rDNA chromatin.
Considerations when testing transcription factor-rDNA regulation
Since rRNA transcription is highly sensitive to a variety of cellular stresses and stimuli, it is critical to take into account multiple alternate explanations when trying to draw a direct mechanistic link between the manipulation of a TF and a change in rRNA transcription (Fig. 4). These alternate explanations may range from indirect effects on cell cycle or apoptosis, to effects on ribonucleotide availability, to changes in abundance of the core rDNA machinery. We urge investigators dissecting TF-rDNA regulation to bear these considerations in mind in the course of their studies.
FIGURE 4.
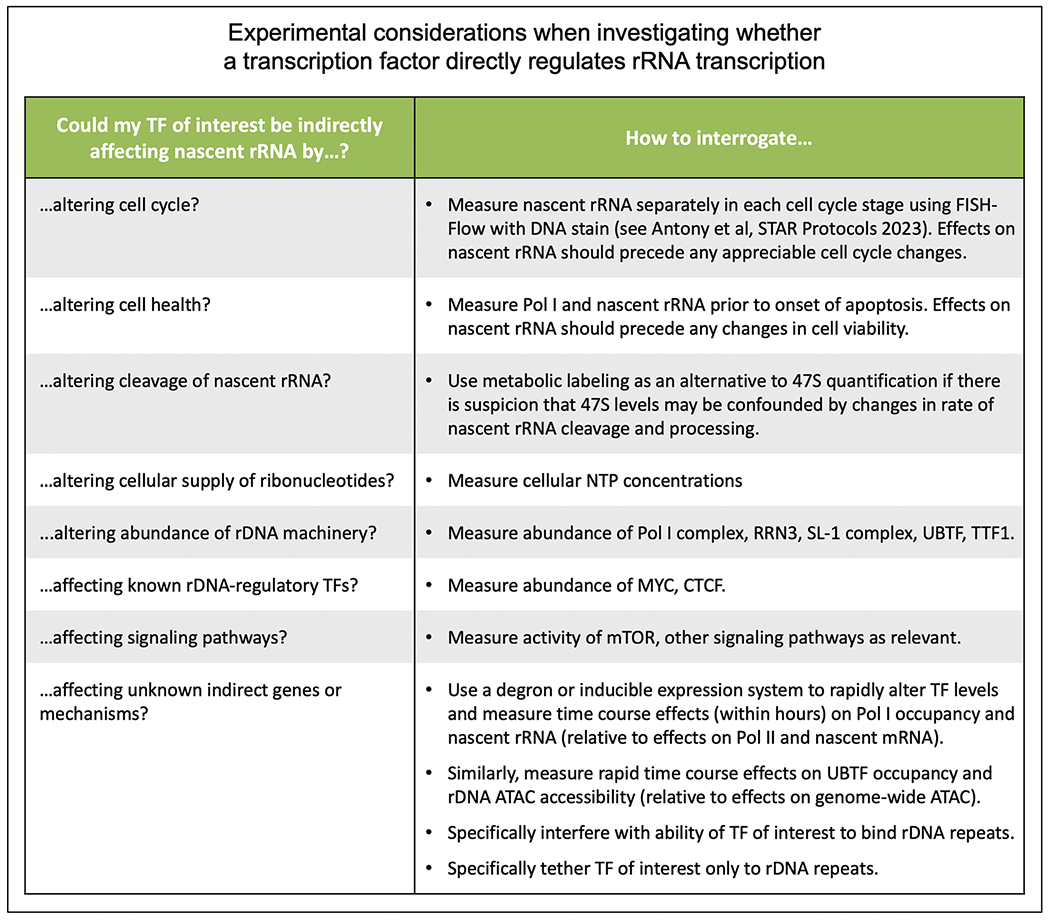
Considerations for TF-rDNA studies. We list several experimental considerations to be borne in mind while investigating whether a TF of interest directly regulates the transcription of rDNA into rRNA.
CONCLUSION
We propose that the rate of rRNA transcription is fine-tuned in each hematopoietic cell type by multiple mechanisms, one of which is through direct rDNA binding and control by some of the same master transcription factors that control the cell’s coding transcriptome. This model has implications for normal and malignant hematopoiesis, given the diversity of transcription factors that control cellular identity and fate, and whose mutations or dysregulation lead to cancer. We hope that recent tools and approaches will make systematic interrogation of rRNA transcription more accessible in hematopoietic biology and beyond.
KEY POINTS.
Ribosomal RNAs are transcribed from hundreds of rDNA repeats, and their transcription rate varies in different hematopoietic cell types.
Recent mapping pipelines have made rDNA more accessible for high-throughput mapping.
Several hematopoietic transcription factors, including MYC, CEBP, RUNX family, and others, bind rDNA, and varying levels of evidence are available to support their role in regulating rRNA transcription.
We provide a list of experimental considerations for future studies of TF-rDNA regulation, including potential indirect mechanisms that should be taken into account and ruled out.
Overall, we propose a model in which the transcription of rRNA, just like the coding transcriptome, is regulated in a cell-type specific manner in normal and malignant hematopoietic cells by master transcription factors.
Acknowledgements
The authors apologize to authors whose work we could not cite due to word limits. They thank members of the Paralkar Lab for valuable scientific discussion. Illustrations were made using BioRender.
Financial support and sponsorship
V.R.P is supported by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health (NIH) under award number R35-GM138035.
Footnotes
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
■ of special interest
■■ of outstanding interest
- 1.Hori Y, Engel C, Kobayashi T. Regulation of ribosomal RNA gene copy number, transcription and nucleolus organization in eukaryotes. Nat Rev Mol Cell Biol 2023; 24:414–429. [DOI] [PubMed] [Google Scholar]
- 2.■■.Antony C, George SS, Blum J, et al. Control of ribosomal RNA synthesis by hematopoietic transcription factors. Mol Cell 2022; 82:3826–3839; e9. [DOI] [PMC free article] [PubMed] [Google Scholar]; Profile of hematopoietic transcription factor-rDNA binding, and demonstration that CEBPA degradation leads to rapid reduction in rRNA transcription.
- 3.■.George SS, Pimkin M, Paralkar VR. Construction and validation of customized genomes for human and mouse ribosomal DNA mapping. J Biol Chem 2023; 299:104766. [DOI] [PMC free article] [PubMed] [Google Scholar]; Customized genome assemblies for mapping human and mouse high-throughput datasets to rDNA.
- 4.Mills EW, Green R. Ribosomopathies: there’s strength in numbers. Science 2017; 358:eaan2755. [DOI] [PubMed] [Google Scholar]
- 5.Dörner K, Ruggeri C, Zemp I, et al. Ribosome biogenesis factors-from names to functions. EMBO J 2023; 42:e112699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vlachos A. Acquired ribosomopathies in leukemia and solid tumors. Hematology Am Soc Hematol Educ Program 2017; 2017:716–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moss T, Mars J-C, Tremblay MG, et al. The chromatin landscape of the ribosomal RNA genes in mouse and human. Chromosome Res 2019; 27:31–40. [DOI] [PubMed] [Google Scholar]
- 8.Daiß JL, Griesenbeck J, Tschochner H, et al. Synthesis of the ribosomal RNA precursor in human cells: mechanisms, factors and regulation. Biol Chem 2023; 404:1003–1023. [DOI] [PubMed] [Google Scholar]
- 9.Girbig M, Misiaszek AD, Müller CW. Structural insights into nuclear transcription by eukaryotic DNA-dependent RNA polymerases. Nat Rev Mol Cell Biol 2022; 23:603–622. [DOI] [PubMed] [Google Scholar]
- 10.Bartsch I, Schoneberg C, Grummt I. Purification and characterization of TTFI, a factor that mediates termination of mouse ribosomal DNA transcription. Mol Cell Biol 1988; 8:3891–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Németh A, Guibert S, Tiwari VK, et al. Epigenetic regulation of TTF-I-mediated promoter-terminator interactions of rRNA genes. EMBO J 2008; 27:1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolznig H, Bartunek P, Nasmyth K, et al. Terminal differentiation of normal chicken erythroid progenitors: shortening of G1 correlates with loss of D-cyclin/cdk4 expression and altered cell size control. Cell Growth Differ 1995; 6:1341–1352. [PubMed] [Google Scholar]
- 13.Le Goff S, Boussaid I, Floquet C, et al. p53 activation during ribosome biogenesis regulates normal erythroid differentiation. Blood 2021; 137:89–102. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi Y, Kuroda T, Kishimoto H, et al. Downregulation of rRNA transcription triggers cell differentiation. PLoS One 2014; 9:e98586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarzebowski L, Le Bouteiller M, Coqueran S, et al. Mouse adult hematopoietic stem cells actively synthesize ribosomal RNA. RNA 2018; 24:1803–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.■.Antony C, Somers P, Gray EM, et al. FISH-Flow to quantify nascent and mature ribosomal RNA in mouse and human cells. STAR Protoc 2023; 4:102463. [DOI] [PMC free article] [PubMed] [Google Scholar]; Protocol for per-cell quantification of nascent and mature rRNA in rare and heterogenous populations, and in different stages of cell cycle.
- 17.Sayers EW, Beck J, Bolton EE, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2024; 52:D33–D43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J-H, Dilthey AT, Nagaraja R, et al. Variation in human chromosome 21 ribosomal RNA genes characterized by TAR cloning and long-read sequencing. Nucleic Acids Res 2018; 46:6712–6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grozdanov P, Georgiev O, Karagyozov L. Complete sequence of the 45-kb mouse ribosomal DNA repeat: analysis of the intergenic spacer. Genomics 2003; 82:637–643. [DOI] [PubMed] [Google Scholar]
- 20.Zentner GE, Saiakhova A, Manaenkov P, et al. Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res 2011; 39:4949–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zentner GE, Balow SA, Scacheri PC. Genomic characterization of the mouse ribosomal DNA locus. G3 2014; 4:243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stengel KR, Ellis JD, Spielman CL, et al. Definition of a small core transcriptional circuit regulated by AML1-ETO. Mol Cell 2021; 81:530–545; e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harada T, Kalfon J, Perez MW, et al. Leukemia core transcriptional circuitry is a sparsely interconnected hierarchy stabilized by incoherent feed-forward loops. bioRxiv 2023. doi: 10.1101/2023.03.13.532438. [DOI] [Google Scholar]
- 24.Harada T, Perez MW, Kalfon J, et al. Rapid-kinetics degron benchmarking reveals off-target activities and mixed agonism-antagonism of MYB inhibitors. bioRxiv 2023. doi: 10.1101/2023.04.07.536032. [DOI] [Google Scholar]
- 25.Comai L, Tanese N, Tjian R. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell 1992; 68:965–976. [DOI] [PubMed] [Google Scholar]
- 26.van de Nobelen S, Rosa-Garrido M, Leers J, et al. CTCF regulates the local epigenetic state of ribosomal DNA repeats. Epigenetics Chromatin 2010; 3:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 2010; 10:301–309. [DOI] [PubMed] [Google Scholar]
- 28.Arabi A, Wu S, Ridderstråle K, et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol 2005; 7:303–310. [DOI] [PubMed] [Google Scholar]
- 29.Grandori C, Gomez-Roman N, Felton-Edkins ZA, et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 2005; 7:311–318. [DOI] [PubMed] [Google Scholar]
- 30.Poortinga G, Hannan KM, Snelling H, et al. MAD1 and c-MYC regulate UBF and rDNA transcription during granulocyte differentiation. EMBO J 2004; 23:3325–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poortinga G, Wall M, Sanij E, et al. c-MYC coordinately regulates ribosomal gene chromatin remodeling and Pol I availability during granulocyte differentiation. Nucleic Acids Res 2011; 39:3267–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lourenco C, Resetca D, Redel C, et al. MYC protein interactors in gene transcription and cancer. Nat Rev Cancer 2021; 21:579–591. [DOI] [PubMed] [Google Scholar]
- 33.Tsukada J, Yoshida Y, Kominato Y, et al. The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors is a multifaceted highly-regulated system for gene regulation. Cytokine 2011; 54:6–19. [DOI] [PubMed] [Google Scholar]
- 34.Zhang D-E, Zhang P, Wang N-D, et al. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein-deficient mice. Proc Natl Acad Sci U S A 1997; 94:569–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang P, Iwasaki-Arai J, Iwasaki H, et al. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity 2004; 21:853–863. [DOI] [PubMed] [Google Scholar]
- 36.Müller C, Bremer A, Schreiber S, et al. Nucleolar retention of a translational C/EBPalpha isoform stimulates rDNA transcription and cell size. EMBO J 2010; 29:897–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taube F, Georgi JA, Kramer M, et al. CEBPA mutations in 4708 patients with acute myeloid leukemia: differential impact of bZIP and TAD mutations on outcome. Blood 2022; 139:87–103. [DOI] [PubMed] [Google Scholar]
- 38.Tamura T, Yanai H, Savitsky D, et al. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol 2008; 26:535–584. [DOI] [PubMed] [Google Scholar]
- 39.Burda P, Laslo P, Stopka T. The role of PU.1 and GATA-1 transcription factors during normal and leukemogenic hematopoiesis. Leukemia 2010; 24:1249–1257. [DOI] [PubMed] [Google Scholar]
- 40.Young DW, Hassan MQ, Pratap J, et al. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature 2007; 445:442–446. [DOI] [PubMed] [Google Scholar]
- 41.Pande S, Ali SA, Dowdy C, et al. Subnuclear targeting of the Runx3 tumor suppressor and its epigenetic association with mitotic chromosomes. J Cell Physiol 2009; 218:473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bakshi R, Zaidi SK, Pande S, et al. The leukemogenic t(8;21) fusion protein AML1-ETO controls rRNA genes and associates with nucleolar-organizing regions at mitotic chromosomes. J Cell Sci 2008; 121:3981–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai X, Gao L, Teng L, et al. Runx1 deficiency decreases ribosome biogenesis and confers stress resistance to hematopoietic stem and progenitor cells. Cell Stem Cell 2015; 17:165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]


