Graphical abstract
KEYWORDS: 3D reconstruction, Cleavage stage, Embryology, Cell arrangement, Cell contact, Network analysis
Highlights
-
•
Increased cell contact is associated with greater developmental potential in embryos.
-
•
This association is strongest at the eight-cell stage.
-
•
Three-dimensional embryo analysis may provide new insights into embryo development.
Abstract
Research question
What can three-dimensional cell contact networks tell us about the developmental potential of cleavage-stage human embryos?
Design
This pilot study was a retrospective analysis of two Embryoscope imaging datasets from two clinics. An artificial intelligence system was used to reconstruct the three-dimensional structure of embryos from 11-plane focal stacks. Networks of cell contacts were extracted from the resulting embryo three-dimensional models and each embryo's mean contacts per cell was computed. Unpaired t-tests and receiver operating characteristic curve analysis were used to statistically analyse mean cell contact outcomes. Cell contact networks from different embryos were compared with identical embryos with similar cell arrangements.
Results
At t4, a higher mean number of contacts per cell was associated with greater rates of blastulation and blastocyst quality. No associations were found with biochemical pregnancy, live birth, miscarriage or ploidy. At t8, a higher mean number of contacts was associated with increased blastocyst quality, biochemical pregnancy and live birth. No associations were found with miscarriage or aneuploidy. Mean contacts at t4 weakly correlated with those at t8. Four-cell embryos fell into nine distinct cell arrangements; the five most common accounted for 97% of embryos. Eight-cell embryos, however, displayed a greater degree of variation with 59 distinct cell arrangements.
Conclusions
Evidence is provided for the clinical relevance of cleavage-stage cell arrangement in the human preimplantation embryo beyond the four-cell stage, which may improve selection techniques for day-3 transfers. This pilot study provides a strong case for further investigation into spatial biomarkers and three-dimensional morphokinetics.
Introduction
In the earliest stages of life, communication is key; nowhere is this more evident than in the world of embryonic development. Across many biological systems, the establishment of cellular polarity, which, in the human embryo takes place between the eight- and 16-cell stages, depends on cell-to-cell communication via intercellular contacts (Ajduk and Zernicka-Goetz, 2016). The amount of contact between blastomeres is determined by the spatial arrangement of blastomeres within the embryo, to which several factors, including cleavage patterns (Ebner et al., 2012; Cauffman et al., 2014; Ajduk and Zernicka-Goetz, 2016; Desai and Gill, 2019), fragmentation (Alikani et al., 1999a) and ploidy (Moayeri et al., 2008) may contribute.
In a clinical setting, the assessment of four- and eight-cell embryos is relatively commonplace, especially in clinics that carry out cleavage-stage transfers. Typical criteria for a cleavage-stage embryo to be considered for transfer include a good number of evenly sized blastomeres, a good degree of symmetrical division and minimal fragmentation (Rijnders and Jansen, 1998; Dennis et al., 2006; Moayeri et al., 2008). Many studies have specifically assessed cell arrangement at the four-cell stage, with most agreeing that embryos with tetrahedral cell arrangements have greater developmental potential than those without (Ebner et al., 2012; 2017; Paternot et al., 2012; Cauffman et al., 2014; Desai and Gill, 2019). To our knowledge, however, only one study has investigated cell arrangement and contact in cleavage-stage embryos beyond the four-cell stage in a clinical setting (Paternot et al., 2012). By modelling blastomeres in embryos as spheres, the investigators found an association between culture conditions, the number of cell contacts and the total surface area of contact in day-3 embryos.
This paucity of research stems from a key difficulty: visualizing embryos with many blastomeres in three dimensions. All previous work in four-cell embryo cell arrangement has involved human assessment of embryos; such an approach is challenging with eight cells owing to the exponential increase in possible contacts between blastomeres to keep track of. Moreover, although the use of confocal microscopy for understanding three-dimensional structure is standard practice in basic research, their use is infeasible in clinical practice because of concerns around staining, phototoxicity and cost (Iyer et al., 2021). Recently, He et al. (2022) described preliminary work towards a method for recovering the three-dimensional structure of cleavage-stage embryos using an artificial intelligence system on focal stacks obtained using the Hoffman modulation contrast microscopes widely integrated into modern time-lapse incubators. Their approach involves automatically detecting individual blastomeres and using the detected outlines to generate three-dimensional meshes for each blastomere in an embryo.
In this pilot study, an artificial intelligence system proposed by He et al. (2022) is used to investigate the effect of cell arrangement on clinical cleavage-stage human embryos at the four- and eight-cell stages. Networks of blastomere contacts are derived from three-dimensional visualizations of embryos and perform simple network analyses. From these analyses, we propose a simple, autonomously assessable quantitative biomarker for the developmental potential of clinical human embryos: the mean number of intercellular contacts per blastomere.
Materials and methods
Study population, setting and clinical data collection
This pilot study was a retrospective analysis of two imaging datasets from two different clinics. All data were captured at 11 focal planes on Embryoscope incubators between 2018 and 2020 and standardized using the Super-Focus method (He et al., 2022).
The first dataset (DS1) consisted of 303 t4 embryos from the first clinic with information on blastulation (blastulated and non-blastulated) and Gardner grade (Gardner and Schoolcraft, 1999). The second dataset (DS2) consisted of 217 embryos from the second clinic at t4 and t8 with information on Gardner grade, biochemical pregnancy, live birth, kinetics (annotated by embryologists) and preimplantation genetic testing for aneuploidy (PGT-A) (which was conducted in-house on all embryos on day 5).
The study was approved by HRA and Health and Care Research Wales (HCRW) on 12 January 2021 (IRAS 287428).
Three-dimensional cell contact analysis
Three-dimensional reconstructions were obtained from each focal stack using the system proposed by He et al. (2022). The reconstructions were visualized in the Unity game engine (v2021.3.7f1) and manually validated with incorrect reconstructions being excluded. After manual validation, DS1 was reduced to 238 four-cell embryos. DS2 was reduced to 115 eight-cell embryos and 201 four-cell embryos owing to factors such as the embryos being too compacted for cell boundaries to be visible, cells not being detected, excessive fragmentation and the embryo being cut off by the well. A total of 101 embryos had reconstructions for both the four- and eight-cell stages. Further details on the datasets after manual validation are presented in Table 1.
TABLE 1.
THE MANUALLY VALIDATED DATASET
| Dataset | Stage | Parameter | n |
|---|---|---|---|
| DS1 | t4 | 238 | |
| Blastulation (blastulated/non-blastulated) | 203/35 | ||
| Blastocyst quality (good/poor) | 106/97 | ||
| DS2 | t4 | 201 | |
| Blastocyst quality (good/poor/unknown) | 125/75/1 | ||
| Blastocyst transfers | 131 | ||
| Biochemical pregnancies | 92 | ||
| Live births | 68 | ||
| Miscarriages | 11 | ||
| Ploidy (euploid/aneuploid) | 151/50 | ||
| t8 | 115 | ||
| Blastocyst quality (good/poor) | 74/41 | ||
| Blastocyst transfers | 80 | ||
| Biochemical pregnancies | 59 | ||
| Live births | 44 | ||
| Miscarriages | 7 | ||
| Ploidy (euploid/aneuploid) | 88/27 |
The existence of cell contacts was determined by scaling the meshes of each blastomere by a small factor (in our case, 1.05, determined empirically over a subset of 10 embryos) and detecting the presence of any resulting overlap. From these calculations, networks of cell contacts were constructed for each embryo, with nodes in the networks representing each blastomere and links between nodes representing cell contacts between the linked nodes. The cell contact networks were then assessed for the mean number of cell contacts per blastomere. An overview of the process is presented in Figure 1. The reconstruction process was carried out on a server running Ubuntu 20.04 machine with an Intel i7-9700K CPU, 64 GB RAM and an NVIDIA Titan X GPU. A Windows 10 desktop machine with an Intel i3-6100 CPU and 36 GB RAM were used for all visualization and further analysis.
Figure 1.
The three-dimensional analysis process: focal stacks (left) were used to generate three-dimensional models of embryos (middle) from which cell contact networks (right) were obtained.
Characterization of cell arrangement
The cell arrangement in each embryo was characterized using Hickman vectors, a mathematical representation of cell contact originally developed for the characterization of four-cell embryos (Hickman et al., 2021). A Hickman vector representing an N-cell embryo is a list of numbers in which the nth number indicates the number of blastomeres with exactly n cell contacts (for 0≤n <N). The frequency distribution of the Hickman vectors observed across the datasets was computed.
The Pandas (v1.4.1) package in Python (v3.10.3) was used for all analyses.
Statistical analysis
Unpaired t-tests were used to compare the mean number of cell contacts per blastomere with blastulation, blastocyst quality, biochemical pregnancy and live birth outcomes. Blastocyst quality was measured as a binary variable (good/poor) derived from the Gardner grade. A ‘good’ embryo was defined as having a Gardner grade with EXP>2, ICM>C and TE>C. Analyses of biochemical pregnancy, live birth and miscarriage rates only included transferred embryos (here, miscarriage is defined as the loss of a clinical pregnancy within the first 20 weeks). The Spearman rank correlation coefficient was used to evaluate the association between the mean number of cell contacts per blastomere at the four- and eight-cell stages for a given embryo.
All statistical analysis was carried out using the Pandas (v1.4.1) and Correlation (v1.0.0) packages in Python (v3.10.3). Correlation coefficient confidence intervals were computed using the bootstrapping method implemented in the Correlation (v1.0.0) package with 10000 samples at 95% confidence.
Evaluation of predictive power
The power of the mean number of contacts per blastomere as a predictor of biochemical pregnancy and live birth was evaluated using five-fold cross-validation on eight-cell embryos in DS2. For each fold, a simple threshold-based model was fitted to the training folds and evaluated on the held-out fold using the accuracy, precision, sensitivity, F1 and area under the receiver operating characteristic curve (AUC) metrics. The threshold-based models took the following form:
where is the input feature, is the outcome prediction based on , and is a cut-off threshold to be calibrated during training. Training involved an exhaustive search of potential thresholds with the goal of maximizing AUC on the training folds. The performance of the model was benchmarked against the KIDScore D3 (Petersen et al., 2016), a well-known biomarker for cleavage-stage embryo viability based on kinetic parameters (tPNf, t2, t3, t5, t8), using the same evaluation protocol. Model AUCs were compared using unpaired t-tests.
All evaluations were carried out using the scikit-learn (v1.2.1) package in Python (v3.10.3).
Results
Comparisons between the mean number of blastomere contacts and embryo outcome are presented in Table 2. At t4, a higher mean number of contacts per cell was associated with greater rates of blastulation in DS1 (P = 0.007) and blastocyst quality in both DS1 (P = 0.003) and DS2 (P = 0.014). At t8, a higher mean number of contacts was associated with increased blastocyst quality (P = 0.017), biochemical pregnancy (P = 0.003) and live birth (P < 0.001).
TABLE 2.
COMPARISON OF MEAN NUMBER OF CELL CONTACTS ACROSS DIFFERENT OUTCOMES
| Dataset | Stage | Comparison (outcome 1 versus outcome 2) | Outcome 1 (mean ± SD) | Outcome 2 (mean ± SD) | t-statistic | P-value |
|---|---|---|---|---|---|---|
| DS1 | t4 | Blastulated versus non-blastulated | 2.54 ± 0.46 | 2.30 ± 0.61 | 2.713 | 0.007a |
| Good versus poor blastocyst quality | 2.61 ± 0.40 | 2.46 ± 0.52 | 3.046 | 0.003a | ||
| DS2 | t4 | Good versus poor blastocyst quality | 2.52 ± 0.45 | 2.35 ± 0.48 | 2.475 | 0.014a |
| Pregnantb versus not pregnant | 2.49 ± 0.46 | 2.41 ± 0.57 | 0.830 | 0.408 | ||
| Live birth versus no live birth | 2.44 ± 0.46 | 2.45 ± 0.55 | −0.095 | 0.925 | ||
| Live birth versus miscarriage | 2.44 ± 0.46 | 2.59 ± 0.44 | −1.005 | 0.318 | ||
| Euploid versus aneuploid | 2.44 ± 0.49 | 2.49 ± 0.42 | −0.607 | 0.545 | ||
| t8 | Good versus poor blastocyst quality | 3.36 ± 0.61 | 3.08 ± 0.55 | 2.423 | 0.017a | |
| Pregnantb versus not pregnant | 3.32 ± 0.56 | 2.87 ± 0.63 | 3.041 | 0.003a | ||
| Live birth versus no live birth | 3.40 ± 0.53 | 2.90 ± 0.59 | 3.684 | <0.001a | ||
| Live birth versus miscarriage | 3.40 ± 0.53 | 3.00 ± 0.50 | 1.842 | 0.072 | ||
| Euploid versus aneuploid | 3.21 ± 0.60 | 3.44 ± 0.57 | 1.725 | 0.087 |
P < 0.05.
Biochemical pregnancy.
DS1, dataset 1; DS2, dataset 2.
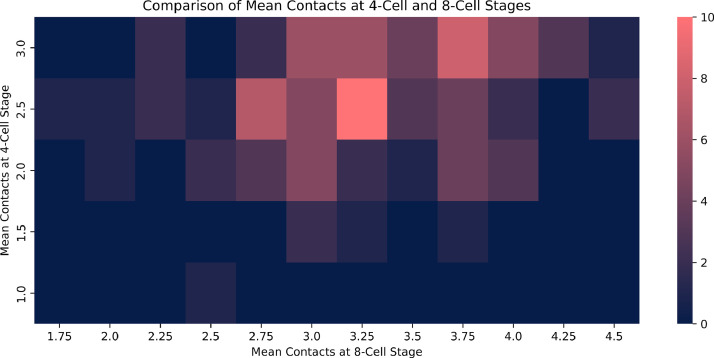
Mean contacts at t4 weakly correlated with those at t8 (ρ = 0.24, 95% CI 0.04 to 0.42). A plot is presented in Figure 2.
Figure 2.
Heatmap of mean contacts at the t4 stage versus mean contacts at the t8 stage. The axes represent the number of cell contacts at the four- and eight-cell stages. The colour of each grid square represents the frequency of embryos with a specific combination of four- and eight- cell contacts. A slight upward trend can be seen.
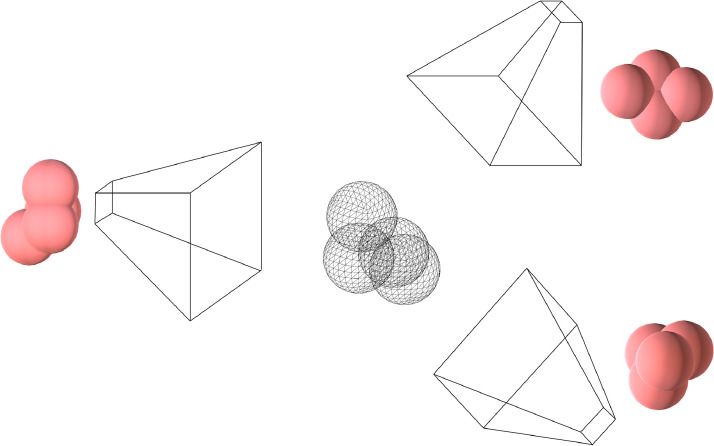
At the four-cell stage, it was observed that embryos fell into nine distinct cell arrangements (Table 3). The most common of these were (pseudotetrahedral), (tetrahedral) and (planar). Examples of each cell arrangement can be found in Figure 3A. At the eight-cell stage, embryos displayed a great degree of variation with 59 distinct cell arrangements, the most common of these, , representing only seven embryos. Examples of the most common eight-cell arrangements are presented in Figure 3B.
TABLE 3.
FREQUENCIES OF FOUR-CELL BLASTOMERE ARRANGEMENTS
| Arrangement | Arrangement nickname | Frequency in DS1 | Frequency in DS2 |
|---|---|---|---|
| [0, 0, 2, 2] | Pseudotetrahedral | 95 | 80 |
| [0, 0, 0, 4] | Tetrahedral | 86 | 60 |
| [0, 0, 4, 0] | Planar | 19 | 30 |
| [0, 1, 2, 1] | Closed Y | 17 | 16 |
| [0, 2, 2, 0] | Linear | 11 | 10 |
| Other | NA | 10 | 5 |
DS1, dataset 1; DS2, dataset 2; NA, not applicable.
Figure 3.
Examples of the most common cell arrangements at the (A) four-cell and (B) eight-cell stages.
A comparison of the mean number of blastomere contacts at the eight-cell stage and the KIDScore D3 as predictive markers of biochemical pregnancy and live birth can be found in Table 4. The models achieved similar AUC scores when predicting biochemical pregnancy suggesting comparable predictive power (P = 0.533) and our method outperformed the KIDScore D3 when predicting live birth (P = 0.029). For predicting biochemical pregnancy, the mean threshold values across all folds (mean ± SD) were 2.4 ± 0.8 for the KIDScore D3 and 3.2 ± 0.2 for the mean number of blastomere contacts. Similarly, for predicting live birth, the mean threshold values were 1.8 ± 0.4 for the KIDScore D3 and 2.9 ± 0.2 for the mean number of contacts.
TABLE 4.
PERFORMANCE COMPARISON BETWEEN THE MEAN NUMBER OF CELL CONTACTS AND KIDSCORE D3 AS PREDICTORS OF BIOCHEMICAL PREGNANCY AND LIVE BIRTH
| Outcome | Metric | Mean cell contacts | KIDScore D3 |
|---|---|---|---|
| Biochemical pregnancy | Accuracy | 0.59 ± 0.11 | 0.74 ± 0.08 |
| Precision | 0.89 ± 0.14 | 0.77 ± 0.06 | |
| Sensitivity | 0.52 ± 0.13 | 0.91 ± 0.11 | |
| F1 | 0.65 ± 0.11 | 0.83 ± 0.06 | |
| AUC | 0.64 ± 0.18 | 0.58 ± 0.10 | |
| Live birth | Accuracy | 0.68 ± 0.08 | 0.64 ± 0.08 |
| Precision | 0.73 ± 0.04 | 0.63 ± 0.05 | |
| Sensitivity | 0.74 ± 0.20 | 0.95 ± 0.10 | |
| F1 | 0.72 ± 0.13 | 0.76 ± 0.07 | |
| AUC | 0.65 ± 0.06 | 0.54 ± 0.07 |
All metrics were evaluated on dataset 2 (D3) blastocysts transferred on day 5 after preimplantation genetic testing for aneuploidy. Values are shown as mean ± SD for all folds in the five-fold cross validation.
AUC, area under the receiver operating characteristic curve; F1, the harmonic mean of the precision and sensitivity.
Discussion
The results of our study echo findings in previous studies that a greater degree of cell contact in the four-cell embryo is conducive to greater developmental potential. We further demonstrated that these findings translate to eight-cell embryos. In this section, we first compare our study to previous works before examining our findings from biological perspective. We then discuss the novel computational aspects of our work followed by the limitations of this study.
Comparison with previous studies on cell arrangement
Four-cell arrangement
Previous studies (Ebner et al., 2012; 2017; Cauffman et al., 2014; Paternot et al., 2014; Desai and Gill, 2019) on the arrangement of blastomeres in four-cell human embryos manually assessed cell arrangement by eye. Our study, in contrast, used a computational method. The advantages of taking this approach include faster and more scalable assessment as well as a reduction in the inter- and intra-observer variability inherent in manual embryological annotation (Baxter Bendus et al., 2006; Khosravi et al., 2019). Notwithstanding, the results of our computational analysis strengthen the argument that increased cell contact in four-cell embryos is associated with greater developmental potential.
Eight-cell arrangement
At the eight-cell stage, an increase in the number of blastomeres present, as well as the possible interactions between them, rule out the feasibility of quick manual assessment. To address this, Paternot et al. (2012) used a semi-automated approach, manually labelling the diameters of blastomeres and using these annotations to generate spheres representing each blastomere. Cell contacts were obtained by detecting overlapping spheres. Although similar in concept, the system used in our work takes a more complex approach, making use of automatically generated cell membrane annotations to represent blastomeres as opposed to spherical approximations. This allowed for a more complex modelling approach which may explain why we detected a correlation between eight-cell stage blastomere contacts and live birth, whereas Paternot et al. (2012) did not.
Characterization of blastomere arrangement in embryos
Over the past years, several systems for the characterization of blastomere arrangement in four-cell embryos have been forwarded. Among these are the tetrahedral/non-tetrahedral system (Desai and Gill, 2019), the tetrahedral/planar system (Ebner et al., 2012; 2017; Milewski and Ajduk, 2017) and Hickman vectors (Hickman et al., 2021). Such systems are useful as they provide a standardized way to categorize and communicate relevant information about embryos encountered in clinical practice, and can thus simplify clinical decision-making.
To our knowledge, no studies have attempted to characterize blastomere arrangement in eight-cell embryos. While simply extending Hickman's system to eight-cell embryos may seem a straightforward approach to fill this gap, our results demonstrate that the large increase in possible blastomere arrangements at the eight-cell stage preclude this: the system was so fine-grained that even the largest group consisted of too few embryos to draw conclusions on clinical outcomes in relation to group membership or the relative prevalence of different groups. Our work overcame this issue by condensing networks into a single numerical descriptor corresponding to the mean degree across all nodes. Although straightforward, such an approach proved sufficient to achieve, albeit with several major caveats discussed in the Limitations section, predictive performance comparable to a well-established existing method based on multiple kinetic parameters. Future works may investigate the use of cluster analysis and graph embedding techniques to identify more coarse-grained, clinically usable characterizations of eight-cell embryos.
Blastomere contact and developmental potential
The development of an embryo into a blastocyst depends on a complex series of events that involve the interaction of multiple signalling pathways, genetic and epigenetic factors, and physical properties of the embryo. Our results suggest that more contact between the blastomeres throughout early embryo development is associated with greater developmental potential.
One explanation for this association may be that a greater degree of blastomere contact enables greater communication between blastomeres. Many of the proteins involved in cell adhesion serve a dual role in enabling cell signalling (Klezovitch and Vasioukhin, 2015; Shawky and Davidson, 2015). Cell signalling is especially important for self-organization and fate determination in the early embryo (Andrzej and Tarkowski, 1959; Rossant, 1976; Strumpf et al., 2005; Ralston et al., 2010; Klezovitch and Vasioukhin, 2015; Saiz et al., 2016; Menchero et al., 2019). It can, therefore, be reasoned that greater levels of contact facilitates improved communication and thus greater developmental potential. Indeed, this notion forms part of the rationale behind the practice of embryo defragmentation, as fragments often interfere with cell junctions (Keltz et al., 2006). Another possible explanation may stem from the fact that early compaction is associated with activation of the embryonic genome and greater developmental potential (Desai et al., 2000; Öztürk et al., 2022; Skiadas et al., 2006). More blastomere contact may, therefore, as opposed to being a causal factor, just be a marker for greater levels of compaction and thus the transcription and activation of the embryonic genome that increases an embryo's developmental potential.
We also observed stronger associations between improved outcomes and blastomere contact at the eight-cell stage than the four-cell stage. This may be explained by the fact that totipotency is gradually lost from four-cell stage onwards as lineage transcription factors change gene expression across the different blastomeres, eventually committing them to different developmental fates (Tarkowski and Wróblewska, 1967; Strumpf et al., 2005; Ralston et al., 2010; de Paepe et al., 2014; Wicklow et al., 2014; Goolam et al., 2016). By the eight-cell stage, this process is more advanced, and thus arguably more representative of the embryo at transfer and implantation. In contrast, four-cell stage embryos are still totipotent and may rely more heavily on maternal mRNA.
It has also been suggested that cell arrangement in an embryo may reflect the presence of cleavage anomalies at the four-cell stage (Piotrowska-Nitsche and Zernicka-Goetz, 2005; Ebner et al., 2012; Ajduk and Zernicka-Goetz, 2016). It is, however, unclear how this may translate to the eight-cell stage and the multitude of possible arrangements we identified. Additional research efforts are necessary to draw any definitive conclusions.
Representation matters
The past few years have seen a surge of interest in the use of artificial intelligence tools for embryo selection. Many recent studies have been based on image analysis techniques from computer vision, with a great number of these trying to predict embryo viability directly from images and videos (Khosravi et al., 2019; Ver Milyea et al., 2021; Berntsen et al., 2022; Kragh et al., 2022). A key difference between the present study and previous studies is that we operate on cell contact networks derived from embryo images as opposed to the images themselves. Taking such an approach carries an important advantage: it gives us a representation of cell organization in an embryo that is ‘invariant’ with respect to the embryo's orientation.
Roughly speaking, ‘orientation invariance’ refers to the fact that even when an embryo is orientated differently under a microscope, its representation as a cell contact network remains the same. This cannot be said about representing embryo cell arrangement using raw image data: the same embryo orientated in different ways will yield different focal stacks (Figure 4). As a result, when training computer vision systems, a large quantity of data is needed to establish that the different focal stacks correspond to the same arrangement, a problem that existing data augmentation methods cannot overcome.
Figure 4.
A three-dimensional model of an embryo (shown as a wireframe) viewed from different angles (illustrated using frustums). The resulting view from each angle is shown next to its respective frustrum.
Moreover, our approach explicitly encodes the ideas that a cell affects the other cells in its neighbourhood and that the overall spatial arrangement of the embryo is of importance. Representing embryos as cell contact networks can thus provide a more biologically grounded basis for automated embryo assessment than purely image-based works. Future works may augment the nodes with further information derived from the embryo, such as features derived from analyses of blastomere shape and cytoplasm texture to produce predictive models for clinical outcomes. They may also integrate temporal information to understand how interactions between cells change over time, giving rise to a new field of three-dimensional morphokinetics.
Limitations
The biggest limitation of our study lies in the data used: this was a small-scale analysis of two very different embryo datasets from two clinics. This limitation stems from the need for manual validation of reconstructions given the newness of the technique as well as the resource constraints of a pilot study. DS2 harbours several biases as it was collected with a view to maximizing data on transferred embryos and PGT-A. It is, therefore, not a representative sample of the general embryo population, which is not transferred. In addition, the fact that PGT-A was conducted on all DS2 embryos necessarily implies they survived to blastocyst stage, which creates a survivorship bias against embryos that arrest in the cleavage or morula stages.
Furthermore, the applicability of our findings to cleavage stage transfers is limited as the study comprised only embryos that were cultured to and transferred at the blastocyst stage. It is, therefore, possible that the associations observed in our study may not hold true for embryos transferred on day 3. A similar limitation is important to consider when reviewing the comparison between our method and the KIDScore D3. While our classifier was trained to predict biochemical pregnancy after PGT-A and blastocyst transfer, the KIDScore D3 was trained to predict implantation from cleavage-stage transfers. Both algorithms, however, were tested on the former population, which raises the possibility that results might diverge when applied to day-3 transfers. As a result, larger collaborative studies with more embryos and appropriate end points are needed to comprehensively evaluate the utility of the method proposed here.
Finally, the three-dimensional reconstruction system was not applicable to all cleavage-stage embryos, especially those obscured by the edge of the well, severely fragmented or having undergone substantial compaction. Nevertheless, a failure to carry out three-dimensional reconstruction does not automatically indicate that nothing can be learnt about the embryo, especially in the latter two cases. As previously mentioned, early compaction has been associated with greater developmental potential (Aslan Öztürk et al., 2022; Skiadas et al., 2006). Moreover, in theory, these embryos have the most blastomere contact of all; therefore, the exclusion of these embryos may mean that we underestimated the strength of the relationship between cell contact and developmental potential (Skiadas et al., 2006). Similarly, severe fragmentation has been associated with negative clinical outcomes, including lower implantation rates (Alikani et al., 1999b; Keltz et al., 2006). Future studies may explore the potential for automated detection of these conditions before attempting reconstruction; however, our findings suggest that, in most cases falling outside these two extremes, three-dimensional analysis has the potential to be a valuable tool.
In conclusion, in this pilot study, we have provided evidence to support the clinical significance of cleavage-stage cell arrangement in the human preimplantation embryo extending beyond the four-cell stage. Our results demonstrate significant correlations between the amount of cell contact in an embryo and developmental outcomes, with the strongest associations seen in eight-cell embryos. Moreover, we highlight that cell contact at the eight-cell stage may be competitive with more traditional morphokinetic scoring algorithms when predicting biochemical pregnancy and live birth, which could be helpful for improving embryo selection and cycle planning, particularly in situations in which a day-3 transfer may be necessary. Overall, our findings suggest that three-dimensional morphokinetics may represent a promising avenue for further investigation.
Acknowledgments
Acknowledgements
The authors would like to thank Lydia Firminger and Neave Croft for helping to provide annotations. The authors are eternally indebted to Chris-Alexandre Pena for looking after the GPU server the research was conducted on. Finally, the authors thank Jules Sales for his creative support on figures and Alexis Gkantiragas for proofreading the manuscript.
Authors’ roles
CH, NK, CJ, JC, RH, FV and Christina Hickman (Chi) conceived the study and designed the methodology; KW, JM, NZ and RM provided raw data and annotations; NK, RH and CHi validated the data and provided further annotations; CJ, TF and JC contributed significant technical resources; CH performed the computational research; CH, LL, NK, RH and CHi sanity-checked the outputs of the three-dimensional reconstruction system; CH conducted the statistical analysis; FV and CHi supervised the research activity; CH and NK wrote the article; all authors contributed to the review and editing of the final manuscript.
Funding
This project was supported by Apricity and the Wellcome/EPSRC Centre for Interventional and Surgical Sciences (WEISS) [203145/Z/16/Z].
Biography

Chloe He completed her undergraduate training in the Department of Computing at Imperial College London before embarking on a PhD at University College London and Apricity. Her research focuses on devising methods to better understand preimplantation embryo development in humans as well as designing inclusive and equitable reproductive healthcare services.
Key message.
Increased cell contact in cleavage-stage embryos is associated with greater developmental potential. Our results provide preliminary evidence for the utility of spatial biomarkers derived from three-dimensional embryo reconstruction. A network representation of embryos carries several advantages over the image-based representations used by many artificial intelligence embryo selection tools.
Alt-text: Unlabelled box
Declaration: CH, NK, RH, LL, CJ, TF, JC and Cristina Hickman (Chi) are employed by Apricity. CHi is also the founder of Avenues, Ovom Care and IVF Professionals, and chief clinical officer of Fairtility. CHi owns shares in TMRW, Apricity, Fairtility, Aria Fertility, Avenues, Ovom Care and IVF Professionals. CHi has received consulting fees for from Gynii Me, Ferring, Vitrolife, Hamilton Thorne, McKinsey and Company, Mouwasat, Salve and TMRW. CHi has received travel support from Vitrolife, CooperSurgical, Mellowood and Kitazato. CHi has received payment or honoraria for lectures, presentations, speakers’ bureaus, manuscript writing or educational events from Merck Serono, Vitrolife, Ferring, Gynii Me, Hamilton Thorne, Cook Medical, Kitazato and CooperSurgical.
Data availability
The data that has been used is confidential.
References
- Ajduk A., Zernicka-Goetz M. Polarity and cell division orientation in the cleavage embryo: From worm to human. Mol. Hum. Reprod. 2016 doi: 10.1093/molehr/gav068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alikani M., Cohen J., Tomkin G., Garrisi G.J., Mack C., Scott R.T. Human embryo fragmentation in vitro and its implications for pregnancy and implantation. Fertil. Steril. 1999;71:836–842. doi: 10.1016/S0015-0282(99)00092-8. [DOI] [PubMed] [Google Scholar]
- Alikani M., Cohen J., Tomkin G., Garrisi G.J., Mack C., Scott R.T. Human embryo fragmentation in vitro and its implications for pregnancy and implantation. Fertil. Steril. 1999;71:836–842. doi: 10.1016/S0015-0282(99)00092-8. [DOI] [PubMed] [Google Scholar]
- Andrzej D.R., Tarkowski K. Experiments on the Development of Isolated Blastomeres of Mouse Eggs. Nature. 1959;184:1286–1287. doi: 10.1038/1841286a0. 4695 184. [DOI] [PubMed] [Google Scholar]
- Aslan Öztürk S., Cincik M., Donmez Cakil Y., Sayan S., Selam B. Early Compaction Might Be a Parameter to Determine Good Quality Embryos and Day of Embryo Transfer in Patients Undergoing Intracytoplasmic Sperm Injection. Cureus. 2022;14 doi: 10.7759/cureus.23593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter Bendus A.E., Mayer J.F., Shipley S.K., Catherino W.H. Interobserver and intraobserver variation in day 3 embryo grading. Fertil. Steril. 2006;86:1608–1615. doi: 10.1016/j.fertnstert.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Berntsen J., Rimestad J., Lassen J.T., Tran D., Kragh M.F. Robust and generalizable embryo selection based on artificial intelligence and time-lapse image sequences. PLoS One. 2022;17 doi: 10.1371/journal.pone.0262661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauffman G., Verheyen G., Tournaye H., Van De Velde H. Developmental capacity and pregnancy rate of tetrahedral-versus non-tetrahedral-shaped 4-cell stage human embryos. J. Assist. Reprod. Genet. 2014;31 doi: 10.1007/s10815-014-0185-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paepe C., Krivega M., Cauffman G., Geens M., van de Velde H. Totipotency and lineage segregation in the human embryo. Mol. Hum. Reprod. 2014;20:599–618. doi: 10.1093/MOLEHR/GAU027. [DOI] [PubMed] [Google Scholar]
- Dennis S.J., Thomas M.A., Williams D.B., Robins J.C. Embryo morphology score on day 3 is predictive of implantation and live birth rates. J. Assist. Reprod. Genet. 2006;23:171–175. doi: 10.1007/s10815-006-9027-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai N., Gill P. Blastomere cleavage plane orientation and the tetrahedral formation are associated with increased probability of a good-quality blastocyst for cryopreservation or transfer: a time-lapse study. Fertil. Steril. 2019;111 doi: 10.1016/j.fertnstert.2019.02.019. [DOI] [PubMed] [Google Scholar]
- Desai, N., Goldstein, J., Rowland, D., Goldfarb, J., 2000. Morphological evaluation of human embryos and derivation of an embryo quality scoring system specific for day 3 embryos: a preliminary study. Hum. Reprod. 15, 2190–2196. 10.1093/humrep/15.10.2190. [DOI] [PubMed]
- Ebner T., Höggerl A., Oppelt P., Radler E., Enzelsberger S.H., Mayer R.B., Petek E., Shebl O. Time-lapse imaging provides further evidence that planar arrangement of blastomeres is highly abnormal. Arch. Gynecol. Obstet. 2017;296 doi: 10.1007/s00404-017-4531-5. [DOI] [PubMed] [Google Scholar]
- Ebner T., Maurer M., Shebl O., Moser M., Mayer R.B., Duba H.C., Tews G. Planar embryos have poor prognosis in terms of blastocyst formation and implantation. Reprod. Biomed. Online. 2012;25 doi: 10.1016/j.rbmo.2012.05.007. [DOI] [PubMed] [Google Scholar]
- Gardner D.K., Schoolcraft W.B. Culture and transfer of human blastocysts. Curr. Opin. Obstet. Gynecol. 1999;11:307–311. doi: 10.1097/00001703-199906000-00013. [DOI] [PubMed] [Google Scholar]
- Goolam M., Scialdone A., Graham S.J.L., MacAulay I.C., Jedrusik A., Hupalowska A., Voet T., Marioni J.C., Zernicka-Goetz M. Heterogeneity in Oct4 and Sox2 Targets Biases Cell Fate in 4-Cell Mouse Embryos. Cell. 2016;165:61–74. doi: 10.1016/J.CELL.2016.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Hariharan R., Karpavičiūtė N., Croft N., Firminger L., Chambost J., Jacques C., Saravelos S., Wouters K., Fréour T., Zaninovic N., Malmsten J., Vasconcelos F., Hickman C. O-177 Towards 3D Reconstructions of Human Preimplantation Embryo Development. Human Reproduction. 2022;37 doi: 10.1093/humrep/deac105.091. [DOI] [Google Scholar]
- He C, Jacques C., Chambost J., Malmsten J., Wouters K., Fréour T., Zaninovic N., Hickman C., Vasconcelos F. Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) Springer Science and Business Media Deutschland GmbH; 2022. Super-Focus: Domain Adaptation for Embryo Imaging via Self-supervised Focal Plane Regression; pp. 732–742. [DOI] [Google Scholar]
- Hickman C., He C., Hariharan R., Jacques C., Chambost J. P–267 Characterising a novel embryo grading system for 4-cell embryos including symmetry, fragmentation, cell configuration, cell contacts per cell, distance between cells, and cell adhesion strength. Human Reproduction. 2021;36 doi: 10.1093/humrep/deab130.266. [DOI] [Google Scholar]
- Iyer S., Mukherjee S., Kumar M. Watching the embryo: Evolution of the microscope for the study of embryogenesis. BioEssays. 2021;43 doi: 10.1002/bies.202000238. [DOI] [PubMed] [Google Scholar]
- Keltz M.D., Skorupski J.C., Bradley K., Stein D. Predictors of embryo fragmentation and outcome after fragment removal in in vitro fertilization. Fertil. Steril. 2006;86:321–324. doi: 10.1016/j.fertnstert.2006.01.048. [DOI] [PubMed] [Google Scholar]
- Khosravi P., Kazemi E., Zhan Q., Malmsten J.E., Toschi M., Zisimopoulos P., Sigaras A., Lavery S., Cooper L.A.D., Hickman C., Meseguer M., Rosenwaks Z., Elemento O., Zaninovic N., Hajirasouliha I. Deep learning enables robust assessment and selection of human blastocysts after in vitro fertilization. NPJ Digit. Med. 2019;2:1–9. doi: 10.1038/s41746-019-0096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klezovitch O., Vasioukhin V. Cadherin signaling: keeping cells in touch. F1000Res. 2015;4 doi: 10.12688/F1000RESEARCH.6445.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragh M.F., Rimestad J., Lassen J.T., Berntsen J., Karstoft H. Predicting Embryo Viability Based on Self-Supervised Alignment of Time-Lapse Videos. IEEE Trans. Med. Imaging. 2022;41:465–475. doi: 10.1109/TMI.2021.3116986. [DOI] [PubMed] [Google Scholar]
- Menchero S., Rollan I., Lopez-Izquierdo A., Andreu M.J., De Aja J.S., Kang M., Adan J., Benedito R., Rayon T., Hadjantonakis A.K., Manzanares M. Transitions in cell potency during early mouse development are driven by notch. Elife. 2019;8 doi: 10.7554/ELIFE.42930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewski R., Ajduk A. Time-lapse imaging of cleavage divisions in embryo quality assessment. Reproduction. 2017 doi: 10.1530/REP-17-0004. [DOI] [PubMed] [Google Scholar]
- Moayeri S.E., Allen R.B., Brewster W.R., Kim M.H., Porto M., Werlin L.B. Day-3 embryo morphology predicts euploidy among older subjects. Fertil. Steril. 2008;89:118–123. doi: 10.1016/j.fertnstert.2007.01.169. [DOI] [PubMed] [Google Scholar]
- Öztürk, S., Cincik, M., Cakil, Y., Sayan, S., Selam, B., 2022. Early Compaction Might Be a Parameter to Determine Good Quality Embryos and Day of Embryo Transfer in Patients Undergoing Intracytoplasmic Sperm Injection. Cureus 14, e23593. 10.7759/cureus.23593. [DOI] [PMC free article] [PubMed]
- Paternot G., Debrock S., De Neubourg D., D'Hooghe T.M., Spiessens C. The spatial arrangement of blastomeres at the 4-cell stage and IVF outcome. Reprod. Biomed. Online. 2014;28:198–203. doi: 10.1016/j.rbmo.2013.10.008. [DOI] [PubMed] [Google Scholar]
- Paternot G., Spiessens M., Verstreken D., Van Bauwel J., Debrock S., D'Hooghe T., Spiessens C. Is there a link between blastomere contact surfaces of day 3 embryos and live birth rate? Reproductive Biology and Endocrinology. 2012;10 doi: 10.1186/1477-7827-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen B.M., Boel M., Montag M., Gardner D.K. Development of a generally applicable morphokinetic algorithm capable of predicting the implantation potential of embryos transferred on Day 3. Human Reproduction. 2016;31:2231–2244. doi: 10.1093/humrep/dew188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowska-Nitsche K., Zernicka-Goetz M. Spatial arrangement of individual 4-cell stage blastomeres and the order in which they are generated correlate with blastocyst pattern in the mouse embryo. Mech. Dev. 2005;122:487–500. doi: 10.1016/j.mod.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Ralston A., Cox B.J., Nishioka N., Sasaki H., Chea E., Rugg-Gunn P., Guo G., Robson P., Draper J.S., Rossant J. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 2010;137:395–403. doi: 10.1242/DEV.038828. [DOI] [PubMed] [Google Scholar]
- Rijnders P.M., Jansen C.A.M. The predictive value of day 3 embryo morphology regarding blastocyst formation, pregnancy and implantation rate after day 5 transfer following in-vitro fertilization or intracytoplasmic sperm injection. Human Reproduction. 1998;13 doi: 10.1093/humrep/13.10.2869. [DOI] [PubMed] [Google Scholar]
- Rossant J. Postimplantation development of blastomeres isolated from 4- and 8-cell mouse eggs. Development. 1976;36:283–290. doi: 10.1242/DEV.36.2.283. [DOI] [PubMed] [Google Scholar]
- Saiz N., Williams K.M., Seshan V.E., Hadjantonakis A.K. Asynchronous fate decisions by single cells collectively ensure consistent lineage composition in the mouse blastocyst. Nat. Commun. 2016;7 doi: 10.1038/NCOMMS13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawky J.H., Davidson L.A. Tissue Mechanics and Adhesion during Embryo Development. Dev Biol. 2015;401:152. doi: 10.1016/J.YDBIO.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiadas C.C., Jackson K.V., Racowsky C. Early compaction on day 3 may be associated with increased implantation potential. Fertil. Steril. 2006;86:1386–1391. doi: 10.1016/j.fertnstert.2006.03.051. [DOI] [PubMed] [Google Scholar]
- Strumpf D., Mao C.A., Yamanaka Y., Ralston A., Chawengsaksophak K., Beck F., Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/DEV.01801. [DOI] [PubMed] [Google Scholar]
- Tarkowski A.K., Wróblewska J. Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. Development. 1967;18:155–180. doi: 10.1242/DEV.18.1.155. [DOI] [PubMed] [Google Scholar]
- Ver Milyea M., Hall J.M.M., Diakiw S.M., Johnston A., Nguyen T., Perugini D., Miller A., Picou A., Murphy A.P., Perugini M. Development of an artificial intelligence-based assessment model for prediction of embryo viability using static images captured by optical light microscopy during IVF. Human Reproduction. 2021;35:770–784. doi: 10.1093/HUMREP/DEAA013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicklow E., Blij S., Frum T., Hirate Y., Lang R.A., Sasaki H., Ralston A. HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst. PLoS Genet. 2014;10 doi: 10.1371/JOURNAL.PGEN.1004618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that has been used is confidential.