Abstract
Intracranial aneurysms (IAs) are present in 2–6% of the global population and can be catastrophic upon rupture with a mortality rate of 30–50%. IAs are commonly detected through time-of-flight magnetic resonance angiography (TOF-MRA), however, this data is rarely available for research and training purposes. The provision of imaging resources such as TOF-MRA images is imperative to develop new strategies for IA detection, rupture prediction, and surgical training. To support efforts in addressing data availability bottlenecks, we provide an open-access TOF-MRA dataset comprising 63 patients, of which 24 underwent interval surveillance imaging by TOF-MRA. Patient scans were evaluated by a neuroradiologist, providing aneurysm and vessel segmentations, clinical annotations, 3D models, in addition to 3D Slicer software environments containing all this data for each patient. This dataset is the first to provide interval surveillance imaging for supporting the understanding of IA growth and stability. This dataset will support computational and experimental research into IA dynamics and assist surgical and radiology training in IA treatment.
Subject terms: Brain, Brain imaging, Magnetic resonance imaging, Cerebrovascular disorders, Aneurysm
Background & Summary
Intracranial aneurysms (IAs) are an outpouching of the arterial wall, caused by a weakening and degradation of arterial tissue layers1–3. IAs are present in 2–6% of the global population, and their rupture causes 85% of subarachnoid haemorrhages with a 30–50% mortality rate4–7. IAs are also a leading cause of stroke in adults below 65 years of age8,9.
Intra-arterial digital subtraction angiography (DSA), computed tomography angiography (CTA) and time-of-flight magnetic resonance angiography (TOF-MRA) are widely implemented for clinical identification of IAs10. TOF-MRA is less invasive and thus more frequently used as it does not require the use of an intravenous contrast material10. Although, some radiologists may still administer contrast material to enhance image quality.
With the widespread use of these imaging modalities for a variety of clinical presentations, the incidental identification of IAs is increasing11,12. However, IA detection relies on specialised radiologists or neuroradiologists, and as implementation of these imaging modalities increases so-to does the workload of neuroradiologists, making it challenging to reach reporting demand on all cranial angiograms13,14. In addition, these specialised radiologists are rarely available outside of specialised facilities and in rural areas, reducing rates of detection.
Furthermore, once an IA has been detected, clinicians are currently reliant on experience and known patient risk factors to determine whether surgical treatment is required. Existing methods to evaluate rupture risk include use of the PHASES risk prediction score (based on Population, Hypertension, Age, Size, Earlier haemorrhage and Site of IA) and the unruptured intracranial aneurysm treatment score (UIATS) which in some cases remain insufficient in providing a clear recommendation for treatment15–17.
To improve IA detection and our understanding of pathophysiology and rupture tendency, various in silico and in vitro models have been developed. These include, but are not limited to: the development of automated algorithms to aid clinicians in detecting aneurysms5,12,13; evaluation of the effect of haemodynamic stress through computational fluid dynamics (CFD) models18,19; and investigating cell behaviour in in vitro aneurysm models with patient-specific geometries20–22.
However, many of these studies are limited to analysis of small patient cohorts (n < 15) and/or do not provide their datasets for public use14. While several publicly available datasets of healthy brain vasculature exist, including the IXI (http://brain-development.org) database, there is a lack of large and publicly available datasets with IAs, with none containing patients with interval surveillance imaging. This hinders research efforts into IA detection and eventual rupture by limiting collaboration, evaluation, validation, and reproducibility of in silico and in vitro models. Additionally, given the multitude of IA morphologies that can arise and inter-individual variability, the generation of large publicly available datasets with expert clinical measurements and segmentations are imperative for a complete understanding of risk factors related to IA formation and rupture.
The AneurRisk Project (http://ecm2.mathcs.emory.edu/aneuriskweb/index) thus arose in 2008 to unite researchers in the goal to uncover the complex pathology around aneurysms, providing DSA scans of 65 patients with IAs. More recently, Di Noto et al.14 published an open-access dataset comprising 284 subjects, with 127 healthy controls and 157 patients with 198 aneurysms. They demonstrated the use of this dataset in developing an automated deep learning algorithm to detect IAs14. To the best of our knowledge, this is currently the only TOF-MRA aneurysm dataset available.
To further support this effort to address bottlenecks in data availability, we provide an open-access annotated TOF-MRA dataset comprising 63 subjects containing IAs. Importantly, this work differs to the existing databases by providing interval surveillance imaging on 24 subjects, ranging from 1–12 years of surveillance. This data is useful for investigating changes in size and morphology of IAs over time, which may help validate the predictive risk of IA geometry and fluid dynamics in rupture. These surveillance images could also highlight changes in vessel geometry and fluid dynamics after treatment to inform surgical planning and aid in the development of new treatment options.
We also provide IA characteristics including size and location, with 3D aneurysm models in standard tessellation language (STL) file format, clinical annotations, labelled maps with the IA and parent vessels, and 3D slicer files comprising all masks within a single workspace (one example patient dataset depicted in Fig. 1). This dataset will therefore support ongoing collaborative research to ultimately improve clinical detection, understanding and treatment of IAs.
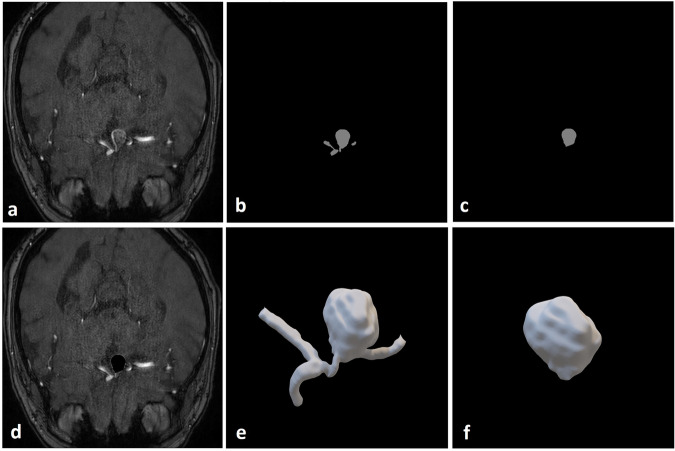
Fig. 1.
One transverse cross-section of a Time-Of-Flight Magnetic Resonance Angiography (TOF-MRA) scan for one representative patient (sub-042) with a right A1/anterior communicating artery (ACOM) aneurysm with clinician-annotated size of 12 × 10 × 12 mm. (a) High-resolution TOF-MRA; (b) Threshold-segmented mask of aneurysm with parent artery; (c) Threshold-segmented mask of aneurysm only; (d) TOF-MRA with aneurysm overlay in black; (e) 3D model of aneurysm with parent vessel smoothed using Taubin’s algorithm in 3D Slicer; (f) 3D model of aneurysm smoothed using Taubin’s algorithm in 3D Slicer. Brightness for (a) and (d) were linearly adjusted.
Methods
Data overview
The TOF-MRA scans were acquired through Metro North Hospital and Health Service (MNHHS) at various institutions between 2008 and 2022, totalling 63 de-identified patients. Interval surveillance imaging was conducted on 24 of the 63 patients through follow-up appointments, as depicted in Fig. 2. The research protocol used for de-identified data collection and provision in a public open-access repository under a waiver of patient consent was approved by the Royal Brisbane and Women’s Hospital (RBWH) Human Research Ethics Committee (Ref: LNR/2019/QRBW/49363) and the Queensland Department of Public Health (Ref: Public Health Agreement #49363).
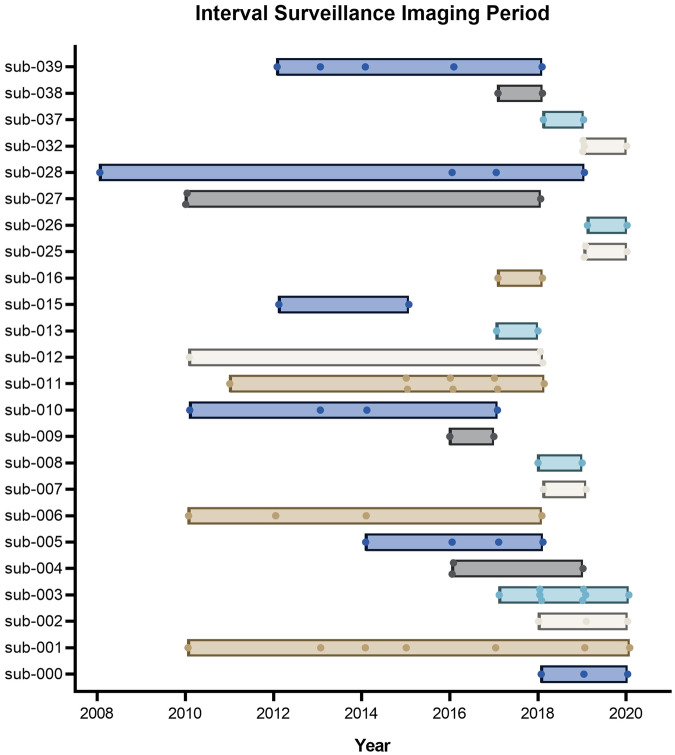
Fig. 2.
Imaging period for patients with interval surveillance (n = 24). Each data point represents one imaging session. Sub-008, sub-009 and sub-013 each had surveillance images taken within one year of the first appointment.
Image acquisition
MR images were acquired using GE, Philips and Siemens MR scanners with varying technical specifications and acquisition parameters, as summarised in Table 1. In some cases, longitudinal imaging was conducted on different MR scanners across the surveillance period. These details are provided in the supporting Excel spreadsheet, ‘Clinical Annotations_MR Specifications_Demographics_Image Quality Analysis’ uploaded with the dataset for each individual scan.
Table 1.
Magnetic Resonance scanners, technical specifications and acquisition parameters of TOF-MRA scans. RT = Repetition time, TE = time to echo.
| Vendor | Model | # of Scans | Avg. Field Strength [T] | Avg. RT [ms] | Avg. TE [ms] | Avg. Voxel Spacing [mm3] |
|---|---|---|---|---|---|---|
| GE | Discovery MR750w | 1 | 3 | 24.0 | 5.7 | 1.00 × 1.00 × 0.50 |
| Signa HDxt | 3 | 1.5 | 22.0 | 3.5 | 1.20 × 1.16 × 0.60 | |
| Unknown | 3 | 2 | 28.0 | 3.8 | 1.40 × 1.36 × 0.70 | |
| Philips | Ingenia | 25 | 3 | 18.6 | 3.5 | 0.66 × 0.91 × 0.82 |
| Unknown | 6 | 3 | 21.7 | 3.5 | 1.13 × 1.10 × 0.57 | |
| Siemens | Avanto | 8 | 1.5 | 164.7 | 6.6 | 2.04 × 1.94 × 15.56 |
| Magnetom Vida | 19 | 3 | 24.1 | 4.0 | 0.41 × 0.41 × 0.42 | |
| Skyra | 12 | 3 | 21.2 | 3.6 | 0.44 × 0.44 × 14.66 | |
| TrioTim | 26 | 3 | 22.7 | 4.1 | 0.48 × 0.48 × 20.49 | |
| Unknown | 34 | 3 | 22.7 | 4.0 | 0.47 × 0.47 × 7.53 | |
| Verio | 6 | 3 | 22.0 | 3.7 | 0.49 × 0.49 × 18.63 |
Data was then de-identified by radiologists using PixelMedTM DicomCleanerTM to remove all health information identifiers. De-identified data was provided for research purposes through established governance processes at the RBWH and data distributed to researchers as per data sharing agreements approved by the Queensland Department of Public Health (Ref: Public Health Agreement #49363). Scans were further cropped to remove the front portion of the skull and facial features while maintaining visibility of the Circle of Willis, with all data visually inspected to ensure appropriate de-identification before release.
Scans were acquired in DICOM (Digital Imaging and Communications in Medicine) format and converted to NIFTI (Neuroimaging Informatics Technology Initiative) files. Where files contained multiple scanning sequences, these were stripped to comprise only de-identified TOF-MRA scanning sequences for each patient.
Segmentation and annotation
Angiographic image sequences were imported into 3D Slicer, with segmentation and annotation of aneurysms and parent vessels performed by a neuroradiologist. Aneurysms were first manually detected by scrolling through the slices of the scan, and threshold segmentation applied to extract the vascular tree. The aneurysm and parent vessels were then isolated by cropping the larger vascular tree. Some aneurysms that were too large or contained clots required manual segmentation of focal areas due to the lack of contrast. Voxel-wise annotations of the aneurysm and parent vessels were then created, where the neuroradiologist manually delineated the aneurysm from the parent vessel, guided by changes in vessel curvature. Annotations were smoothed using Taubin’s algorithm in 3D Slicer, and surface meshes produced using the Flying Edges algorithm in 3D Slicer before exporting as 3D STL mesh files. Original segmentations and binary label maps are provided as raw voxel-wise annotations, allowing the user of the dataset to apply different smoothing settings as necessary.
Data Records
63 patients with 85 IAs were imaged using TOF-MRA. 39 patients comprise singular scans while 24 patients include interval surveillance images. Patient images were organised according to the Brain Imaging Data Structure (BIDS) and were made publicly available through the OpenNeuro repository 10.18112/openneuro.ds005096.v1.0.023.
All information relating to MR acquisition parameters, clinical annotations and patient age and gender has been uploaded in the supporting Excel spreadsheet, ‘Clinical Annotations_MR Specifications_Demographics_Image Quality Analysis’ uploaded with the dataset. A summary of aneurysm characteristics is provided in Table 2, with location and size grouped according to the PHASES score due to its clinical use.
Table 2.
Summary of patient scans, aneurysm locations and size grouped according to the PHASES score currently used for clinical interpretation. ICA = Internal Carotid Artery, MCA = Middle Cerebral Artery, ACA = Anterior Cerebral Arteries, Pcom = Posterior Communicating Artery, Posterior = posterior circulation (including vertebral, basilar, cerebellar, and posterior cerebral arteries).
| Count | ||
|---|---|---|
| Total Patients | 63 | |
| Patient Scans | Patients with 1 aneurysm | 47 |
| Patients with > 1 aneurysm | 16 | |
| Patients with single scans | 39 | |
| Patients with follow-up scans | 24 | |
| Aneurysm Location | MCA and M1 | 30 |
| ICA | 24 | |
| ACA/Pcom/Posterior | 31 | |
| Aneurysm Maximum Diameter | <7.0 mm | 46 |
| 7–9.9 mm | 23 | |
| 10–19.9 mm | 10 | |
| >20 mm | 2 | |
| Difficult or previously treated aneurysm | 4 | |
Patients having undergone interval surveillance imaging have been further analysed for increases in size, daughter sac formation, aneurysm recurrence, new aneurysm formation, and treatment. This has been summarised in Fig. 3 below. Importantly, due to the poor spatial resolution offered by TOF-MRA scans, a shape change is only indicated for an increase in diameter of more than 2 mm. If aneurysm shape change is suspected from a TOF-MRA in the clinic, subsequent CTA or DSA is typically performed to provide more accurate analyses of aneurysm morphology and confirm any suspected shape change.
Fig. 3.
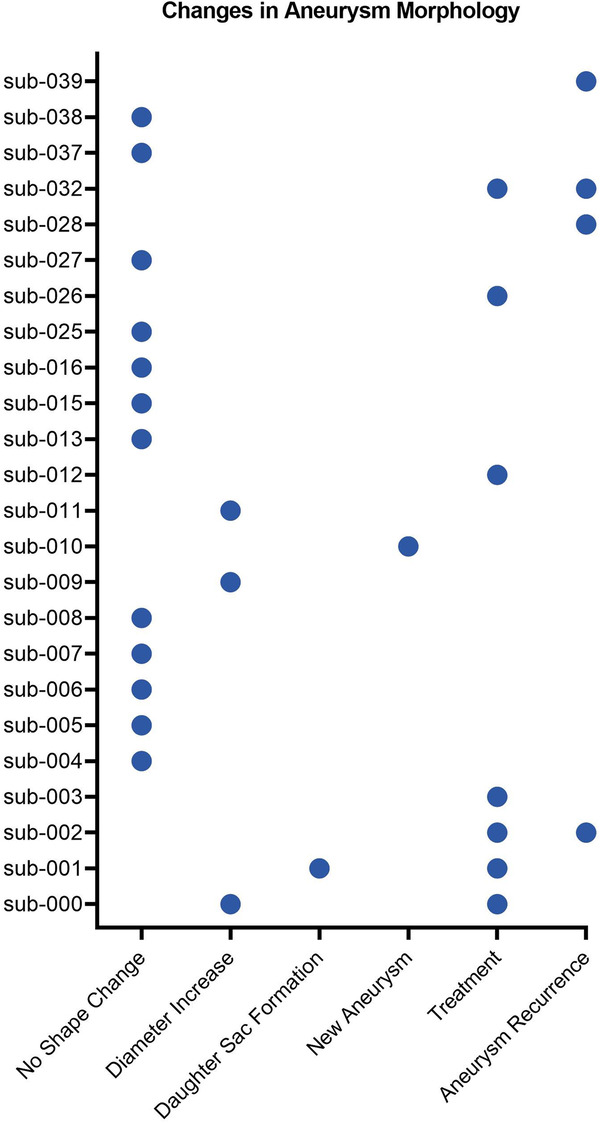
Summary of changes in aneurysm morphology across subjects with interval surveillance imaging. A ‘diameter increase’ was indicated if an aneurysm exhibited more than a 2 mm increase. A ‘new aneurysm’ indicates initiation of a new aneurysm at a different location during imaging. ‘Treatment’ refers to scans taken before- and after- aneurysm treatment. Aneurysms that have recurred after treatment are also indicated.
Each patient comprises at least one angiography scan in NIFTI format with accompanying JSON (JavaScript Object Notation) file organised according to the BIDS standard (Fig. 4a). Some patients include multiple angiographic scans for a single scanning session, with some scans including both single or multi-slab imaging, or with varying resolution. These have been indicated through corresponding acquisition labels (e.g. ‘acq-singleslab’ for single slab imaging or ‘acq-highres’ for high resolution).
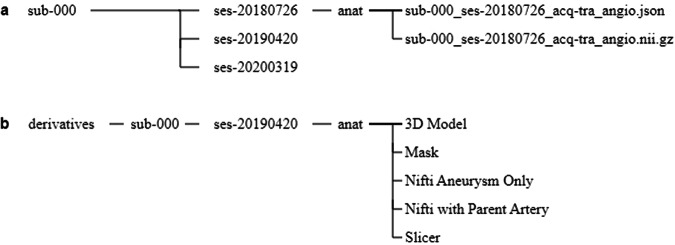
Fig. 4.
Folder structure of the dataset for sub-000. (a) Original Time-of-Flight Magnetic Resonance Angiography (TOF-MRA) scans are provided for each individual subject with all scanning sessions available. (b) A derivatives folder provides all segmentations and annotations performed for a single scanning session for each subject. For sub-000, the TOF-MRA from ses-20190420 was used for segmentation and annotation by a neuroradiologist as this was the most recent high-quality scan conducted before aneurysm coiling.
In addition to the original patient data, segmentations and annotations performed by a neuroradiologist using 3D Slicer are provided for a single scanning session. Where several scanning sessions are available for 24 patients, segmentations were performed on scans before treatment if applicable, on the best quality scan, or on the most recent scan provided as determined by the neuroradiologist. These annotations are contained in a derivatives folder using a similar folder structure to the BIDS specification.
Specifically, for each patient a mask has been created for the segmented aneurysm and parent vessels (Fig. 4b – ‘Nifti Aneurysm Only’ and Fig. 4b – ‘Nifti with Parent Artery’ respectively) overlayed onto the original NIFTI scan (Fig. 4b – ‘Mask’), along with the segmented 3D STL aneurysm models (Fig. 4b – ‘3D Model’), and the 3D Slicer scene where segmentation and annotations were performed (Fig. 4b – ‘Slicer’). For patients where characterisations and annotations were performed using DSA or CTA scans, only the segmented masks and 3D STL models are provided of the aneurysm, parent vessels and vascular tree.
An example of the data provided for a single patient is summarised in Fig. 4.
Technical Validation
Segmented aneurysm morphology and resulting CFD analyses can be influenced by imaging technique and image quality, as well as segmentation and post-processing methods. Paritala et al. (2023) has previously published an inter-user variability study on sub-001, with interval surveillance over a decade. They found the level of smoothing on segmented models to be a significant contributor to variations in aneurysm morphology, causing a 20% variation in CFD results between analysts19. This inter-user variability is consistent with existing studies on variations in intracranial aneurysm segmentation24.
While establishing universal segmentation procedures is essential to reduce variability in aneurysm morphology and subsequent analyses using this dataset, the provision of image quality metrics can support these efforts. Image quality and suitability of this dataset for identification and segmentation of IAs was thus verified through evaluation of the signal-to-noise ratio (SNR) and contrast-to-noise ratio (CNR).
Using the annotated masks provided by a neuroradiologist, pixel locations for the aneurysm and parent vessel were extracted to determine corresponding pixel intensities in the original scans. To evaluate contrast between the vasculature and surrounding brain tissue, pixel intensities in a user-selected small brain region devoid of vasculature was also extracted. Noise was determined based on signal intensity in the air outside of the scan. SNR, CNR and coefficients of variation (CV) were then calculated for the aneurysm-only, and aneurysm with parent vessels using equations (1) to (3) below, with results depicted in Fig. 5. Regions of interest used for calculations were sized based on the number of pixels to account for changes in imaging resolution. Where some patient scans did not include any ‘air’ regions or were annotated using DSA or CTA images (n = 5), these were excluded from analysis of all quality metrics.
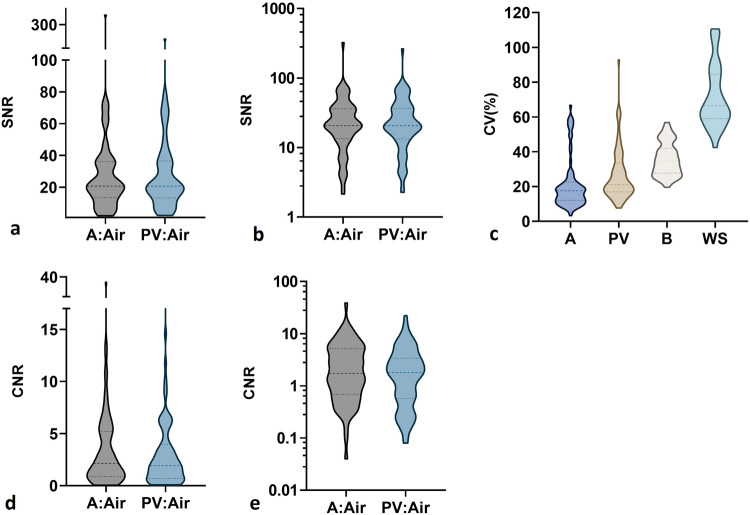
Fig. 5.
Analysis of image quality metrics across TOF-MRA patient scans (n = 73 aneurysms in 58 patients). Some patients scans (n = 5) did not include any ‘air’ region suitable for calculations, or did not include MRA annotations, and thus were excluded from analysis for all image quality metrics. (a) Signal-to-noise ratio (SNR) of the region of interest relative to noise in the air; (b) SNR presented on a logarithmic (base 10) scale; (c) Coefficient of variation (CV) of pixel intensity in region of interest; (d) Contrast-to-noise ratio (CNR) between the region of interest and a user-selected small brain region devoid of vasculature, relative to noise in the air; (e) CNR presented on a logarithmic (base 10) scale. A = Aneurysm, PV = Parent Vessel with Aneurysm, WS = Whole Scan, B = Brain region. Dotted lines in the violin plots indicate quartiles, with the middle being the median value.
SNR, signal-to-noise ratio
| 1 |
CNR, contrast-to-noise ratio
| 2 |
CV, coefficient of variation (%)
| 3 |
Where is the mean signal intensity for the aneurysm-only or the aneurysm with parent vessel, is the mean signal intensity for the neighbouring brain region, is the standard deviation of pixel intensity in the air outside of the head, and is the standard deviation of pixel intensity across the aneurysm-only or the aneurysm with parent vessel.
As can be seen in Fig. 5a,d, there is a wide range of SNR and CNR values across patients, which should be considered when selecting suitable images for modelling or segmentation purposes. These have been further presented on a logarithmic scale (base 10) in Fig. 5b,c respectively for clarity due to the wide range of values.
A similar range of SNR values was previously observed across existing databases of the healthy Circle of Willis (n = 15) when comparing the signal in vessels to manual segmentations25. A comparable SNR range is also seen in databases containing T1-weighted images as opposed to angiograms26. It is uncertain as to how the quality of these images compare to those provided by Di Noto et al.14 as SNR and CNR statistics were not published alongside the dataset.
A previous study evaluated the SNR of brain arteries in the Circle of Willis using 1.5 T, 3 T and 7 T TOF-MRA scanners, finding 1.5 T scanners resulted in the lowest SNR of 23.8 which remained excellent for visualising these major arteries27. It has also been shown through computational simulations that for an SNR ≥ 10, vessel cross-sectional area can be accurately determined with an error of <5% for images with a sampling matrix down to 256 × 256 pixels28. It can be clearly seen from Fig. 5b that the SNR of most scans is greater than 10. In general, for accurate segmentation of aneurysms from the TOF-MRA scans, a high SNR is desired to ensure vasculature can be clearly differentiated from noise to produce well-defined vessel edges29.
A similar range of CNR values across TOF-MRA has also been reported across limited studies30,31. However, there is no clear guideline or evidence for a CNR range that clearly supports accurate segmentation of vasculature from TOF-MRA. From equation (2) above, a CNR > 1 indicates the contrast between the vessels and neighbouring tissue is greater than the noise itself, which is desirable for vessel segmentation. In general, a high CNR will ensure vessels are well-contrasted against the neighbouring tissue of the brain to ease segmentation32,33.
As illustrated in Fig. 5e, the CNR of almost half of the scans in this dataset is less than 1, implying the signal difference between the aneurysm and brain is less than the noise from the background ‘air’. Segmentation and modelling from these ‘noisy’ scans could therefore be difficult. Meanwhile, sub-004 had high CNR values of 39 for the aneurysm and 22 for the aneurysm with parent vessel, likely due to the contrast agent administered for this scan in combination with the TOF-MRA.
The IA region in most patient scans in this dataset exhibited an SNR > 10, while half had a CNR > 1, and therefore should facilitate accurate segmentations. However, many scans obtained an SNR < 10 and CNR < 1, and thus care should be taken when using these patient scans of variable image quality for further modelling and analysis. To ensure appropriate comparisons between studies utilising multiple datasets, it is important to consider these SNR and CNR results, as models developed on patient scans of high SNR may be challenging to adapt to those with a low SNR34.
Usage Notes
While the free and open-source 3D Slicer software can be used for viewing and processing of raw images and clinician-segmented vascular trees, parent vessels, and IA models, the NIFTI format provided ensures various commercial software suites can also utilise the de-identified TOF-MRAs for further processing. This imaging format is also consistent with existing aneurysm repositories previously described, including the IXI database and the recently published aneurysm dataset by Di Noto et al. (2023). This ensures similar data-processing workflows can be used across available datasets.
Patient scans within this dataset have been collected with comparable acquisition parameters to existing datasets, albeit with some scanners using significantly larger spacing between slices, thereby increasing voxel size and reducing image resolution, as summarised in Table 1. In some cases, the voxel size is comparable to arterial diameters across the Circle of Willis, thus decreasing spatial resolution and detail. Through investigating the effect of voxel size on IA morphological and haemodynamic parameters, Berg et al. (2018) found a larger voxel size to result in inaccurate aneurysmal neck representation in segmentations, with the inability to capture smaller side branches, small perforators and potentially smaller aneurysms35. Therefore, while TOF-MRA is often used for monitoring aneurysms due to the lack of ionising radiation required and greater safety for patients, the poor spatial resolution can limit analysis of aneurysm progression and shape change. Figure 3 thus provides only an indication for whether aneurysm shape change has been observed.
Furthermore, sub-001 from this dataset has been previously investigated for inter-user variability and the effect of segmentation morphology on fluid dynamics simulations19. Paritala et al. (2023) found that minor variations in threshold segmentation parameters and the level of smoothing resulted in a 20% variation in computational fluid dynamics results related to wall shear stress and derived parameters.
Similarly, SNR and CNR is variable across the dataset as summarised in Fig. 5. and can influence segmentation accuracy and the ability to distinguish vessels from neighbouring tissue. Therefore, caution must be exercised when utilising low resolution patient scans for research necessitating segmentation and consider the inter-user segmentation variability previously described19. As such, voxel spacing, SNR and CNR results are provided for each scan in the supporting documentation for the dataset and can be consulted when selecting appropriate patient images to incorporate into modelling and analysis.
This broad distribution of image quality represents realistic TOF-MRA images from a major tertiary hospital, and thus is valuable for the development of robust automated patient screening and aneurysm detection algorithms. To ensure widespread use of IA models, including in frugal, remote, or austere clinical settings, it is important that clinicians and clinical research be educated and developed using TOF-MRA images of varying quality.
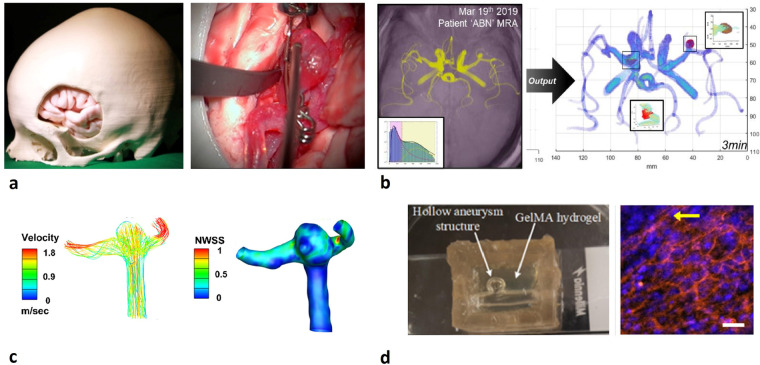
This dataset aims to support the effort to reduce bottlenecks in data availability which currently limits research and modelling into IA growth and rupture dynamics. As depicted in Fig. 6 this dataset can be, and has already been, useful for developing surgical training models and surgical guides, automated detection algorithms, in CFD studies and for evaluating cell behaviour and dynamics in in vitro IA models. Specifically, Fig. 6b5 and Fig. 6c19 have made use of this dataset.
Fig. 6.
Potential applications of dataset in intracranial aneurysm research. (a) Patient-specific 3D printed skull, brain and middle cerebral artery aneurysm model for surgical clipping training36; (b) Automated intracranial aneurysm identification algorithm using MATLAB applied to sub-03713; (c) Computational fluid dynamics analyses of sub-001 as part of the inter-user variability study yielding velocity and wall shear stress contour plots in a middle cerebral artery aneurysm19; (d) Gelatine Methacrylate aneurysm model with open lumen, seeded with endothelial cells and smooth muscle cells22. Figures have been reused from referenced articles with permission and under approved licenses.
Acknowledgements
The authors would like to thank Dr. John Clouston for connecting the authors involved in this project, and Dr. David Alonso-Caneiro for his assistance in image processing and interpretation. Dr. Karen Dobeli from the Department of Medical Imaging at the RBWH and Dr. Katie McMahon from the Herston Imaging Research Facility also shared their knowledge in medical imaging and de-identification requirements. The authors would like to thank the Royal Brisbane and Women’s Hospital (RBWH) Foundation and the RBWH Department of Medical Imaging for their support, including RBWH Foundation grants in 2020 and 2021. The authors would also like to thank Metro North Health who funded some of the researchers through the CranioFacial program of the Herston Biofabrication Institute. This work was supported by the Australian Research Council (DE220100757) and Advance Queensland (AQIRF1312018) fellowships to MCA as well as a collaborative research agreement between CMD, JIN, CDW, ARM and MCA with the Herston Biofabrication Institute.
Author contributions
Research conception: M.C.A., C.D.W. Data collection: M.P., E.L. and C.D.W. Data processing and organisation: C.M.D., E.L. Data analysis and annotation: C.M.D., E.L. Manuscript preparation: C.M.D. All authors reviewed the manuscript.
Code availability
The described dataset was manually organised according to BIDS (v1.8.0). DICOM files were converted to NIFTI format using MRIcroGL dcm2niix software (NITRC, v1.0.20220720). Masks and annotations were prepared using 3DSlicer software (v5.2.1). Python (Spyder, v5.4.3) was used to order scans by imaging modality and scanning dates, and for the stripping of unnecessary NIFTI slices in some patients where portions of the skull not related to the Circle of Willis were also imaged. Code used for these purposes is not published as it is not required for generation or processing of the published dataset.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chloe M. de Nys, Ee Shern Liang.
References
- 1.Vlak MH, Algra A, Brandenburg R, Rinkel GJ. Prevalence of unruptured intracranial aneurysms, with emphasis on sex, age, comorbidity, country, and time period: a systematic review and meta-analysis. Lancet Neurol. 2011;10:626–636. doi: 10.1016/s1474-4422(11)70109-0. [DOI] [PubMed] [Google Scholar]
- 2.Imaizumi Y, Mizutani T, Shimizu K, Sato Y, Taguchi J. Detection rates and sites of unruptured intracranial aneurysms according to sex and age: an analysis of MR angiography–based brain examinations of 4070 healthy Japanese adults. Journal of Neurosurgery JNS. 2019;130:573–578. doi: 10.3171/2017.9.JNS171191. [DOI] [PubMed] [Google Scholar]
- 3.Wiebers DO. Unruptured intracranial aneurysms: natural history, clinical outcome, and risks of surgical and endovascular treatment. The Lancet. 2003;362:103–110. doi: 10.1016/S0140-6736(03)13860-3. [DOI] [PubMed] [Google Scholar]
- 4.Zhou S, Dion PA, Rouleau GA. Genetics of Intracranial Aneurysms. Stroke. 2018;49:780–787. doi: 10.1161/strokeaha.117.018152. [DOI] [PubMed] [Google Scholar]
- 5.Allenby MC, et al. Detection of clustered anomalies in single-voxel morphometry as a rapid automated method for identifying intracranial aneurysms. Computerized Medical Imaging and Graphics. 2021;89:101888. doi: 10.1016/j.compmedimag.2021.101888. [DOI] [PubMed] [Google Scholar]
- 6.Zuurbier CCM, et al. Sex Difference and Rupture Rate of Intracranial Aneurysms: An Individual Patient Data Meta-Analysis. Stroke. 2022;53:362–369. doi: 10.1161/STROKEAHA.121.035187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nieuwkamp DJ, et al. Changes in case fatality of aneurysmal subarachnoid haemorrhage over time, according to age, sex, and region: a meta-analysis. The Lancet Neurology. 2009;8:635–642. doi: 10.1016/S1474-4422(09)70126-7. [DOI] [PubMed] [Google Scholar]
- 8.Bevan H, Sharma K, Bradley W. Stroke in young adults. Stroke. 1990;21:382–386. doi: 10.1161/01.STR.21.3.382. [DOI] [PubMed] [Google Scholar]
- 9.Vlak MHM, Algra A, Brandenburg R, Rinkel GJE. Prevalence of unruptured intracranial aneurysms, with emphasis on sex, age, comorbidity, country, and time period: a systematic review and meta-analysis. The Lancet Neurology. 2011;10:626–636. doi: 10.1016/S1474-4422(11)70109-0. [DOI] [PubMed] [Google Scholar]
- 10.Keedy A. An overview of intracranial aneurysms. Mcgill J Med. 2006;9:141–146. [PMC free article] [PubMed] [Google Scholar]
- 11.The Natural Course of Unruptured Cerebral Aneurysms in a Japanese Cohort. New England Journal of Medicine366, 2474–2482, 10.1056/NEJMoa1113260 (2012). [DOI] [PubMed]
- 12.Duan Z, et al. Morphological parameters and anatomical locations associated with rupture status of small intracranial aneurysms. Scientific Reports. 2018;8:6440. doi: 10.1038/s41598-018-24732-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Din M, et al. Detection of cerebral aneurysms using artificial intelligence: a systematic review and meta-analysis. J Neurointerv Surg. 2023;15:262–271. doi: 10.1136/jnis-2022-019456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Noto T, et al. Towards Automated Brain Aneurysm Detection in TOF-MRA: Open Data, Weak Labels, and Anatomical Knowledge. Neuroinformatics. 2023;21:21–34. doi: 10.1007/s12021-022-09597-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Etminan N, et al. The unruptured intracranial aneurysm treatment score. Neurology. 2015;85:881. doi: 10.1212/WNL.0000000000001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neulen, A. et al. Comparison of Unruptured Intracranial Aneurysm Treatment Score and PHASES Score in Subarachnoid Hemorrhage Patients With Multiple Intracranial Aneurysms. Frontiers in Neurology12, 10.3389/fneur.2021.616497 (2021). [DOI] [PMC free article] [PubMed]
- 17.Stumpo V, et al. Retrospective Application of UIATS Recommendations to a Multicenter Cohort of Ruptured Intracranial Aneurysms: How It Would Have Oriented the Treatment Choices? World Neurosurgery. 2021;147:e262–e271. doi: 10.1016/j.wneu.2020.12.041. [DOI] [PubMed] [Google Scholar]
- 18.Murayama Y, Fujimura S, Suzuki T, Takao H. Computational fluid dynamics as a risk assessment tool for aneurysm rupture. Neurosurg Focus. 2019;47:E12. doi: 10.3171/2019.4.Focus19189. [DOI] [PubMed] [Google Scholar]
- 19.Paritala PK, et al. Reproducibility of the computational fluid dynamic analysis of a cerebral aneurysm monitored over a decade. Scientific Reports. 2023;13:219. doi: 10.1038/s41598-022-27354-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levitt MR, et al. Genetic correlates of wall shear stress in a patient-specific 3D-printed cerebral aneurysm model. J Neurointerv Surg. 2019;11:999–1003. doi: 10.1136/neurintsurg-2018-014669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaneko N, et al. A patient-specific intracranial aneurysm model with endothelial lining: a novel in vitro approach to bridge the gap between biology and flow dynamics. J Neurointerv Surg. 2018;10:306–309. doi: 10.1136/neurintsurg-2017-013087. [DOI] [PubMed] [Google Scholar]
- 22.Yong KW, et al. Engineering a 3D human intracranial aneurysm model using liquid-assisted injection molding and tuned hydrogels. Acta Biomaterialia. 2021;136:266–278. doi: 10.1016/j.actbio.2021.09.022. [DOI] [PubMed] [Google Scholar]
- 23.de Nys CM, 2024. Royal Brisbane_TOFMRA_Intracranial Aneurysm_Database. OpenNeuro. [DOI]
- 24.Voß S, Beuing O, Janiga G, Berg P. Multiple Aneurysms AnaTomy CHallenge 2018 (MATCH)—Phase Ib: Effect of morphology on hemodynamics. PLOS ONE. 2019;14:e0216813. doi: 10.1371/journal.pone.0216813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dumais F, et al. eICAB: A novel deep learning pipeline for Circle of Willis multiclass segmentation and analysis. NeuroImage. 2022;260:119425. doi: 10.1016/j.neuroimage.2022.119425. [DOI] [PubMed] [Google Scholar]
- 26.Isherwood SJS, Bazin P-L, Alkemade A, Forstmann BU. Quantity and quality: Normative open-access neuroimaging databases. PLOS ONE. 2021;16:e0248341. doi: 10.1371/journal.pone.0248341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heverhagen, J. T., Bourekas, E., Sammet, S., Knopp, M. V. & Schmalbrock, P. Time-of-Flight Magnetic Resonance Angiography at 7 Tesla. Investigative Radiology43 (2008). [DOI] [PubMed]
- 28.Jiang J, Haacke EM, Dong M. Dependence of vessel area accuracy and precision as a function of MR imaging parameters and boundary detection algorithm. J Magn Reson Imaging. 2007;25:1226–1234. doi: 10.1002/jmri.20918. [DOI] [PubMed] [Google Scholar]
- 29.Bizino, M. B. et al. High Spatial Resolution Coronary Magnetic Resonance Angiography at 7 T: Comparison With Low Spatial Resolution Bright Blood Imaging. Investigative Radiology49 (2014). [DOI] [PubMed]
- 30.Zhang Z, et al. Visualization of the lenticulostriate arteries at 3T using black-blood T1-weighted intracranial vessel wall imaging: comparison with 7T TOF-MRA. European Radiology. 2019;29:1452–1459. doi: 10.1007/s00330-018-5701-y. [DOI] [PubMed] [Google Scholar]
- 31.von Morze C, et al. Intracranial time-of-flight MR angiography at 7T with comparison to 3T. Journal of Magnetic Resonance Imaging. 2007;26:900–904. doi: 10.1002/jmri.21097. [DOI] [PubMed] [Google Scholar]
- 32.Yang Q, et al. Whole-brain vessel wall MRI: A parameter tune-up solution to improve the scan efficiency of three-dimensional variable flip-angle turbo spin-echo. Journal of Magnetic Resonance Imaging. 2017;46:751–757. doi: 10.1002/jmri.25611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luu HM, Klink C, Moelker A, Niessen W, van Walsum T. Quantitative evaluation of noise reduction and vesselness filters for liver vessel segmentation on abdominal CTA images. Physics in Medicine & Biology. 2015;60:3905. doi: 10.1088/0031-9155/60/10/3905. [DOI] [PubMed] [Google Scholar]
- 34.Pham, W. et al. A critical guide to the automated quantification of perivascular spaces in magnetic resonance imaging. Frontiers in Neuroscience16, 10.3389/fnins.2022.1021311 (2022). [DOI] [PMC free article] [PubMed]
- 35.Berg, P. et al. in 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). 1327–1330.
- 36.Joseph FJ, Weber S, Raabe A, Bervini D. Neurosurgical simulator for training aneurysm microsurgery-a user suitability study involving neurosurgeons and residents. Acta Neurochir (Wien) 2020;162:2313–2321. doi: 10.1007/s00701-020-04522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- de Nys CM, 2024. Royal Brisbane_TOFMRA_Intracranial Aneurysm_Database. OpenNeuro. [DOI]
Data Availability Statement
The described dataset was manually organised according to BIDS (v1.8.0). DICOM files were converted to NIFTI format using MRIcroGL dcm2niix software (NITRC, v1.0.20220720). Masks and annotations were prepared using 3DSlicer software (v5.2.1). Python (Spyder, v5.4.3) was used to order scans by imaging modality and scanning dates, and for the stripping of unnecessary NIFTI slices in some patients where portions of the skull not related to the Circle of Willis were also imaged. Code used for these purposes is not published as it is not required for generation or processing of the published dataset.