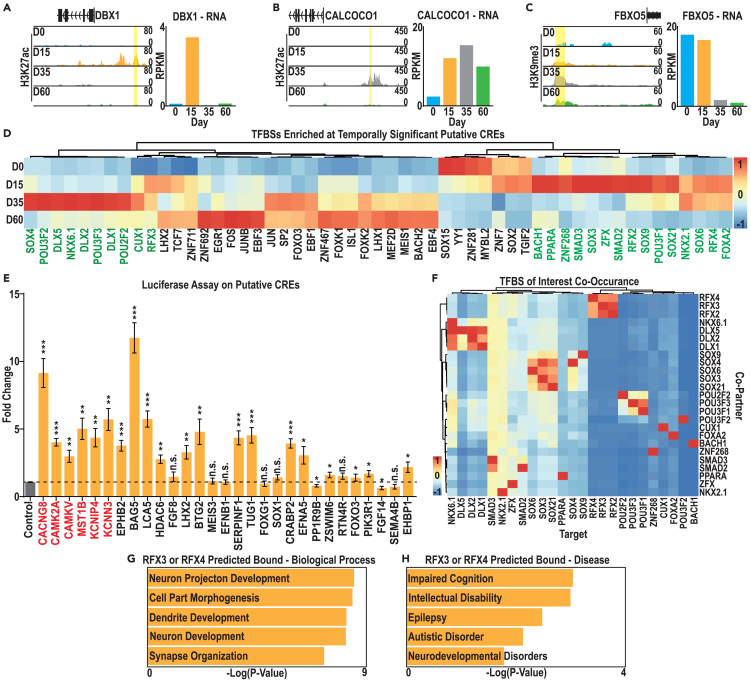
Figure 4.
Analysis of putative CREs and transcription factors with binding site enrichment under these peaks
(A–C) Examples from each set of putative CREs: (A) D15 enriched H3K27ac associated with high D15 gene expression, (B) D35 enriched H3K27ac associated with high D35 gene expression, and (C) D35 enriched H3K9me3 associated with high expression at early (D0 and D15) time points.
(D) Expression, during hESC-mature cIN differentiation, of TFs that recognize TFBS enriched in putative CREs. TFs with enriched expression at D15 and D35 are indicated in green text.
(E) Luciferase assay measures luminescence in D15 NPCs transfected with putative CRE-luciferase reporter plasmids, normalized to the unmodified (no CRE) luciferase vector. Dotted line represents basal expression from the no CRE control. CREs indicated in red are associated with an RFX binding motif. Data were analyzed with Student’s t test n = 4, ∗p value < 0.05, ∗∗p value < 0.01, ∗∗∗p value < 0.001 and ∗∗∗∗p value < 0.0001.
(F) Co-enrichment for target and co-occuring TFBSs within peak sets of interest, averaged across all peaks as a percentage of peak numbers with corresponding TFBS.
(G and H) Biological process and disease GO term enrichment for genes associated with a putative RFX3/4 TFBS-containing CRE within our peak set of interest.