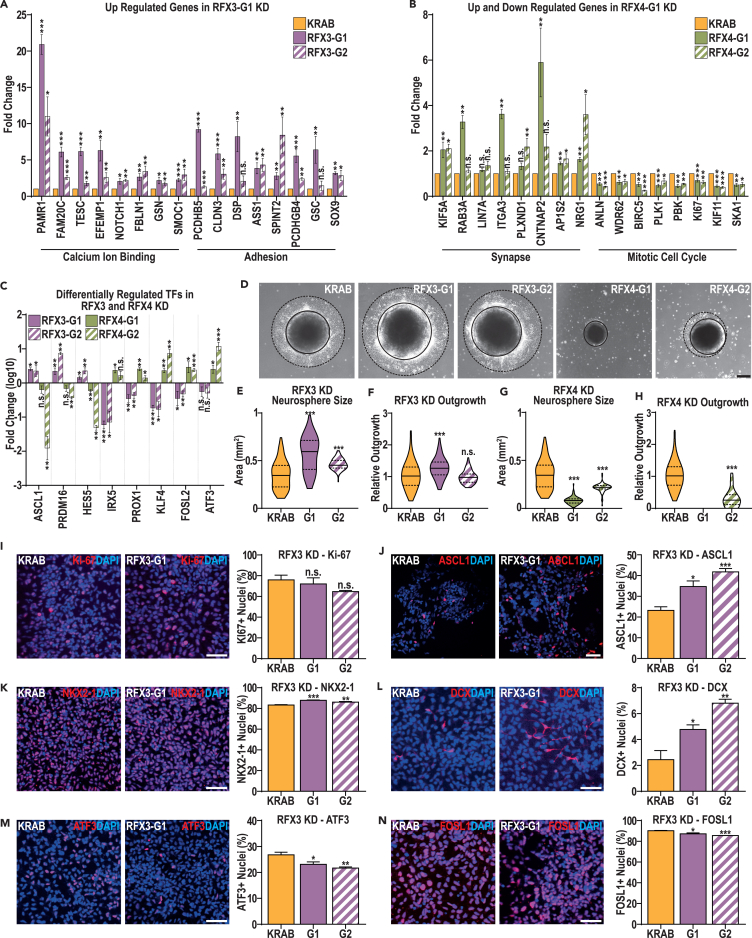
Figure 6.
Both RFX3 and RFX4 KD models exhibit deficits in human cortical interneuron differentiation
(A and B) RT-qPCR was used to confirm gene expression changes identified in the (A) RFX3 KD and (B) RFX4 KD models, using both the G1 and G2 KD models to test each gene at D15 of cortical interneuron differentiation.
(C) RT-qPCR was also used to investigate the reciprocal regulation of a selection of TFs previously identified as DEGs in the RFX3 KD and RFX4 KD models. Data are represented as mean fold change ± SEM.
(D–H) (D) Representative images indicating sphere size (marked by solid black lines) and outgrowth (marked by dotted black lines) and quantification of neurosphere size and relative outgrowth (outgrowth minus neurosphere size, relative to neurosphere size) from neurospheres at D12 of cortical interneuron differentiation in the RFX3 (E and F) and RFX4 (G and H) KD models. Data are represented as distributions, using all measures taken with quartiles (dotted lines) and means (black line) marked (scale bar: 100 μm).
(I–N) Representative images and quantification of the fraction of cells immunopositive for (I) Ki-67, (J) ASCL1, (K) NKX2.1, (L) DCX, (M) ATF3, and (N) FOSL1 in the KRAB and RFX3 KD models. Data are represented as mean ± SEM; data were analyzed by Student’s t-test, versus the paired KRAB controls. n = 4 biological replicate experiments for all conditions, ∗p value < 0.05, ∗∗p value < 0.01, ∗∗∗p value < 0.001 and ∗∗∗p value < 0.0001. Scale bars: 50 μm.